Abstract
Error-free chromosome segregation requires stable attachment of sister kinetochores to the opposite spindle poles (amphitelic attachment). Exactly how amphitelic attachments are achieved during spindle assembly remains elusive. We employed photoactivatable GFP and high-resolution live-cell confocal microscopy to visualize for the first time complete 3-D movements of individual kinetochores throughout mitosis in non-transformed human cells. Combined with electron microscopy, molecular perturbations, and immunofluorescence analyses, this approach reveals unexpected new details of chromosome behavior. Our data demonstrate that unstable lateral interactions between kinetochores and microtubules dominate during early prometaphase. These transient interactions lead to the reproducible arrangement of chromosomes in an equatorial ring on the surface of the nascent spindle. A computational model predicts that this toroidal distribution of chromosomes exposes kinetochores to a high-density of microtubules which facilitates subsequent formation of amphitelic attachments. Thus, spindle formation involves a previously overlooked stage of chromosome prepositioning which promotes formation of amphitelic attachments.
Keywords: mitosis, spindle assembly, chromosome congression, kinetochore
Introduction
The goal of mitosis is to ensure that daughter cells inherit identical genetic information transmitted in the form of duplicated chromosomes. To achieve this goal cells employ a microtubule-based molecular machine termed the ‘spindle’. Chromosomes attach to spindle microtubules via kinetochores, discrete macromolecular assemblies that reside at the chromosome’s centromere. The two kinetochores on each chromosome must stably attach to the opposite spindle poles (amphitelic attachment, reviewed in Walczak et al., 2010).
The general principle of mitotic spindle assembly is described as microtubule ‘search & capture’ (Kirschner and Mitchison, 1986). In this model dynamic plus ends of microtubules grow and shrink until they are captured and stabilized by a kinetochore. Modern computational models predict that for unbiased search and capture would require hours before each of the 200-nm small kinetochores on 46 chromosomes present in a typical human cell encounters a single microtubule (Wollman et al., 2005). Nevertheless, mitosis takes less than 30 minutes in diploid human cells (Yang et al., 2008). This discrepancy implies that additional mechanisms facilitate mitotic spindle assembly by guiding microtubules growth toward kinetochores (O'Connell et al., 2009; Wollman et al., 2005) and/or positioning chromosomes to the areas with high density of microtubules (Kapoor et al., 2006; Lenart et al., 2005; Paul et al., 2009). To which extent various accessory pathways are harnessed by chromosomes during normal mitosis remains unknown.
One feature of mitosis that must be considered in the analysis of spindle assembly is that the spindle forms in 3-D space. Yet, owing to technical limitations, most recordings of spindle assembly and chromosome movements are limited to single focal planes. Here we report a 3-D analysis of centrosome and kinetochore movements in non-transformed diploid human cells RPE1. Our data reveal that spindle assembly is facilitated by a transient arrangement of chromosomes in a ring surrounding the central part of the spindle during early prometaphase. Formation of the chromosome ring is driven by the combination of labile lateral kinetochore/microtubule interactions and spindle ejection forces. As a result, centromeres become prepositioned near the spindle equator where kinetochores are exposed to the high density of microtubules which promotes formation of stable amphitelic attachments.
Results
The pattern of spindle elongation and orientation
The length and orientation of the spindle are determined by spatial separation of the duplicated centrosomes. This separation can occur during prophase or after nuclear envelope breakdown (NEB), during prometaphase (Roos, 1973). In the latter case the spindle was reported to form as a monopolar structure that subsequently bipolarizes. The ‘prophase’ and ‘prometaphase pathways’ (Whitehead et al., 1996) were observed in a variety of cell types (Roos, 1973; Toso et al., 2009) and these different routes of centrosome separation may affect the efficiency of spindle assembly (Rosenblatt, 2005; Toso et al., 2009).
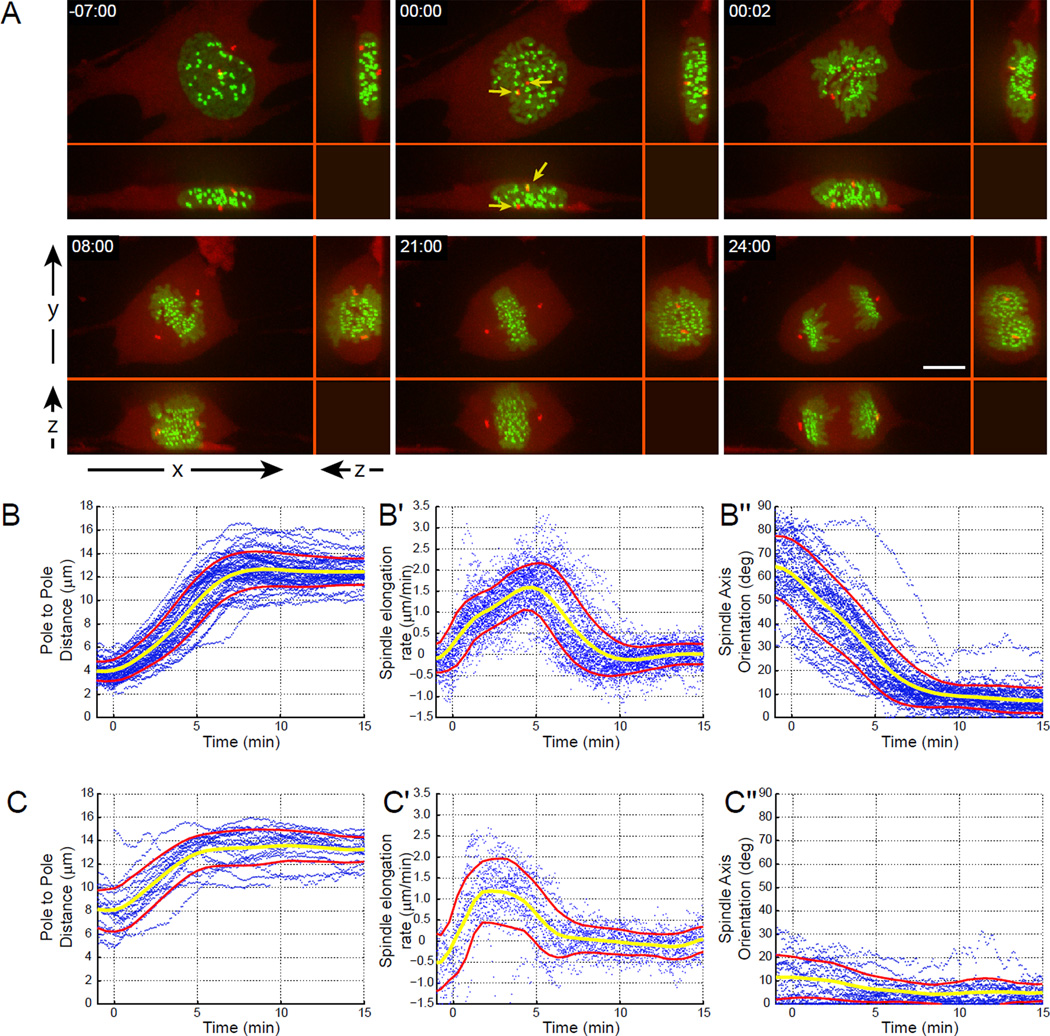
Our 3-D analyses of centrosome movements reveal that centrosomes always separate to the opposite sides of the nucleus prior to NEB in RPE1 cells (Fig. 1). In the majority of late-prophase cells (~73%, 49/67) one centrosome resides above and one below the nucleus so that upon NEB the forming spindle is initially oriented vertically (the angle between the spindle axis and the surface of the coverslip exceeds 30°). Hereafter we refer to these cells as ‘V-cells’. In the remaining ~27% (18/67) of cells, centrosomes are separated to the opposite sides of the nucleus horizontally so that spindle axis at NEB is tilted less than 30° with respect to the coverslip (hereafter ‘H-cells’). In planar XY view, vertical separation of centrosomes in V-cells may create an impression that the centrosomes form a common complex. However, as evident from 3-D microscopy the centrosomes in V-cells are in fact physically separated by the intervening nucleus (Fig. 1A; Movie S1).
Figure 1. The pattern of spindle elongation and orientation in RPE1 cells.
(A) An RPE1 cell expressing CENP-A-GFP (green) to label the kinetochores and centrin1-tdTomato (red) to label the centrosomes is shown. Although in XY view the centrosomes appear to reside in a common complex just before NEB (arrows in 00:00), XZ and YZ views demonstrate that the centrosomes are actually positioned on the opposite sides of the nucleus (above and below). (B–C) Numeric characterization of spindle elongation and orientation in 67 RPE1 cells co-expressing centrin-GFP and CENP-A-PAGFP. Each plot presents individual trajectories (blue dots), the average value (yellow line), and standard deviation (red lines). Spindle length (B, C), rate of spindle elongation (B’, C’), and spindle orientation (B”, C”) in V- (B–B”) vs. H-cells (C–C”). Note the remarkable reproducibility of spindle elongation and rotation pattern.
Due to the disk-like shape of the nucleus inter-centrosome distances at NEB are much greater in H- than in V-cells and this distance begins to increase immediately after NEB (Fig. 1B, C). The rate of spindle elongation is not linear with the velocity increasing gradually to 2.2±0.5 µm/min which is generally consistent with the velocity of antiparallel sliding of microtubules driven by kinesin-5 (Kapitein et al., 2005; Uteng et al., 2008). Although maximal rate of spindle elongation is similar between V- and H-cells (2.3±0.4 and 1.9±0.5 µm/min, Fig. 1B’, C’), the peak velocity is reached ~5 min after NEB in V-cells and ~2.5 min in H-cells when the spindle length is ~10 µm in both cases. This suggests that the initial slower phase of spindle elongation is not due to a gradual activation of mitotic kinesins (Blangy et al., 1995; Cahu et al., 2008; Goshima and Vale, 2005). Instead, the elongation rate is likely to reflect changes in the region of antiparallel microtubule overlap.
Since at NEB the centrosomes are already further apart in H-cells (7.9±2.3 µm), the spindle reaches its full length (13.4±1.2 µm) more rapidly in H- vs. V-cells (~5 min vs. ~8 min in V-cells; Fig. 1 B and C). In V-cells spindle elongation is concurrent with spindle rotation at the average rate of 6–7 degrees per min, so that ~8 min after NEB the spindle is oriented parallel to the coverslip surface (Fig. 1 B”, Movie S1). The final orientation of the spindle is identical in V- and H-cells (81.8±6.6° and 82.4±6.9° respectively; Fig. 1 B” and C”). Therefore, we conclude that RPE1 cells rely exclusively on the prophase pathway of centrosome separation and the efficiency of spindle assembly does not depend on the direction of the initial centrosome separation. Our data also reveal the remarkable consistency of spindle assembly in RPE1 cells – in all cells the spindle is fully elongated and properly oriented ~8 min after NEB.
Chromosomes reproducibly arrange in a ring around the spindle during early prometaphase
We used 3-D time-lapse movies of RPE1 cell co-expressing centrin1-GFP and CENP-A-GFP fusions to explore whether there is a specific pattern in the spatial arrangement of chromosomes during the initial stages of spindle formation. Restricting image acquisition to a single channel allowed us to avoid significant photodamage while the centrioles and kinetochores could still be easily discerned in the recordings due to their dramatically different behaviors.
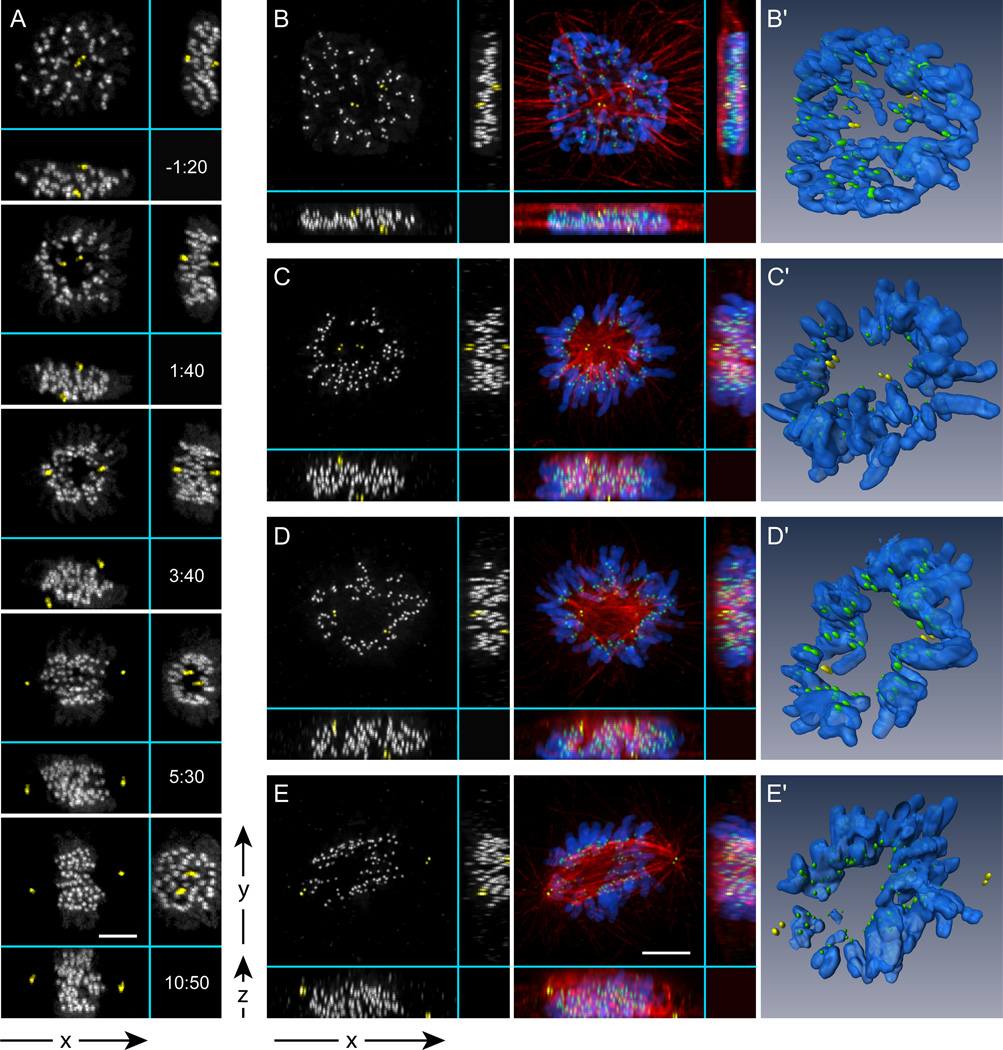
Shortly after nuclear envelope breakdown (NEB) kinetochores residing in the inner parts of the nucleus are rapidly expelled from the central part of the early spindle. This outward movement of the centrally-located kinetochores, combined with the inward movement of more peripheral kinetochores, leads to the arrangement of the chromosomes in a ring with the arms pointing outwards and the centromeres inwards towards the spindle axis (Fig. 2A, 1:40; Movie S2). This ring forms in both V- and H-cells although it can be easily overlooked in the conventional XY view due to unfavorable spindle orientation. The ring becomes apparent when viewed along the spindle axis (Fig. 2A, 5:30; also see Fig. S2). Fixed-cell immunofluorescence analysis confirms that the space inside the chromosome ring is filled with microtubules comprising the compact spindle that forms following NEB (Fig. 2B–C). The effects of occlusion by the dense network of microtubules are clearly seen in 3-D reconstructions (Fig 2B'–E' and Movie S3). At later stages of spindle formation chromosomes move into the central part of the spindle so that the toroidal distribution of kinetochores gradually converts into a typical metaphase plate with evenly spaced kinetochores (Fig. 2A 10:50). The chromosome ring is not unique to RPE1 cells. Similar patterns form during mitosis in transformed human cells (HeLa; Fig. S1) as well as in cells originating from other species (rat NRK-52E; Fig. S1).
Figure 2. Multi-dimensional analysis of spindle assembly.
(A) Selected frames from a high-resolution 4-D time-lapse movie of a cell labeled with centrin1-GFP and CENP-A-GFP. For clarity, centrioles are pseudo-colored yellow. Notice that one centrosome is positioned above and the other – below the nucleus (V-cell). In less than 2 min after NEB a clear zone, void of chromosomes, develops between the separating centrosomes (1:40). As the spindle rotates, the zone persists as evident from the YZ view (5:30). Later, the chromosomes re-populate the central part of the spindle (10:50). Time shown relative to NEB in min:sec. (B–E) Immunofluorescence images and computer generated surface renderings (B'–E') of fixed RPE1 cells during early-to-mid prometaphase. The volume between the poles that is void of chromosomes is filled with high-density of microtubules (C–D; C'–D'). Once the spindle rotates to a vertical position a typical prometaphase morphology becomes apparent in the conventional XY view (E, E’). Bars, 5 µm.
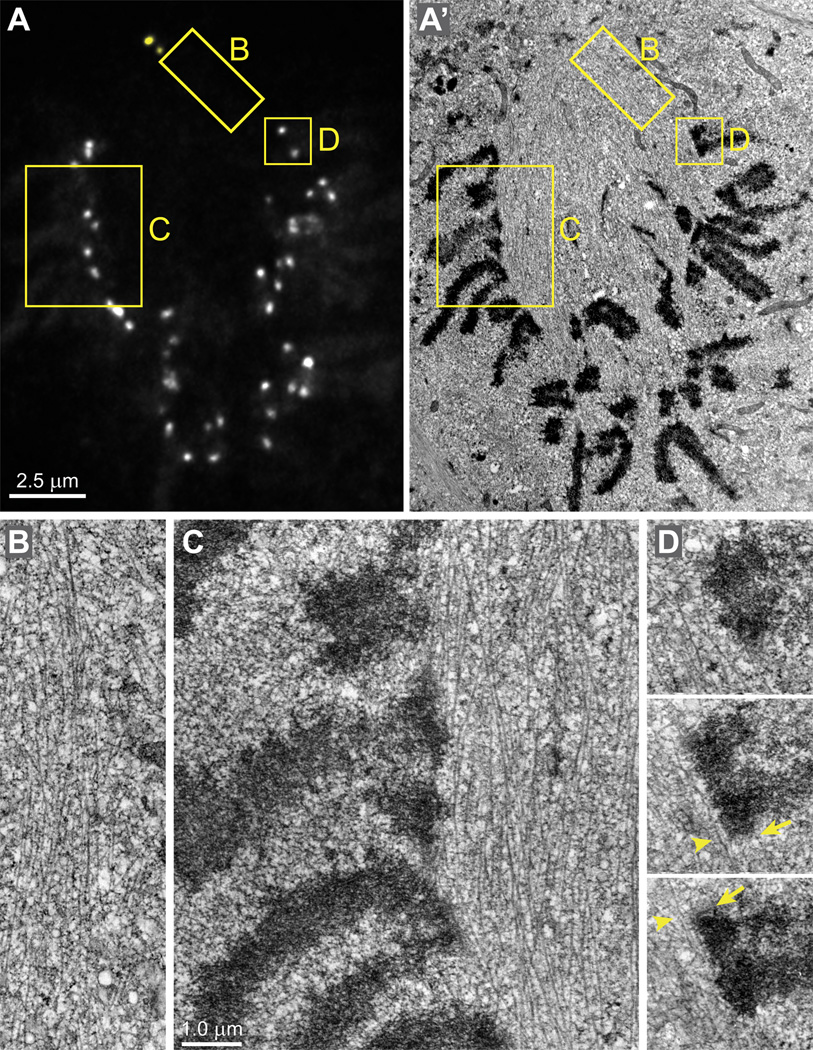
To gain deeper insight into the organization of the chromosome ring we employed correlative light/electron microscopy (Figs.3 and S2). Serial-section reconstruction of an RPE1 cell fixed during early prometaphase reveals that spindle microtubules densely populate the central part of the nascent spindle between the centrosomes. Interestingly, there is a sharp demarcation in the density of microtubules with only few microtubules protruding beyond the spindle proper (Fig. 3 B). Most centromeres reside at the boundary of the spindle with their kinetochores interacting with microtubules in a lateral fashion (Fig. 3C). Surprisingly, centromeres can be markedly stretched even when both sister kinetochores lack proper end-on microtubule attachments (Fig. 3D). This observation is surprising as it is generally assumed that stable amphitelic attachments are required for centromere stretching (reviewed in Maresca and Salmon, 2010; Nezi and Musacchio, 2009).
Figure 3. Architecture of the early-prometaphase spindle.
(A–A’) A single GFP-fluorescence focal plane (A) and the corresponding EM section (A’) selected from complete 3-D datasets. Chromosomes are excluded from the spindle and the centromeres reside on the spindle surface. Insets denote the areas presented at higher magnification in (B–D). (B) A view of the sharp demarcation between the spindle and the rest of the cytoplasm showing the high density of microtubules inside the spindle and their absence in the cytoplasm. (C) The centromeres reside on the surface of the spindle. Note that only few microtubules can be found outside the spindle between the chromosome arms. (D) Serial sections through a centromere on the surface of the spindle. Both sister kinetochores (arrows) lack end-on microtubule attachments but laterally interact with individual microtubules (arrowheads) that run parallel to the centromere. The distance between sister kinetochores is ~1 µm in spite the lack of end-on attachments. See Fig.S3 for 3-D data on the kinetochore distribution in this cell. Scale bars are 2.5 µm for (A–B) and 1 µm for (B–D).
The chromosome ring accelerates mitotic spindle assembly
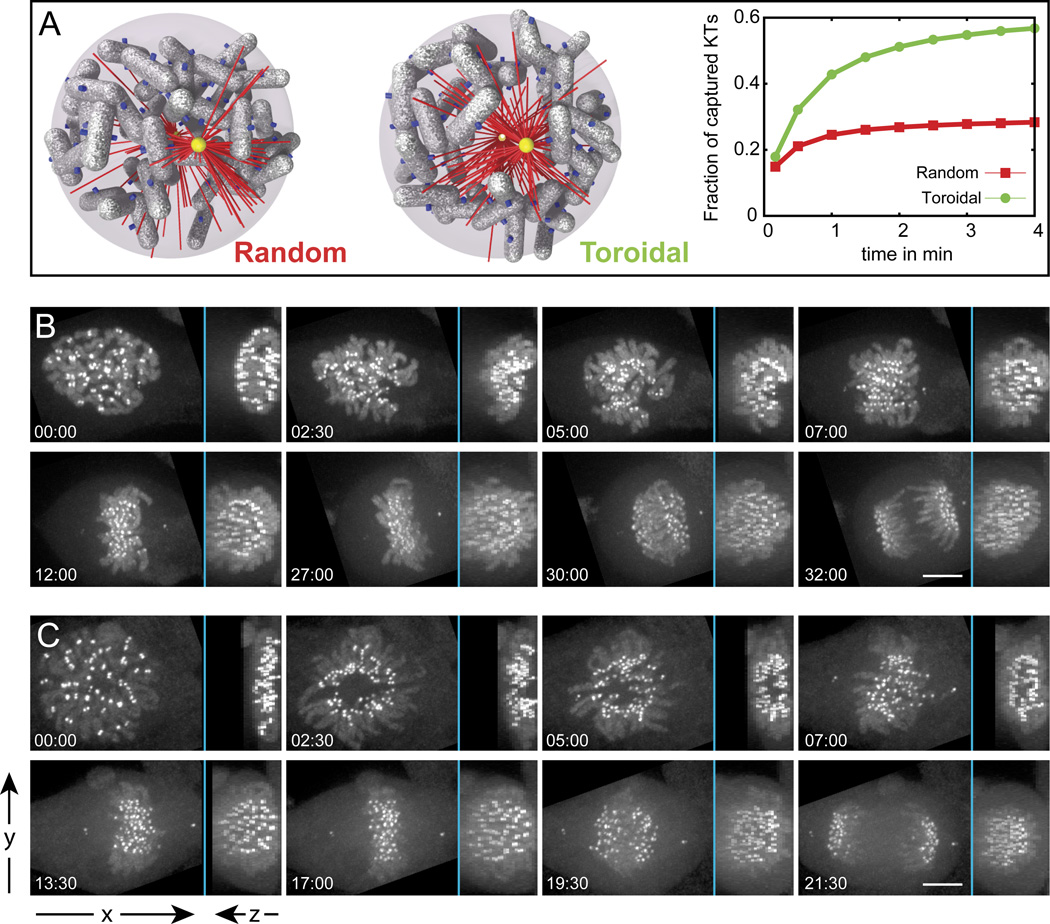
Having observed reproducible formation of the chromosome ring during mitosis we sought to establish whether this pattern bears a functional significance for spindle assembly. To this end, we harnessed the computational model constructed by Paul and co-workers (2009) which predicted that only a few kinetochores would be initially exposed to microtubules in the crowded environment of a human cell with 46 chromosomes. To estimate whether formation of the chromosome ring would facilitate S&C within the constraints of the Paul model, two types of simulations were conducted. The chromosomes were assumed to either be spread uniformly and randomly throughout the nuclear space (oblate spheroid with 14 ×14 × 7 µm) or form a toroid with the dimensions extracted from our live- and fixed-cell observations (inner radius 4 µm and outer radius 7 µm). Although the difference between these two types of chromosome distribution is visually subtle (cf. Fig. 4A Random vs. Toroidal), the simulation predicts that the efficiency of S&C is significantly improved by the chromosome ring. The number of kinetochores exposed to microtubules increases from ~30% in the case of uniformly distributed chromosomes to ~70% in the ring configuration. As a result, within a 3-min long search, ~60% of the chromosomes would be captured and incorporated into the spindle in the ring configuration, which is a dramatic improvement over the randomly distributed chromosomes. Thus, formation of the chromosome ring at the onset of mitosis is advantageous for S&C and is predicted to accelerate mitotic spindle assembly by approximately 6–8 minutes.
Figure 4. The chromosome ring facilitates spindle assembly.
(A) Two types of initial chromosome distribution (Random and Toroidal) and corresponding dynamics of kinetochore capture predicted in our computer simulations. The toroidal distribution provides a clear kinetic advantage. (B–C) Mitosis in chromokinesin Kid-depleted (B) vs. control (C) cells. Depletion of Kid inhibits formation of the central clear zone. In contrast, chromosomes in control cells are excluded from the center of the spindle during early prometaphase (C; 02:30 – 07:00). Notice that to generate consistent perspective, both sequences are illustrated by maximal-intensity projections that are perpendicular (left part of each frame) and parallel (right part) to the spindle axis during metaphase.
To experimentally test this prediction we followed the dynamics of mitosis in cells depleted of the chromokinesin Kid (kinesin-10) (Tokai et al., 1996). We reasoned that expulsion of chromosomes from the central part of the spindle is likely to be driven by the spindle ejection force (Rieder et al., 1986) which is primarily generated by Kid (Levesque and Compton, 2001). Previous studies have established that inactivation of Kid does not prevent formation of a functional bipolar spindle although several aspects of chromosome movement are affected and the duration of mitosis is increased (Levesque and Compton, 2001; Tokai-Nishizumi et al., 2005). Our 3-D recordings reveal, that in fact, formation of the chromosome ring is inhibited upon siRNA depletion of Kid (Fig. 4B) and the duration of mitosis increases by approximately 6 min, (from 19.4±2.9 min; in control [N=8] to 25±3.3 min in Kid-depleted cells [N=10]), which is in excellent agreement with the model. The delay is due to slower formation of the metaphase plate (cf. Fig. 4B and C; Movies S4 and S5). We also observed similar inhibition of the ring formation and prolongation of prometaphase in cells microinjected with an antibody raised against the Kid DNA-binding domain which was previously used by Levesque and Compton (2001) (N=4, data not shown). Thus, experimental perturbation of chromosome ring formation decreases the efficiency of spindle assembly as predicted by the computational model.
The pattern of chromosome movements
3-D recordings of cells with GFP-tagged kinetochores and centrioles allowed us to visualize the general pattern of spindle assembly. However, due to the large number of chromosomes and complexity of their movements in 3-D space, we were unable to continuously follow trajectories of individual chromosomes from NEB through anaphase in these recordings. To overcome this limitation we developed an assay in which one or two pairs of sister kinetochores were photoactivated in cells expressing CENP-A-PAGFP (Fig. S3A). With this approach 3-D positions of both sister kinetochores and centrosomes can be reliably tracked and analyzed (Fig. S3B–C, Movie S6). Comparative and averaging analyses of 81 chromosome trajectories (50 from NEB through anaphase and 31 from NEB through metaphase) obtained in 67 cells allowed us to identify features that are common (Fig. S3D), which in turn helps to reveal the pathways that are prevalent during normal spindle assembly in diploid human cells.
Consistent with data obtained in cells with all kinetochores labeled, individual-kinetochore tracking reveals that most centromeres remain near the spindle equator from NEB through anaphase onset (AO). Typically, the distance between kinetochores and spindle poles increases gradually during prometaphase (Figs. 5A and S3) until it reaches its maximum of 6.7±1.6 µm ~8 min after NEB when the prometaphase centrosome separation is completed (Fig. S3D). Thus, somewhat counterintuitive, during spindle assembly the total displacement of centrosomes from their positions at NEB is greater than the total displacement of a typical chromosome.
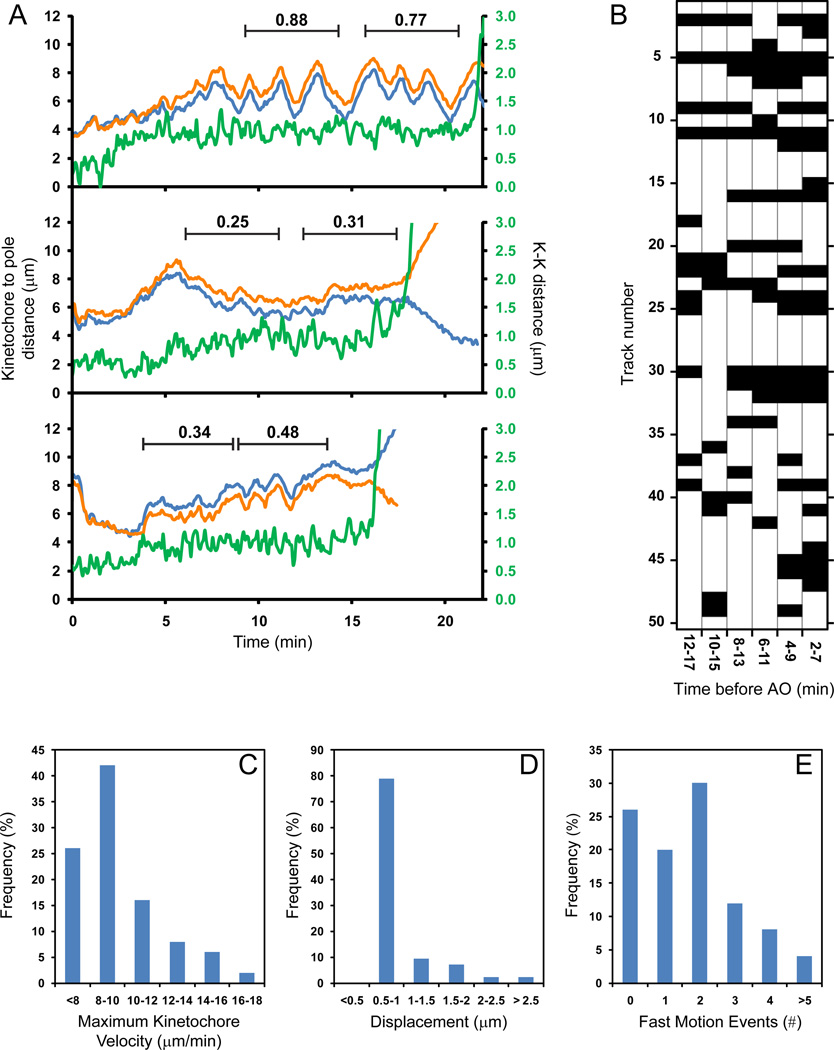
Figure 5. Chromosome movements during prometaphase and metaphase.
(A) Examples of individual-chromosome behavior. The plots present changes in the distance between one spindle pole and each photoactivated kinetochore in a sister pair (orange and blue lines) as well as centromere stretch (green) from NEB through AO. One chromosome (top) exhibits oscillatory behavior; another chromosome remains relatively motionless during metaphase (middle) while the third chromosome switches between periods of oscillation and irregular movements (bottom). Deviation from Average Position (DAP) values are shown for periods marked by black lines. (B) Summary of oscillatory behavior for 50 individual chromosomes. Black blocks represent DAP <0.4 (non-oscillating behavior), white blocks correspond to DAP values exceeding 0.4 (oscillation). (C) Histogram of maximum velocity reached by kinetochores. (D) Displacements resulting from rapid (>8 µm/min) kinetochore movements. (E) Number of rapid kinetochore movements exhibited by individual chromosomes.
At the spindle equator some chromosomes undergo continuous oscillations throughout metaphase; other chromosomes remain motionless, and some switch between periods of oscillations and irregular movements (Fig. 5A). To characterize these behaviors numerically we used the DAP criterion (deviation from average position) developed by Stumpff and co-authors (2008). We determined DAP for every chromosome in a series of 5-min windows that span from late prometaphase to AO. Chromosomes with DAP >0.4 were considered oscillating (Stumpff et al., 2008). As shown in Fig. 5B, 28% chromosomes oscillate continuously, 68% undergo transitions between periods of oscillations and relative motionless, and 4% remain motionless throughout metaphase. The reason(s) for this variable behavior of congressed chromosomes, which are all expected to continuously maintain amphitelic attachments, remain unknown. We noticed that regular oscillations always begin after the centromere becomes stretched to ~1 µm which is consistent with the notion that oscillating chromosomes are stably attached to microtubules in the end-on fashion (Jaqaman et al., 2010). However, achieving the full stretch of the centromere is not sufficient to induce oscillation (Fig. 5A).
Descriptions of mitosis in newt lung cells demonstrate that microtubule capture leads to a rapid (~18 µm/min) gliding of the kinetochore along the captured microtubule towards the centrosome (Rieder and Alexander, 1990) (Skibbens et al., 1993). Chromosome gliding displaces the chromosome by ~10 µm on average (Skibbens et al., 1993). Rapid centromere gliding (~10 µm/min) was also observed in human U2-OS cells, although it appeared to affect less than 20% of chromosomes. The average displacement generated by the fast movement was not reported for human cells (Yang et al., 2008).
Our 4-D recordings reveal that ~75% kinetochores in RPE1 cells exceed momentous velocity of 8 µm/min at least once during the course of prometaphase and metaphase. Higher velocities up to 18 µm/min are also observed, but at a progressively lower frequencies (Fig. 5C). The periods of rapid movement are brief (5–15 sec) resulting in the average displacement of 0.93±0.44 µm, although in rare cases the centromere displaces up to 3-µm (Fig. 5D). An individual chromosome can undergo several rapid movements (Fig. 5E) which indicates that the initial interactions with microtubules often do not result in a stable attachment of the kinetochore. While the majority of the rapid movements (~60%) are observed within 5 min after NEB, some (~10%) can occur 5–10 min before AO when the metaphase plate is already fully formed. These late occurrences of rapid kinetochore movements generally correlate with centromere disorientation (see next chapter).
Surprisingly, most rapid kinetochore movements are not directed toward one of the centrosomes. As evident from the plot presented in Fig. S3C (arrows), fast movement can lead to a simultaneous decrease of the distances between the centromere and both centrosomes to a similar extent indicating that the chromosome moves to a position located near the middle of the nascent spindle. We used the ratio of kinetochore displacement toward different centrosomes to characterize the predominant direction of fast movement. This ratio is negative when the movement is directed toward one centrosome and away from the other. For centromeres that move towards both centrosomes to the same extent the ratio is 1. This metric reveals that ~50% of fast kinetochore movements (N=65) during early prometaphase are directed to center of the spindle with the ratios between 0.5 and 1.5 (SD = 0.25).
Together, these observations suggest that during spindle formation unattached kinetochores in RPE1 cells experience frequent albeit transient lateral interactions with spindle microtubules. These interactions do not result in a significant repositioning of the chromosome and only some of these interactions lead to a stable attachment.
Lateral interactions between kinetochores and microtubules pre-position and orient centromeres to foster formation of stable end-on attachments
Thus far our experiments reveal that during early prometaphase centromeres become positioned on the surface of the nascent spindle where the high density of microtubules results in numerous lateral interactions with the unattached kinetochores. To identify the aspects of spindle assembly that depend upon these lateral interactions during normal mitosis we compared the behavior of chromosomes in normal and Nuf2-depleted RPE1 cells. siRNA depletion of Nuf2, an NDC80-complex protein, has been shown to preclude formation of end-on microtubule attachments without significantly affecting lateral interactions (DeLuca et al., 2005) In fact, chromosomes can congress to a typical metaphase plate in cells co-depleted for Nuf2 and HSET (Cai et al., 2009). 3-D recordings in RPE1 cells with all kinetochores labeled via CENP-A-GFP expression demonstrate that the chromosome ring forms in Nuf2 depleted cells and it tends to persist longer than in untreated cells (Fig. S4).
Tracking individual photoactivated kinetochore pairs demonstrates that immediately prior to NEB the average distances between sister kinetochores is somewhat smaller in Nuf2-depleted (0.33±0.14 µm) than in control RPE1 cells (0.45±0.22 µm). During prometaphase these distances increase gradually until they reach plateaus approximately 10 min after NEB in both control and Nuf2-depleted cells (Fig. 6A–B). In agreement with previous studies (Cai et al., 2009; DeLuca et al., 2002) on average centromere stretching is greater by ~40% in controls (0.96±0.21 µm) than in Nuf2 depleted cells (0.62±0.2 µm) during late prometaphase. However, an important outcome of our time-resolved analysis is that even in the absence of end-on attachments interkinetochore distances progressively increase during early prometaphase. We also find the distribution of interkinetochore distances in Nuf2-depleted cells to be similar to that in control cells during early prometaphase when the chromosome ring is most prominent (Fig. S4C). This similarity is consistent with the notion that lateral interactions dominate during chromosome ring formation in control cells. Another interesting feature evident in trajectories of individual chromosomes is that transition from the low-stretch to high-stretch state usually occurs gradually over a period of several minutes both in control (Figs. 6C, S3C) and Nuf2 depleted cells (Fig. 6D) and this transition does not strictly correlate with achieving a stable orientation of the centromere.
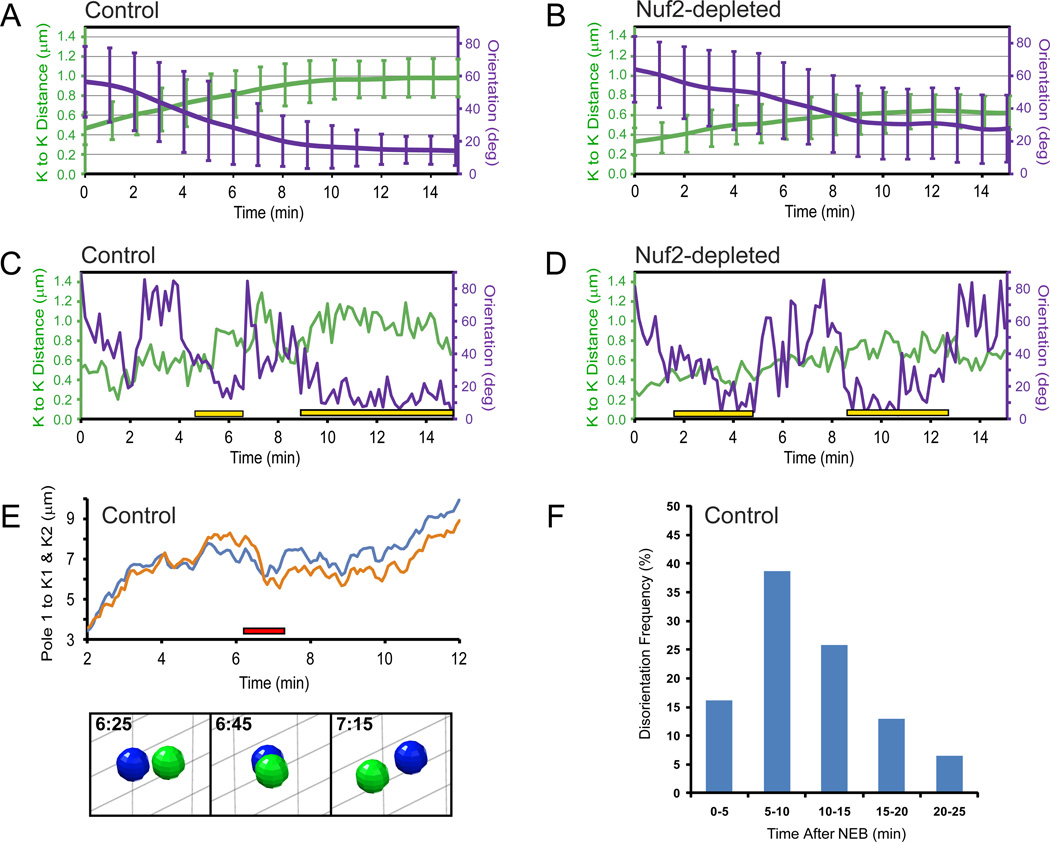
Figure 6. Centromere stretch and orientation during prometaphase.
(A–B) Changes in the average value of interkinetochore distance (green lines) and centromere orientation with respect to the spindle axis (violet lines) during first 15 min after NEB in control (A) and Nuf2-depleted (B) cells. (C–D) Examples of the changes in the interkinetochore distance and centromere orientation in control (C) and Nuf2-depleted (D) cells. Yellow bars denote periods when persistent, proper alignment of the centromere has been achieved. Notice that interkinetochore distances do not change when centromeres become disoriented. (E) An example of centromere re-orientation during normal prometaphase (same kinetochore pair as in (C)). The kinetochore oriented to the left at 6:25 becomes oriented to the right at 7:15 (images; also see Movie S7). Note that the re-orientation occurs when the centromere resides close to the spindle equator. (F) Frequency of centromere disorientations at different stages of spindle assembly.
In both control and Nuf2-depleted cells, centromeres are randomly oriented with respect to the axis of the forming spindle at NEB. Within the first 10 min of prometaphase the average angle between the lines connecting the centrosomes and the line connecting sister kinetochores decreases to ~15° in control and ~30° in Nuf2-depleted cells (Fig. 6A–B, violet). Thus, even in the absence of end-on microtubule attachments, centromeres become roughly oriented with respect to the spindle. However, analysis of chromosome trajectories demonstrates that the orientation of individual centromeres in Nuf2-depleted cells continues to fluctuate between periods of relative stability and ‘wobbling’ (Fig. 6D). These fluctuations are reflected in the standard deviation from the average angle that remains wide even as the average values gradually improve (Fig. 6B). Similar fluctuations are consistently observed during earlier prometaphase in control cells (cf. Fig 6A and C). In severe cases centromeres are seen to undergo a complete revolution so that the kinetochore that initially faces one centrosome becomes oriented toward the other centrosome (Fig. 6E and Movie S7). In other instances the original centromere orientation is restored after a period of ‘wobbling’. To quantify the frequency of centromere disorientations we determined the number of events when a centromere that has remained stably oriented for at least one minute (12 frames) lost its orientation by tilting more than 45° with respect to the spindle axis. By this criterion, ~42% of chromosomes (21/50) transiently lose their initial orientation while ~33% (7/21) of these chromosomes become disoriented more than once. Centromere disorientations are most frequent during early- to mid-prometaphase (Fig. 6F) although a significant number of them (~20%) occur later in mitosis when the metaphase plate is already fully formed (Fig. 6F).
Presence of laterally-attached kinetochores in a fully congressed metaphase plate
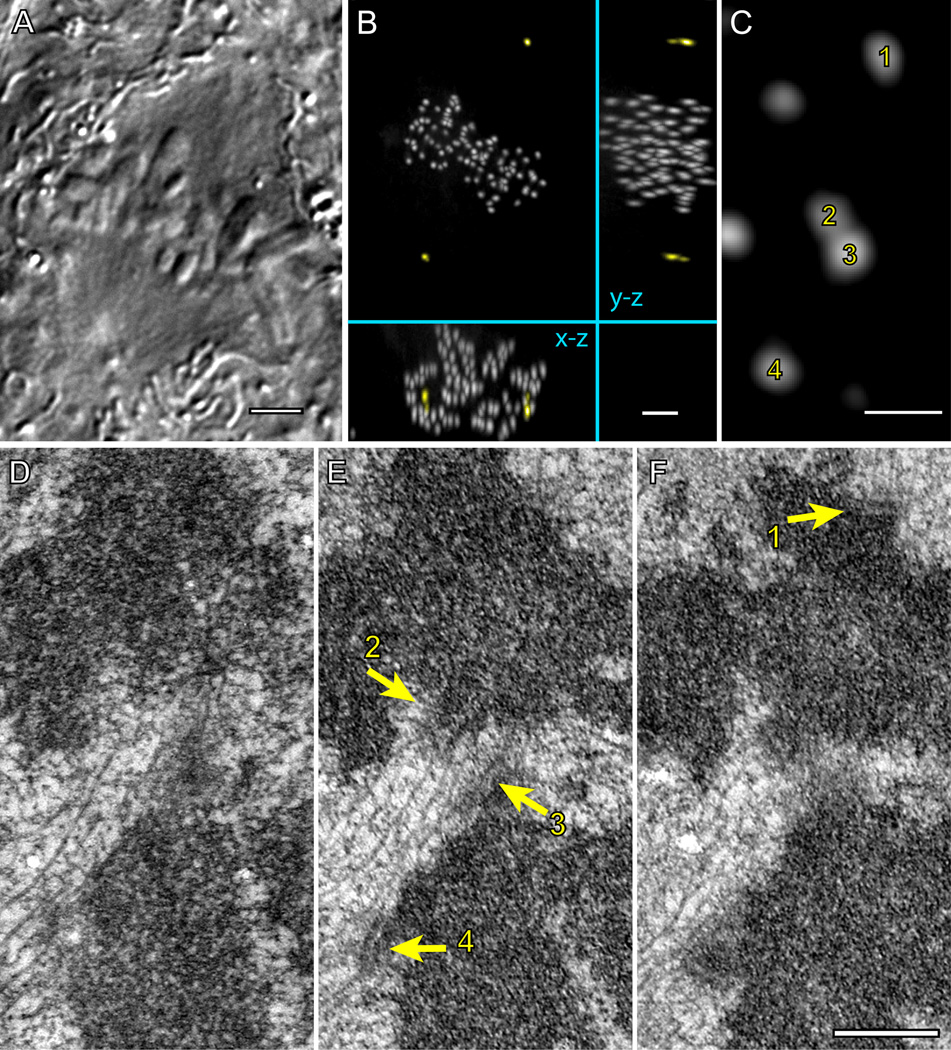
Our analysis of centromere stretch and orientation support that amphitelic attachment is not required for positioning the chromosome at the spindle equator. To investigate whether all chromosomes inside completely congressed metaphase plates are in fact attached to microtubules in amphitelic fashion we employed serial-section electron microscopy (EM). By correlating complete 3-D LM and EM datasets we were able to locate each of the 92 kinetochores in a metaphase cell.
The reconstructed cell is in late metaphase with all chromosomes fully congressed (Fig. 7A) and the kinetochores are uniformly distributed in the central part of the spindle characteristic of late metaphase. Expectedly, most of the 92 kinetochores are properly attached to prominent K-fibers with microtubules terminating within the kinetochore plate (e.g., kinetochores 1, 2, and 4; Fig. 7 D–F). However, three chromosomes lack amphitelic attachment. In each of these instances one sister kinetochore is attached to microtubules in an end-on fashion, while the other kinetochore only laterally interacts with microtubules of a K-fiber that terminates in a kinetochore on a different chromosome (kinetochore 3; Fig. 7 D–F). This configuration has been previously observed only during congression of monooriented chromosomes but not inside the metaphase plate (Kapoor et al., 2006). It is noteworthy that in two cases the laterally-attached kinetochores are completely shielded from one of the spindle poles by arms of other chromosomes. This steric impediment prevents a direct microtubule connection to the spindle pole.
Figure 7. Fully congressed chromosomes can lack amphitelic attachment.
DIC image (A) and maximal-intensity XY, XZ, and YZ projections of GFP fluorescence (B) of a fixed metaphase RPE1 cell expressing centrin1-GFP and CENP-A-GFP. (C) A higher-magnification view (XY projection) showing two pairs (1–2 and 3–4) of sister chromosomes positioned within the metaphase plate. (D–F) Serial 70-nm thin sections through the area presented in (C) demonstrate that kinetochores 1, 2, and 4 are attached to microtubule in the end-on fashion, which implies that the chromosome in the top half of the image is amphitelic. In contrast, kinetochore 3 lacks end-on attachment and it is shielded from the top spindle pole by a mass of chromatin positioned in front of the kinetochore. This kinetochore laterally interacts with microtubules of the K-fiber that terminates within kinetochore 2. Bars in A and B, 5 µm. Bars in C–F, 0.5 µm.
Discussion
An emerging theme in the field of cell division is that severe mitotic abnormalities, such as formation of persistently multipolar spindle (Ganem et al., 2009) or significant prolongation of mitosis (Uetake and Sluder, 2010) lead to the formation of non-viable progeny. In contrast, seemingly mild deficiencies in the mechanisms facilitating mechanisms profoundly affect the fidelity of chromosome segregation and the fate of the daughter cells (Chandhok and Pellman, 2009; Thompson et al., 2010). For example, transient deviations from the bipolar spindle geometry during prometaphase or subtle changes in the stability of kinetochore microtubules have been shown to cause chromosomal instability (Bakhoum et al., 2009; Ganem et al., 2009; Silkworth et al., 2009). Thus, it is critical to reveal the exact pathways responsible for the timely assembly of the spindle and accurate establishment of proper kinetochore attachments. Our approach of tracking individual spindle components in 3-D throughout mitosis allows us shed new light on this issue. The rationale is that different spindle assembly mechanisms results in distinct chromosome behavior. Thus, contributions of the mechanisms governing normal spindle assembly can be inferred from the analyses of the unique route taken by each chromosome during mitosis.
A major finding of our work presented here is that the great majority of chromosomes in normal human cells become instantaneously bioriented (positioned close to the spindle equator) from the onset of mitosis and they remain in this locale until anaphase. Interestingly, centromeres of these bioriented chromosomes frequently ‘wobble’ indicating that they have not achieved stable amphitelic attachment. This notion gains support from the EM data that many kinetochores in the middle of the spindle interact with microtubules only in a lateral fashion. Some of these laterally-attached kinetochores can even be found in a mature metaphase plate. While it has been shown that chromosomes can in principle congress in the absence of end-on attachments (Cai et al., 2009) the functional significance of this mechanism remained ambiguous. We find that most chromosomes normally achieve biorientation prior to formation of stable amphitelic attachments lateral interactions make a major contribution during normal spindle assembly.
Instantaneous biorientation can only be achieved if both kinetochores reside in an area with extremely high microtubule density and are not shielded by other chromosomes. Such a condition is not possible when chromosomes are randomly distributed in the relatively small space formerly occupied by the nucleus (Paul et al., 2009). The reproducible pattern of chromosome and centrosome movement observed during early prometaphase provides a straightforward explanation how the chromosome shielding constraint is overcome.
Arrangement of chromosomes in a ring around the spindle during prometaphase has long been known to exist in a variety of cell types (Chaly and Brown, 1988; Mosgoller et al., 1991) although the functional significance of this distribution remained ambiguous. The ring has been suggested to provide a means for non-random distribution of chromosomes into daughter cells (Nagele et al., 1995). However, data indicating chromosomes are arranged randomly within the ring do not support this hypothesis (Allison and Nestor, 1999).
We find that formation of the ring depends on the spindle ejection force (Rieder and Salmon, 1994) which is mediated by plus-end-directed motor activity of kinesin-10 (Levesque and Compton, 2001). Until now the role of the spindle ejection force remained poorly understood. Originally thought to provide spatial cues for chromosome congression (Khodjakov et al., 1999; Rieder et al., 1986) the spindle ejection force was left without a clear function due to demonstration of normal chromosome congression upon experimental inhibition of the spindle ejection force (Levesque and Compton, 2001). Our data suggest that the spindle ejection force functions to efficiently expel the chromosome arms from the center of the nascent spindle. This, combined with centripetal forces acting on the centromeres, positions the kinetochores on the surface of the nascent spindle where they are exposed to a high density of microtubules from both spindle poles. Consistent with the notion that early prometaphase is dominated by lateral interactions between kinetochores and microtubules we find that the chromosome ring forms in Nuf2-depleted cells where kinetochores are not capable of stable end-on microtubule attachments.
Assembly of a compact spindle densely packed with microtubules appears to be the key for efficient spindle assembly. The high density of microtubules between the centrosomes is likely to be established initially by the preferential growth of microtubules towards the high concentration of RanGTP inside the volume formerly occupied by the nucleus (O'Connell et al., 2009). In this mechanism, microtubule density would be particularly high within the spindle if at NEB the centrosomes reside on the opposite sides of the nucleus which according to our 3-D recordings occurs in the great majority of RPE1 cells. It would be extremely interesting to determine whether the efficiency of spindle assembly and/or the fidelity of chromosome segregation are compromised in cells that naturally fail to separate the centrosome prior to NEB (see Toso et al., 2009).
In summary, our work reveals a new mechanism that facilitates S&C by pre-positioning spindle components so that kinetochores can more easily establish end-on microtubule attachments. This was made possible by two technological breakthroughs: 1) Continuous tracking of an individual chromosome from the onset of mitosis to anaphase; 2) Following spindle formation in true 3-D space at high temporal and spatial resolutions. These advancements allowed us for the first time to reconstruct the path taken by a typical chromosome during spindle assembly by averaging the unique trajectories of randomly selected chromosomes. The data presented here establish the baseline of normal chromosome behavior which will be invaluable in the future examinations of pathological conditions arising from the deficiencies in key proteins involved in mitosis.
Methods
Cell culture and generation of stable cell lines
RPE1 cells (Clontech) were grown in DMEM supplemented with 10% FCS (Invitrogen) at 37°, 5% CO2. To generate cells with fluorescent kinetochores and centrosomes cells were first transfected with CENP-A-PAGFP in LentiLox 3.1. Individual clones selected for the desired expression level were subsequently transfected with centrin1-GFP. This approach allowed us to ensure that the intensity of individual kinetochores after photoactivation is comparable with the intensity of GFP-labeled centrioles. A similar strategy was used to construct RPE1 cells co-expressing GFP-CENP-A + centrin1-GFP, and GFP-CENP-A + centrin1-tdTomato. For high-resolution imaging, cells were grown on glass coverslips to sub-confluence and mounted in Rose chambers containing CO2-independent medium (Invitrogen) supplemented with 10% FCS and antibiotics.
Protein inactivation
Oligofectamine (Invitrogen) was used for siRNA transfections according to the manufacturer’s protocol. Cells were analyzed 36–72h after transfection. Target sequences are described elsewhere (Tokai-Nishizumi et al., 2005 for Kid) and (DeLuca et al., 2002 for Nuf2). Efficiency of siRNA depletions was confirmed by antibody staining (anti-Hec1 antibody was used for Nuf2 depletion). In Nuf2 experiments only cells that failed to form a tight metaphase plate were analyzed.
Alternatively, Kid was inactivated via microinjection of a function-blocking antibody raised against the DNA-binding domain of the molecule (Levesque and Compton, 2001). The antibody was purified and injected into the nucleus as in Levesque and Compton (2001) except the injections were conducted during prophase and the antibody concentration in the needle was 16.8 mg/ml.
Photoactivation and analysis of kinetochore/centrosome trajectories
Individual kinetochore pairs were photoactivated with 405-nm diode laser (OZ-2000, Oz Optics, Ottawa, Ontario). Details of the microscopy workstation and laser coupling are described elsewhere (Magidson et al., 2007). Briefly, the collimated beam was steered through a dedicated epi-port of a Nikon TE-2000E PFS microscope and focused by a 100X Plan Apo, N.A. 1.4 oil immersion objective lens. Images were recorded in spinning-disk confocal mode (CSU-10, Yokogawa, Tokyo, Japan) on a back-illuminated Cascade 512B EM CCD camera (Photometrics). Kinetochores were activated during late G2 or prophase and the recordings were initialized shortly before nuclear envelope breakdown. 17 focal planes at 0.5-µm Z-steps were recorded at each time point.
To decrease unnecessary exposure of cells to light we introduced a shutter override into automatic image acquisition. In those instances when centrosomes and kinetochores were positioned at similar depth, excitation light was blocked once in-focus images of all objects had been recorded. Details of this approach are presented elsewhere (Schilling et al., manuscript under review).
Determining complete 3-D coordinates requires that the objects do not overlap in two of the three possible orthogonal projections (XY, XZ, and YZ). Due to low number of objects this condition was always satisfied in our datasets. Centroids of each mother centriole and each kinetochore were determined manually in ImageJ (NIH, Bethesda, MA) and the 3-D coordinates extracted in MatLab (Mathworks, Natick, MA). The results were validated by superimposing the final 3-D trajectories over the original time-lapse movies in an in-house written MatLab viewer. MatLab code used for visualization and analysis is available upon request.
Deviation from Average Position (DAP) was calculated as described in Stumpff et al., (2008). For each chromosome DAP was calculated in a series of 5-min windows that span from −17 to −2 min prior to AO. To classify oscillating and non-oscillating chromosomes we used a threshold DAP value of 0.4. This threshold was chosen based on the demonstration that overexpression of the kinesin Kif18A in HeLa cells abrogates chromosome oscillation and changes DAP from 0.46±0.02 to 0.31±0.01(Stumpff et al., 2008).
Correlative electron microscopy
Cells were fixed in 2.5% glutaraldehyde (Sigma). DIC and fluorescence images were acquired at 0.1-µm Z steps through the entire cell volume immediately after fixation. Post-fixation, embedding, and sectioning were done as previously described (Rieder and Cassels, 1999). 80 nm-thin sections were imaged on a Zeiss 910 microscope operated at 80 kV. Scaling and alignment of LM and EM images were done manually using Photoshop. Correlation of conspicuous morphological features between DIC and EM images was used to match the orientation and Z-positions for individual focal planes and then fluorescence images were overlaid on the EM reconstruction to determine exact positions of kinetochores.
Fixation and Immunofluorescence
Cells were pre-extracted in warm PEM buffer (100 mM PIPES pH 6.9, 2.5 mM EGTA, 5 mM MgCl2) supplemented with 0.5% Triton-X100 for 1 min and fixed with 1–2% glutaraldehyde for 10 minutes in PEM. Microtubules were visualized with DM1A monoclonal anti-α-tubulin antibody (Sigma). Hoechst 33343 was used to stain DNA (chromosomes).
Amira software (Visage Imaging) was used for surface rendering. To display centrioles and kinetochores in different colors, it was necessary to separate them in the imported images by masking either centrin1-GFP or CENP-A-GFP containing structures.
Computational modeling
We considered the nuclear space to be an oblate spheroid with dimensions 14 µm ×14 µm × 7 µm (based on dimensions gleaned from experimental images), with 2 centrosomes at the poles of the spheroid and 46 chromosomes (92 kinetochores) inside. The chromosomes were either distributed in the nuclear space uniformly, or concentrated in the ring (toroid) with inner radius 4 µm and outer radius 7 µm. The chromosome arms were allowed to slightly overlap (due to their elasticity). In the course of the simulations, the chromosomes neither moved nor rotated. Chromosomes and kinetochores were cylindrical objects with dimensions given below. During the search, each centrosome nucleated 150 microtubules in random directions undergoing dynamic instability with the growth and shortening rates shown below. There were neither rescues, nor spontaneous catastrophe events. The microtubules were undergoing a catastrophe immediately if growing outside the nuclear space, or when hitting a chromosome arm; the microtubules did not turn. When a microtubule encountered a kinetochore, the microtubule was stabilized and the capture took place. Stochastic Monte Carlo simulations using this algorithm and parameters below were performed as described by Paul and coworkers (2009). The results of the simulations were obtained from running each search for 4 min (of physical, not computer time), for 100 times, and then by averaging.
Parameters used in the simulations
Number of chromosomes = 46; Number of MTs from each pole = 150; KT length = 0.35 µm; KT diameter = 0.35 µm; Chromosome diameter = 1.5 µm; Chromosome length = 4 µm; MT growth rate = 0.35 µm/s; MT shortening rate = 1 µm/s.
Highlights.
-
➢
During early prometaphase kinetochores are arranged on the surface of the spindle
-
➢
This arrangement is driven by chromokinesin-mediated ejection of chromosome arms
-
➢
Chromosome pre-positioning increases the efficiency of microtubule search & capture
-
➢
Stable amphitelic attachments are formed during late prometaphase and metaphase
Supplementary Material
Acknowledgements
This work was supported by NIH grants GM059363 (A.K.), GM068952 (A.M.), and a Kirschstein National Research Service Award GM077911 to C.B.O. We acknowledge use of the Wadsworth Center’s electron microscopy core facility and the Ordway Research Institute’s Flow Cytometry Core. A special thanks to Dr. Duane Compton (Dartmouth Medical School) for his generous donation of anti-KID serum.
Footnotes
Author Contributions
V.M., C.B.O., and A.K. designed the experiments. V.M., CBO, and J.L. performed the experiments. R.P and A.M developed the computational model. V.M. designed the MatLab tools to visualize and numerically analyze kinetochore/centrosome trajectories. All authors analyzed and interpreted the data. The manuscript was written primarily by C.B.O and A.K. with significant input from V.M. and other authors.
References
- Allison DC, Nestor AL. Evidence for a relatively random array of human chromosomes on the mitotic ring. J Cell Biol. 1999;145:1–14. doi: 10.1083/jcb.145.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Genovese G, Compton DA. Deviant kinetochore-microtubule dynamics underlie chromosomal instability. Current Biology. 2009;19:R1032–R1034. doi: 10.1016/j.cub.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy A, Lane HA, d'Herin P, Harper M, Kress M, Nigg EA. Phosphorilation by p34 cdc2 regulates spindle assosiation of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- Cahu J, Olichon A, Hentrich C, Schek H, Drinjakovic J, Zhang C, Doherty-Kirby A, Lajoie G, Surrey T. Phosphorylation by Cdk1 increases the binding of Eg5 to microtubules in vitro and in Xenopus egg extract spindles. PLoS ONE. 2008;3:e3936. doi: 10.1371/journal.pone.0003936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, O'Connell CB, Khodjakov A, Walczak CE. Chromosome congression in the absence of kinetochore fibers. Nature Cell Biology. 2009;11:832–838. doi: 10.1038/ncb1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaly N, Brown DL. The prometaphase configuration and chromosome order in early mitosis. Journal of Cell Science. 1988;91:325–335. doi: 10.1242/jcs.91.3.325. [DOI] [PubMed] [Google Scholar]
- Chandhok NS, Pellman D. A little CIN may cost a lot: revisiting aneuploidy and cancer. Current Opinion in Genetics & Development. 2009;19:74–81. doi: 10.1016/j.gde.2008.12.004. [DOI] [PubMed] [Google Scholar]
- DeLuca JG, Dong Y, Hergert P, Strauss J, Hickey JM, Salmon ED, McEwen BF. Hec1 and Nuf2 Are Core Components of the Kinetochore Outer Plate Essential for Organizing Microtubule Attachment Sites. Molecular Biology of the Cell. 2005;16:519–531. doi: 10.1091/mbc.E04-09-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol. 2002;159:549–555. doi: 10.1083/jcb.200208159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Vale RD. Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Molecular Biology of the Cell. 2005;16:3896–3907. doi: 10.1091/mbc.E05-02-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon ED, McEwen BF, Khodjakov A. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006;311:388–391. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Gabashvili IS, Rieder CL. "Dumb" versus "smart" kinetochore models for chromosome congression during mitosis in vertebrate somatic cells. Cell Motility & the Cytoskeleton. 1999;43:179–185. doi: 10.1002/(SICI)1097-0169(1999)43:3<179::AID-CM1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Kirschner M, Mitchison T. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- Lenart P, Daigle N, Hand AR, Eils R, Terasaki M, Ellenberg J. A contractile nuclear actin network drives chromosome congression in oocytes. Nature. 2005;436:812–818. doi: 10.1038/nature03810. [DOI] [PubMed] [Google Scholar]
- Levesque AA, Compton DA. The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J Cell Biol. 2001;154:1135–1146. doi: 10.1083/jcb.200106093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V, Loncarek J, Hergert P, Rieder CL, Khodjakov A. Laser microsurgery in the GFP era: A cell biologist's perspective. In: Berns MW, Greulich KO, editors. Laser Manipulations of Cells and Tissues. Elsevier; 2007. pp. 237–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca TJ, Salmon ED. Welcome to a new kind of tension: translating kinetochore mechanics into a wait-anaphase signal. Journal of Cell Science. 2010;123:825–835. doi: 10.1242/jcs.064790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosgoller W, Leitch AR, Brown JK, Heslop-Harrison JS. Chromosome arrangements in human fibroblasts at mitosis. Human Genetics. 1991;88:27–33. doi: 10.1007/BF00204924. [DOI] [PubMed] [Google Scholar]
- Nagele R, Freeman T, McMorrow L, Lee HY. Precise spatial positioning of chromosomes during prometaphase: evidence for chromosomal order. Science. 1995;270:1831–1835. doi: 10.1126/science.270.5243.1831. [DOI] [PubMed] [Google Scholar]
- Nezi L, Musacchio A. Sister chromatid tension and the spindle assembly checkpoint. Current Opinion in Cell Biology. 2009;21:785–795. doi: 10.1016/j.ceb.2009.09.007. [DOI] [PubMed] [Google Scholar]
- O'Connell CB, Loncarek J, Kalab P, Khodjakov A. Relative contributions of chromatin and kinetochores to mitotic spindle assembly. J Cell Biol. 2009;187:43–51. doi: 10.1083/jcb.200903076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Wollman R, Silkworth WT, Nardi IK, Cimini D, Mogilner D. Computer simulations predict that chromosome movements and rotations accelerate mitotic spindle assembly without compromising accuracy. Proceedings of National Academy of Sciences USA. 2009;106:15708–15713. doi: 10.1073/pnas.0908261106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Alexander SP. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J Cell Biol. 1990;110:81–95. doi: 10.1083/jcb.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Cassels G. Correlative light and electron microscopy of mitotic cells in monolayer cultures. Methods in Cell Biology. 1999;61:297–315. doi: 10.1016/s0091-679x(08)61987-1. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Davison EA, Jensen LC, Cassimeris L, Salmon ED. Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half-spindle. J Cell Biol. 1986;103:581–591. doi: 10.1083/jcb.103.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos UP. Light and electron microscopy of rat kangaroo cells in mitosis. I. Formation and breakdown of the mitotic apparatus. Chromosoma. 1973;40:43–82. doi: 10.1007/BF00319836. [DOI] [PubMed] [Google Scholar]
- Rosenblatt J. Spindle assembly: asters part their separate ways. Nature Cell Biology. 2005;7:219–222. doi: 10.1038/ncb0305-219. [DOI] [PubMed] [Google Scholar]
- Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS ONE. 2009;4:e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens RV, Rieder CL, Salmon ED. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J Cell Biol. 1993;122:859–875. doi: 10.1083/jcb.122.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Developmental Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Bakhoum SF, Compton DA. Mechanisms of chromosomal instability. Current Biology. 2010;20:R285–R295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokai-Nishizumi N, Ohsugi M, Suzuki E, Yamamoto T. The chromokinesin Kid is required for maintenance of proper metaphase spindle size. Mol Biol Cell. 2005;16:5455–5463. doi: 10.1091/mbc.E05-03-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokai N, Fujimoto-Nishiyama A, Toyoshima Y, Yonemura S, Tsukita S, Inoue J, Yamamota T. Kid, a novel kinesin-like DNA binding protein, is localized to chromosomes and the mitotic spindle. EMBO J. 1996;15:457–467. [PMC free article] [PubMed] [Google Scholar]
- Toso A, Winter JR, Garrod AJ, Amaro AC, Meraldi P, McAinsh AD. Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. J Cell Biol. 2009;184:365–372. doi: 10.1083/jcb.200809055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y, Sluder G. Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Current Biology. 2010;20:1666–1671. doi: 10.1016/j.cub.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uteng M, Hentrich C, Miura K, Bieling P, Surrey T. Poleward transport of Eg5 by dynein-dynactin in Xenopus laevis egg extract spindles. J Cell Biol. 2008;182:715–726. doi: 10.1083/jcb.200801125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE, Cai S, Khodjakov A. Mechanisms of chromosome behaviour during mitosis. Nature Reviews Molecular Cell Biology. 2010;11:91–102. doi: 10.1038/nrm2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead CM, Winkfein RJ, Rattner JB. The relationship of HsEg5 and the actin cytoskeleton to centrosome separation. Cell Motility & the Cytoskeleton. 1996;35:298–308. doi: 10.1002/(SICI)1097-0169(1996)35:4<298::AID-CM3>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Wollman R, Cytrynbaum EN, Jones JT, Meyer T, Scholey JM, Mogilner A. Efficient chromosome capture requires a bias in the 'search-and-capture' process during mitoticspindle assembly. Current Biology. 2005;15:828–832. doi: 10.1016/j.cub.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Yang Z, Loncarek J, Khodjakov A, Rieder CL. Extra centrosomes and/or chromosomes prolong mitosis in human cells. Nature Cell Biology. 2008;10:748–751. doi: 10.1038/ncb1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.