SYNOPSIS
Ubiquitin-like protein, containing PHD and RING finger domains-1 (UHRF1) is required for cell cycle progression and epigenetic regulation. In this study, we show that depleting cancer cells of UHRF1 causes activation of the DNA damage response pathway, cell cycle arrest in G2/M and apoptosis dependent on caspase-8. The DNA damage response in cells depleted of UHRF1 is illustrated by: phosphorylation of histone H2AX on serine 139, phosphorylation of CHK2 on threonine 68, phosphorylation of CDC25 on serine 216 and phosphorylation of CDK1 on tyrosine 15. Moreover, we find that UHRF1 accumulates at sites of DNA damage suggesting that the cell cycle block in UHRF1 depleted cells is due to an important role in damage repair. The consequence of UHRF1 depletion is apoptosis: cells undergo activation of caspases 8 and 3 and depletion of caspase-8 prevents cell death induced by UHRF1 knock-down. Interestingly, the cell cycle block and apoptosis occurs in p53 containing and deficient cells. From these studies we conclude that UHRF1 links epigenetic regulation with DNA replication.
Keywords: Cell cycle, UHRF1, apoptosis, caspase, phosphorylation, methylation
INTRODUCTION
Following genotoxic injury, the regulated process of mammalian cell cycle progression is often halted. This arrest occurs through the activation of checkpoints that result in cell cycle blocks, allowing for damage repair or cell death if the damage is irreparable [1]. The consequence of abrogating such checkpoints includes proliferation of cells with defective DNA content, a frequent cause of carcinogenesis.
UHRF1 (ubiquitin-like protein, containing PHD and RING finger domains-1) was identified as a factor that binds the inverted CCAAT box in the topoisomerase IIα promoter and regulate its expression [2]. However, subsequent studies have not demonstrated that UHRF1 functions as a transcription factor. Instead, UHRF1 functions to regulate gene expression through epigenetic mechanisms including DNA methylation [3, 4], histone deacetylation [5], histone methylation [6] and possibly, histone ubiquitination [7]. UHRF1 is a multi-domained protein (Figure 1A) that contains 1) an amino-terminal ubiquitylation-like domain 2) a plant homeodomain , through which it interacts with methylated histones [8, 9], retinoblastoma protein [10] and DNA methyltransferase 1 (DNMT1) [3], 3) a set and ring finger associated domain (SRA) that interacts with hemimethylated DNA [11–13], and histone deacetylase 1[5] and 4) a ring finger motif that has an E3 ubiquitin-ligase activity [7, 14]. Therefore, UHRF1 is thought to regulate gene expression through epigenetic mechanisms.
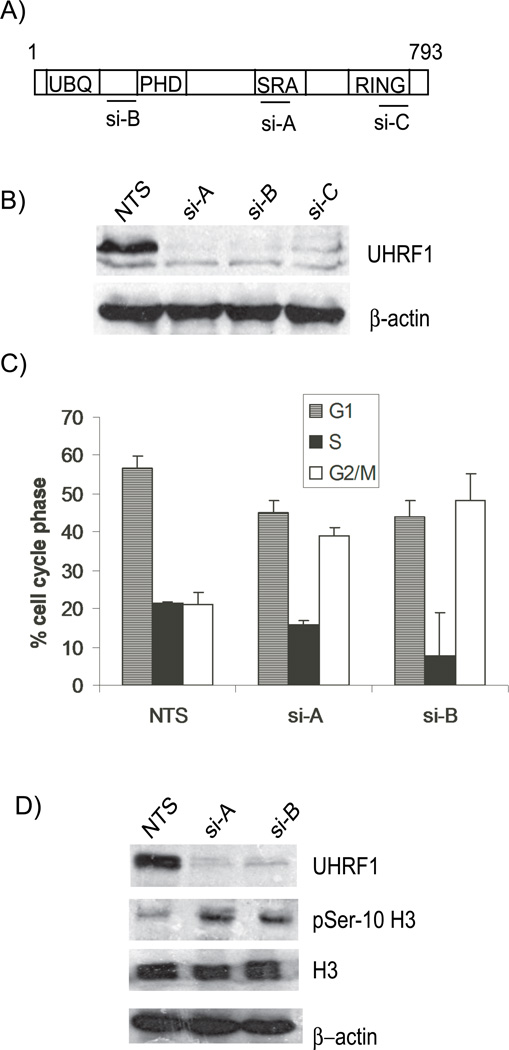
Figure 1. Depletion of UHRF1 results in a G2/M Block.
A). UHRF1 is a multi-domained protein with ubiquitin, PHD, SRA and RING finger domains. The location of UHRF1 siRNAs: si-A, si-B, and si-C target are shown B). Western immunoblot of HCT116 protein lysates from cells transfected with non-targeting siRNA (NTS) or UHRF1 targeting siRNA (si-A, si-B and si-C), show efficient knockdown of UHRF1 with targeting siRNAs. C). Depletion of UHRF1 leads to a G2/M arrest. Percentage of cells in G1 (hatched), S (black) and G2/M (white) in cells transfected with NTS, si-A or si-B. Error bars represent standard deviations from the mean of three experiments. D) Enhanced serine 10 phospho histone H3 levels in UHRF1 knockdown cells. Depletion of UHRF1 (lanes 2 and 3) leads to increased levels of phosphorylated histone H3 (panel 2). Total histone H3 levels are constant. E) There is no concurrent G1 arrest in UHRF1 depleted cells. The remnant G1 population in UHRF1 containing and deficient cells (top histograms) progress to the nocodazole-induced mitotic block (bottom histograms). F) The level of G1 and S cyclins: D2, E and A remain stable in UHRF1 depleted cells (top three panels) while cyclin B1 level is elevated (panel 4). β-actin is used as a loading control for all experiments.
UHRF1 is required for cell cycle progression of non-cancerous cells. UHRF1 mRNA and protein fluctuate with the cell cycle [15, 16], depletion of UHRF1 abrogates S phase entry [17] and zebrafish with a loss of function mutation in uhrf1 have defects in hepatocyte proliferation and increased apoptosis [15]. In cancer cells, UHRF1 levels are high and the protein is equally expressed in all phases of the cell cycle [10, 16, 18]. However, reports on the effects of UHRF1 depletion in cancer cells have been varied. For example, siRNA mediated knockdown of UHRF1 in HeLa cells concurrently treated with adriamycin causes a small percentage of cells to arrest in G1[18]. However, in H1299 cells a modest two fold knockdown of UHRF1 by shRNA causes cells to arrest in either G1 or G2/M [14]. Regardless of these differences, it is clear that cell cycle progression requires UHRF1 [10, 18]. These data establish the possibility that depleting cancer cells of UHRF1 may lead to cell death.
Recent studies show that UHRF1 functions to conserve epigenetic inheritance [3, 4]. UHRF1 interacts with DNMT1which methylates cytosines on CpG islands of hemimethylated DNA. UHRF1 also interacts with hemimethylated DNA allowing the methyl cytosine of the parent strand to “flip out” of the double helix so that DNMT1 can access the unmethylated cytosine on the daughter strand [11–13]. Indeed, depletion of UHRF1 prevents the association of DNMTI with the chromatin leading to hypomethylation of many genes [3]. A role for UHRF1 in maintaining genomic integrity has been suggested in experiments that show that cells lacking UHRF1 are hypersensitive to DNA damage by genotoxic agents [19]. Furthermore, DNMT1, which interacts with UHRF1, accumulates at sites of DNA damage [20]. Lastly, the inactivation of DNMT1 in HCT116 cells leads to activation of the DNA damage response pathway and a G2/M block [21]. These studies support the hypothesis that proper UHRF1 function is required for genomic fidelity.
In this study, we test this hypothesis by depleting UHRF1 from cancer cells and investigating the effects on the cell cycle. We show that UHRF1 depleted cells undergo a caspase-8 mediated apoptosis and that cell cycle arrest and cell death in response to UHRF1 knock-down cells does not require p53. Moreover, we find that UHRF1 accumulates rapidly at sites of DNA injury. Together, these data support a model in which UHRF1 is required for genomic fidelity and its loss causes activation of the DNA damage response and cell death.
EXPERIMENTAL PROCEDURES
Materials and additional experimental procedures are provided in the accompanying supplementary information.
Fluorescent activated-cell sorting (FACS) analysis
Cell cycle analysis was performed as previously described [22]. Briefly, following fixation and propidium iodide (PI) staining, PI positive cells were sorted and histograms were analyzed using Modfit LT (version 3.0, Verity Softaware House, Inc). For nocodazole treatment, cells were transfected with control or UHRF1 targeting siRNA for 24 hours and then incubated with or without nocodazole (40 ng/ml) for an additional 24 hours. Cells were then collected for FACS analysis.
UVA-Laser-scissors induced DNA injury
HeLa cells were treated with 10 µM 5-iodo-2-deoxyuridine (Sigma; St. Louis, MO) for 24h prior to laser irradiation. LabTek chambers were mounted on a Zeiss Axiovert 200 microscope integrated with the P.A.L.M Microlaser workstation (P.A.L.M. Laser Technologies, Bernried, Germany). Cells were visualized under visible light and laser targeted nuclei selected using the supplied software. A pulsed UVA-laser (30 Hz, 337 nm) coupled to the bright field path of the microscope was focused through a LD 40x; NA 0.6 Zeiss Achroplan objective to yield a spot size of approximately 1 µm. Nuclei were subsequently irradiated with a pulsed solid-state UVA-laser (30 Hz, 337 nm) with following settings a) Energy: 35, b) Focus: 57 and c) Cut speed between 10–15 with laser output set to 50%. An average of 100 cells were micro-irradiated within 2–5 min, and cell nucleus exposed to the laser beam for less than 500-ms.
RESULTS
UHRF1 depletion arrests cells in G2/M
UHRF1 is expressed at high levels in many cancer cell lines and primary tumors [14, 16]. Studies have suggested that loss of UHRF1 prevents cell cycle progression and thus UHRF1 may be a potential target for decreasing tumor growth in vivo. To characterize the cell cycle behavior of colorectal cancer cells (HCT116) depleted of UHRF1, we used small interfering RNA (siRNA) to knockdown UHRF1. Three siRNAs (si-A, si-B, and si-C) targeting distinct regions of UHRF1 (Figure 1A) effectively depleted UHRF1 as assessed by Western blot analysis (Figure 1B). Similar UHRF1 depletion was achieved in liver tumor cells cell lines (Huh7 and Hep3B) and breast cancer cell lines (MCF7) transfected with these siRNAs (not shown). To control for potential off target effects, the majority of our studies were carried out using two different UHRF1 targeted siRNAs (si-A and si-B).
We evaluated the relationship between UHRF1 depletion and cell cycle progression. Cells harvested 48 hours post-transfection were processed for FACS analysis to assess cell cycle stage. As shown in Figure 1C, depletion of UHFR1 in HCT116 cells led to a doubling in the percentage of cells in the G2/M phase. Western immunoblots (Figure 1D), show a corresponding increase of serine 10 phosphorylated histone H3, confirming a block in mitosis. In Hep3B cells depleted of UHRF1 we see a similar pattern (Figure S1A, supplementary information); indicating the G2/M cell cycle arrest is not cell type specific.
Previous studies in HeLa cells with reduced UHRF1 levels showed that a small percentage of cells remain blocked in G1 following DNA damage [18]. We reasoned that if loss of UHRF1 causes a G1 arrest in HCT116, some UHRF1 depleted cells should be retained in G1 when treated with nocodazole, a drug that induces cell arrest in prometaphase. FACS analyses revealed no difference between UHRF1 depleted and control cells: in both cases, the G1 population progressed to G2/M where they arrested (Figure 1E). Consistent with this, Western immunoblots showed no changes in the levels of G1 or S phase cyclins, indicating that there is no cell cycle block in G1 or S. Instead, we saw an increase in the mitotic cyclin B1 (Figure 1F), suggesting that UHRF1 depletion caused a block after cyclin B synthesis, but before its destruction in mid-mitosis [23, 24]. These experiments confirm that there is no G1 arrest in UHRF1 depleted HCT116 cells. This observation is reminiscent of the phenotype observed in these same cells depleted of DNMT1 [21].
Loss of UHRF1 activates the DNA damage and response pathway
Cyclin dependent kinases are regulators of cell cycle progression [23]. Because cyclin dependent kinase 1 (CDK1, also called Cdc2) is required for progression of cells from G2 into and through mitosis, we questioned whether the G2/M block was associated with evidence of CDK1 inhibition. Figure 2A shows that inhibitory phosphorylation of CDK1 on tyrosine-15 is enhanced in HCT116 cells depleted of UHRF1, while total CDK1 remains constant (compare panel 2 to 3, Figure 2A). Similar elevation of tyrosine-15 CDK1 phosphorylation was also seen in Hep3B cells transfected with UHRF1 siRNA (Figure S1B, supplementary information). Taken together, these data indicate that in at least two different cancer cell lines depletion of UHRF1 caused cell cycle arrest in the G2/M phase.
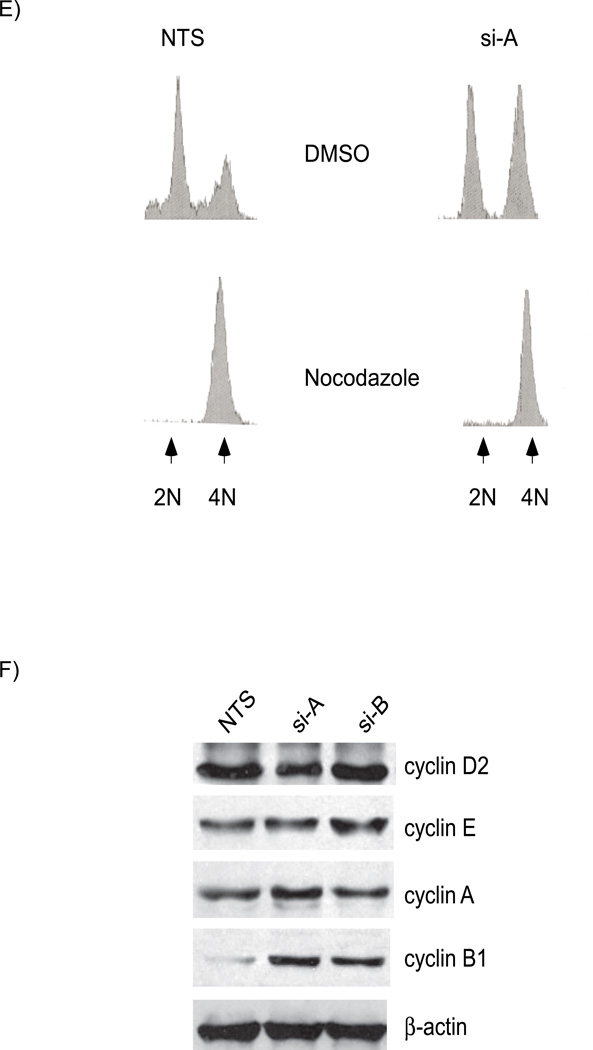
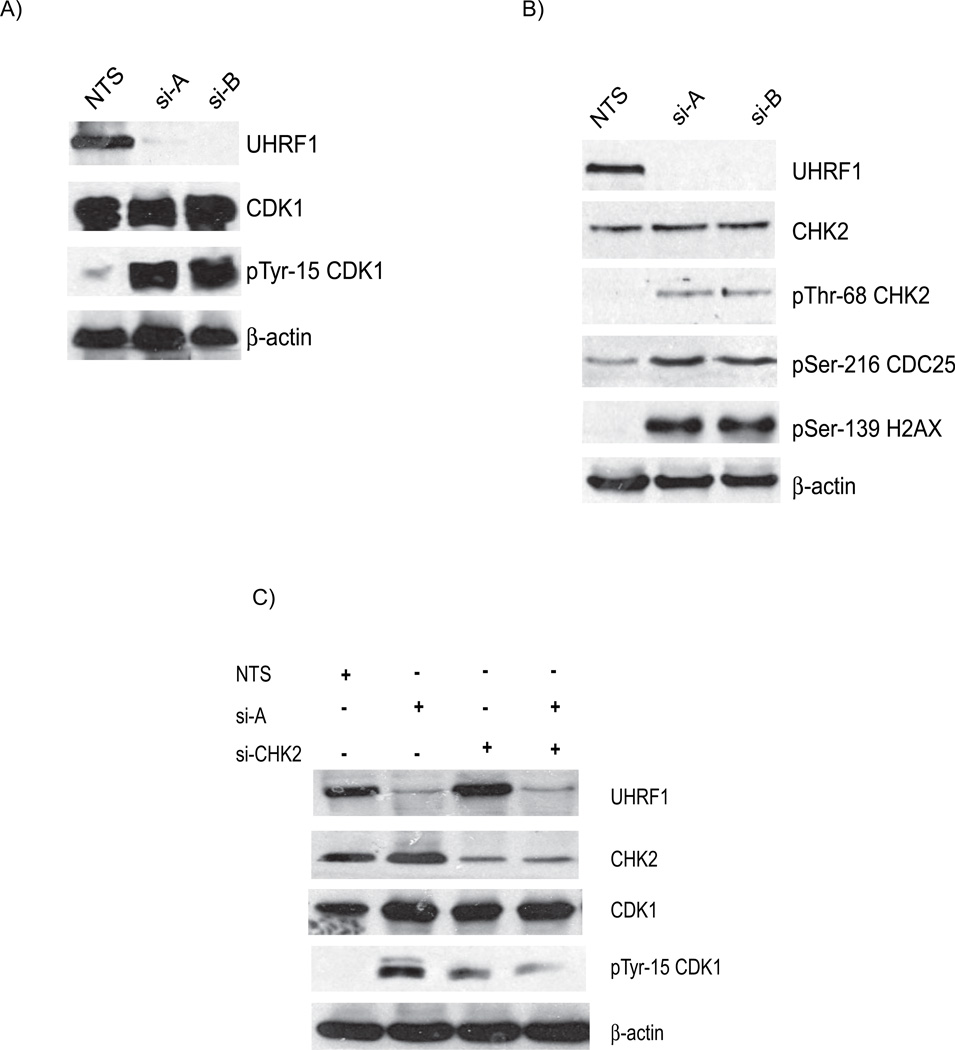
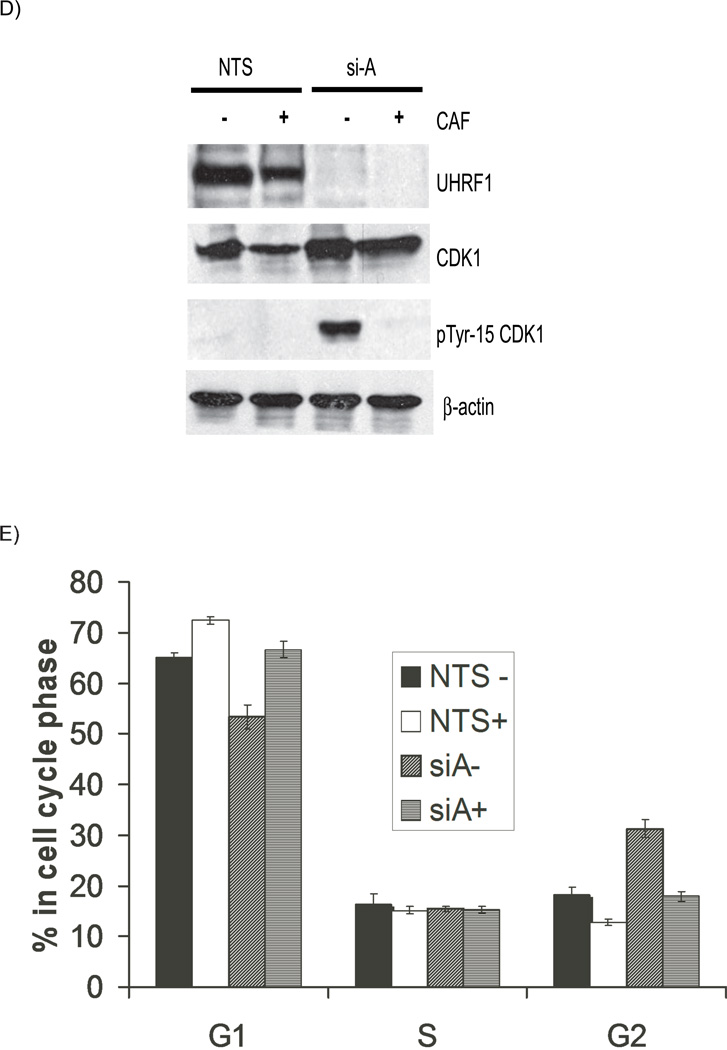
Figure 2. Depletion of UHRF1 activates the DNA damage response pathway.
A). Western immunoblots show similar levels of total cellular CDK1 (panel 2) but there are enhanced levels of tyrosine 15 phosphorylated CDK1 in UHRF1 depleted cells (panel 3, compare lanes 2 and 3 with 1). B). Additional markers of DNA damage response are present in UHRF1 depleted cells. Total CHK2 is unchanged (panel 2) but CHK2 is phosphorylated on threonine-68 (panel 3), CDC25C is phosphorylated on serine-216 (panel 4) and histone H2AX is phosphorylated on serine-139 (panel 5). β-actin is used as a loading control for the experiments above. C) Knockdown of CHK2 in UHRF1 depleted cells reduces tyrosine-15 phosphorylation. The increased tyrosine-15 phosphorylation of CDK1 seen in UHRF1 depleted cells (lane 2, panel 4) is reduced when CHK2 is concurrently knocked down (lane 4, panel 4). si-CHK2 represents siRNA against CHK2. D). Caffeine abrogates the G2/M checkpoint in UHRF1 depleted cells. The marked tyrosine-15 phosphorylation of CDK1 in UHRF1 depleted cells (panel 3, lane 3) is absent in UHRF1 deficient cells treated with caffeine (CAF). CDK1 levels for NTS transfected cells are similar (panel 2, lanes 1 and 2) as well as for si-A transfected cells (panel 2, lanes 3 and 4). E) Representative FACS analysis of caffeine treated cell. Percentage of cells at various stages of the cell cycle following caffeine treatment. NTS represents UHRF1 containing cells while si-A represents UHRF1 depleted cells. + and − represent the presence or absence of caffeine. Error bars represent standard deviations from the mean of replicates. Note that caffeine abrogates the increase in G2/M population seen with UHRF1 depletion.
The G2/M checkpoint is activated as a response to DNA damage. For example, ultraviolet and ionizing irradiation or genotoxic drug treatment of cells causes cells to arrest in order to repair the damage [25]. Because UHRF1 depletion renders cells more sensitive to DNA damaging agents [18, 19], we hypothesized that the G2/M block observed in UHRF1 depleted cells was associated with activation of the DNA damage pathway. To address this we evaluated several markers of the DNA damage response in HCT116 cells depleted of UHRF1. (i) Checkpoint 1 (CHK1) and Checkpoint 2 (CHK2) kinases are activated through phosphorylation by ataxia telangiectasia mutated (ATM) and/or ATM-Rad3 related (ATR) kinase mediated pathways in response to DNA damage [1]. We did not see any phosphorylation of CHK1, but did find increased threonine-68 phosphorylation of CHK2 in UHRF1 depleted cells (Figure 2B, compare lanes 2 and 3 to lane 1 in panel 3). (ii) CDC25 is the phosphatase responsible for removing the inhibitory tyrosine-15 phosphorylation of CDK1. CDC25 is inactivated when it is phosphorylated on serine-216 by CHK1 and/or CHK2. We found increased phosphorylation of serine-216 on CDC25 in cells depleted of UHRF1, confirming the downstream effect of CHK2 phosphorylation (Figure 2B, panel 4). (iii) Activation of the DNA damage pathway causes phosphorylation of the histone variant, H2AX on serine-139. Western blot (Figure 2B) shows enhanced H2AX phosphorylation and immunofluorescence shows positive phospho-H2AX cells (Figure S2, supplementary information) in UHRF1 depleted cells. These data demonstrate that the DNA damage response system is activated in response to UHRF1 depletion.
We rationalized that if the G2/M block is due to the activation of DNA damage repair pathway, then the loss of CHK2 in UHRF1 depleted cells should affect phosphotyrosine-15 CDK1 levels. Figure 2C shows that increased tyrosine-15 phosphorylation of CDK1 in UHRF1 depleted cells is reduced when CHK2 is concurrently knocked down (panel 4, compare lane 2 and 4). The G2/M checkpoint can be abolished in cells following caffeine treatment, likely through inhibition of ATM and ATR [26]. We treated transfected cells with caffeine and then harvested cells for Western blots. Figure 2D shows that there is no change in total CDK1 levels whereas tyrosine-15 phosphorylation of CDK1 is markedly increased in UHRF1 depleted cells (Figure 2D, compare lane 1 to lane 3 in panels 2 and 3). However, CDK1 phosphorylation is abolished when the cells are treated with caffeine (Figure 2D, compare lane 3 to 4 in panel 3). In support of the role of this checkpoint in the observed G2/M block, FACS analysis of UHRF1 containing and depleted cells treated with or without caffeine shows that caffeine treatment abolishes the increase in the population of G2/M cells seen with UHRF1 depletion (Figure 2E). These data suggest that loss of UHRF1 activates DNA damage signals initiated through ATM and/or ATR leading to the inhibitory phosphorylation of CDK1 and the subsequent cell cycle block.
Cell cycle arrest is not p53 dependent
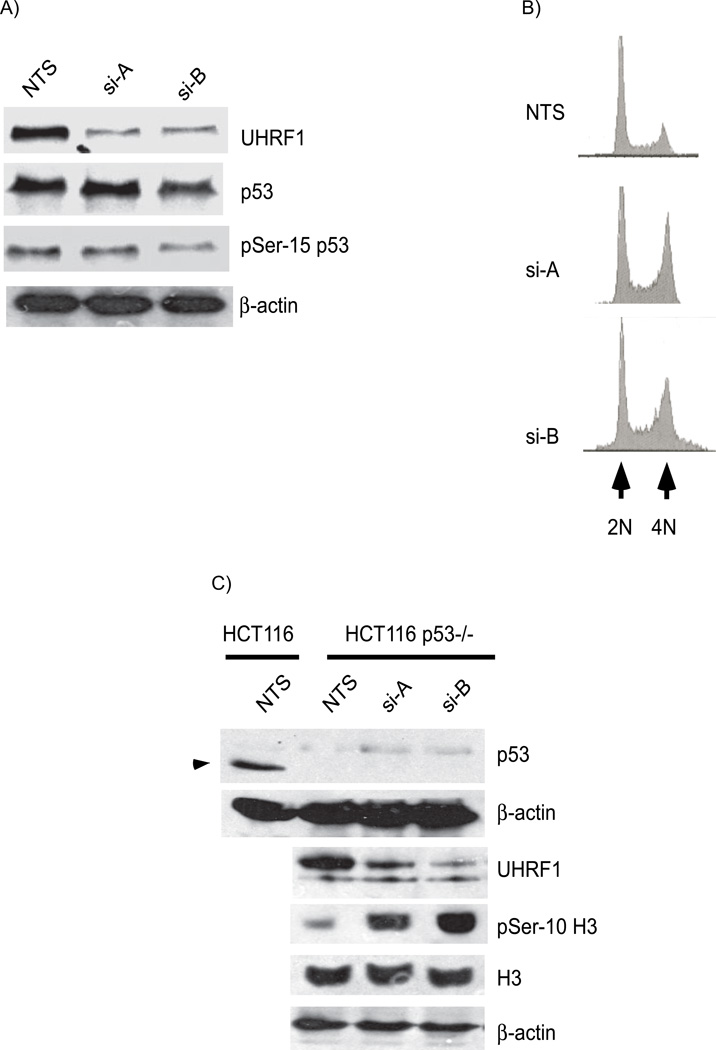
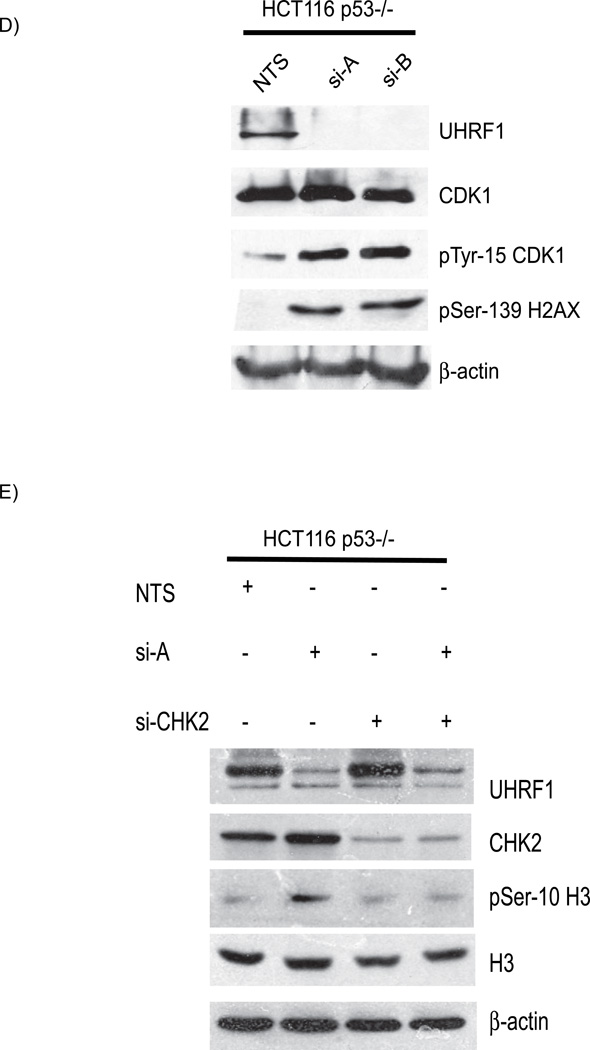
In a previous study, adriamycin treatment caused a small population of UHRF1 depleted cells to arrest in G1, in a p53 dependent manner [18]. p53 is a downstream effector of ATM-mediated DNA damage signaling and coordinates DNA repair with cell cycle progression [27]. Activation of ATM/ATR, CHK1 and/or CHK2, leads to p53 phosphorylation and stabilization [28, 29]. Interestingly, DNMT1 inactivation also activates and stabilizes p53 [21]. We asked whether the cell cycle arrest and DNA damage response activation in cells depleted of UHRF1 is p53 dependent. We found no difference in total p53 levels in cells depleted of UHRF1 (Figure 3A; panel 2, compare lanes 2 and 3 to lane 1) and there was no corresponding increase in the serine-15 phosphorylated moiety (panel 3, compare lanes 2 and 3 to lane 1). Next, we assessed whether the cell cycle arrest due to UHRF1 depletion requires p53 by knocking down UHRF1 in HCT116 cells devoid of p53 (HCT116 p53−/−). FACS analysis revealed the same increase in the G2/M population in response to UHRF1 depletion (Figure 3B). In support of the FACS analysis data, Western immunoblots of UHRF1 depleted p53−/− HCT cells showed that serine 10 phosphorylation of histone H3 is increased in UHRF1 knockdown cells (Figure 3C, compare lane 1 to lanes 2 and 3 in panel 4). Western blot analysis of HCT116 p53−/− cells also showed that UHRF1 depletion led to inhibitory phosphorylation of CDK1 and phosphorylation of histone H2AX on serine-139 (Figure 3D, panels 3 and 4). In addition, the abrogation of the G2/M checkpoint by knockdown of CHK2 leads to the alleviation of the cell cycle arrest as illustrated by the decrease in phospho Histone H3 levels (Figure 3E). These experiments demonstrate that independent of p53 status, the same DNA damage pathway components are activated in response to UHRF1 depletion.
Figure 3. The cell cycle block in UHRF1 depleted cells is p53 independent.
A). Western immunoblot of total p53 (panel 2) and serine-15 phophorylated p53 (panel 3). Total p53 and serine-15 p53 proteins are not increased in HCT116 cells depleted or sufficient for UHRF1. B). Representative histograms of HCT116 p53−/− cells depleted of UHRF1. Note the increased 4N DNA content in cells transfected with si-A and si-B (panel 2 and 3). C) Increased phosphohistone H3 levels in UHRF1 depleted cells. Top panel: p53 is absent in p53−/− cells (compare lanes 2, 3 and 4 with lane 1, arrow head indicates p53 in WT HCT116 cells). In p53−/− HCT116 cells, knockdown of UHRF1 increase phosphohistone H3 levels (panel 4 compare lanes 2 and 3 to 1). D) UHRF1 knockdown enhances phosphorylation of CDK1 and enhances phospho-H2AX levels (panels 3 and 4). E) CHK2 depletion in UHRF1 knockdown cells affects phosphohistone H3 levels. UHRF1 depletion increases phosphohistone H3 levels (panel 3, compare lane 1 and 2). Loss of CHK2 in UHRF1 depleted cells abrogates histone H3 phosphorylation (panel 3, compare lanes 2 and 4). si-CHK2 represents siRNA against CHK2. β-actin is used as a loading control.
UHRF1 depleted cells undergo a p53 independent apoptosis
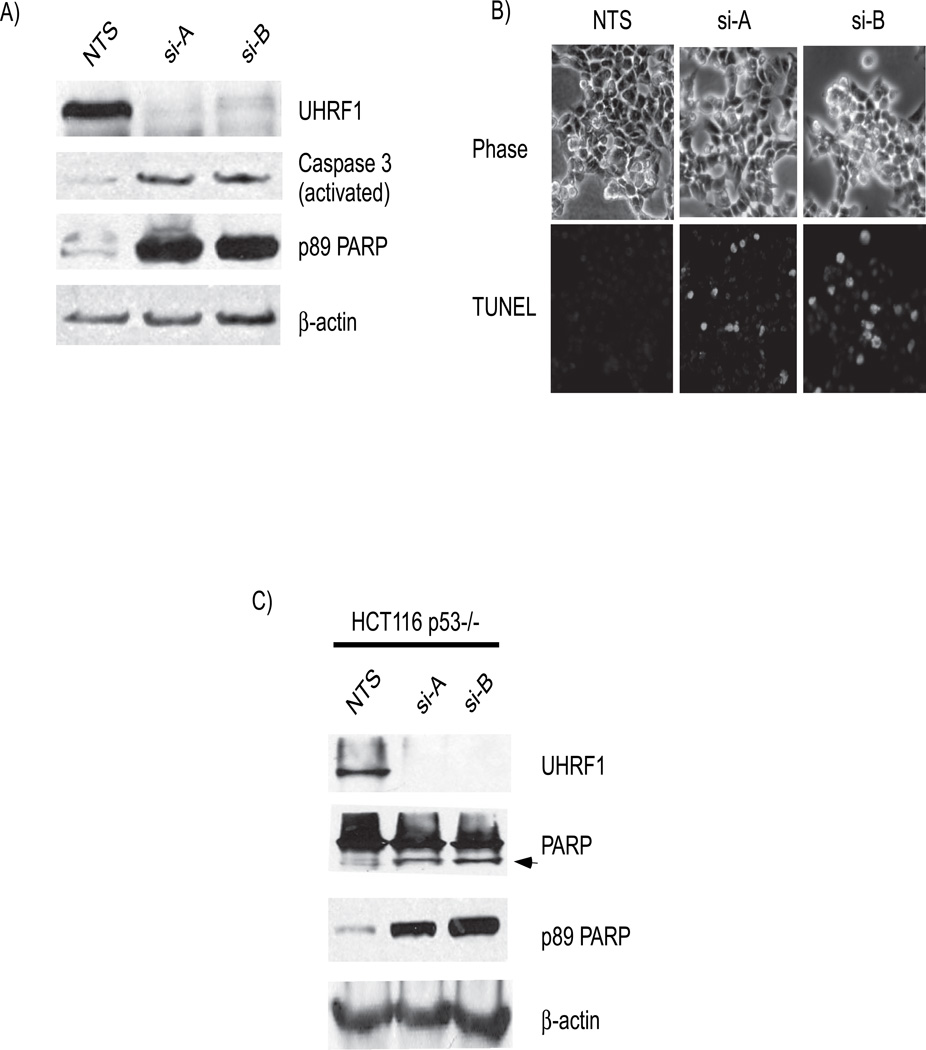
We noted that some cells became non-adherent after UHRF1 knockdown. The ATM/ATR-mediated DNA damage signaling pathways can promote apoptosis and thus we examined whether loss of UHRF1 causes apoptosis in cancer cells, as it does in embryonic cells [15]. Apoptosis can be detected biochemically by assaying for caspase activation through detecting their cleavage products. Caspase-3, an effector caspase downstream of multiple apoptotic stimuli, was detected in its active (cleaved) form in lysates from HCT116 cells depleted of UHRF1 (Figure 4A). Additionally, cleavage of the 116 kDa protein, poly ADP ribose polymerase-1 (PARP-1) to an 89 kDa proteolytoic fragment by activated caspase-3 was detected in cells depleted of UHRF1 (Figure 4A). Fluorescence microscopy of UHRF1 depleted cells also showed abundant TUNEL positive cells (Figure 4B and Figure S3, supplementary information) as well as numerous cells positive for another marker of apoptosis, annexin-V (not shown). Similar to p53 containing cells, p53 deficient HCT116 cells depleted of UHRF1 also expressed markers of active apoptosis with detection of cleaved PARP-1 (Figure 4C). We conclude that loss of UHRF1 in human cancer cells results in apoptosis, similar to the finding in non cancerous embryonic zebrafish liver devoid of uhrf1 [15]. It is also interesting that similar findings were reported in DNMT1 depleted cells, however, unlike the loss of DNMT1, where p53 is stabilized and maybe required; apoptosis in response to UHRF1 depletion is at least partly independent of p53.
Figure 4. UHRF1 cells undergo a p53-independent apoptosis.
A). Markers of active apoptosis are present in UHRF1 depleted cells. Caspase-3 is cleaved to generate activated caspase-3 (panel 2, compares lanes 2 and 3 to 1) and PARP is cleaved (panel 4; compare lanes 2 and 3 with 1). B). TUNEL assays shows the presence of TUNEL positive cells (bottom) in UHRF1 depleted cells. Phase photographs are shown for comparison (top). C) Apoptosis also occurs in HCT116 p53−/− cells. The second panel shows both the 116 kDa and 89 kDa (arrow) moieties of PARP in UHRF1 depleted cells. Only the 116 kDa PARP is seen in the lysate from NTS transfected cells (panel 2, lane 1). Blots with an antibody that recognizes the 89 kDa fragment confirms that it is generated from PARP (panel 3).
We did not find evidence that other p53 family members (p63 and p73) which share structural homology and overlapping functions with p53 [30] were activated in response to UHRF1 depletion. Neither p63 and p73 RNA (Figure S4A, supplementary information) nor protein (Figure S4B, supplementary information) levels were enhanced with UHRF1 depletion. Consistent with this, cells depleted of UHRF1 did not upregulate the p53 upregulated modulator of apoptosis (PUMA) gene or protein (Figure S4, supplementary information), a downstream target of p53, p63 and p73 and a factor upstream of caspase-9 [31]. Taken together results suggest that neither p53, p63 nor p73 are absolute requirements for apoptosis in UHRF1 depleted cells.
UHRF1 depletion causes apoptosis in a caspase-8 dependent manner
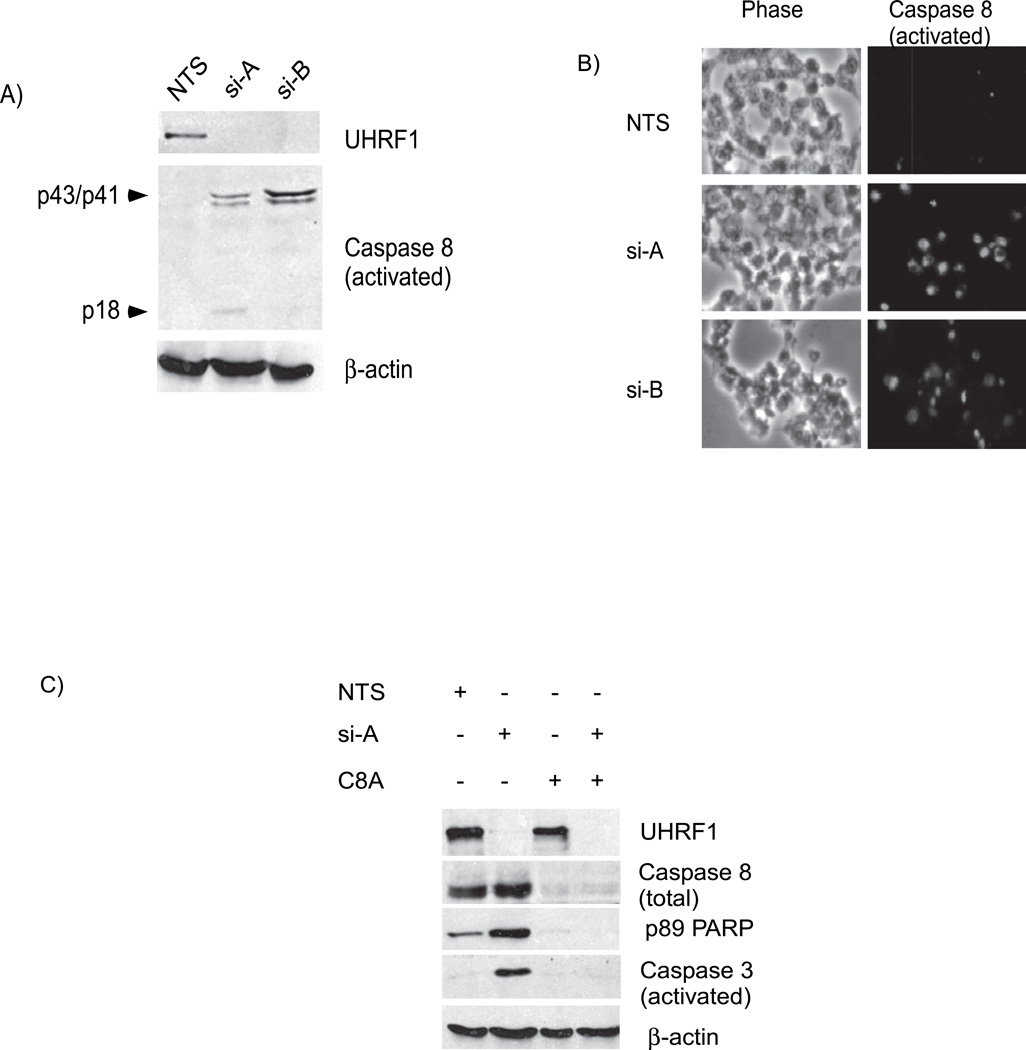
Caspase-8, an initiator caspase, can directly activate caspase-3 bypassing the mitochondrial mediated death pathway [31]. A p53 independent pathway that regulates caspase-8 activity has been described to account for apoptosis seen following irradiation of p53 deficient glioma cells [32]. The presence of activated and cleaved caspase-8 was detected in lysates from UHRF1 depleted cells, but not controls (Figure 5A). Immunofluorescence with antibodies to activated caspase-8 show greater than a 10-fold increase in the number of caspase-8 positive cells when UHRF1 depleted cells are compared to UHRF1 containing cells (Figure 5B). We depleted caspase-8 from UHRF1 knockdown cells and evaluated whether apoptosis still occurred. As expected, the p89 fragment of PARP-1 (Figure 5C, compare lane 1 to lane 2 of panel 3) and activated caspase-3 (Figure 5C, compare lane 1 to lane 2 of panel 4) increases with knockdown of UHRF1 in caspase-8 containing cells. However in UHRF1 depleted cells, loss of caspase-8 blocks PARP-1 cleavage (Figure 5C, compare lane 2 to lane 4 of panel 3) and caspase-3 activation (Figure 5C, compare lane 2 to lane 4 of panel 4). These data suggest that loss of UHRF1 causes a cell cycle arrest that is irreparable, causing activation of caspase-8 which leads to activation of caspase-3 and cell death.
Figure 5. Apoptosis in UHRF1 depleted cells occurs through the activation of caspase-8.
A) Proteolytic fragments of caspase-8 representing its activated form (p43/p41 and p18) are detected in UHRF1 depleted cells but not in UHRF1 containing cells (panel 2, compare lanes 2 and 3 with 1). B). Immunofluoresence photographs (right panel, green) of UHRF1 containing and depleted cells show that activated caspase-8 is present in cells lacking UHRF1. Phase photographs are also shown for comparison (left) C) Depletion of total caspase-8 prevents PARP and caspase-3 cleavage. p89 PARP and activated caspase-3 levels increase in UHRF1 depleted but caspase-8 containing cells (panels 3 and 4, lane 2) while low in caspase-8 depleted (C8A) but UHRF1 containing cells (panels 3 and 4, lane 3). When caspase-8 in depleted in cells lacking UHRF1, PARP and caspase-3 cleavage are suppressed (panels 3 and 4, lane 4). Note that “Caspase 8” represents total caspase-8 levels while “Caspase 8 (activated)” represents proteolyzed forms of caspase-8.
5-azacytidine treatment reproduces some of the effects of UHRF1 depletion
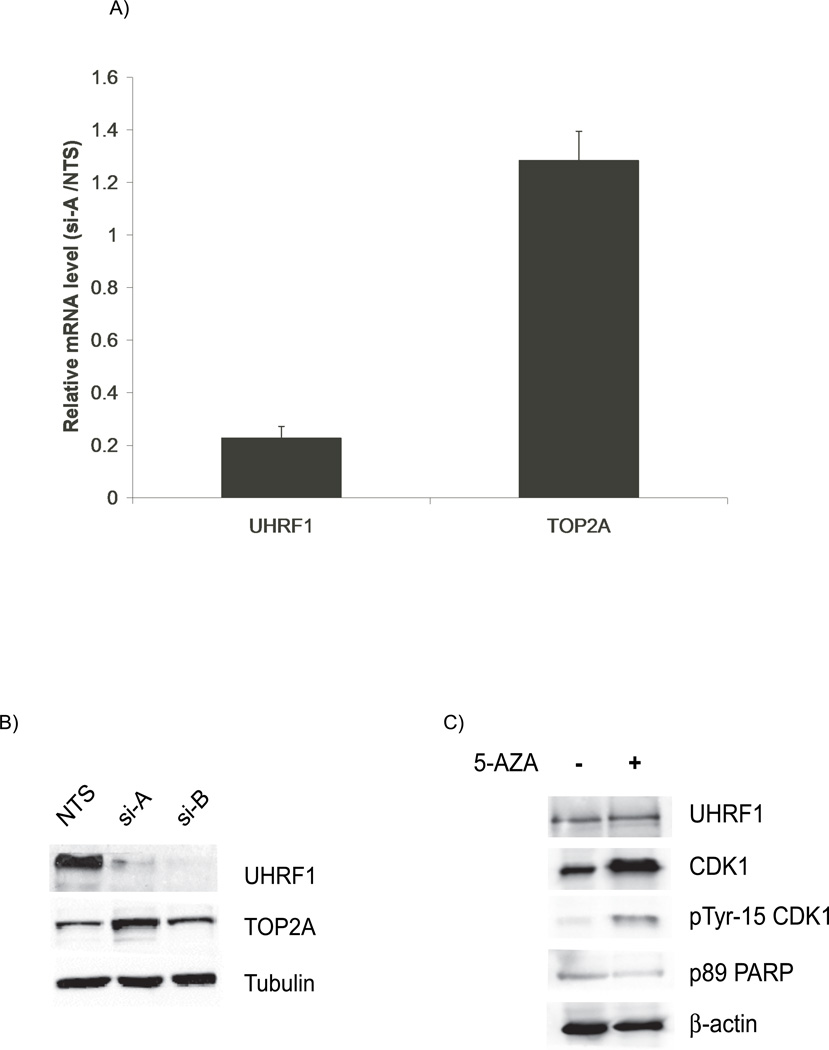
We turned to studies designed to begin unraveling the mechanisms governing how UHRF1 interacts with the cell cycle. UHRF1 has been reported to bind the inverted CCAAT box in the topoisomerase 2A (TOP2A) promoter and activate its expression [2]. Given that TOP2A plays important roles in DNA replication and chromosomal segregation, events whose disruption can elicit cell cycle blocks, we investigated whether the effects of UHRF1 depletion could be explained through decreased TOP2A levels. Neither the mRNA nor the protein levels of TOP2A are decreased in UHRF1 depleted HCT116 cells (Figure 6A and B), nor in HuH7, MCF7 or zebrafish embryos lacking uhrf1 (not shown). In fact, cells transfected with si-A have a modest increase TOP2A mRNA and protein. These data suggest that TOP2A is unlikely to play a direct role in the cell cycle arrest seen in UHRF1 depleted cells and support the notion that UHRF1 does not directly regulate topoisomerase 2A expression.
Figure 6. Topoisomerase 2A levels are unchanged with UHRF1 depletion.
A). Quantitative PCR assays shows that TOP2A mRNA levels are not decreased in UHRF1 depleted cells. UHRF1 mRNA levels are decreased by 80% for this experiment. Error bars represent standard deviations from the mean of three replicates.
B). TOP2A protein levels are not decreased in UHRF1 knockdown cells. Tubulin is used as a loading control. C) UHRF1 levels remain unchanged in 5-azacytidine treated cells (top panel). Total CDK1 and Tyr 15-CDK1 also increase (panel 2 and 3) but p89 levels are not increased in 5-azacytidine treated cells (panel 4).
Loss of UHRF1 may lead to increased concentration of hemimethylated DNA in UHRF1 depleted cells. We suspected that such improperly modified DNA maybe viewed as defective by the cell cycle machinery leading to a cell cycle block. If true, treatment of cells with 5-azacytidine, a drug that inhibits DNA methylation would recapitulate the phenotype of UHRF1 loss. Our findings partially support defective methylation as a variable. In support of a role for methylation, there is an increase in the G2/M population of cells (Figure S5) as well as an increase in tyrosine-15 CDK1 content of cells treated with 5-azacytidine (Figure 6C). However, unlike UHRF1 depletion, there is a parallel increase in total CDK1 levels which is not seen in UHRF1 depleted cells. Secondly, there is no increase in p89 fragment of PARP-1 in 5-azacytidine treated cells (Figure 6C). Thus, failed DNA methylation is one possible mechanism by which UHRF1 induces the DNA damage response and cell cycle arrest, however, it is not likely that this alone accounts for the phenotype seen with UHRF1 knockdown.
UHRF1 is recruited to sites of DNA injury
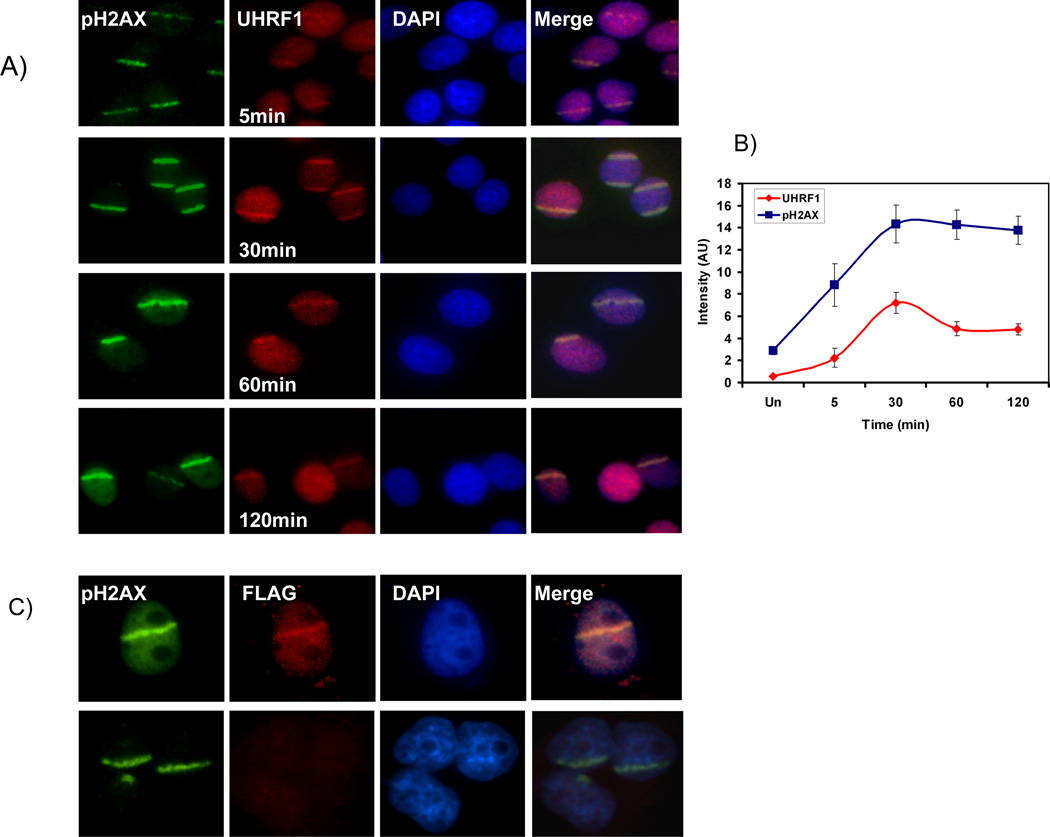
Previous studies have shown that UHRF1 interacting protein DNMT1 is recruited to sites of DNA injury[20]. Because DNMTI is dependent on UHRF1 for interaction with chromatin [3], we questioned whether UHRF1 is recruited to these sites as a possible participant in the repair process. It is also known that the DNA damage response pathway is frequently activated when constituents of the pathway are inactivated. For example, inactivation of Hsu1, Claspin 1, Chk1 all result in DNA damage repair [33–35]. Thus, it is possible that UHRF1 depleted cells activate the DNA damage response because UHRF1 participates in this pathway. To address this, we used laser scissors to introduce DNA damage at discrete sites which can be marked by the presence of phospho-H2AX (Figure 7A). We show that UHRF1 is recruited to these sites and is seen within 5 minutes with peak intensity 30 minutes after injury. The dynamic nature of UHRF1 in the DNA damage stripes and the difference with that of phospho-H2AX, suggest that this is a specific interaction of UHRF1 and the damage site (Figure 7B). To confirm the specificity of UHRF1 for the DNA damage sites, we transfected cells with cDNA encoding FLAG tagged UHRF1 or empty vector and repeated the experiment with FLAG antibody. Once again we find that UHRF1 (as detected with the FLAG antibody) is seen at the sites of injury in FLAG-UHRF1 expressing cells (Figure 7C top panel) but not in vector transfected cells (Figure 7C, bottom panel). These data suggest that UHRF1 senses DNA damage and we speculate that it is recruited to these sites for essential DNA modification linked to repair.
Figure 7.
UHRF1 localizes to UV laser scissor induced DNA damage. A) phospho- H2AX accumulates at site of DNA damage after targeted injury (first row, green). UHRF1 is recruited to these sites within 5 minutes (second row, red), peaks in intensity at 30 minutes and then begins to fade. B) Quantitation of UHRF1 localization with pH2AX. The ratio of intensity of UHRF1 at region of laser scissors induced damage (marked by pH2AX) compared to undamaged region is plotted. Note that the change in intensity of UHRF1 staining is dynamic and differs from that of phospho-H2AX. C) Flag-UHRF1 localizes to DNA damage. Cells were transfected with Flag-UHRF1 (top panel) or empty vector (bottom panel). Cells were subjected to UV laser and processed for immunofluorescence 30 min post DNA damage with pH2AX and Flag antibodies. UHRF1 is present in the stripe in transfected cells (top, column 2) but not in control cells (bottom, column 2).
DISCUSSION
In this study, we identify a link between UHRF1, cell cycle progression and DNA damage response. Cancer cells depleted of UHRF1 show an increase in the population of cells with 4N DNA, activation of the DNA damage response and caspase-8 dependent apoptosis. The cell cycle arrest and apoptosis induced by loss of UHRF1 does not appear to require any p53 family members. Lastly, we show that UHRF1 accumulates at sites of DNA injury. These studies suggest a direct link between genomic integrity and UHRF1 function.
A previous report found that cancer cells depleted of UHRF1 arrest in G1 only when treated with adriamycin [18], whereas another showed that reduction of UHRF1 causes arrest in both G1 and G2 [14]. In our study, we find only a G2/M arrest. If UHRF1 is required for G1/S transition in these cells, then the G1 arrested cells would be revealed when nocodazole is used to trap cells in mitosis. Instead, we find that cells in G1 progressed to the mitotic block in UHRF1 sufficient and deficient cells. Thus no G1 block is present in HCT116 cells depleted of UHRF1. A number of reasons may account for the differences in the cell cycle arrest in our studies when compared to previous studies. 1) The requirement for UHRF1 in cell cycle progression may vary in different cancer cell lines due to differences in mechanisms to bypass cell cycle arrest 2) The G1 arrest seen in cells with low levels of UHRF1, occurred when cells were concurrently treated with adriamycin, raising the possibility that the arrest may be an indirect effect [18] 3) Variations in degree UHRF1 knockdown may result in elaboration of different cell cycle phenotypes. Such differences have been seen with DNMT1 depletion, where different levels of DNMT1 loss result in varying cell cycle effects [36].
It is noteworthy that we do not see a G2/M block in a higher fraction of our cells. This is partly due to the fact that only adherent cells are harvested, and thus cells that have died and floating are not analyzed. A second reason maybe as a result of the limitations of siRNA mediated knockdown and possible variation in cell to cell levels of UHRF1. Third, it may be that only a small population of cells that have gone through the S phase and contain inadequately repaired DNA are candidates for cell cycle block and death.
The results presented here have a number of similarities with the consequences of DNMT1 inactivation in HCT116 cells [21]. In both cases, cells arrest with a 4N DNA content and the DNA damage response system is activated. In addition, both UHRF1 depleted cells and DNMT1 inactivated cells undergo apoptosis. The similarities between DNMT1 inactivation and UHRF1 depletion however differ in one aspect. In cells with devoid of DNMT1, p53 levels are increased and phosphoserine-15 p53 level is elevated [21]. These data suggest that p53 may be integral to the DNA damage activation and the subsequent cell cycle block. However, we do not find any evidence that the response to low levels of UHRF1 involves the stabilization of p53 nor do we find any qualitative evidence that p53 is required for the cell cycle effects of UHRF1 loss. This difference may be attributed to the fact that in our experiments, UHRF1 levels are substantially reduced but not abolished, as is the case for the experiments on DNMT1 in which cells are completely devoid of DNMT1. Alternatively, since UHRF1 has multiple activities, that include but are not limited to driving DNMT1 dependent DNA methylation, loss of UHRF1 may abrogate both DNA methylation as well as other processes. Indeed, insights from the recent crystal structure of the SRA domain in complex with DNA suggest that in addition to methylation, UHRF1 may also allow other enzymes access to the double helix [12]. Lastly, it remains to be tested whether the apoptosis observed due to DNMT1 depletion depends on p53.
The apoptosis in UHRF1 depleted cells supports our data that p53 is dispensable given a known role for caspase-8 in a pathway that bypasses p53 [32]. As noted, cell death also occurs in DNMT1 deficient cells [21]. Given the functional parallels between two proteins, it is tempting to speculate that cell death in DNMT1 deficient cells may also occur through a caspase-8 dependent process. In addition, the data here suggest a relationship between UHRF1 and caspase-8 activity and the mechanism of this regulatory effect needs to be examined.
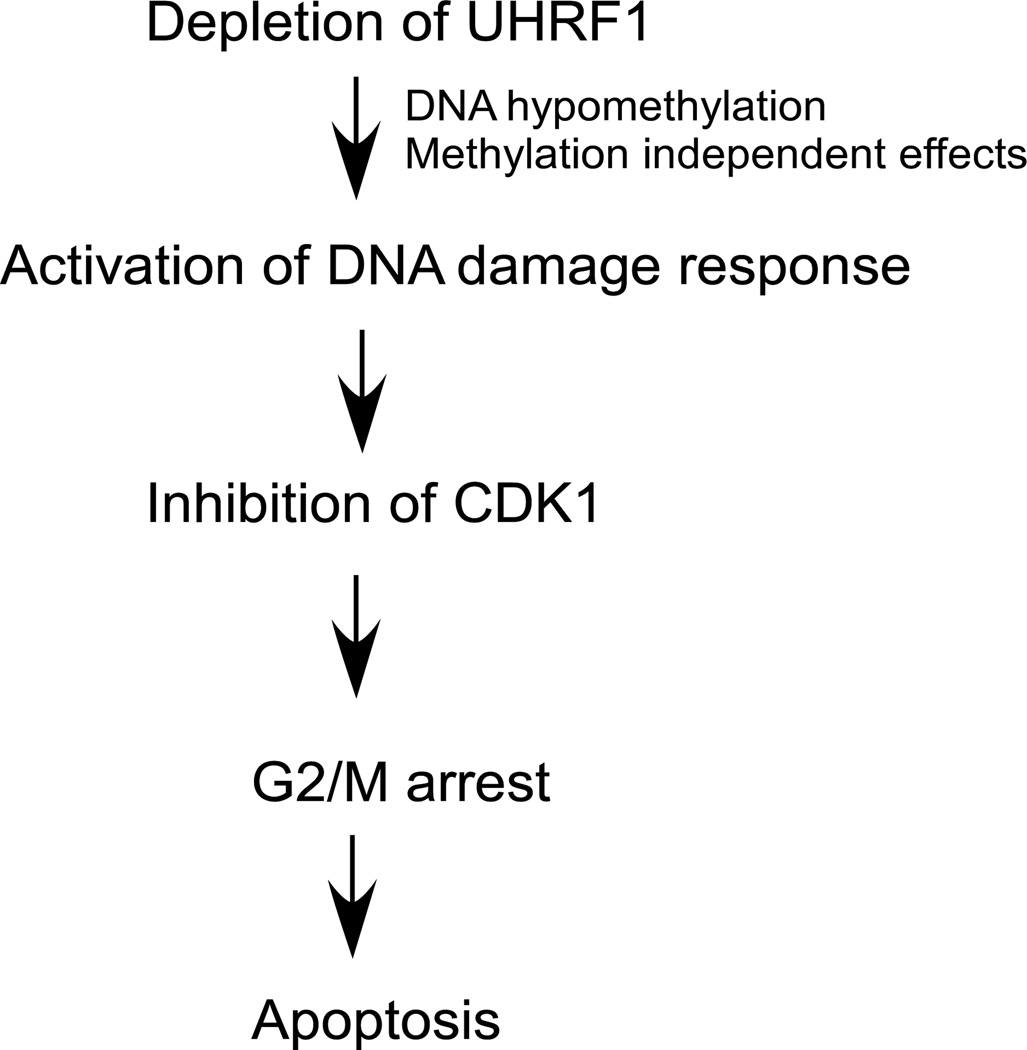
How then does UHRF1 depletion lead to activation of DNA damage response and the subsequent arrest and apoptosis? We do not believe that this effect is related to the putative effects of UHRF1 on TOP2A expression, since we find no evidence that UHRF1 directly regulates TOP2A expression. We reasoned that high concentrations of hemimethylated DNA which likely accumulate in UHRF1 depleted cells may be viewed by the cell as a loss of genomic fidelity. We show that treatment of cells with 5-azacytidine recapitulates some but not all of the phenotypes of UHRF1 loss and suggests that DNA methylation defects may play at least a partial role. We find that UHRF1 is recruited to sites of DNA injury enhancing our argument that it plays a role in DNA damage repair. This observation is supported by reports linking UHRF1 to DNA damage. In one study, repair factors including PARP-1, XRCC1, TopBP1, PCNA were found in complex with UHRF1 [4]. Additionally, Eme1 a component of an endonuclease required for damage repair is reported to associate with UHRF1 [37]. Therefore, loss of UHRF1 may lead to a defective replication product by any of these mechanisms, activating the damage response pathway and arresting the cell cycle in G2/M (Figure 8). While this is an intriguing finding we are cognizant of the fact that it begs the question of what UHRF1 does at the damage site. This will be the focus of further studies.
Figure 8.
Proposed model of the effects of UHRF1 depletion in HCT116 cells.
In summary we provide molecular evidence linking UHRF1 and DNA damage repair. Our results suggest that UHRF1 loss leads to genomic defects that are sensed by the cell cycle apparatus and result in cell cycle block and cell death. Our data combined with existing evidence that cells lacking UHRF1 are more sensitive to DNA damaging agents [19] provides a basis for investigating UHRF1 as a possible target in cancer.
Supplementary Material
Acknowledgements
AT was supported by a T32 grant (Gastroenterology Division, Brigham and Women’s Hospital). BG was supported by the Four Directions Summer Research Program. Grant support: 1R01DK080789 to CU and KS, DFCI/ Harvard Cancer Center, NCI Cancer Center Support Grant and Sidney A. Swensrud Foundation grants to CU.
Abbreviations
- UHRF1
Ubiquitin-like protein, containing PHD and RING finger domains-1
- DNMT1
DNA methyltrnasferase 1
- CDK1
Cyclin dependent kinase 1
- CHK1
Checkpoint kinase 1
- CHK2
Checkpoint kinase 2
- ATM
Ataxia telangectasia related
- ATR
ataxia telangectasia mutated and Rad 3 related
- CDC25
Cell division control 25
- PARP-1
Poly ADP ribose polymerase-1
- TOP2A
Topoisomerase 2A
Footnotes
Author Contributions: AT, SS, AK, RM, BG and CU performed experiments. AT and CU prepared manuscript. CU, KSE and SG edited manuscript.
References
- 1.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 2.Hopfner R, Mousli M, Jeltsch JM, Voulgaris A, Lutz Y, Marin C, Bellocq JP, Oudet P, Bronner C. ICBP90, a novel human CCAAT binding protein, involved in the regulation of topoisomerase IIalpha expression. Cancer Res. 2000;60:121–128. [PubMed] [Google Scholar]
- 3.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 4.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K, Okano M, Koseki H. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 5.Unoki M, Nishidate T, Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23:7601–7610. doi: 10.1038/sj.onc.1208053. [DOI] [PubMed] [Google Scholar]
- 6.Kim JK, Esteve PO, Jacobsen SE, Pradhan S. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res. 2009;37:493–505. doi: 10.1093/nar/gkn961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Citterio E, Papait R, Nicassio F, Vecchi M, Gomiero P, Mantovani R, Di Fiore PP, Bonapace IM. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol. Cell. Biol. 2004;24:2526–2535. doi: 10.1128/MCB.24.6.2526-2535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hashimoto H, Horton JR, Zhang X, Cheng X. UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications. Epigenetics. 2009;4:8–14. doi: 10.4161/epi.4.1.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karagianni P, Amazit L, Qin J, Wong J. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol. Cell. Biol. 2008;28:705–717. doi: 10.1128/MCB.01598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeanblanc M, Mousli M, Hopfner R, Bathami K, Martinet N, Abbady AQ, Siffert JC, Mathieu E, Muller CD, Bronner C. The retinoblastoma gene and its product are targeted by ICBP90: a key mechanism in the G1/S transition during the cell cycle. Oncogene. 2005;24:7337–7345. doi: 10.1038/sj.onc.1208878. [DOI] [PubMed] [Google Scholar]
- 11.Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 14.Jenkins Y, Markovtsov V, Lang W, Sharma P, Pearsall D, Warner J, Franci C, Huang B, Huang J, Yam GC, Vistan JP, Pali E, Vialard J, Janicot M, Lorens JB, Payan DG, Hitoshi Y. Critical role of the ubiquitin ligase activity of UHRF1, a nuclear RING finger protein, in tumor cell growth. Mol. Biol. Cell. 2005;16:5621–5629. doi: 10.1091/mbc.E05-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sadler KC, Krahn KN, Gaur NA, Ukomadu C. Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, uhrf1. Proc. Natl. Acad. Sci. U.S.A. 2007;104:1570–1575. doi: 10.1073/pnas.0610774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mousli M, Hopfner R, Abbady AQ, Monte D, Jeanblanc M, Oudet P, Louis B, Bronner C. ICBP90 belongs to a new family of proteins with an expression that is deregulated in cancer cells. Br. J. Cancer. 2003;89:120–127. doi: 10.1038/sj.bjc.6601068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonapace IM, Latella L, Papait R, Nicassio F, Sacco A, Muto M, Crescenzi M, Di Fiore PP. Np95 is regulated by E1A during mitotic reactivation of terminally differentiated cells and is essential for S phase entry. J. Cell Biol. 2002;157:909–914. doi: 10.1083/jcb.200201025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arima Y, Hirota T, Bronner C, Mousli M, Fujiwara T, Niwa S, Ishikawa H, Saya H. Down-regulation of nuclear protein ICBP90 by p53/p21Cip1/WAF1-dependent DNA-damage checkpoint signals contributes to cell cycle arrest at G1/S transition. Genes Cells. 2004;9:131–142. doi: 10.1111/j.1356-9597.2004.00710.x. [DOI] [PubMed] [Google Scholar]
- 19.Muto M, Kanari Y, Kubo E, Takabe T, Kurihara T, Fujimori A, Tatsumi K. Targeted disruption of Np95 gene renders murine embryonic stem cells hypersensitive to DNA damaging agents and DNA replication blocks. J. Biol. Chem. 2002;277:34549–34555. doi: 10.1074/jbc.M205189200. [DOI] [PubMed] [Google Scholar]
- 20.Mortusewicz O, Schermelleh L, Walter J, Cardoso MC, Leonhardt H. Recruitment of DNA methyltransferase I to DNA repair sites. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8905–8909. doi: 10.1073/pnas.0501034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen T, Hevi S, Gay F, Tsujimoto N, He T, Zhang B, Ueda Y, Li E. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat. Genet. 2007;39:391–396. doi: 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
- 22.Ukomadu C, Dutta A. Inhibition of cdk2 activating phosphorylation by mevastatin. J. Biol. Chem. 2003;278:4840–4846. doi: 10.1074/jbc.M208658200. [DOI] [PubMed] [Google Scholar]
- 23.Morgan DO. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 24.Glotzer M. Cell cycle. The only way out of mitosis. Curr. Biol. 1995;5:970–972. doi: 10.1016/s0960-9822(95)00190-4. [DOI] [PubMed] [Google Scholar]
- 25.Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge SJ. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc. Natl. Acad. Sci. U.S.A. 2000;97:10389–10394. doi: 10.1073/pnas.190030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu W, Chen Y, Dutta A. Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol. Cell. Biol. 2004;24:7140–7150. doi: 10.1128/MCB.24.16.7140-7150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 28.Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge SJ, Mak TW. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 29.Shieh SY, Taya Y, Prives C. DNA damage-inducible phosphorylation of p53 at N-terminal sites including a novel site, Ser20, requires tetramerization. EMBO J. 1999;18:1815–1823. doi: 10.1093/emboj/18.7.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deyoung MP, Ellisen LW. p63 and p73 in human cancer: defining the network. Oncogene. 2007;26:5169–5183. doi: 10.1038/sj.onc.1210337. [DOI] [PubMed] [Google Scholar]
- 31.Sidi S, Sanda T, Kennedy RD, Hagen AT, Jette CA, Hoffmans R, Pascual J, Imamura S, Kishi S, Amatruda JF, Kanki JP, Green DR, D'Andrea AA, Look AT. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell. 2008;133:864–877. doi: 10.1016/j.cell.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Afshar G, Jelluma N, Yang X, Basila D, Arvold ND, Karlsson A, Yount GL, Dansen TB, Koller E, Haas-Kogan DA. Radiation-induced caspase-8 mediates p53-independent apoptosis in glioma cells. Cancer Res. 2006;66:4223–4232. doi: 10.1158/0008-5472.CAN-05-1283. [DOI] [PubMed] [Google Scholar]
- 33.Zhu M, Weiss RS. Increased common fragile site expression, cell proliferation defects, and apoptosis following conditional inactivation of mouse Hus1 in primary cultured cells. Mol. Biol. Cell. 2007;18:1044–1055. doi: 10.1091/mbc.E06-10-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu S, Bekker-Jensen S, Mailand N, Lukas C, Bartek J, Lukas J. Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol. Cell. Biol. 2006;26:6056–6064. doi: 10.1128/MCB.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puc J, Parsons R. PTEN loss inhibits CHK1 to cause double stranded-DNA breaks in cells. Cell Cycle. 2005;4:927–929. doi: 10.4161/cc.4.7.1795. [DOI] [PubMed] [Google Scholar]
- 36.Brown KD, Robertson KD. DNMT1 knockout delivers a strong blow to genome stability and cell viability. Nat. Genet. 2007;39:289–290. doi: 10.1038/ng0307-289. [DOI] [PubMed] [Google Scholar]
- 37.Mistry H, Gibson L, Yun JW, Sarras H, Tamblyn L, McPherson JP. Interplay between Np95 and Eme1 in the DNA damage response. Biochem. Biophys. Res. Commun. 2008;375:321–325. doi: 10.1016/j.bbrc.2008.07.146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.