Abstract
We identified a short-grain mutant (Short grain1 (Sg1) Dominant) via phenotypic screening of 13,000 rice (Oryza sativa) activation-tagged lines. The causative gene, SG1, encodes a protein with unknown function that is preferentially expressed in roots and developing panicles. Overexpression of SG1 in rice produced a phenotype with short grains and dwarfing reminiscent of brassinosteroid (BR)-deficient mutants, with wide, dark-green, and erect leaves. However, the endogenous BR level in the SG1 overexpressor (SG1:OX) plants was comparable to the wild type. SG1:OX plants were insensitive to brassinolide in the lamina inclination assay. Therefore, SG1 appears to decrease responses to BRs. Despite shorter organs in the SG1:OX plants, their cell size was not decreased in the SG1:OX plants. Therefore, SG1 decreases organ elongation by decreasing cell proliferation. In contrast to the SG1:OX plants, RNA interference knockdown plants that down-regulated SG1 and a related gene, SG1-LIKE PROTEIN1, had longer grains and internodes in rachis branches than in the wild type. Taken together, these results suggest that SG1 decreases responses to BRs and elongation of organs such as seeds and the internodes of rachis branches through decreased cellular proliferation.
Grain size is one of the major determinants of crop yield in the cereals. Moreover, grain size is also a major determinant of cooking and edibility characteristics in crops such as rice (Oryza sativa). Since rice grains are usually cooked as intact grains, without milling or processing, preferences for various grain characteristics depend on local cultures and cuisines. For example, long and slender grains are favored in Chinese, Indian, and European cuisine, whereas medium-length and round grains are preferred in Japanese cuisine. Therefore, grain size has been an important target in rice breeding.
In recent years, several genes (GRAIN SIZE3 [GS3], GW2, QTL for seed width on chromosome 5 [qSW5]) that control grain size have been isolated from rice (Fan et al., 2006; Song et al., 2007; Shomura et al., 2008; Takano-Kai et al., 2009). Rice grain length is largely determined by a single-nucleotide polymorphism in GS3, which encodes a protein containing a phosphatidyl-ethanolamine-binding protein-like domain and a domain in the tumor necrosis factor receptor–nerve growth factor receptor family (Fan et al., 2006; Takano-Kai et al., 2009). The C165A mutation at the GS3 locus causes truncation of the C-terminal region of the GS3 protein, and is strongly associated with increased grain length in both indica and japonica cultivars, as well as in wild-type accessions (Takano-Kai et al., 2009). On the other hand, rice grain width is mainly controlled by two major quantitative trait loci (QTL), GW2 and qSW5 (Song et al., 2007; Shomura et al., 2008). GW2 encodes a putative RING-type E3 ubiquitin ligase, and its loss of function widens the spikelet hull, resulting in increased grain weight (Song et al., 2007). The deletion allele of the other QTL for grain width, qSW5, which encodes a protein with unknown function, is tightly linked with the wide-grain phenotype and is likely to have been selected during the rice domestication process (Shomura et al., 2008).
On the other hand, genes that confer an erect panicle (DENSE AND ERECT PANICLE1 [DEP1]/QTL for panicle erectness on chromosome 9-1 [qPE9-1], ERECT PANICLE2 [EP2], EP3) have also been isolated recently (Huang et al., 2009; Piao et al., 2009; Zhou et al., 2009; Zhu et al., 2010). Interestingly, these genes also affect grain shape, although they were originally isolated because of their effect on panicle traits. DEP1/qPE9-1 encodes a putative transmembrane-domain protein that resembles GS3 (Huang et al., 2009; Zhou et al., 2009). The mutant alleles of qep9-1 that encode a truncated protein that lacks its C-terminal domain confer a dense and erect panicle phenotype as well as a short-grain phenotype (Zhou et al., 2009). Likewise, loss-of-function mutations of EP2 and EP3, which encode a protein with unknown function and a putative F-box protein, respectively, caused an erect-panicle phenotype as well as a short-grain phenotype (Piao et al., 2009; Zhu et al., 2010). These results suggested that some common mechanisms are likely to control both panicle erectness and grain shape, at least to some extent, during rice reproductive development.
In addition, some dwarf mutants whose phytohormone signaling or metabolism was affected also showed effects on grain and panicle sizes. For example, brassinosteroid (BR)-related dwarf mutants such as d61, brassinosteroid-dependent1/brassinosteroid-deficient dwarf1 (brd1), d2, and d11 (Yamamuro et al., 2000; Mori et al., 2002; Hong et al., 2003; Tanabe et al., 2005) have shorter grains combined with altered panicle length. The loss-of-function mutant of the gene for rice G-PROTEIN ALPHA1 (RGA1), d1, affects multiple signaling pathways such as those for BRs and gibberellins, and produces plants with short and round seeds, dense panicles, and a dwarf phenotype (Ashikari et al., 1999; Oki et al., 2009a).
In this study, we identified SHORT GRAIN1 (SG1), a gene that controls the elongation of both grains and internodes in rachis branches, using a modified activation-tagging system in rice (Mori et al., 2007). We described the expression pattern of SG1 and characterized the phenotypes of SG1 overexpressor (SG1:OX) plants and RNA interference (RNAi) knockdown plants that strongly down-regulated both SG1 and a gene for a related protein, SG1-LIKE PROTEIN1 (SGL1). Based on these results, we discuss the roles of SG1 in BR signaling and its control of organ length. Further, we will discuss the potential of SG1 in molecular breeding of rice and related cereals to let breeders engineer the lengths of grains and internodes in rachis branches.
RESULTS
Isolation of the Sg1 Dominant Mutant by Activation Tagging
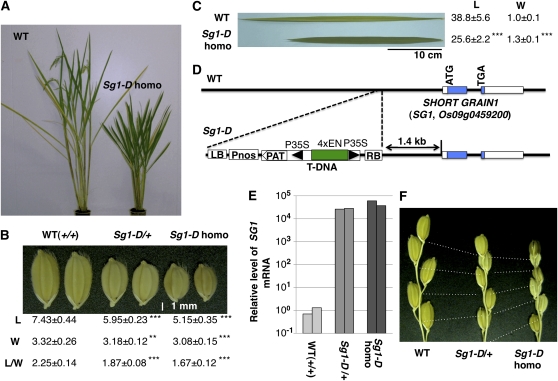
Previously, we reported the isolation of a lesion-mimic mutant, Spotted leaf18, from 13,000 activation-tagging lines (Mori et al., 2007). In the course of phenotypic screening of visible phenotypes in the activation-tagging population, we also isolated a dominant semidwarf mutant with a short-grain phenotype, and named the mutant Sg1 Dominant (Sg1-D; Fig. 1, A and B). The Sg1-D mutant also had short, wide, dark-green leaves reminiscent of a BR-deficient mutant (Fig. 1C). We cloned the T-DNA 3′-flanking fragment, corresponding to the probe B in Supplemental Figure S1, by thermal asymmetric interlaced PCR (Liu et al., 1995). Sequence analysis of the fragment revealed that a gene, Os09g0459200, is located 1.4-kb downstream of the T-DNA insertion (Fig. 1D). The short-grain phenotype cosegregated with the T-DNA insertion (Supplemental Fig. S1). In addition, the expression level of Os09g0459200 increased dramatically (to 20,000–50,000 times the level in the wild-type plants) in a dose-dependent manner in the mutants (Fig. 1E). We also examined the expression of another T-DNA flanking gene (Os09g0458900), which encodes an EF-hand protein located 10.3-kb upstream of the T-DNA. The expression level of Os09g0458900 in the Sg1-D mutant was only 1.3 times that in the wild type. On this basis, Os09g0459200 appears to be the gene responsible for the Sg1-D mutant, and we have tentatively designated this gene as SG1.
Figure 1.
Identification of the rice SG1 gene by activation tagging. A to C, Phenotypes of the Sg1-D rice mutant. A, Heading stage: In comparison with the wild-type plants (WT), the Sg1-D homozygote showed a semidwarf phenotype. B, The short-grain phenotype cosegregated with the T-DNA insertion in a dose-dependent manner. Seeds of WT (+/+), Sg1-D heterozygotes (Sg1-d/+), and Sg1-D homozygotes (Sg1-D homo) are shown. Means and sd from at least 30 measurements of lengths (L, mm), widths (W, mm), and the ratio of length to width (L/W) are shown below the seeds. C, Leaf blades of the Sg1-D homozygote were dark green and were shorter and wider than those of wild type. Means and sd from at least nine measurements of lengths (L, cm) and widths (W, cm) of second uppermost leaves are shown. D, Schematic representation of the T-DNA flanking region of the Sg1-D mutant. A candidate for the ORF responsible for the phenotype (Os09g0459200, which encodes a novel protein with unknown function, colored blue), referred to as SG1, is located 1.4-kb downstream of the T-DNA insertion. The cauliflower mosaic virus 35S minimal promoter (P35S) and a tetramer of the 35S enhancer (4×EN) are shown by the black triangles and the green box, respectively. LB and RB are left and right borders of the T-DNA. Pnos, Nopaline synthase promoter sequence; PAT, ORF-containing region of the phosphinothricin acetyltransferase gene. E, The levels of SG1 mRNA in the WT, Sg1-D heterozygote, and Sg1-D homozygote plants. For each genotype, RNA from two independent plants was used for qPCR, and values were normalized using RUBQ2 as a standard. F, The Sg1-D mutant shows reduced panicle internode elongation in a dose-dependent manner. Bases of the terminal spikelets and nodes of the primary rachis branches are connected with dotted lines. Asterisks show that significantly different from WT. **, P < 0.01; ***, P < 0.001 (Student’s t test).
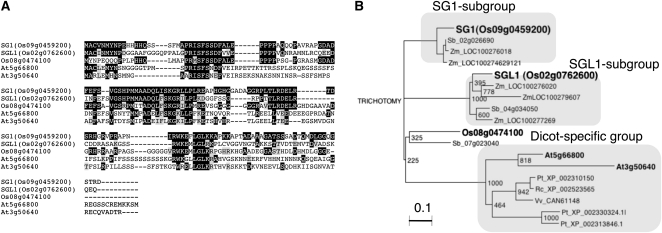
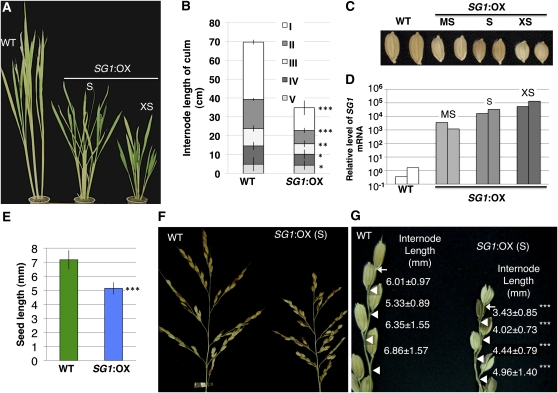
The full-length cDNA (AK110733) for SG1 has already been isolated from panicles by the Japanese full-length rice cDNA project (Kikuchi et al., 2003). The SG1 cDNA (AK110733) is 1,139-bp long and contains a 474-bp open reading frame (ORF) that encodes 157 amino acids (Fig. 2A). Figure 2B shows the phylogenetic relationship between the encoded protein and related proteins from other species (discussed in the section “SG1 Encodes a Novel Protein That Is Conserved among Angiosperms”). To confirm whether overexpression of SG1 is necessary for the short-grain phenotype, we transformed wild-type rice with an expression vector that drives SG1 cDNA (AK110733) under the control of the maize (Zea mays) ubiquitin promoter. As shown in Figure 3, the SG1:OX plants showed various degrees of dwarfism and of the short-grain phenotype (Fig. 3, A–C). The magnitude of the change in the phenotypes was apparently correlated with the levels of SG1 expression (Fig. 3D). We therefore concluded that SG1 is the gene responsible for the Sg1-D dominant mutant.
Figure 2.
Amino acid sequences and phylogenetic analysis of the SG1 and SGL1 proteins. A, Sequence alignment of the SG1, SGL1, and SG1-like proteins in rice and Arabidopsis. Amino acid residues identical and similar to those of SG1 are shaded in black and gray, respectively. The multiple sequence alignment was performed using the CLUSTAL W analysis tool provided by the DNA Data Bank of Japan. B, Phylogenetic relationships among the SG1, SGL1, and SG1-like proteins in plants. The corresponding sequence alignment is shown in Supplemental Figure S2. Bootstrap values from 1,000 replicates are indicated at each node. At, Arabidopsis; Os, O. sativa; Pt, Populus trichocarpa; Rc, Ricinus communis; Sb, S. bicolor; Vv, Vitis vinifera; Zm, Z. mays. The bar corresponds to 0.1 amino acid substitutions per site.
Figure 3.
Phenotype of the SG1:OX rice plants. A, Mature plants at the heading stage. Wild-type (WT) and SG1:OX plants with the short-grain phenotype (S) and the extremely short-grain phenotype (XS) are shown. B, Patterns of internode elongation of the WT and SG1:OX plants with the short-grain phenotype (S). More than 10 culms of the wild-type and SG1:OX plants were measured. I to V represent the internode number, with I representing the uppermost internode. C, Seeds of wild-type plants, and of SG1:OX plants with moderately short (MS), short (S), and extremely short (XS) length are shown. D, qPCR analysis of SG1 mRNA in the wild-type and SG1:OX plants. Symbols (MS, S, XS) correspond to those in A to C. E, Comparison of seed length in the WT and SG1:OX plants. Values are means ± sd for more than 250 seeds. F, Panicles of WT plants and SG1:OX plants with the short-grain phenotype. G, Close-up view of the primary rachis branches. Arrows and triangles indicate the bases of the terminal spikelets and of the pedicels, respectively. Values are means ± sd for the length of each internode (mm). At least 30 internodes were measured for each line of plants. Asterisks show that significantly different from WT. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student’s t test).
SG1 Is Preferentially Expressed in the Developing Panicle and in the Roots
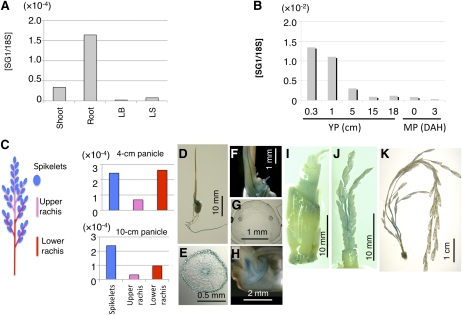
We examined the organ specificity of SG1 expression using quantitative real-time reverse transcription (RT)-PCR (qPCR; Fig. 4, A and B). During vegetative growth, SG1 was preferentially expressed in the roots. During reproductive growth, SG1 was strongly expressed in the young panicles (0.3–1 cm in length), and expression then gradually decreased as the panicles matured. In the developing panicles, SG1 was expressed most strongly in the spikelets and the basal parts of rachis and rachis branches (Fig. 4C). The organ specificity of SG1 expression was confirmed by experiments using SG1::GUS reporter plants (Fig. 4, D–K). During vegetative growth, GUS activity was detected in the primary root, the vascular bundle of the coleoptile, and the embryo of 7-d-old seedlings, as well as in the vegetative nodes (Fig. 4, D–I). In the developing panicles, GUS activity was observed most strongly in the basal parts of rachis and rachis branches (Fig. 4, J and K). However, grain hulls did not show GUS activity.
Figure 4.
Expression analysis of SG1 using qPCR and SG1::GUS transgenic plants. A and B, qPCR analysis of SG1 in various organs of wild-type plants. Expression of SG1 was normalized using 18S rRNA. A, Vegetative organs. Shoots and roots of 10-d-old seedlings and leaf blades (LB) and leaf sheaths (LS) of 2-month-old plants were used. B, Reproductive organs. Young panicles (YP) at 0.3, 1, 5, 15, and 18 cm and mature panicles (MP) at 0 and 3 d after heading (DAH) were used. C, qPCR analysis of SG1 in various parts of the young panicle. Schematic representation of the rice panicle (left). Young panicles were dissected into three parts: spikelets, upper rachis, and rachis branches, and lower rachis and rachis branches. These parts were then used for qPCR analysis (right). qPCR analyses are shown for SG1 in the young panicles at the 4-cm stage and the 10-cm stage. Expression of SG1 was normalized using 18S rRNA. D to K, Histochemical staining of transgenic plants harboring the SG1::GUS construct. D to H, Seven-day-old seedlings: Views are of the whole plant (D), a cross section of the seminal root (E), a close-up of the coleoptile (F), a cross section of the coleoptile (G), and a cross section of the embryo (H). I, Vegetative node of the 2-month-old plant. J and K, Young panicles: at the 2-cm stage (J) and 10-cm stage (K) are shown.
SG1 Encodes a Novel Protein That Is Conserved among Angiosperms
A BLAST search using the SG1 amino acid sequence revealed that SG1 does not share significant identity with any proteins of known function. However, SG1 and related proteins comprise a family that is conserved among monocots and dicots (Fig. 2, A and B; Supplemental Fig. S2). The rice genome encodes two SG1-related proteins (Os02g0762600 and Os08g0474100) that have 49% to 53% sequence identity with SG1. We designated Os02g0762600 as SGL1, since it was expressed at levels similar to those of SG1 in the rice panicle (Supplemental Fig. S3). The full-length cDNA for SGL1 has also been isolated by the full-length rice cDNA project (AK110321). Interestingly, the cDNAs for both SG1 and SGL1 have relatively long 3′-untranslated regions (UTRs): 540 bp for SG1 and 1,016 bp for SGL1. On the other hand, Os08g0474100 is likely to be produced at lower levels than SG1 and SGL1, since no corresponding full-length cDNA or EST have been cloned. In fact, the spatial and temporal profiles of Os08g0474100 expression retrieved from the Rice Expression Profile Database (Sato et al., 2011) were similar to those of SG1 and SGL1, although the signal intensity was weak.
Phylogenetic analysis revealed that SG1-like proteins can largely be classified into three subgroups: SG1, SGL1, and dicot-specific groups (Fig. 2B). Members of the dicot-specific group are more distant from the SG1 subgroup than from the SGL1 subgroup. Therefore, the SG1 and SGL1 subgroups are likely to have diverged from a common ancestor after diversification of the dicot and monocot species groups. In addition to the SG1 and SGL1 subgroups, Os08g0474100 and its putative ortholog (SORBIDRAFT_07g023040) in sorghum (Sorghum bicolor Moench) form another small subgroup.
The SG1 protein contains no putative sequence motifs that define a specific subcellular localization or extracellular secretion. In fact, fusion proteins of SG1 with GFP that are transiently expressed in rice seedlings were uniformly distributed throughout the bombarded cells (Supplemental Fig. S4).
SG1 Overexpression Confers a BR-Insensitive Phenotype
Phenotypes of the SG1:OX plants and the Sg1-D mutant were similar to those of BR-deficient mutants with respect to the degree of dwarfing and their erect, short, wide, and dark-green leaves. In addition, the Sg1-D mutant showed a pattern of culm compression that was most obvious in the second internode (Supplemental Fig. S5). These two phenotypes, erect leaf, and the second internode-specific culm compression, are common and specific features of BR-related dwarf mutants in rice (Yamamuro et al., 2000; Hong et al., 2005; Oki et al., 2009a). In contrast, these phenotypes have not been reported from other classes of dwarf mutants including GA-defective mutants (Sato et al., 1999; Sasaki et al., 2002; Itoh et al., 2004). Therefore, we hypothesized that biosynthesis of BRs or BR signaling may be impaired in the SG1:OX plants.
First, we compared levels of endogenous BR intermediates in the SG1:OX plants with those in wild-type plants. We found no major differences in BR levels between wild-type and SG1:OX plants that depended on the magnitude of the change in the phenotype (Supplemental Fig. S6A). In addition, the transcriptional level of a BR biosynthetic gene (OsBR6ox/BRD1) that encodes BR-6-oxidase was not significantly affected in the SG1:OX plants (Supplemental Fig. S6, B and C).
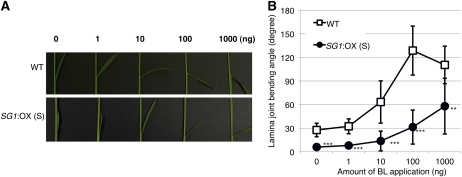
Next, we tested the response of the SG1:OX plants to exogenous brassinolide (BL) using the lamina-joint inclination assay (Fujioka et al., 1998; Fig. 5). Compared with the wild-type plants, the SG1:OX plants showed reduced bending in response to various concentrations of BL. The decrease in the BR response of lamina-joint inclination was similar to that of BR-insensitive mutants (Oki et al., 2009b). Therefore, SG1 may play an inhibitory role in BR signaling or in the response to BRs.
Figure 5.
SG1:OX plants showed a BR-insensitive lamina-joint phenotype. A, Typical response of the lamina joint of the second leaf of wild-type plants (WT) and SG1:OX plants (overexpressor) treated with 0, 1, 10, 100, and 1,000 ng of BL. B, The response of the bending angle to BL as a function of dose in the WT and SG1:OX plants. Data represent means ± sd of the results from at least six plants. Asterisks show that significantly different from WT. **, P < 0.01; ***, P < 0.001 (Student’s t test).
We also tested the possibility whether GA signaling might be down-regulated in SG1:OX plants. The SG1:OX plants responded to the exogenous GA3 in a dose-dependent manner similar to wild type (Supplemental Fig. S7). Therefore GA signaling is not responsible for the dwarf phenotype of SG1:OX.
SG1 Decreases the Elongation of Rice Seeds and of Internodes in Rachis Branches
Next, we characterized the short-grain and short-panicle phenotypes of SG1:OX plants in detail. SG1:OX plants have a shorter panicle than the wild-type plants (Fig. 3F). Since SG1 is expressed in the spikelets and in the rachis and rachis branches (Fig. 4), we compared the lengths of these organs in SG1:OX and wild-type plants (Fig. 3, F and G). The lengths of the seeds and internodes in the rachis branches were measured from images of rice panicles using custom-developed software (Supplemental Fig. S8). The average seed length of the SG1:OX plants was 29% shorter than those of the wild-type plants (Fig. 3E). On the other hand, internode lengths in the rachis branches of the SG1:OX plants were 27% to 43% shorter than those in wild-type plants (Fig. 3, F and G). The internodes in the rachis branches in Sg1-D mutants were also consistently and dose-dependently shorter than those of the wild-type plants (Fig. 1F). Considering its expression in the rice panicle and the overexpression phenotype, SG1 may therefore play an inhibitory role in the growth of seeds and of the internodes in rachis branches.
SGL1 Also Decreases Elongation of Rice Seeds and of the Internodes in Rachis Branches
The SGL1 protein shows moderate sequence identity (49%) with SG1. In addition, SGL1 is also preferentially expressed in the developing panicles (Supplemental Fig. S3, B and C). However, the specificity of SGL1 expression to various developmental stages differed from that of SG1. SG1 is strongly expressed in young panicles, and its expression gradually decreases as they mature (Fig. 4B), whereas SGL1 expression increased toward panicle maturity (Supplemental Fig. S3B). As in the SG1::GUS plants, GUS expression was detected in the rachises and branches in developing panicles of the SGL1::GUS plants (Supplemental Fig. S3C). In addition, GUS staining was also detectable in the grain hulls (Supplemental Fig. S3, D and E). Considering SGL1’s structural similarity and overlapping expression with SG1 in the panicles, SGL1 may also play a role in the regulation of seed and panicle development in rice. To address this possibility, we created and characterized transgenic rice plants that overexpressed SGL1 under the control of the maize ubiquitin promoter (Supplemental Fig. S9). We obtained SGL1:OX plants that express SGL1 at 38 to 78 times the level in the wild-type plants (Supplemental Fig. S9, A and B). As expected, the SGL1:OX plants showed dwarfing (Supplemental Fig. S9A), short grains (Supplemental Fig. S9A), and dense panicles (Supplemental Fig. S9, D and E), similar to the changes observed in the SG1:OX plants (Fig. 3). In addition, SGL1:OX plants showed a phenotype with erect leaves (Supplemental Fig. S9D), similar to that of BR-deficient mutants. In comparison with the wild-type plants, the SGL1:OX plants showed a 45% decrease in plant height due to a uniform decrease of culm internode elongation (Supplemental Fig. S9F). Lengths of seeds, panicles, and internodes in the rachis branches were 17%, 39%, and 31% shorter than those of the wild-type plants, respectively, and all differences were statistically significant (Supplemental Fig. S9, G to I; P < 0.001). Further, we tested BR response of SGL1:OX plants by lamina-joint inclination assay (Supplemental Fig. S10). The SGL1:OX plants showed a partially reduced response of lamina-joint inclination to BR, which suggests that SGL1 also down-regulates signaling or response of BR in rice.
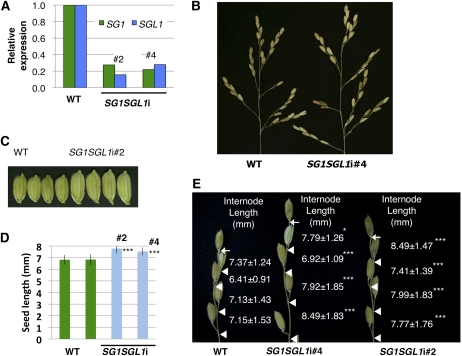
Down-Regulation of Both SG1 and SGL1 Caused Elongation of Rice Seeds and of the Internodes between Spikelets in the Rachis Branches
To explore the intrinsic functions of SG1 and SGL1, we analyzed the phenotypes produced by loss of function of these genes. Since we could not find T-DNA- or transposon-tagged mutants of SG1 and SGL1, we created RNAi-knockdown plants for SG1 and SGL1 using an inverted-repeat-mediated RNAi vector (the pANDA vector; Miki and Shimamoto, 2004; Miki et al., 2005). As expected, the SG1SGL1-RNAi double-knockdown plants showed the opposite phenotype of the overexpressor plants in terms of the lengths of the seeds and of the internodes of the rachis branches (Fig. 6). These RNAi lines showed SG1 and SGL1 expression at 18% to 28% of the levels in the wild-type plants (Fig. 6A). The lengths of seeds and of internodes in the rachis branches were 11% to 14% and 6% to 20% longer, respectively, than those of the wild-type plants (Fig. 6, B–E). On the other hand, overall plant morphology and size did not appear to be altered in these SG1SGL1-RNAi lines.
Figure 6.
Phenotypes of RNAi knockdown plants that down-regulated expression of both SG1 and SGL1. A, Expression of SG1 and SGL2 in the wild-type (WT) and SG1SGL1i RNAi knockdown lines. B, Panicles of WT and SG1SGL1-RNAi plants. C, Seeds of WT and SG1SGL1-RNAi lines. D, Comparison of seed lengths of WT and SG1SGL1-RNAi lines. Values represent means ± sd of at least 25 measurements. E, Close-up view of the primary rachis branches of the wild-type and SG1SGL1-RNAi lines. Bases of the terminal spikelets and of the pedicels are indicated by arrows and triangles, respectively. Values are shown for means ± sd based on at least 30 rachis internodes for each plant. Asterisks show that significantly different from WT. *, P < 0.05; ***, P < 0.001 (Student’s t test).
We also created RNAi-knockdown plants that down-regulated only SG1 (Supplemental Fig. S11). The SG1-RNAi lines decreased the expression of SG1 in panicles to 25% to 35% of the level in the wild-type plants (Supplemental Fig. S11A). Lengths of seeds and of the internodes between spikelets were 5% to 8% and 3% to 20% longer, respectively, than those of wild-type plants (Supplemental Fig. S11, B and C). We also created RNAi-knockdown plants that down-regulate only SGL1. The SGL1-RNAi lines decrease the expression of SGL1 in panicles to 19% to 35% of the level in the wild-type plants (Supplemental Fig. S12A). Lengths of seeds and of the internodes between spikelets were 3% to 7% and 4% to 20% longer, respectively, than those of wild-type plants (Supplemental Fig. S12B). The elongation of seeds and internodes in the SG1SGL1-RNAi double-knockdown lines was greater than in the single RNAi lines that down-regulate either SG1 or SGL1. Therefore, SG1 and SGL1 redundantly decrease the elongation of seeds and internodes during panicle development.
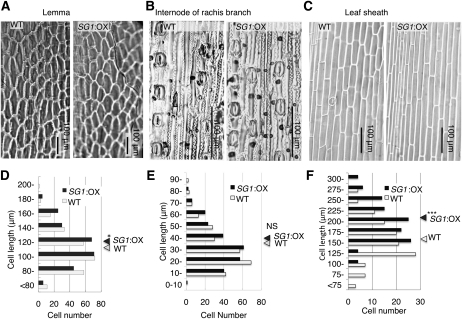
SG1 Decreases Organ Elongation without Decreasing Cellular Elongation
To clarify whether a reduction of cell number or cellular elongation in comparison with the wild-type plants was responsible for the reduction of organ size in the SG1:OX plants, we compared the cellular morphology of various organs in the two groups of plants (Fig. 7, A–C). The inner epidermis of the lemma of the wild-type and SG1:OX plants was observed (Fig. 7A). Even though the seeds of SG1:OX plants were 29% shorter than those of the wild-type plants, the length of the inner epidermal cell of the lemma of the SG1:OX plants was slightly longer than those of the wild-type plants (Fig. 7D, P < 0.05). Internodes of the rachis branches were also examined (Fig. 7B). There was no significant difference between the SG1:OX and wild-type plants with respect to the lengths of the epidermal cells between the stomata (Fig. 7E). In the case of the leaf sheaths, the epidermal cells of SG1:OX were 20% longer than those of the wild type (Fig. 7, C and F, P < 0.001). These results suggest that a decrease in the number of cells and not in cell length was mainly responsible for the reduction of organ size in the SG1:OX plants.
Figure 7.
Cellular sizes of the SG1:OX plants are comparable to those of the wild-type (WT) plants. A to C, Comparisons of epidermal imprint images of the WT and SG1:OX plants. A, Adaxial surface of the lemma. B, An internode of the rachis branch. C, The adaxial surface of the leaf sheath. D to F, Size distribution of epidermal cell lengths. Triangles at the right of each plot show the mean cellular length of the WT and SG1:OX plants. D, Lower epidermis of the lemma. E, Epidermal cells between the stomata in the rachis branch. F, Adaxial surface of the leaf sheath. For both types of plants, 250 cells (D and E) or 120 cells (F) from at least five independent organs were measured. Asterisks show that significantly different from WT. *, P < 0.05; ***, P < 0.001 (Student’s t test). NS, Nonsignificant (P > 0.05).
DISCUSSION
Several rice genes that control grain size (Fan et al., 2006; Song et al., 2007; Shomura et al., 2008; Takano-Kai et al., 2009; Kitagawa et al., 2010) and panicle traits (Huang et al., 2009; Piao et al., 2009; Zhou et al., 2009; Zhu et al., 2010) have been isolated by means of conventional map-based cloning approaches. These conventional approaches are only available when allelic differences or mutation phenotypes are apparent. In contrast, we isolated SG1 by means of a gain-of-function approach (i.e. activation tagging). To identify gene function, gain-of-function approaches such as activation tagging (Kakimoto, 1996) and the full-length cDNA overexpressor gene (FOX) hunting system (Ichikawa et al., 2006; Nakamura et al., 2007) have advantages over other genetic approaches, especially when redundant genes play the same or overlapping roles (Tsuchida-Mayama et al., 2010). In our case, single-knockdown SG1 or SGL1 plants showed only minor phenotypic differences compared with the wild type due to the functional redundancy of these genes. However, because SG1 does not contain any known functional domain or organelle-targeting signal, it is difficult to estimate the function of its protein from its amino acid sequence. Therefore, the role of SG1 in the control of organ length is likely to be identified only by using gain-of-function approaches.
SG1 and SGL1 are strongly expressed in roots and developing panicles (Fig. 4; Supplemental Fig. S3). In developing panicle, activities of SG1::GUS and SGL1::GUS were detected in rachis and rachis branches (Fig. 4; Supplemental Fig. S3), where organ length increased in the SG1SGL1 RNAi plants. In the SG1SGL1 RNAi plants, length of spikelet hulls also increased (Fig. 6, C and D). However, we couldn’t detect SG1::GUS activity in the spikelet hulls (Fig. 4, I and J). In contrast, expression of SG1 was detected in spikelet by qPCR analysis (Fig. 4C). In the case of promoter fusion analysis, 5′-upstream region is often insufficient to accurately reflect the in vivo expression of the gene of interest. The SG1::GUS construct contained only a 2-kb 5′-upstream region, which may not be enough to confer spikelet hull expression of SG1.
Moreover SG1::GUS activity was also detected in the coleoptile and vegetative stem. These results suggested that SG1 and SGL1 may also serve some unidentified function in these organs. However, we could not find major differences between the wild-type and SG1/SGL1 double-knockdown plants in terms of the sizes of these organs. Residual activities of SG1, SGL1, or both in the knockdown lines may be sufficient to allow normal development of these organs. Otherwise, these genes may play only minor roles in the control of the size of these organs.
We found that overexpression of SG1 decreased both the response to BRs and organ elongation. SG1:OX plants were similar to rice BR-related mutants with respect to their short grains, semidwarf phenotype, and dark-green erect leaves. Since the SG1:OX plants were insensitive to BRs, the decrease of organ elongation may be caused by a decrease in the response to BRs. However, the explanation does not seem to be so simple because the dwarf phenotype of SG1:OX differed from those of BR-related rice mutants at a cellular level. In the case of SG1:OX plants, a decrease in the number of cells was mainly responsible for the dwarf phenotype. In contrast, the dwarf phenotypes of a BR-insensitive mutant that had a defect in the OsBRI1 gene, d61 (Yamamuro et al., 2000), and of a BR-deficient mutant, brd1 (Hong et al., 2002; Mori et al., 2002), were mainly caused by decreased cellular elongation. It has also been reported that BRs control both the expansion and the proliferation of leaf cells in Arabidopsis (Arabidopsis thaliana; Nakaya et al., 2002). Therefore, it is probable that SG1 only decreases cell proliferation via a mechanism that occurs downstream of the response to BRs.
The SG1:OX phenotype is similar to that of d1, a rice mutant that is deficient in the heterotrimeric RGA1 (Ashikari et al., 1999). Like BR-related dwarf mutants, the d1 mutant is insensitive to BRs and shows an abnormal phenotype, with dwarfism, small and round seeds, and dark-green erect leaves (Wang et al., 2006; Oki et al., 2009a). SG1:OX is more similar to the d1 mutant than to other BR-related mutants such as d61, since the dwarf phenotype of the d1 mutant is mainly caused by a reduction in the number of cells in various organs (Izawa et al., 2010). The phenotypic similarity between SG1:OX plants and the two BR-insensitive dwarf mutants, d61 and d1, raises the possibility that either OsBRI1 or RGA1 may control the expression of SG1, SGL1, or both in developing panicles. However, the expression levels of SG1 and SGL1 were not altered in loss-of-function mutants for OsBRI1 or RGA1 (Supplemental Fig. S13). The expression levels of OsBRI1 and RGA1 were also not significantly affected by overexpression of SG1 (Supplemental Fig. S14).
Besides BR-related mutants and the d1 mutant, several other mutants or loci that affect grain length have been reported. GS3, a major QTL that controls grain length (Takano-Kai et al., 2009), also acts as a negative regulator of organ size (Mao et al., 2010). DEP1/qPE9-1 is a QTL used for breeding lines with a dense and erect panicle (Huang et al., 2009; Zhou et al., 2009) and also shows pleiotropic effects on grain length (Zhou et al., 2009). DEP2/SMALL AND ROUND SEED1 (SRS1) encodes a protein with unknown function that controls the elongation of panicles and grains via regulation of both cell number and cell elongation (Abe et al., 2010; Li et al., 2010). SRS3 encodes a kinesin 13 protein (Kitagawa et al., 2010) and EP3 (Piao et al., 2009) encodes an F-box protein; both of these are also involved in the control of grain length. SG1 may regulate organ elongation by controlling expression levels of these genes. To test this possibility, we compared expression levels of genes that are involved in development of the panicles and spikelets (Supplemental Fig. S15). However, the expression levels of these genes were not significantly affected in the SG1:OX plants. Therefore SG1 and SGL1 do not appear to regulate the expression of these panicle- and grain-related genes.
Because spatial and temporal expression patterns of these grain-related genes overlap with those of SG1 and SGL1, these genes may act cooperatively to control the lengths of grains and of internodes in panicles. Studies of the consequences of RNAi-based down-regulation and ectopic expression of SG1 and SGL1 in the plants containing mutant alleles of these panicle and grain-related genes would be helpful to reveal any genetic interactions between SG1 and SGL1 and these genes in the control of organ size.
To understand the molecular mechanism by which SG1 decreases cell proliferation, we compared the expression levels of six genes related to the cell cycle, CYCLIN B1;1 (CycB1;1), CycB2;1, CycB2;2, CycD3;1, CYCLIN-DEPENDENT KINASE B;1(CDKB;1), and CDKB;2 (La et al., 2006; Guo et al., 2007) in SG1:OX plants with those in wild-type plants. Unfortunately, we could not find obvious difference between expression levels in the SG1:OX and wild-type plants (data not shown).
As mentioned above, we have not found any structural clue that would elucidate the molecular functions of SG1. Proteomic approaches, such as a yeast (Saccharomyces cerevisiae) two-hybrid experiment or copurification of protein complexes using TAP-tagged SG1 protein, might identify the partners of SG1 and provide clues that would reveal the biochemical function of SG1.
Orthologs of SG1 and SGL1 exist in other cereals, such as maize and sorghum (Fig. 2). In addition, moderately similar proteins are conserved among several dicotyledonous plants, including Arabidopsis, Populus, Ricinus, and Vitis (Fig. 2; Supplemental Fig. S2). Expression patterns of these SG1-related proteins have not yet been investigated. However, the EST profile of an SG1 ortholog in maize (LOC100276018) can be retrieved from the UniGene database (http://www.ncbi.nlm.nih.gov/UniGene, ID Zm.4194). Consistent with our observations for SG1 expression, the EST sequences for the maize SG1 ortholog were preferentially isolated from inflorescence tissues (ears and tassels) and from roots. Therefore, the roles of SG1 and SGL1 orthologs in the control of organ length are likely to be conserved among other cereals. Similar approaches can be applied to other cereals to modify the lengths of grains and of the internodes in panicles by modulating the functions of these homologous genes. However, to guide such efforts, it will be necessary to clarify the functions of SG1-related proteins in dicotyledonous species in future research.
MATERIALS AND METHODS
Plant Materials
Rice (Oryza sativa) wild-type plants (japonica cultivars: Nipponbare and T65), BR-insensitive mutants (d61-2 [Yamamuro et al., 2000] and d1 [Ashikari et al., 1999]), and transgenic plants were grown under greenhouse conditions at 27°C to 30°C. The Sg1-D mutant was isolated from a previously described library of rice activation-tagging lines (Mori et al., 2007).
Isolation and Analysis of Nucleic Acids
Isolation, purification, and Southern-blot analysis of rice genomic DNA were performed as previously described (Mori et al., 2007). The 3′-flanking region of the T-DNA insertion was isolated by means of thermal asymmetric interlaced PCR (Liu et al., 1995) and sequenced as previously described, except that the RB3 primer (5′-AACGTGACTCCCTTAATTCTCCG-3′) was used instead of RBR80R (Mori et al., 2007).
Total RNA was isolated and purified from rice tissues as previously described (Tanaka et al., 2009). Using the RNA, cDNA synthesis and qPCR were performed as previously described (Tanaka et al., 2009) using the primers shown in Supplemental Table S1.
Plasmid Construction and Rice Transformation
For overexpression of SG1 and SGL1 under the control of maize (Zea mays) ubiquitin promoter in rice, we constructed the expression vectors as follows: Full-length cDNA clones for SG1 (AK110733) and SGL1 (AK110321), which were provided by the Rice Genome Resource Center of the National Institute of Agrobiological Sciences, were cloned into an entry vector (pDONR/ZEO; Invitrogen) using the BP-clonase reaction according to the manufacturer’s instructions. The full-length cDNA clones in the entry vector were then cloned into pRiceFOX-GateA, a modified version of the pRiceFOX vector (Nakamura et al., 2007) that is compatible with the gateway recombination system, using the LR-clonase (Invitrogen) reaction.
We constructed a gene-silencing vector to down-regulate both SG1 and SGL1 as follows: The 3′-UTR fragments of SG1 and SGL1 were amplified by means of PCR using the primers in Supplemental Table S1 and their full-length cDNA as templates (the first PCR). We then constructed chimeric fragments of the 3′-UTR fragments of SGL1 and SG1 by means of nested PCR to fuse the first PCR products (the second PCR). The chimeric PCR fragments were cloned into the pENTR/D-TOPO vector (Invitrogen) and used to make an inverted-repeat construct in the pANDA vector (Miki and Shimamoto, 2004; Miki et al., 2005).
We constructed a gene-silencing vector specific to SG1 as follows: The 3′-UTR of SG1 was amplified by means of PCR using the primers in Supplemental Table S1 and its full-length cDNA as templates. The PCR fragment was then cloned into the pENTR/D-TOPO vector and used to make an inverted-repeat construct in the pANDA vector.
A gene-silencing vector specific to SGL1 was constructed following substantially the same approach used for the SG1-specific silencing plasmid, except that SGL1-cDNA and SGL1-specific primers in Supplemental Table S1 were used for the PCR.
To construct the SG1::GUS reporter plasmid, we amplified a 2-kb promoter region of SG1 from rice genomic DNA by means of nested PCR using the primers shown in Supplemental Table S1. We then digested the SG1-promoter fragment with SbfI and HindIII, and ligated it into pSMAHdN632L-M2GUS (Hakata et al., 2010) between its compatible sites. The SGL1::GUS reporter plasmid was constructed following substantially the same approach used for the SG1::GUS plasmid, except that the BglII and NcoI sites were used to introduce the SGL1-promoter fragment into the GUS vector.
For construction of 35S::eGFP:SG1, a region of SG1 containing the ORF was amplified from the SG1 full-length cDNA using the primers in Supplemental Table S1. The PCR product was then digested with the restriction enzymes BamHI and BglII, then was ligated into the expression vector pSAT6-EGFP-C1 (Tzfira et al., 2005) at the BamHI site. Transformation of the rice was performed as previously described (Toki et al., 2006).
Phylogenetic Analysis of SG1-Related Protein
Protein sequences showing similarity to SG1 protein were retrieved using the public BLAST server on the National Center for Biotechnology Information Web site (http://blast.ncbi.nlm.nih.gov/). The phylogenetic analysis was performed using the DNA Data Bank of Japan CLUSTAL W analysis tool (http://clustalw.ddbj.nig.ac.jp/) using the neighbor-joining method, and the resulting phylogenetic tree was visualized using the Dendroscope software (Huson et al., 2007; http://ab.inf.uni-tuebingen.de/software/dendroscope/).
Microscopic Observations
GUS histochemical staining and observation was performed as previously described (Tanaka et al., 2009). Transient expression of the GFP-fusion protein and observation of fluorescence signals of the GFP-fusion protein was performed as previously described (Tanaka et al., 2009).
To observe epidermal morphology, we obtained imprints of the organ surface as follows: Translucent nail polish was spread on the surface of the rice organ. After drying, the resulting replica of the epidermis was transferred to a slide using adhesive tape and examined using an optical microscope at ×100 to ×400.
Quantification of Endogenous BRs
Quantification of endogeneous BRs was performed as previously described (Fujioka et al., 2002) using 5 to 10 g of fresh shoot tissue.
Lamina-Joint Inclination Assay
Seeds of wild type and the SG1:OX plants were dehusked, sterilized, and grown on half-strength Murashige and Skoog agar medium at 25°C. After 7 d, 1 μL of ethanol:dimethyl sulfoxide solution (9:1, v/v) containing 0, 10, 100, or 1,000 ng of BL (Fuji Chemical) was spotted on the lamina joint of second leaf of the seedling. After incubation for 3 d, the angle of the lamina joint was measured (Fujioka et al., 1998).
Measurement of Lengths of Grains and Internodes in Rachis Branches
To measure the lengths of rice grains and of the internodes in the rachis branches, mature panicles that had been air dried for more than 1 week were mounted on a clear sheet in a single layer and photographed with a digital camera system α-100 (Sony). The lengths of grains and internodes in the rachis branches were then measured from the images of the rice panicles using rice panicle measurement software that we developed for this work (Supplemental Fig. S8). Details of the measurement procedure are provided in the caption of Supplemental Figure S8. After measurements from the digital image were complete, we calculated statistical data for organ length using the Microsoft Excel software. Quantitative difference was tested for the significance of difference using Student’s t test.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AK110733 (SG1) and AK110321 (SGL1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Analysis of the T1 progeny of the Sg1-D dominant mutant.
Supplemental Figure S2. Sequence alignment of SG1, SGL1, and related protein sequences from various plants.
Supplemental Figure S3. Organ specificity of SGL1 expression.
Supplemental Figure S4. Subcellular localization of the SG1:eGFP fusion protein.
Supplemental Figure S5. Sg1-D dominant mutant plants show an aberrant pattern of culm internode elongation.
Supplemental Figure S6. SG1:OX did not affect BR biosynthesis.
Supplemental Figure S7. Elongation of the second leaf sheath in response to various concentration of GA treatment in wild-type and SG1:OX plants.
Supplemental Figure S8. Measurements of the lengths of seeds and of the internodes in rachis branches using custom-developed software.
Supplemental Figure S9. Phenotypes of the SGL1:OX rice.
Supplemental Figure S10. SGL1:OX plants show a reduced response to BR in lamina-joint inclination assay.
Supplemental Figure S11. Phenotype of RNAi knockdown plants that down-regulate the expression of SG1.
Supplemental Figure S12. Phenotype of RNAi knockdown plants that down-regulate the expression of SGL1.
Supplemental Figure S13. Expression of SG1 and SGL1 in developing panicles of wild-type plants and BR-insensitive mutants.
Supplemental Figure S14. Expression of OsBRI1 and RGA1 in wild-type and SG1:OX seedlings.
Supplemental Figure S15. Expression of genes that may control the sizes of panicles and grains.
Supplemental Table S1. Primer sequences used in this study.
Acknowledgments
We thank Dr. Ko Shimamoto (Nara Institute of Science and Technology, Japan) and Dr. Tzvi Tzfira (State University of New York) for providing pANDA and pSAT6-EGFP, respectively. We thank Dr. Motoyuki Ashikari (Nagoya University, Japan) and Dr. Hidemi Kitano (Nagoya University) for providing seeds of the d1 mutant and the d61 mutant, respectively. We also thank Dr. Suguru Takatsuto (Joetsu University of Education, Japan) for supplying deuterium-labeled BRs. We thank Dr. Chang-Jie Jiang (National Institute of Agrobiological Sciences, Japan) for helpful advice on the observation of GFP fluorescence signals. We thank Lois Ishizaki, Chiyoko Umeda, and Tomiko Senba (National Institute of Agrobiological Sciences) for their technical assistance and maintenance of the transgenic plants, and Satoru Maeda (National Institute of Agrobiological Sciences) for helpful discussion.
References
- Abe Y, Mieda K, Ando T, Kono I, Yano M, Kitano H, Iwasaki Y. (2010) The SMALL AND ROUND SEED1 (SRS1/DEP2) gene is involved in the regulation of seed size in rice. Genes Genet Syst 85: 327–339 [DOI] [PubMed] [Google Scholar]
- Ashikari M, Wu J, Yano M, Sasaki T, Yoshimura A. (1999) Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the alpha-subunit of GTP-binding protein. Proc Natl Acad Sci USA 96: 10284–10289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Xing Y, Mao H, Lu T, Han B, Xu C, Li X, Zhang Q. (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet 112: 1164–1171 [DOI] [PubMed] [Google Scholar]
- Fujioka S, Noguchi T, Takatsuto S, Yoshida S. (1998) Activity of brassinosteroids in the dwarf rice lamina inclination bioassay. Phytochemistry 49: 1841–1848 [Google Scholar]
- Fujioka S, Takatsuto S, Yoshida S. (2002) An early C-22 oxidation branch in the brassinosteroid biosynthetic pathway. Plant Physiol 130: 930–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Song J, Wang F, Zhang XS. (2007) Genome-wide identification and expression analysis of rice cell cycle genes. Plant Mol Biol 64: 349–360 [DOI] [PubMed] [Google Scholar]
- Hakata M, Nakamura H, Iida-Okada K, Miyao A, Kajikawa M, Imai-Toki N, Pang JH, Amano K, Horikawa A, Tsuchida-Mayama T, et al. (2010) Production and characterization of a large population of cDNA-overexpressing transgenic rice plants using Gateway-based full-length cDNA expression libraries. Breed Sci 60: 575–585 [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Fujioka S, Takatsuto S, Yoshida S, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M. (2005) The rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell 17: 2243–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Shimizu-Sato S, Inukai Y, Fujioka S, Shimada Y, Takatsuto S, Agetsuma M, Yoshida S, Watanabe Y, et al. (2002) Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J 32: 495–508 [DOI] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Umemura K, Uozu S, Fujioka S, Takatsuto S, Yoshida S, Ashikari M, Kitano H, Matsuoka M. (2003) A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15: 2900–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, Xia G, Chu C, Li J, Fu X. (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet 41: 494–497 [DOI] [PubMed] [Google Scholar]
- Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. (2007) Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics 8: 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa T, Nakazawa M, Kawashima M, Iizumi H, Kuroda H, Kondou Y, Tsuhara Y, Suzuki K, Ishikawa A, Seki M, et al. (2006) The FOX hunting system: an alternative gain-of-function gene hunting technique. Plant J 48: 974–985 [DOI] [PubMed] [Google Scholar]
- Itoh H, Tatsumi T, Sakamoto T, Otomo K, Toyomasu T, Kitano H, Ashikari M, Ichihara S, Matsuoka M. (2004) A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol Biol 54: 533–547 [DOI] [PubMed] [Google Scholar]
- Izawa Y, Takayanagi Y, Inaba N, Abe Y, Minami M, Fujisawa Y, Kato H, Ohki S, Kitano H, Iwasaki Y. (2010) Function and expression pattern of the alpha subunit of the heterotrimeric G protein in rice. Plant Cell Physiol 51: 271–281 [DOI] [PubMed] [Google Scholar]
- Kakimoto T. (1996) CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science 274: 982–985 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, et al. (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301: 376–379 [DOI] [PubMed] [Google Scholar]
- Kitagawa K, Kurinami S, Oki K, Abe Y, Ando T, Kono I, Yano M, Kitano H, Iwasaki Y. (2010) A novel kinesin 13 protein regulating rice seed length. Plant Cell Physiol 51: 1315–1329 [DOI] [PubMed] [Google Scholar]
- La H, Li J, Ji Z, Cheng Y, Li X, Jiang S, Venkatesh PN, Ramachandran S. (2006) Genome-wide analysis of cyclin family in rice (Oryza Sativa L.). Mol Genet Genomics 275: 374–386 [DOI] [PubMed] [Google Scholar]
- Li F, Liu W, Tang J, Chen J, Tong H, Hu B, Li C, Fang J, Chen M, Chu C. (2010) Rice DENSE AND ERECT PANICLE 2 is essential for determining panicle outgrowth and elongation. Cell Res 20: 838–849 [DOI] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Mao H, Sun S, Yao J, Wang C, Yu S, Xu C, Li X, Zhang Q. (2010) Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc Natl Acad Sci USA 107: 19579–19584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D, Itoh R, Shimamoto K. (2005) RNA silencing of single and multiple members in a gene family of rice. Plant Physiol 138: 1903–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D, Shimamoto K. (2004) Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol 45: 490–495 [DOI] [PubMed] [Google Scholar]
- Mori M, Nomura T, Ooka H, Ishizaka M, Yokota T, Sugimoto K, Okabe K, Kajiwara H, Satoh K, Yamamoto K, et al. (2002) Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol 130: 1152–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Tomita C, Sugimoto K, Hasegawa M, Hayashi N, Dubouzet JG, Ochiai H, Sekimoto H, Hirochika H, Kikuchi S. (2007) Isolation and molecular characterization of a Spotted leaf 18 mutant by modified activation-tagging in rice. Plant Mol Biol 63: 847–860 [DOI] [PubMed] [Google Scholar]
- Nakamura H, Hakata M, Amano K, Miyao A, Toki N, Kajikawa M, Pang J, Higashi N, Ando S, Toki S, et al. (2007) A genome-wide gain-of function analysis of rice genes using the FOX-hunting system. Plant Mol Biol 65: 357–371 [DOI] [PubMed] [Google Scholar]
- Nakaya M, Tsukaya H, Murakami N, Kato M. (2002) Brassinosteroids control the proliferation of leaf cells of Arabidopsis thaliana. Plant Cell Physiol 43: 239–244 [DOI] [PubMed] [Google Scholar]
- Oki K, Inaba N, Kitagawa K, Fujioka S, Kitano H, Fujisawa Y, Kato H, Iwasaki Y. (2009a) Function of the alpha subunit of rice heterotrimeric G protein in brassinosteroid signaling. Plant Cell Physiol 50: 161–172 [DOI] [PubMed] [Google Scholar]
- Oki K, Inaba N, Kitano H, Takahashi S, Fujisawa Y, Kato H, Iwasaki Y. (2009b) Study of novel d1 alleles, defective mutants of the α subunit of heterotrimeric G-protein in rice. Genes Genet Syst 84: 35–42 [DOI] [PubMed] [Google Scholar]
- Piao R, Jiang W, Ham TH, Choi MS, Qiao Y, Chu SH, Park JH, Woo MO, Jin Z, An G, et al. (2009) Map-based cloning of the ERECT PANICLE 3 gene in rice. Theor Appl Genet 119: 1497–1506 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, et al. (2002) Green revolution: a mutant gibberellin-synthesis gene in rice. Nature 416: 701–702 [DOI] [PubMed] [Google Scholar]
- Sato Y, Antonio BA, Namiki N, Takehisa H, Minami H, Kamatsuki K, Sugimoto K, Shimizu Y, Hirochika H, Nagamura Y. (2011) RiceXPro: a platform for monitoring gene expression in japonica rice grown under natural field conditions. Nucleic Acids Res (Database issue) 39: D1141–D1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Sentoku N, Miura Y, Hirochika H, Kitano H, Matsuoka M. (1999) Loss-of-function mutations in the rice homeobox gene OSH15 affect the architecture of internodes resulting in dwarf plants. EMBO J 18: 992–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Yano M. (2008) Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet 40: 1023–1028 [DOI] [PubMed] [Google Scholar]
- Song XJ, Huang W, Shi M, Zhu MZ, Lin HX. (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet 39: 623–630 [DOI] [PubMed] [Google Scholar]
- Takano-Kai N, Jiang H, Kubo T, Sweeney M, Matsumoto T, Kanamori H, Padhukasahasram B, Bustamante C, Yoshimura A, Doi K, et al. (2009) Evolutionary history of GS3, a gene conferring grain length in rice. Genetics 182: 1323–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe S, Ashikari M, Fujioka S, Takatsuto S, Yoshida S, Yano M, Yoshimura A, Kitano H, Matsuoka M, Fujisawa Y, et al. (2005) A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17: 776–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Nakagawa H, Tomita C, Shimatani Z, Ohtake M, Nomura T, Jiang CJ, Dubouzet JG, Kikuchi S, Sekimoto H, et al. (2009) BRASSINOSTEROID UPREGULATED1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol 151: 669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H. (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47: 969–976 [DOI] [PubMed] [Google Scholar]
- Tsuchida-Mayama T, Nakamura H, Hakata M, Ichikawa H. (2010) Rice transgenic resources with gain-of-function phenotypes. Breed Sci 60: 493–501 [Google Scholar]
- Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V. (2005) pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57: 503–516 [DOI] [PubMed] [Google Scholar]
- Wang L, Xu YY, Ma QB, Li D, Xu ZH, Chong K. (2006) Heterotrimeric G protein alpha subunit is involved in rice brassinosteroid response. Cell Res 16: 916–922 [DOI] [PubMed] [Google Scholar]
- Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M. (2000) Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12: 1591–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhu J, Li Z, Yi C, Liu J, Zhang H, Tang S, Gu M, Liang G. (2009) Deletion in a quantitative trait gene qPE9-1 associated with panicle erectness improves plant architecture during rice domestication. Genetics 183: 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K, Tang D, Yan C, Chi Z, Yu H, Chen J, Liang J, Gu M, Cheng Z. (2010) Erect panicle2 encodes a novel protein that regulates panicle erectness in indica rice. Genetics 184: 343–350 [DOI] [PMC free article] [PubMed] [Google Scholar]