Abstract
The plasma membrane proton gradient is an essential feature of plant cells. In Arabidopsis (Arabidopsis thaliana), this gradient is generated by the plasma membrane proton pump encoded by a family of 11 genes (abbreviated as AHA, for Arabidopsis H+-ATPase), of which AHA1 and AHA2 are the two most predominantly expressed in seedlings and adult plants. Although double knockdown mutant plants containing T-DNA insertions in both genes are embryonic lethal, under ideal laboratory growth conditions, single knockdown mutant plants with a 50% reduction in proton pump concentration complete their life cycle without any observable growth alteration. However, when grown under conditions that induce stress on the plasma membrane protonmotive force (PMF), such as high external potassium to reduce the electrical gradient or high external pH to reduce the proton chemical gradient, aha2 mutant plants show a growth retardation compared with wild-type plants. In this report, we describe the results of studies that examine in greater detail AHA2’s specific role in maintaining the PMF during seedling growth. By comparing the wild type and aha2 mutants, we have measured the effects of a reduced PMF on root and hypocotyl growth, ATP-induced skewed root growth, and rapid cytoplasmic calcium spiking. In addition, genome-wide gene expression profiling revealed the up-regulation of potassium transporters in aha2 mutants, indicating, as predicted, a close link between the PMF and potassium uptake at the plasma membrane. Overall, this characterization of aha2 mutants provides an experimental and theoretical framework for investigating growth and signaling processes that are mediated by PMF-coupled energetics at the cell membrane.
Plants cannot move to soil with sufficient minerals or water; therefore, their growth and development are highly influenced by the availability of solutes and moisture in their immediate environment. Plasma membrane proteins play a fundamental role in ensuring that the nutritional needs of plant cells are met. Unlike the sodium-coupled system of animal cells, most plasma membrane solute transport systems in plants are proton coupled, and an ATP-coupled proton pump creates the protonmotive force (PMF) that energizes these systems (Bush, 1993; Sussman, 1994). The plasma membrane of higher plants is unique because it does not produce the PMF directly by light or sugar metabolism, as found in organelles and prokaryotes (Jagendorf, 1967; Kocherginsky, 2009). Instead, the plasma membrane PMF is energized by ATP hydrolysis via a P-type H+-ATPase consisting of a single polypeptide of 100,000 D (Sussman, 1994). The amino acid sequence and three-dimensional structure indicate that this is a unique type of H+-ATPase, different from those found in organelles and endomembranes (Schaller and Sussman, 1988; Harper et al., 1989; Pedersen et al., 2007).
In Arabidopsis (Arabidopsis thaliana) root cells, a membrane potential in excess of approximately 200 mV and a ΔpH (proton chemical concentration gradient) of approximately 2 (equivalent to approximately 120 mV) both contribute to the large PMF, which provides the driving force for ion channels like those for potassium and calcium and for secondary symporters transporting amino acids and sugars (Buckhout, 1989; Hirsch et al., 1998; Fasano et al., 2001; Fig. 1A). In Arabidopsis, there are 11 plasma membrane proton pump genes, denoted AHA1 to -11 (for Arabidopsis H+-ATPase; Baxter et al., 2003). Throughout the Arabidopsis life cycle, the plasma membrane PMF is maintained by the two most predominantly expressed isoforms, AHA1 and AHA2, and homozygous T-DNA insertions in both genes create embryonic lethality, indicating a collectively essential function (Haruta et al., 2010). The mRNA expression patterns and protein abundances of AHA1 and AHA2 indicate that, together, these proteins generate up to 80% of the overall ATPase activity in Arabidopsis seedlings and in vegetative tissues of adult plants.
Figure 1.
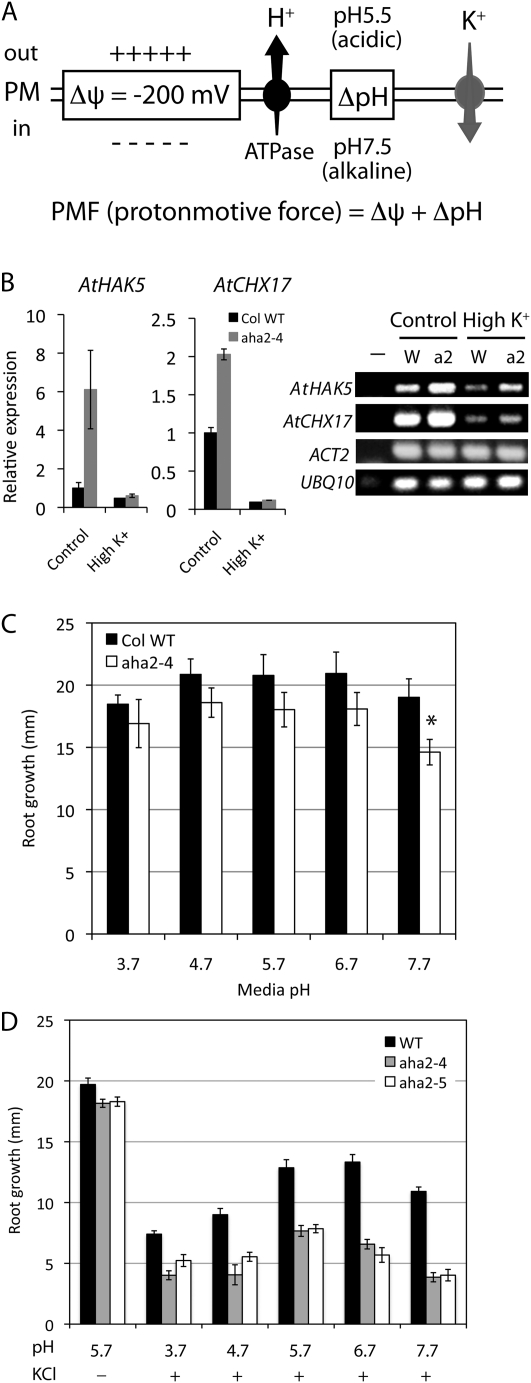
Role of the plasma membrane H+-ATPase in maintaining the PMF. A, Plasma membrane PMF composed of a membrane potential (Δψ) and a proton chemical concentration gradient (ΔpH). The proton gradient (outside high), generated by the H+-ATPase, contributes to both a charge gradient (positive outside) and a pH gradient (acidic outside). High concentrations of potassium entering the cytoplasm result in a reduction of the charge gradient and membrane depolarization. B, Relative expression of K+ transporter genes, AtHAK5 and AtCHX17, in wild-type or aha2 mutant seedlings grown in the presence or absence of 100 mm KCl. The endogenous reference genes, ACT2 and UBQ10, were used to normalize and calculate the relative expression of the K+ transporter genes. The left panels show the quantitative analysis of gene expression, and the right panel shows images of agarose gels presenting PCR products of AtHAK5, AtCHX17, ACT2, and UBQ10. −, Negative control; W, wild type; a2, aha2-4 mutant. C, Root growth of the wild type (WT) and aha2-4 at various pH values. The asterisk indicates a statistical significance between the wild type and the aha2 mutant (P = 0.0036). D, Hypersensitivity of aha2 mutant root growth to reduced PMF. High external pH (reduced ΔpH) and high external potassium (reduced membrane potential) have an additive effect on the impaired growth of aha2 mutants. Plants were grown under various pH values in the presence or absence of 100 mm KCl. Data are shown as means of 14 seedlings ± se.
Plants and fungi have one of the largest membrane potentials of any cell in nature, and a major function of the purely electrical component (the membrane potential) of the PMF is to ensure that a high enough cytoplasmic concentration of potassium is maintained for turgor pressure control (Rodríguez-Navarro, 2000). Since potassium is the major osmolyte in plant cells (Humble and Raschke, 1971), which helps to create the outward force for cell expansion and stomatal opening, turgor pressure can also be very sensitive to changes in membrane potential (Merlot et al., 2007). According to the Nernst potential, each 60 mV of membrane potential is equivalent to a 10-fold gradient of potassium. For example, when the membrane potential is −240 mV, even if the external potassium concentration is low (10 μm), the membrane potential provides enough energy to drive the concentrative uptake of potassium to a much higher level (0.1 m, 104-fold; Maathuis and Sanders, 1994; Hirsch et al., 1998; Spalding et al., 1999). Due to the high permeability of potassium via channels, the cytoplasmic potassium level is very sensitive to changes in the membrane potential (Maathuis and Sanders, 1997). Conversely, when the external potassium level is high, a large influx of this cation will cause a reduction in the membrane potential, which, if not corrected by the proton pump, can cause deleterious effects on other transporters driven by membrane potential.
One can understand the energetic aspects of transport processes at the plasma membrane by thinking of proton and/or charge-coupled transporters as devices that tap into and drain the PMF. The proton pump is the only means to recharge the system by converting the chemical energy of ATP to the PMF; thus, it is thought to play a fundamental role in diverse processes, including pathogen perception, hormone-mediated cell elongation, nutrient acquisition, root gravitropism, stomatal regulation, and salinity stress resistance (Michelet and Boutry, 1995; Schaller and Oecking, 1999; Palmgren, 2001; Hager, 2003; Fuglsang et al., 2007; Liu et al., 2009). Consistent with these predicted roles, mutant tobacco (Nicotiana tabacum) or Arabidopsis plants with altered H+-ATPase expression exhibit several phenotypes related to transport processes (Zhao et al., 2000; Vitart et al., 2001; Gévaudant et al., 2007).
To elucidate the role of plasma membrane energetics on cell elongation, we have focused on measurements of root elongation in knockdown mutants of AHA1 and AHA2. We recently showed that aha2 mutants display phenotypic differences from the wild type only when grown in the presence of toxic ions whose cellular uptake is strongly coupled with the plasma membrane PMF. For example, root growth of the aha2 mutant was more resistant to lithium-, cesium-, and hygromycin-induced inhibition compared with the wild type, similar to observations in yeast proton pump mutants (McCusker et al., 1987; Perlin et al., 1989; Goossens et al., 2000; Williams-Hart et al., 2002). These results are consistent with the hypothesis that the aha2 mutant is resistant to these toxic cations because it has a lower membrane potential and, thus, a smaller driving force for the uptake of these positively charged toxins.
In this report, we describe a transcriptome study of aha1 and aha2 mutants that helps identify molecular changes in the mutants and separates the isoform-specific roles during seedling growth. We further use the aha2 mutant in conditions of high external potassium and high external pH to explore in greater detail the consequences of a reduced PMF on the physiology and growth of Arabidopsis. With this experimental system, we have observed PMF-dependent differences in root and hypocotyl growth, ATP-induced cytoplasmic calcium elevation, and the tropic response of roots. In addition, our observations suggest that the bioenergetics underlying cell expansion in hypocotyls may be fundamentally different from that in roots. In summary, we demonstrate an experimental and theoretical framework by which aha2 knockdown mutants can be used to delineate the role of the plasma membrane PMF in the life of higher plants.
RESULTS
Transcriptome Analysis of aha1-6 and aha2-4 Mutants
We previously demonstrated that the ability of aha1 and aha2 knockdown mutants to grow normally was not due to compensation at the mRNA or protein level of other AHA members, as quantitative RNA and protein measurements did not show a significant increase in the concentration of mRNA or protein derived from any of the 10 other AHA genes (Haruta et al., 2010). We did observe a possible compensation mechanism at the posttranslational level, with an increase in the level of phosphorylation of the penultimate Thr of the wild-type AHA1 or AHA2 proteins in both mutants. Phosphorylation at this residue causes activation of the pump, and this could help maintain the PMF with reduced pump protein. However, it was unclear to what extent this modification would offset the approximately 25% to 50% reduction of overall AHA protein levels.
In order to determine whether other changes in gene expression were present in the homozygous single mutant plants, a transcriptome analysis was performed using the Affymetrix genome array. Seedlings were grown on complete nutrient medium (one-half-strength Murashige and Skoog [MS] medium with 1% [w/v] Suc at pH 5.7), and transcript abundance was compared between the wild type and aha1 or aha2 mutants (Supplemental Fig. S1, A and B). This study demonstrated that aha1 mutants had a 65% reduction in AHA1 expression and that 12 other genes were significantly increased or decreased more than 2-fold (Table I). The proteins encoded by these 12 genes included an oxysterol-binding protein-related protein and lipoxygenase, indicating that lipid metabolism may be altered in the aha1-6 mutant. In the aha2-4 mutant, 27 genes were significantly up- or down-regulated (Table II). We also analyzed the transcriptional profiles of plants carrying other aha1 and aha2 mutant alleles, aha1-7 and aha2-5, with the same results. The very small number of genes that showed a significant change in expression in the two mutants compared with the wild type helps explain the lack of an observable phenotype in these mutants under complete nutrient conditions. However, since the aha2 mutant plants show growth differences such as reduced sensitivity to toxic cations and an enhanced sensitivity to environmental stresses that challenge the plasma membrane PMF, the identities of these 27 genes could provide useful information on the plant’s responses to genetically induced changes in plasma membrane energetics. Among the up-regulated mRNAs observed in aha2 mutants are, notably, a potassium transporter (AtHAK5) and a proton/potassium antiporter (K+/H+ exchanger 17) known to be induced under potassium starvation (Cellier et al., 2004; Gierth et al., 2005; Maresova and Sychrova, 2006). Changes in the expression of potassium transporter genes may provide avenues by which the plant cells compensate for reductions in turgor caused by a reduced membrane potential in the mutant. A gene encoding a d-arabinono-1,4-lactone oxidase family protein that had previously been shown to be up-regulated during potassium starvation (Armengaud et al., 2004) was also up-regulated in the aha2 mutant.
Table I. Up- or down-regulated genes in aha1-6 mutant compared with wild-type plants on the complete nutrient growth condition.
| Fold Change | Pa | Identifier | Annotation |
| Up-regulated genesb | |||
| 3.05 | 0.0498 | At2g43140 | Basic helix-loop-helix (bHLH) DNA-binding superfamily protein |
| 2.77 | 0.0428 | At2g04050 | MATE efflux family protein |
| 2.51 | 0.0373 | At2g04040 | ATDTX1_TX1_MATE efflux family protein |
| 2.26 | 0.0377 | At5g57240 | ORP4C_OSBP (oxysterol-binding protein)-related protein 4C |
| 2.20 | 0.0414 | At3g23150 | ETR2_signal transduction His kinase, hybrid type, ethylene sensor |
| 2.09 | 0.0455 | At5g43450 | 2-Oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
| 2.07 | 0.0020 | At2g39980 | HXXXD-type acyl-transferase family protein |
| 2.06 | 0.0238 | At5g45820 | CIPK20_PKS18_SnRK3.6_CBL-interacting protein kinase 20 |
| 2.02 | 0.0174 | At2g36690 | 2-Oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
| Down-regulated genesc | |||
| 6.14 | 0.0500 | At3g30720 | QQS_qua-quine starch |
| 2.84 | 0.0225 | At2g18960 | AHA1, OST2_PMA_H+-ATPase 1 |
| 2.43 | 0.0036 | At4g03060 | AOP2, alkenyl hydroxyalkyl producing 2 |
| 2.38 | 0.0495 | At3g45140 | ATLOX2_lipoxygenase 2 |
The paired t test was used to calculate the two-tailed P value to determine statistical significance with 95% confidence.
The nine genes that are significantly up-regulated greater than 2-fold in the aha1-6 mutant represent 0.040% of the Arabidopsis genome (22,746 probe sets).
The four genes that are significantly down-regulated greater than 2-fold in the aha1-6 mutant represent 0.018% of the Arabidopsis genome (22,746 probe sets).
Table II. Up- or down-regulated genes in aha2-4 mutant compared with wild-type plants on the complete nutrient growth condition.
| Fold Change (Linear) | Pa | Identifier | Annotation |
| Up-regulated genesb | |||
| 5.29 | 0.0183 | At3g56980 | BHLH039, basic helix-loop-helix (bHLH) DNA-binding protein |
| 4.97 | 0.0043 | At4g36700 | RmlC-like cupins superfamily protein |
| 3.89 | 0.0365 | At4g13420 | ATHAK5, high-affinity K+ transporter 5 |
| 3.30 | 0.0359 | At2g30750 | CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
| 2.96 | 0.0257 | At5g48850 | ATSDI1, tetratricopeptide repeat (TPR)-like superfamily protein |
| 2.86 | 0.0316 | At1g26410 | FAD-binding berberine family protein |
| 2.78 | 0.0208 | At1g47400 | Unknown protein |
| 2.74 | 0.0333 | At2g39510 | Nodulin MtN21, EamA-like transporter family protein |
| 2.72 | 0.0010 | At1g02920/30 | ATGST11, ATGSTF7, GST |
| 2.50 | 0.0082 | At4g37290 | Unknown protein |
| 2.49 | 0.0212 | At2g43590 | Chitinase family protein |
| 2.39 | 0.0134 | At2g02930/2520 | ATGSTF3, GST17, GST |
| 2.34 | 0.0300 | At5g26220 | Cha (cation/H+ antiporter) C-like family protein |
| 2.34 | 0.0358 | At5g57220 | CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
| 2.33 | 0.0300 | At2g46750 | d-Arabinono-1,4-lactone oxidase family protein |
| 2.32 | 0.0453 | At1g12030 | Protein of unknown function (DUF506) |
| 2.27 | 0.0080 | At1g19900 | Glyoxal oxidase-related protein |
| 2.17 | 0.0078 | At3g54040 | PAR1 protein |
| 2.16 | 0.0011 | At4g23700 | ATCHX17, CHX17, cation/H+ exchanger 17 |
| 2.13 | 0.0150 | At1g23020 | ATFRO3, FRO3, ferric reduction oxidase 3 |
| 2.10 | 0.0441 | At5g23220 | NIC3, nicotinamidase 3 |
| 2.01 | 0.0345 | At5g59530 | 2-Oxoglutarate (2OG) and Fe(II)-dependent oxygenase protein |
| Down-regulated genesc | |||
| 17.64 | 0.0074 | At4g30190 | AHA2, HA2, PMA2, H+-ATPase 2 |
| 9.01 | 0.0332 | At3g30720 | QQS, qua-quine starch |
| 2.39 | 0.0440 | AtCg00180 | RPOC1, DNA-directed RNA polymerase family protein |
| 2.12 | 0.0380 | At3g49160 | Pyruvate kinase family protein |
| 2.09 | 0.0016 | At5g01600 | ATFER1, FER1, ferretin 1 |
The paired t test was used to calculate the two-tailed P value to determine statistical significance with 95% confidence.
The 22 genes that are significantly up-regulated greater than 2-fold in the aha2-4 mutant represent 0.097% of the Arabidopsis genome (22,746 probe sets).
The five genes that are significantly down-regulated greater than 2-fold in the aha2-4 mutant represent 0.018% of the Arabidopsis genome (22,746 probe sets).
Other genes up-regulated in the aha2 mutants and previously reported to be induced in nutrient-limited conditions are basic helix-loop-helix transcription factor and ferric reductase (iron starvation; Mukherjee et al., 2006; Vorwieger et al., 2007), tetratricopeptide repeat-like superfamily protein and cation/H+ antiporter-like family protein (sulfate starvation; Nikiforova et al., 2003; Maruyama-Nakashita et al., 2005), and a family of glutathione S-transferases (phosphate starvation; Hammond et al., 2003). We also observed the up-regulation of genes previously found to be induced by pathogen infection (At2g30750; Vellosillo et al., 2007) and by salt stress (At1g12030; Ma et al., 2006). There were five down-regulated genes in the aha2 mutant including, as expected, the AHA2 gene itself. The most greatly affected in this set was a gene encoding a protein involved in starch metabolism, the qua-quine starch protein (Li et al., 2009). This gene was also greatly down-regulated in the aha1-6 mutant; however, there was no causal link between the down-regulation of this starch gene and root gravitropic responses of aha mutants (see below and “Discussion”). The remaining three down-regulated genes showed only small but statistically significant differences.
The transcript abundance of the potassium transporter genes AtHAK5 and AtCHX17 was reexamined with quantitative reverse transcription (RT)-PCR (Fig. 1B). Consistent with the microarray results, we detected the up-regulation of AtHAK5 and AtCHX17 in the aha2-4 mutant compared with wild-type seedlings under normal growth conditions. At high potassium (100 mm KCl), expression of the two genes was suppressed in both the wild type and the aha2-4 mutant. At this high-potassium condition, the differences of AtHAK5 and AtCHX17 transcript abundance between the genotypes were not significant.
Effects of High External pH, High K+, Toxic Cations, and Amino Acids on the Root Growth of aha2 Mutants
Previously, we found that the conditions in which aha2 root growth was reduced compared with the wild type were limited to high potassium (100 mm) and high external pH (pH 8.5; Supplemental Fig. S1D; Haruta et al., 2010). These conditions are expected to reduce the plasma membrane PMF via decreases in the membrane potential (high potassium) or the proton chemical gradient (alkaline medium). In contrast, mutant plants grew better than the wild type in medium containing inhibitory concentrations of positively charged compounds such as aminoglucoside antibiotics (gentamycin and hygromycin), monovalent cations (cesium and lithium), and basic amino acids (Lys and Arg; Supplemental Fig. S1D).
One can predict that if the aha2 mutants have a reduced PMF that does not cause a growth impairment in plants grown under complete nutrient conditions, the effect of reduced pump activity on growth could become evident when the PMF is further challenged. To test this, we grew wild-type and aha2 mutant seedlings under a range of pH values from 3.7 to 7.7 in the presence or absence of 100 mm KCl (Fig. 1, C and D). To eliminate the effect of chemicals on germination and to examine the effect mainly on root elongation, we germinated seeds on control nutrient medium for 3 d before transferring seedlings to stress medium and further incubating for 4 d. The relative growth of aha2 mutants compared with wild-type plants at pH 7.7 in the presence of high potassium decreased significantly in comparison with that at pH 5.7 in the presence of high potassium. This result indicates that reducing both the membrane potential and the proton concentration gradient has additive effects on reducing the root growth of aha2 mutants.
Since the uptake of amino acids across the plasma membrane is known to be mediated in part by proton-coupled cotransporter systems (Li and Bush, 1990), it is likely that aha2 mutants have a reduced uptake of amino acids. Many amino acids externally supplied at high concentrations cause growth retardation in Arabidopsis seedlings (Wu et al., 1994; Voll et al., 2004; Pratelli et al., 2010), possibly by creating metabolic imbalances detrimental to growth. Thus, we predicted that there would be differential growth responses between the wild type and aha2 mutants exposed to high concentrations of amino acids in the growth medium. We first optimized the concentration of each amino acid by finding the concentrations that induced 20% to 80% inhibition in wild-type plants compared with control conditions (one-half-strength MS medium, 1% [w/v] Suc at pH 5.7; Supplemental Fig. S2). The concentration at which a particular amino acid was effective varied, possibly due to differences in the transport and/or catabolic mechanisms for each amino acid or to the endogenous concentrations of those amino acids in Arabidopsis seedlings, as documented by Voll et al. (2004). From these results, we chose the appropriate amino acid concentrations to compare the differential growth responses of the wild type and aha2 mutants.
The aha2 mutant plants showed the most dramatic differences in comparison with the wild type when grown in high concentrations of the basic amino acids, Arg and Lys (Fig. 2, A and B). For example, in [Arg] dose-response analyses, root growth inhibition in wild-type plants was nearly saturated at 5 mm, and the root growth of the aha2-4 plants was over 3-fold greater than in the wild type at 20 mm (Fig. 2C). Only one other amino acid, Val, showed a similar phenotypic difference at high concentrations (Supplemental Fig. S2). Our results thus predict that the uptake of Arg, Lys, and Val is uniquely strongly coupled to the PMF. While this result may seem to be obvious with the positively charged cationic amino acids but not with the neutral amino acid, Val, previous studies have suggested that Val has a unique repertoire of PMF-coupled transporters active in Arabidopsis and pea (Pisum sativum; Fischer et al., 1998; Borstlap and Schuurmans, 2000; de Jong and Borstlap, 2000).
Figure 2.
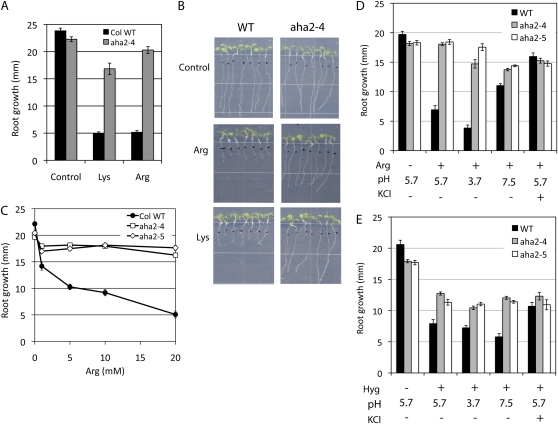
Root growth responses of wild-type (WT) and aha2 mutant plants to high concentrations of basic amino acids. A, Root growth of wild-type and aha2-4 mutant plants in the presence of basic amino acids. Amino acids are supplied to medium at 100 μm Lys or 10 mm Arg. B, Images of root growth on control medium or medium supplied with amino acids. C, Dose-response effect of Arg on the root growth of wild-type and aha2 mutant plants. D, Effects of altered PMF on the root growth of wild-type and aha2 mutant plants on Arg-supplemented medium. Seedlings were grown under various pH values in the presence or absence of 50 mm KCl: the control medium (one-half-strength MS, 1% Suc, pH 5.7), medium supplied with 5 mm Arg, medium adjusted to pH 3.7 and supplied with Arg, medium adjusted to pH 7.5 and supplied with Arg, or medium supplied with Arg and 50 mm KCl. E, Effects of altered PMF on the root growth of wild-type and aha2 mutant plants on hygromycin-supplemented medium. Plants were grown under various pH values in the presence or absence of 50 mm KCl: the control medium (one-half-strength MS, 1% Suc, pH 5.7), medium supplied with 5 μg mL−1 hygromycin, medium adjusted to pH 3.7 and supplied with hygromycin, medium adjusted to pH 7.5 and supplied with hygromycin, or medium supplied with hygromycin and 50 mm KCl. [See online article for color version of this figure.]
The tolerance of aha2 mutants to various species of potentially toxic cations such as hygromycin and Arg suggests that cation uptake is dependent on the PMF, which is composed of both a membrane potential and a chemical gradient of protons. We distinguished the effects of the membrane potential and ΔpH on the growth inhibitory effects of these cations by changing the medium pH to alter the chemical gradient of protons in the presence or absence of high external potassium to reduce the membrane potential. To increase the external pH, the medium pH was adjusted with NaOH to pH 7.5, requiring approximately 1 mm NaOH to reach the final desired pH. The effect of this low concentration of additional sodium on root growth is negligible, and any change in growth will be due to changes in pH rather than sodium concentration. Changing the medium pH from 3.7 to 7.7 caused only a small change in the inhibitory effect of hygromycin on wild-type growth, whereas the same changes in pH had a much larger effect on Arg sensitivity (Fig. 2, compare D and E; Supplemental Fig. S3). In contrast, the Arg and hygromycin effects on root inhibition were both strongly affected by KCl. A possible explanation for these results is that hygromycin uptake is mediated exclusively by membrane potential, while Arg uptake is mediated by both membrane potential and ΔpH.
In order to compare the mechanism of the other toxic cations inducing differential growth between wild-type and aha2 mutant plants, we also examined the effect of the addition of different monovalent cations. Whereas potassium and sodium were inhibitory only at concentrations of 50 mm or higher (Supplemental Fig. S4), lithium and cesium were inhibitory at much lower concentrations (Fig. 3, A and B) for both the wild type and mutants. One possible explanation for the cytotoxicity of these latter elements is a higher affinity for potassium uptake systems, thus preventing the uptake of potassium (Sheahan et al., 1993). However, once inside the cell, these cations may be inhibiting growth by interacting with specific protein targets. For example, cesium has been reported to inhibit the uroporphyrinogen decarboxylase involved in chlorophyll biosynthesis, which is consistent with cesium-induced bleaching of cotyledons (see below). Furthermore, lithium is known to block calcium channel activity that may play a role in growth regulation (Knight et al., 1996; Shalygo et al., 1997; Haruta et al., 2008). More importantly, unlike sodium, the inhibitory effect of cesium and lithium was lower in the mutant compared with wild-type roots. The difference in root elongation in the wild type and aha2 mutants became distinguishable 2 to 3 d after transferring to the stress medium containing 5 mm CsCl or 20 mm LiCl (Supplemental Fig. S5, A and B).
Figure 3.
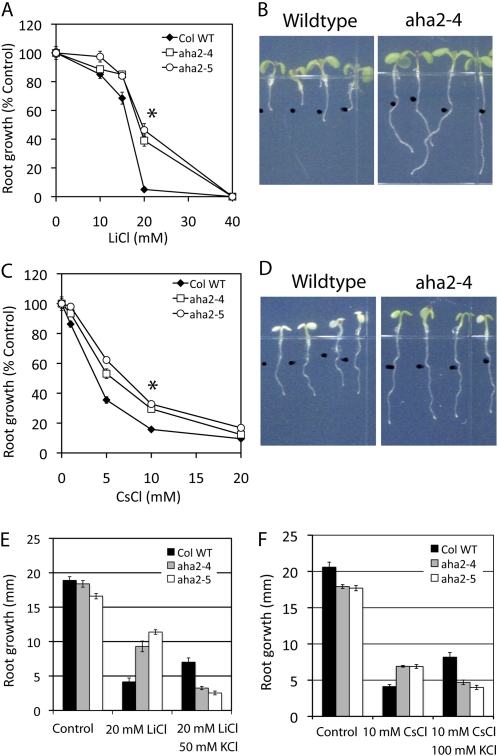
Growth responses of wild-type (WT) and aha2 mutant plants to lithium and cesium. A, Root growth response to LiCl. Seedlings were germinated for 3 d on control medium (one-half-strength MS, 1% Suc, pH 5.7) and further grown on control medium or medium supplemented with LiCl at the concentrations indicated on the x axis. The asterisk indicates statistical significance between the wild type and aha2 mutants (P < 0.0001). B, Images of seedling growth on the medium supplied with LiCl. Seedlings were germinated for 3 d on control medium (one-half-strength MS, 1% Suc, pH 5.7) and further grown on medium supplemented with 20 mm LiCl. C, Root growth responses to CsCl. The asterisk indicates statistical significance between the wild type and aha2 mutants (P < 0.0001). D, Images of seedling growth on medium supplied with CsCl. E, Effect of potassium on lithium-induced root growth inhibition. F, Effect of potassium on cesium-induced root growth inhibition.
Interestingly, the effect of cesium and lithium on wild-type and aha2 mutant root growth was opposite from that of potassium (Fig. 3, A and B; Supplemental Fig. S4). Whereas wild-type roots are less sensitive to growth inhibition by potassium, they are more sensitive to inhibition with cesium and lithium. To gain further insight into the mechanism of inhibition, we compared the effect of 10 mm CsCl and 20 mm LiCl in the presence of high potassium (Fig. 3, E and F). High potassium reverses the mutant-specific resistance to cesium and lithium (Fig. 3, E and F; Supplemental Fig. S5C). When the membrane potential is reduced in the presence of high potassium, the aha2 mutants are inhibited by lithium or cesium to a greater extent than the wild type.
Effect on Hypocotyls of Reduced PMF
While characterizing the cesium-insensitive phenotypes of the aha2 mutant, we observed that the aha2 mutant also exhibited greener cotyledons compared with CsCl-induced bleaching in wild-type seedlings (Fig. 3D). Chlorophyll quantification in seedling extracts from wild-type and aha2 mutant plants showed that aha2 mutants contained 4-fold more chlorophyll than wild-type plants (Fig. 4A). This observation led us to hypothesize that the aerial tissues of the mutant may also have a reduced PMF. To test this, we grew seedlings under low blue light or in the dark to induce hypocotyl elongation and examined the effect of high external potassium on hypocotyl growth (Fig. 4B). The hypocotyl length of the aha2 mutant was not significantly reduced compared with wild-type plants on control medium but was reduced in the presence of 100 mm KCl (Fig. 4C). The reduced hypocotyl length in aha2 mutants was specific to membrane depolarization caused by high potassium and not to osmotic stresses, as we did not detect growth differences in high-sodium or -sorbitol conditions (Fig. 4C).
Figure 4.
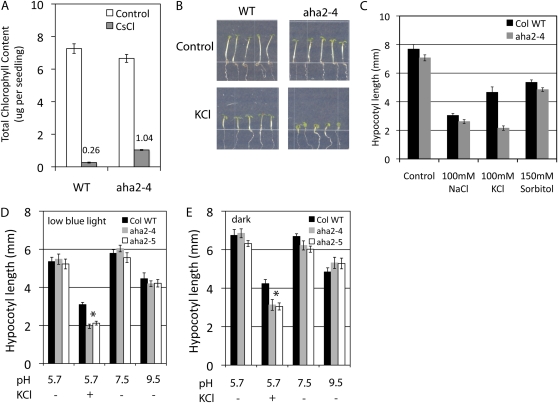
Hypocotyl growth responses of wild-type (WT) and aha2 mutant plants to reduced PMF. A, Total chlorophyll content in seedlings grown under cesium stress. Seedlings were grown as described in Figure 3. Total chlorophyll was extracted from seedlings and quantified. Data shown are from one representative experiment of three experiments. B, Hypocotyl elongation of seedlings grown under low blue light on control medium (top panels) or medium supplemented with 100 mm KCl (bottom panels). C, Hypocotyl length of wild-type and aha2 plants grown under low blue light on control medium or medium supplied with 100 mm NaCl, 100 mm KCl, or 150 mm sorbitol. D, Hypocotyl length of wild-type and aha2 mutant plants grown under low blue light. Plants were grown on control medium (pH 5.7), medium supplied with 100 mm KCl, or medium adjusted to pH 7.5 or 9.5. The asterisk indicates statistical significance between wild-type and aha2 mutant plants (P < 0.001). E, Hypocotyl length of wild-type and aha2 mutant plants grown in the dark. Plants were grown as described in D. The asterisk indicates statistical significance between wild-type and aha2 mutant plants (P < 0.001). [See online article for color version of this figure.]
It has been proposed that medium pH is an important factor determining the cell expansion rate in tissues that can elongate rapidly in response to auxin (Schenck et al., 2010, and refs. therein). We thus investigated whether aha2 mutant hypocotyls behave differently from wild-type hypocotyls at higher external pH, pH 7.5 or 9.5. The seedlings were grown under low blue light or in darkness. As shown in Figure 4, D and E, the hypocotyl length of aha2-4 and aha2-5 mutants at higher pH does not differ from that of the wild-type plants, which is distinct from the strong effect elicited by changes in pH seen in roots (Haruta et al., 2010).
Root Tropic Responses of aha2 Mutants in the Presence of ATP or ADP
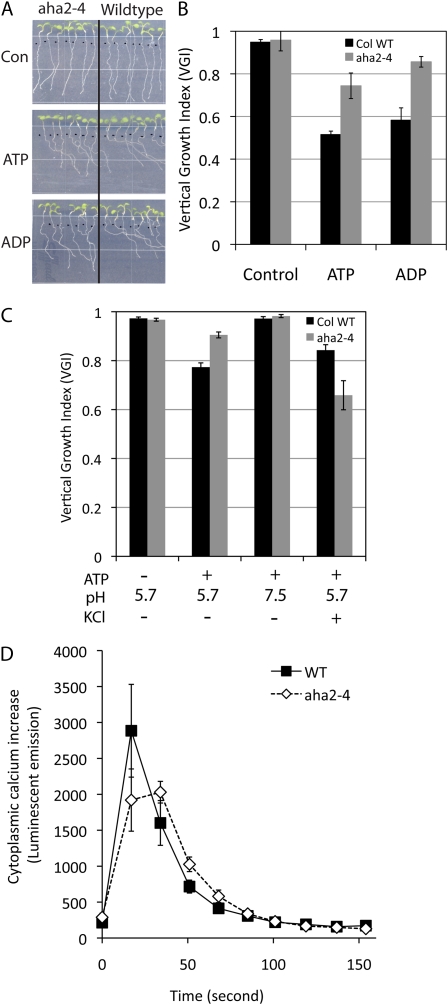
During our root growth assay, plates are placed in an upright manner with respect to gravity, and we noticed that wild-type roots were skewed and grew in a unidirectional pattern in the presence of ATP or ADP. This phenomenon was also previously described by Tang et al. (2003). Both the wild type and aha2 showed normal downward gravitropism on control medium; however, the addition of ATP or ADP to the medium induced skewed root growth in wild-type plants but less so in aha2 mutants (Fig. 5A). The degree of the gravitropic response was quantified by calculating the ratio of root length over vertical downward distance between the growth initiation point and the root tip (Fig. 5B; Supplemental Fig. S6A). Since the aha2 mutant with a reduced PMF is less sensitive to ATP-induced skewed growth, it is likely that the membrane potential and/or the proton chemical gradient are involved in this response. Further experiments with ATP-induced root tropism demonstrated that medium adjusted to pH 7.5 caused the elimination of the ATP effect on the wild type, whereas high potassium had little or no effect on the ATP-induced directional root growth (Fig. 5C; Supplemental Fig. S6B).
Figure 5.
Root tropism of wild-type and aha2 mutant plants in the presence of ATP. A, Root tropism of wild-type and aha2 mutant roots in the presence of ATP or ADP. Seedlings were vertically germinated on control medium (one-half-strength MS, 1% Suc, pH 5.7) for 3 d and transferred to control medium, medium with 200 μg mL−1 ATP, or medium with 200 μg mL−1 ADP. After vertical incubation for an additional 4 d, images were captured and the degree of vertical root growth of wild-type and aha2-4 mutant plants was quantified as shown in B. B, Vertical growth index of wild-type (WT) and aha2-4 mutant plants in response to ATP or ADP. Vertical growth index, described by Vicente-Agullo et al. (2004), is defined as the ratio between a vertical projection of the base-to-tip chord and the root length (Supplemental Fig. S6). C, Effect of high external pH and potassium on ATP-induced root curving. Root tropism in response to ATP was examined with plants grown on medium (one-half-strength MS, 1% Suc) adjusted to pH 7.5 or supplied with 50 mm KCl. D, ATP-induced cytoplasmic calcium increase in wild-type and aha2-4 mutant seedlings. ATP at 1 mm concentration was added to elicit cytoplasmic calcium elevation. Data are shown as averages of 10 seedlings per genotype. [See online article for color version of this figure.]
aha2 Mutants Show an Altered Cytoplasmic Calcium Response Compared with the Wild Type
Exogenous application of ATP or ADP is known to cause membrane depolarization and transient increases in cytoplasmic calcium (Lew and Dearnaley, 2000; Demidchik et al., 2003, 2009). Therefore, we examined whether the ATP-induced calcium response is altered in the aha2 mutants using an aequorin-based cytoplasmic calcium assay (Knight et al., 1991). To this end, we crossed the mutants with transgenic wild-type plants expressing aequorin and reisolated homozygous aha2 plants carrying the calcium reporter. We found that aha2 mutants showed reduced cytoplasmic calcium elevation compared with the wild type when seedlings were stimulated with ATP application (Fig. 5D). Together with root growth data, these results suggest that the PMF mediated by AHA2 function, specifically the pH component, is involved in ATP-induced skewed root growth.
DISCUSSION
In this report, we describe the effects of an altered plasma membrane PMF on plant growth using Arabidopsis mutants with genetic impairments in the two most predominantly expressed plasma membrane proton pump genes. We provide several lines of molecular and physiological evidence that support the role of AHA2 in maintaining the membrane potential and ΔpH, together forming the PMF required for root and hypocotyl elongation and root tropism. The use of potassium to clamp down the membrane potential and external pH to alter the proton chemical gradient allowed us to compare the relative contribution of the two components with the PMF.
Long-Term Changes in Gene Expression in aha1 and aha2 Mutants
In aha1 and aha2 transcriptome analyses, we identified genes showing changes in expression in the mutants. The identities of these differentially expressed genes in the two mutants are somewhat overlapping but also uniquely different (Tables I and II). These results indicate that the reduction of AHA1 expression causes different consequences on cellular activity from the reduction of AHA2 expression and that the regulatory mechanism of the two proton pumps seems to be isoform specific during seedling growth. Whereas the affected genes in aha2 mutants are involved in various nutrient stresses, those in aha1 mutants appear to be related to lipid metabolism, perhaps reflecting lipid-based intracellular signaling. This notion is also consistent with a proposed role of AHA2 in iron transport and that of AHA1 in steroid signaling (Santi and Schmidt, 2009; Caesar et al., 2011).
The localization of AHA1 and AHA2 proteins at the cellular or tissue level is also likely to elucidate the isoform-specific function of these enzymes during seedling growth. A study of the cell type-specific transcriptome of Arabidopsis roots indicates that AHA1 and AHA2 are highly expressed in the entire root, with some isoform-specific expression. While root hair and epidermal cells express AHA2 more than AHA1, the conducting cells, such as phloem companion and xylem cells, express AHA1 more highly than AHA2 (Brady et al., 2007). The impaired growth of aha2 mutants on conditions that are likely to stress the PMF seen in this study thus correlates with AHA2 function in solute uptake in root epidermal cells.
The genes encoding two potassium transporters, AtHAK5 and K+/H+ exchanger 17, are up-regulated in aha2 mutants and are also preferentially expressed in root hair and/or epidermal cells (Cellier et al., 2004; Gierth et al., 2005; Brady et al., 2007). These correlating expression patterns for the potassium transporters and the AHA2 proton pump suggest that they may coordinately function in maintaining membrane energetics and potassium uptake at the root surface.
A Reduction in the Plasma Membrane Electrical Potential and/or the Proton Chemical Gradient Explains Growth Differences Observed in aha2 Mutants
The observation that high external pH and potassium have an additive effect on reducing aha2 mutant root growth supports the concept that a PMF is the sum of both charge differences and proton concentration differences and that this equation holds true at the Arabidopsis plasma membrane in planta. Throughout our experiments, our prediction that high external potassium reduces membrane potential was consistent with seedling growth responses. It is important to note that while membrane depolarization may be the immediate cellular response to high external potassium, long-term effects of high potassium may involve other cellular stress responses as well. The PMF could also be affected by many other variables, such as the intracellular concentration of protons or potassium. Therefore, our interpretation of observed growth differences between mutant and wild-type plants must be confirmed in future experiments in which additional parameters, including intracellular pH and potassium concentration, are controlled and measured.
Insensitivity of the aha2 mutant toward the amino acids Arg and Lys was interpreted as the mutant taking up less of the basic amino acids compared with wild-type plants. H+/amino acid cotransporters have substrate specificities based on side chain charges (Fischer et al., 1995), but they respond to the membrane potential because they transport one proton per one amino acid irrespective of their overall net charge (Boorer and Fischer, 1997). Alternatively, if a particular amino acid is taken up with more than one proton, which was proposed in the case of Val uptake, wild-type and mutant plants will respond differently to such an amino acid (Borstlap and Schuurmans, 2000). The unique insensitivity of the aha2 mutant toward the two basic amino acids, Arg and Lys, and to the neutral amino acid, Val, suggests the presence of transport systems for Arg, Lys, and Val, which are coupled to the PMF to a greater extent than those for the other amino acids.
By chemically changing the membrane potential or proton chemical concentration, we have examined the contributions of these two components to the overall PMF, which was strongly coupled with the uptake of Arg and hygromycin. Reports indicate that the uptake of hygromycin is ubiquitously dependent on the membrane potential in yeast and bacteria (Taber et al., 1987). Hygromycin carries two primary amines with pKa(dissociation constant) values of 7.1 and 8.8 and is thus doubly positively charged in normal growth medium over the pH range tested. This suggests that the inside-negative electrical potential is the inward-driving force for the uptake of this antibiotic compound. The strong effect of high external pH in eliminating the mutant-specific responses to Arg, but not hygromycin, suggests that at least one mechanism of Arg uptake is driven by a proton-coupled cotransporter but that hygromycin uptake is not. These observations agree with the requirements of the ΔpH for H+/amino acid cotransport into plant cells (Li and Bush, 1990; Boorer and Fischer, 1997), although the uptake system for hygromycin is unknown.
Our observation that aha2 mutants with a reduced PMF are more tolerant to cesium toxicity compared with the wild type agrees with previous reports that cesium enters into cells in part through a potassium transporter (Qi et al., 2008; Kanter et al., 2010). The cesium-resistant phenotype of aha2 mutants was observed at a narrow concentration range, 5 to 10 mm, where cesium concentration becomes comparable to potassium concentration in growth medium (one-half-strength MS medium contains approximately 10 mm K+). Cesium is known to induce membrane depolarization like potassium when directly injected into cells (Rubio et al., 1996). However, the cesium toxicity in our root growth test was saturated in both wild-type and aha2 mutant plants at concentrations (approximately 20 mm) that are lower than those capable of eliciting a large membrane depolarization. The resistance of roots to cesium or lithium in the aha2 mutant is eliminated when the membrane is depolarized, supporting our interpretation that the mutants are resistant because their resting potential is lower than that of the wild type. According to this model, the presence of 50 to 100 mm potassium causes the difference in membrane potential between the mutants and the wild type to be reduced and eliminates the sparing effect of reduced membrane potential in the mutants compared with the wild-type plants.
We demonstrated that root tropism in aha2 mutants is less affected compared with wild-type plants when the root tip is subjected to ATP-induced skewed growth on the vertically oriented agar plates. Since root length itself was not significantly different between the two genotypes with ATP treatment (Haruta et al., 2010), we concluded that the differential root tropism was caused by the alteration of root tropic sensing or signaling rather than cell elongation rates in aha2 mutants. This mutant phenotype together with the wild type’s response to ATP under alkaline pH indicates the involvement of the pH component of the PMF in the activation of a signal transduction pathway for ATP-induced tip reorientation (Fig. 5C). A previous study showed that rapid acidification of cell wall pH and alkalinization of cytoplasmic pH were occurring after gravistimulation in root caps (Fasano et al., 2001). In our assays of gravistimulation, however, the aha2 mutant showed a similar response of root-tip orientation as with the wild type (Supplemental Fig. S6). Therefore, the differential tip behavior observed in the mutant is likely unique to the ATP-mediated root tropic response. The lack of a mutant-specific phenotype to gravistimulation may be explained by the fact that the aha2 growth phenotypes revealed here occur only under extreme chemical conditions, including high concentrations of cesium, lithium, potassium, amino acids, and ATP, which may not be common in natural growth environments. On the other hand, plant roots are naturally exposed to conditions causing root tip reorientation, such as mechanostress and obstacle avoidance, and thus, with some inherent adaptive mechanisms, this small reduction of PMF in the mutant may not be sufficient to elicit a differential gravitropic response between the genotypes. Weerasinghe et al. (2009) reported that the exogenous application of ATP mimics the mechanostress-induced release of ATP into extracellular space during root tip reorientation. A plausible explanation of the observed aha2 phenotype is that aha2 mutants with reduced PMF are impaired in the ability to activate an ATP-induced signal pathway for mechanostress response through cytoplasmic calcium elevation.
The Plasma Membrane Bioenergetics Underlying Cell Expansion in Roots and Hypocotyls May Be Fundamentally Different
In this study, we have provided evidence for a critical role of the membrane potential but not a pH gradient for the elongation of Arabidopsis hypocotyls. Surprisingly, an alkaline external pH of 9.5 that gives a nearly 1,000-fold reduction in proton concentration had little effect on both wild-type and aha2 mutant hypocotyls (Fig. 4E). It is possible that the lack of a high pH effect on hypocotyl is caused by structural barriers such as a cuticle or other “unstirred” layers preventing direct contact of the plasma membrane with external solutions rather than by underlying differences in the driving force for cell elongation between the two organs. However, our observation that hypocotyls and roots have similar sensitivity to high external potassium is inconsistent with the possibility of unstirred layers preventing the accessibility of the external solutes to the hypocotyl (Fig. 4, B and C; Supplemental Fig. S4A). The differential response between the two organs could be due to proton-specific effects, which could occur if the two organs have different buffering capacities for pH change in the wall space. Plant roots secrete protons and acidify the root rhizosphere to increase the solubility of minerals such as iron (Palmer and Guerinot, 2009; Santi and Schmidt, 2009). Thus, it is reasonable that roots also have mechanisms to respond to changes in external pH in a manner different from hypocotyls. While the effect of an altered transmembrane pH on intact hypocotyl cells remains unclear, it appears that the membrane potential is an important factor in hypocotyl elongation. From the impaired hypocotyl growth of aha2 mutants under high external potassium conditions, we suggest that the membrane potential regulated by AHA2 and potassium transporters is an essential cellular component for determining maximal hypocotyl elongation in planta and that the proton chemical concentration may play a secondary role.
We described here a detailed comparative analysis of wild-type and aha2 mutant plants grown under various chemical environments to induce changes to the PMF. It is important to note that the only growth phenotypes that we observed with the mutant plants were related to conditions that are closely linked with the plasma membrane PMF. In the traditional model of tissue elongation mechanisms, the PMF, and more specifically, the proton pump, plays a critical role during rapid cell expansion; however, it has yet to be elucidated how endogenous signals or environmental cues are linked to pump-mediated membrane events. Future work using the phenotypes demonstrated by aha mutants for forward genetic studies to search for genetic modifiers or with site-directed mutations may provide some insight into the molecular mechanism by which the proton pump interacts with signaling systems initiated by auxin and other growth regulators.
MATERIALS AND METHODS
Plant Materials and Genotyping
Homozygous Arabidopsis (Arabidopsis thaliana) mutant plants carrying aha1-6, aha1-7, aha2-4, and aha2-5 alleles were previously isolated and characterized (Haruta et al., 2010). Mutant lines are in the Columbia genetic background and were originally obtained from the SALK collection (Alonso et al., 2003). Seeds used for phenotyping assays were harvested from wild-type, aha1-6, aha1-7, aha2-4, or aha2-5 plants propagated simultaneously under identical growth conditions. Plants were grown in soil:perlite (4:1 ratio; Jiffy-Mix [Jiffy Products of America] and horticultural perlite [Schundler Co.]) at 21°C under constant light (32 μmol m–2 s–1). aha2 mutants homozygous for an aequorin transgene insertion were generated by crossing (Lewis et al., 1997). AHA2−/−; AEQ+/+ and AHA2+/+; AEQ+/+ plants were identified in the F2 population by PCR genotyping with aequorin- or aha2-4 gene-specific primers. Primer sequences for the aequorin transgene are 5′-ACTTCGACAACCCAAGATGG-3′ and 5′-ACCGTAGAGCTTTTCGCAAG-3′.
Growth Assay
Protocols for growth tests were described by Weigel and Glazebrook (2002). To sterilize, seeds were suspended in 70% ethanol and 0.1% Triton X-100 for 5 min, rinsed with absolute ethanol, and dried. Seeds were deposited on plates containing one-half-strength MS medium (PhytoTechnology Laboratories) supplied with 1% Suc and adjusted to pH 5.7. The plates were treated at 4°C for 2 d in darkness and then incubated in a vertically oriented position at 22°C under constant white light (Percival Scientific; 25 μmol m–2 s–1). At 3 d old, the seedlings were aseptically transferred to experimental medium and incubated for an additional 4 d. At the end of the assay, seedling images were acquired by scanning (EPSON; Perfection V300 PHOTO), and root growth was measured and quantified using ImageJ (Abramoff et al., 2004) and Microsoft Excel. Hypocotyl elongation assays were carried out by germinating seedlings under constant blue-filtered lights (2.2 μmol m–2 s–1) or complete darkness for 4 or 5 d. Hypocotyl growth was quantified as described above.
For stress tests, various l-amino acids (Sigma), hygromycin (PhytoTechnology Laboratories), ATP, or ADP were filter sterilized prior to adding to autoclaved medium. The other types of salts, NaCl, KCl, CsCl, and LiCl, were supplied to medium before autoclaving.
Quantification of Root Tropism
Seedlings were grown vertically on one-half-strength MS medium for 3 d and further grown on medium supplied with ATP for 4 d. Root tropism was quantified by dividing the root growth length by the vertical downward distance between the growth initiation point and the root tip. This value was defined as the vertical growth index as described previously (Vicente-Agullo et al., 2004).
Chlorophyll Measurements
Seven 1-week-old seedlings were ground in 200 μL of 90% acetone. Chlorophyll extracts were recovered from the supernatant after centrifugation at 13,000 rpm for 5 min. Absorbances at 645 and 663 nm were measured using a photospectrometer (GENESYS 5; Spectronic Unicam). Chlorophyll content was determined using the formula 20.2(A645) + 8.02(A663) (Arnon, 1949).
Cytoplasmic Calcium Assay
Cytoplasmic calcium elevation was measured as described previously (Haruta et al., 2008). Aequorin-expressing transgenic seeds were grown on a one-half-strength MS medium for 4 d. A single seedling was incubated in 200 μL of one-half-strength MS medium supplemented with 2.5 μm coelentrazine cyclopentyl (Sigma) for 16 h. Bioluminescent emission was recorded for 150 s with a Techan plate reader (Tecan). For experiments with reduced PMF, the assay medium was modified by adding 100 mm KCl or adjusting the pH to 7.5.
Microarray Analysis
Five-day-old seedlings grown on one-half-strength MS medium in a vertical orientation were used for the transcriptome study. Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen). RNA sample processing, hybridization, and data extraction for the microarray experiments were carried out at the Gene Expression Center at the University of Wisconsin-Madison. All CEL files were simultaneously normalized with the Robust Multichip Average algorithm, and the transcript intensity of biological replicates was averaged using Microsoft Excel (Bolstad et al., 2003). Fold change of the gene expression level in mutants was calculated by dividing the transcript intensities of the mutants by those of wild-type plants. Affymetrix probe annotation was converted to AtID (Arabidopsis locus identifier) at the Bio-Array Resource for Plant Biology siteand verified at The Arabidopsis Information Resource site.
Quantitative RT-PCR
Seedlings were vertically grown on one-half-strength MS medium for 3 d and transferred to the same medium as the control or that supplied with 100 mm KCl. After additional growth for 4 d, seedlings were harvested. Total RNA was extracted using the RNeasy Plant Kit (Qiagen), and DNase treatment was performed on column as described in the manufacturer’s protocol. Total RNA (1.5 μg) was converted to cDNA using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). Quantitative RT-PCR was carried out using SYBR Green with a real-time thermal cycler (Roche LC480 Light Cycler). One microliter of cDNA was used for the 20-μL reaction in a 96-well format. Cycle conditions were 95°C for 5 min followed by 40 cycles of 95°C for 10 s, 58°C for 7 s, and 72°C for 20s. The expression of reference genes or K+ transporter genes was analyzed with gene-specific pairs of primers: ACT2 (Pischke et al., 2006), 5′-GCATGAAGATCAAGGTGGTTGCAC-3′ and 5′-ATGGACCTGACTCATCGTACTCACT-3′; UBQ10 (Czechowski et al., 2005), 5′-GGCCTTGTATAATCCCTGATGAATAAG-3′ and 5′-AAAGAGATAACAGGAACGGAAACATAGT-3′; AtHAK5 (Qi et al., 2008), 5′-CGTTTTCATTGTTCTTCAGG-3′ and 5′-ATCTTCTGGTTCTTGGTTTG-3′; AtCHX17, 5′-CCGTCCCGATATAATCTGCGTTTTC-3′ and 5′-CAACGCGTGTTCCAGAGGATTC-3′. The sequences of the primers for AtCHX17 were designed using the AtRTPrimer program (Han and Kim, 2006). To confirm clean PCR amplification, product was verified with melt curve analysis and approximate product size was determined by running on a 2% agarose gel. The average value of the crossing point was calculated from three technical replicates of PCR. Biological replicates were composed of wild-type and aha2 mutant seedlings grown on control or high-potassium conditions in three different experiments. Data were analyzed with the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Transcriptional and phenotypic comparisons of wild-type and aha1-6 or aha2-4 mutant plants.
Supplemental Figure S2. Root growth of wild-type, aha1-6, and aha2-4 plants in the presence of various amino acids.
Supplemental Figure S3. Root growth of the wild type and aha2 mutants in response to altered PMF in the presence of 5 mm Arg.
Supplemental Figure S4. Root growth of wild-type and aha2 mutant plants in the presence of KCl or NaCl.
Supplemental Figure S5. Root growth in the presence of LiCl or CsCl.
Supplemental Figure S6. ATP-induced root tip response.
Acknowledgments
We thank Angie Bewell and Gabby Gwaclawik for technical assistance with plant growth, Sandra Splinter BonDurant for assistance with quantitative RT-PCR, and Yoshiro Saimi, Sandra Austin-Phillips, and Rachel Rodrigues for critical reading of the manuscript.
References
- Abramoff MD, Magelhaes PJ, Ram SJ. (2004) Image processing with ImageJ. Biophotonics Int 11: 36–42 [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Armengaud P, Breitling R, Amtmann A. (2004) The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiol 136: 2556–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI. (1949) Copper enzymes in isolated chloroplasts: polyphenol oxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter I, Tchieu J, Sussman MR, Boutry M, Palmgren MG, Gribskov M, Harper JF, Axelsen KB. (2003) Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol 132: 618–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. (2003) A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Boorer KJ, Fischer WN. (1997) Specificity and stoichiometry of the Arabidopsis H+/amino acid transporter AAP5. J Biol Chem 272: 13040–13046 [DOI] [PubMed] [Google Scholar]
- Borstlap AC, Schuurmans JAMJ. (2000) Proton-symport of L-valine in plasma membrane vesicles isolated from leaves of the wild-type and the Val(r)-2 mutant of Nicotiana tabacum L. Plant Cell Physiol 41: 1210–1217 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Buckhout TJ. (1989) Sucrose transport in isolated plasma-membrane vesicles from sugar beet (Beta vulgaris L.): evidence for an electrogenic sucrose-proton symport. Planta 178: 393–399 [DOI] [PubMed] [Google Scholar]
- Bush DR. (1993) Proton-coupled sugar and amino acid transporters in plants. Annu Rev Plant Physiol Plant Mol Biol 44: 513–542 [Google Scholar]
- Caesar K, Elgass K, Chen Z, Huppenberger P, Witthöft J, Schleifenbaum F, Blatt MR, Oecking C, Harter K. (2011) A fast brassinolide-regulated response pathway in the plasma membrane of Arabidopsis thaliana. Plant J 66: 528–540 [DOI] [PubMed] [Google Scholar]
- Cellier F, Conéjéro G, Ricaud L, Luu DT, Lepetit M, Gosti F, Casse F. (2004) Characterization of AtCHX17, a member of the cation/H+ exchangers, CHX family, from Arabidopsis thaliana suggests a role in K+ homeostasis. Plant J 39: 834–846 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong A, Borstlap AC. (2000) Transport of amino acids (L-valine, L-lysine, L-glutamic acid) and sucrose into plasma membrane vesicles isolated from cotyledons of developing pea seeds. J Exp Bot 51: 1663–1670 [DOI] [PubMed] [Google Scholar]
- Demidchik V, Nichols C, Oliynyk M, Dark A, Glover BJ, Davies JM. (2003) Is ATP a signaling agent in plants? Plant Physiol 133: 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidchik V, Shang Z, Shin R, Thompson E, Rubio L, Laohavisit A, Mortimer JC, Chivasa S, Slabas AR, Glover BJ, et al. (2009) Plant extracellular ATP signalling by plasma membrane NADPH oxidase and Ca2+ channels. Plant J 58: 903–913 [DOI] [PubMed] [Google Scholar]
- Fasano JM, Swanson SJ, Blancaflor EB, Dowd PE, Kao TH, Gilroy S. (2001) Changes in root cap pH are required for the gravity response of the Arabidopsis root. Plant Cell 13: 907–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer WN, André B, Rentsch D, Krolkiewicz S, Tegeder M, Breitkreuz K, Frommer WB. (1998) Amino acid transport in plants. Trends Plant Sci 3: 188–195 [Google Scholar]
- Fischer WN, Kwart M, Hummel S, Frommer WB. (1995) Substrate specificity and expression profile of amino acid transporters (AAPs) in Arabidopsis. J Biol Chem 270: 16315–16320 [DOI] [PubMed] [Google Scholar]
- Fuglsang AT, Guo Y, Cuin TA, Qiu Q, Song C, Kristiansen KA, Bych K, Schulz A, Shabala S, Schumaker KS, et al. (2007) Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19: 1617–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gévaudant F, Duby G, von Stedingk E, Zhao R, Morsomme P, Boutry M. (2007) Expression of a constitutively activated plasma membrane H+-ATPase alters plant development and increases salt tolerance. Plant Physiol 144: 1763–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierth M, Mäser P, Schroeder JI. (2005) The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol 137: 1105–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens A, de La Fuente N, Forment J, Serrano R, Portillo F. (2000) Regulation of yeast H(+)-ATPase by protein kinases belonging to a family dedicated to activation of plasma membrane transporters. Mol Cell Biol 20: 7654–7661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager A. (2003) Role of the plasma membrane H+-ATPase in auxin-induced elongation growth: historical and new aspects. J Plant Res 116: 483–505 [DOI] [PubMed] [Google Scholar]
- Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ. (2003) Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol 132: 578–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Kim D. (2006) AtRTPrimer: database for Arabidopsis genome-wide homogeneous and specific RT-PCR primer-pairs. BMC Bioinformatics 7: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JF, Surowy TK, Sussman MR. (1989) Molecular cloning and sequence of cDNA encoding the plasma membrane proton pump (H+-ATPase) of Arabidopsis thaliana. Proc Natl Acad Sci USA 86: 1234–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Burch HL, Nelson RB, Barrett-Wilt G, Kline KG, Mohsin SB, Young JC, Otegui MS, Sussman MR. (2010) Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity. J Biol Chem 285: 17918–17929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Monshausen G, Gilroy S, Sussman MR. (2008) A cytoplasmic Ca2+ functional assay for identifying and purifying endogenous cell signaling peptides in Arabidopsis seedlings: identification of AtRALF1 peptide. Biochemistry 47: 6311–6321 [DOI] [PubMed] [Google Scholar]
- Hirsch RE, Lewis BD, Spalding EP, Sussman MR. (1998) A role for the AKT1 potassium channel in plant nutrition. Science 280: 918–921 [DOI] [PubMed] [Google Scholar]
- Humble GD, Raschke K. (1971) Stomatal opening quantitatively related to potassium transport: evidence from electron probe analysis. Plant Physiol 48: 447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagendorf AT. (1967) The chemiosmotic hypothesis of photophosphorylation. San Pietro A, Greer FA, Army TJ, eds, Harvesting the Sun: Photosynthesis in Plant Life. Academic Press, New York, pp 69–78 [Google Scholar]
- Kanter U, Hauser A, Michalke B, Dräxl S, Schäffner AR. (2010) Caesium and strontium accumulation in shoots of Arabidopsis thaliana: genetic and physiological aspects. J Exp Bot 61: 3995–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Trewavas AJ, Knight MR. (1996) Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 8: 489–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight MR, Campbell AK, Smith SM, Trewavas AJ. (1991) Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352: 524–526 [DOI] [PubMed] [Google Scholar]
- Kocherginsky N. (2009) Acidic lipids, H(+)-ATPases, and mechanism of oxidative phosphorylation: Physico-chemical ideas 30 years after P. Mitchell’s Nobel Prize award. Prog Biophys Mol Biol 99: 20–41 [DOI] [PubMed] [Google Scholar]
- Lew R, Dearnaley JDW. (2000) Extracellular nucleotide effects on the electrical properties of growing Arabidopsis thaliana root hairs. Plant Sci 153: 1–6 [Google Scholar]
- Lewis BD, Karlin-Neumann G, Davis RW, Spalding EP. (1997) Ca2+-activated anion channels and membrane depolarizations induced by blue light and cold in Arabidopsis seedlings. Plant Physiol 114: 1327–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Foster CM, Gan Q, Nettleton D, James MG, Myers AM, Wurtele ES. (2009) Identification of the novel protein QQS as a component of the starch metabolic network in Arabidopsis leaves. Plant J 58: 485–498 [DOI] [PubMed] [Google Scholar]
- Li ZC, Bush DR. (1990) ΔpH-dependent amino acid transport into plasma membrane vesicles isolated from sugar beet leaves. I. Evidence for carrier-mediated, electrogenic flux through multiple transport systems. Plant Physiol 94: 268–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Fuglsang AT, Palmgren MG, Staskawicz BJ, Coaker G. (2009) RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol 7: e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Ma S, Gong Q, Bohnert HJ. (2006) Dissecting salt stress pathways. J Exp Bot 57: 1097–1107 [DOI] [PubMed] [Google Scholar]
- Maathuis FJ, Sanders D. (1994) Mechanism of high-affinity potassium uptake in roots of Arabidopsis thaliana. Proc Natl Acad Sci USA 91: 9272–9276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis FJ, Sanders D. (1997) Regulation of K+ absorption in plant root cells by external K+: interplay of different plasma membrane K+ transporters. J Exp Bot 48: 451–458 [DOI] [PubMed] [Google Scholar]
- Maresova L, Sychrova H. (2006) Arabidopsis thaliana CHX17 gene complements the kha1 deletion phenotypes in Saccharomyces cerevisiae. Yeast 23: 1167–1171 [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Watanabe-Takahashi A, Inoue E, Yamaya T, Takahashi H. (2005) Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J 42: 305–314 [DOI] [PubMed] [Google Scholar]
- McCusker JH, Perlin DS, Haber JE. (1987) Pleiotropic plasma membrane ATPase mutations of Saccharomyces cerevisiae. Mol Cell Biol 7: 4082–4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Leonhardt N, Fenzi F, Valon C, Costa M, Piette L, Vavasseur A, Genty B, Boivin K, Müller A, et al. (2007) Constitutive activation of a plasma membrane H(+)-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J 26: 3216–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelet B, Boutry M. (1995) The plasma membrane H+-ATPase: a highly regulated enzyme with multiple physiological functions. Plant Physiol 108: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee I, Campbell NH, Ash JS, Connolly EL. (2006) Expression profiling of the Arabidopsis ferric chelate reductase (FRO) gene family reveals differential regulation by iron and copper. Planta 223: 1178–1190 [DOI] [PubMed] [Google Scholar]
- Nikiforova V, Freitag J, Kempa S, Adamik M, Hesse H, Hoefgen R. (2003) Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J 33: 633–650 [DOI] [PubMed] [Google Scholar]
- Palmer CM, Guerinot ML. (2009) Facing the challenges of Cu, Fe and Zn homeostasis in plants. Nat Chem Biol 5: 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren MG. (2001) Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annu Rev Plant Physiol Plant Mol Biol 52: 817–845 [DOI] [PubMed] [Google Scholar]
- Pedersen BP, Buch-Pedersen MJ, Morth JP, Palmgren MG, Nissen P. (2007) Crystal structure of the plasma membrane proton pump. Nature 450: 1111–1114 [DOI] [PubMed] [Google Scholar]
- Perlin DS, Harris SL, Seto-Young D, Haber JE. (1989) Defective H(+)-ATPase of hygromycin B-resistant pma1 mutants from Saccharomyces cerevisiae. J Biol Chem 264: 21857–21864 [PubMed] [Google Scholar]
- Pischke MS, Huttlin EL, Hegeman AD, Sussman MR. (2006) A transcriptome-based characterization of habituation in plant tissue culture. Plant Physiol 140: 1255–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli R, Voll LM, Horst RJ, Frommer WB, Pilot G. (2010) Stimulation of nonselective amino acid export by glutamine dumper proteins. Plant Physiol 152: 762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Navarro A. (2000) Potassium transport in fungi and plants. Biochim Biophys Acta 1469: 1–30 [DOI] [PubMed] [Google Scholar]
- Rubio F, Gassmann W, Schroeder JI. (1996) Response: high-affinity potassium uptake in plants. Science 273: 978–979 [DOI] [PubMed] [Google Scholar]
- Qi Z, Hampton CR, Shin R, Barkla BJ, White PJ, Schachtman DP. (2008) The high affinity K+ transporter AtHAK5 plays a physiological role in planta at very low K+ concentrations and provides a caesium uptake pathway in Arabidopsis. J Exp Bot 59: 595–607 [DOI] [PubMed] [Google Scholar]
- Santi S, Schmidt W. (2009) Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol 183: 1072–1084 [DOI] [PubMed] [Google Scholar]
- Schaller A, Oecking C. (1999) Modulation of plasma membrane H+-ATPase activity differentially activates wound and pathogen defense responses in tomato plants. Plant Cell 11: 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Sussman MR. (1988) Isolation and sequence of tryptic peptides from the proton-pumping ATPase of the oat plasma membrane. Plant Physiol 86: 512–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck D, Christian M, Jones A, Lüthen H. (2010) Rapid auxin-induced cell expansion and gene expression: a four-decade-old question revisited. Plant Physiol 152: 1183–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalygo NV, Averina NG, Grimm B, Mock HP. (1997) Influence of cesium on tetrapyrrole biosynthesis in etiolated and greening barley leaves. Physiol Plant 99: 160–168 [Google Scholar]
- Sheahan JJ, Ribeiro-Neto L, Sussman MR. (1993) Cesium-insensitive mutants of Arabidopsis thaliana. Plant J 3: 647–656 [Google Scholar]
- Spalding EP, Hirsch RE, Lewis DR, Qi Z, Sussman MR, Lewis BD. (1999) Potassium uptake supporting plant growth in the absence of AKT1 channel activity: inhibition by ammonium and stimulation by sodium. J Gen Physiol 113: 909–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman MR. (1994) Molecular analysis of proteins in the plant plasma membrane. Annu Rev Plant Physiol Plant Mol Biol 45: 211–234 [Google Scholar]
- Taber HW, Mueller JP, Miller PF, Arrow AS. (1987) Bacterial uptake of aminoglycoside antibiotics. Microbiol Rev 51: 439–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Brady SR, Sun Y, Muday GK, Roux SJ. (2003) Extracellular ATP inhibits root gravitropism at concentrations that inhibit polar auxin transport. Plant Physiol 131: 147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellosillo T, Martínez M, López MA, Vicente J, Cascón T, Dolan L, Hamberg M, Castresana C. (2007) Oxylipins produced by the 9-lipoxygenase pathway in Arabidopsis regulate lateral root development and defense responses through a specific signaling cascade. Plant Cell 19: 831–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Agullo F, Rigas S, Desbrosses G, Dolan L, Hatzopoulos P, Grabov A. (2004) Potassium carrier TRH1 is required for auxin transport in Arabidopsis roots. Plant J 40: 523–535 [DOI] [PubMed] [Google Scholar]
- Vitart V, Baxter I, Doerner P, Harper JF. (2001) Evidence for a role in growth and salt resistance of a plasma membrane H+-ATPase in the root endodermis. Plant J 27: 191–201 [DOI] [PubMed] [Google Scholar]
- Voll LM, Allaire EE, Fiene G, Weber AP. (2004) The Arabidopsis phenylalanine insensitive growth mutant exhibits a deregulated amino acid metabolism. Plant Physiol 136: 3058–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorwieger A, Gryczka C, Czihal A, Douchkov D, Tiedemann J, Mock H-P, Jakoby M, Weisshaar B, Saalbach I, Bäumlein H. (2007) Iron assimilation and transcription factor controlled synthesis of riboflavin in plants. Planta 226: 147–158 [DOI] [PubMed] [Google Scholar]
- Weerasinghe RR, Swanson SJ, Okada SF, Garrett MB, Kim SY, Stacey G, Boucher RC, Gilroy S, Jones AM. (2009) Touch induces ATP release in Arabidopsis roots that is modulated by the heterotrimeric G-protein complex. FEBS Lett 583: 2521–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Williams-Hart T, Wu X, Tatchell K. (2002) Protein phosphatase type 1 regulates ion homeostasis in Saccharomyces cerevisiae. Genetics 160: 1423–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Mourad G, King J. (1994) A valine-resistant mutant of Arabidopsis thaliana displays an acetolactate synthase with altered feedback control. Planta 192: 249–255 [Google Scholar]
- Zhao R, Dielen V, Kinet JM, Boutry M. (2000) Cosuppression of a plasma membrane H+-ATPase isoform impairs sucrose translocation, stomatal opening, plant growth, and male fertility. Plant Cell 12: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]