Abstract
Cellular Na+/K+ ratio is a crucial parameter determining plant salinity stress resistance. We tested the function of plasma membrane Na+/K+ cotransporters in the High-affinity K+ Transporter (HKT) family from the halophytic Arabidopsis (Arabidopsis thaliana) relative Thellungiella salsuginea. T. salsuginea contains at least two HKT genes. TsHKT1;1 is expressed at very low levels, while the abundant TsHKT1;2 is transcriptionally strongly up-regulated by salt stress. TsHKT-based RNA interference in T. salsuginea resulted in Na+ sensitivity and K+ deficiency. The athkt1 mutant lines overexpressing TsHKT1;2 proved less sensitive to Na+ and showed less K+ deficiency than lines overexpressing AtHKT1. TsHKT1;2 ectopically expressed in yeast mutants lacking Na+ or K+ transporters revealed strong K+ transporter activity and selectivity for K+ over Na+. Altering two amino acid residues in TsHKT1;2 to mimic the AtHKT1 sequence resulted in enhanced sodium uptake and loss of the TsHKT1;2 intrinsic K+ transporter activity. We consider the maintenance of K+ uptake through TsHKT1;2 under salt stress an important component supporting the halophytic lifestyle of T. salsuginea.
Next to the Salt Overly Sensitive (SOS) pathway, proteins in the High-affinity K+ Transporter (HKT) families of monovalent ion transporters have emerged as relevant components in the defense of plants against increased salinity (Rus et al., 2001, 2006). Nonetheless, the precise function of plant ion transporters in the HKT family remains elusive. HKT1 was first isolated and described in wheat (Triticum aestivum) as a high-affinity K+ uptake transporter with a Na+/K+ cotransport component (Rubio et al., 1995). In Arabidopsis (Arabidopsis thaliana), however, HKT1 functions as a selective Na+ transporter (Uozumi et al., 2000). This difference becomes important under environmental conditions that change the normal Na+/K+ ratio in the medium or soil. The confusing story about how to place HKT proteins into a general picture of the plant ion uptake menagerie has recently been analyzed by Kronzucker and Britto (2011). In short, after the first description of a wheat K+ transporter, TaHKT1 (Schachtman and Schroeder, 1994), studies in a variety of organisms, most detailed in rice (Oryza sativa) and Arabidopsis, documented characteristics for HKT proteins that seemed irreconcilable. HKTs are represented by small gene families in some species, while the Arabidopsis genome includes only a single HKT1 gene. The multiple genes in some species apparently evolved to perform functions in preferential potassium transport over sodium ions (Mäser et al., 2002; Kato et al., 2007). Other versions transport according to the concentration of monovalent cations in the external medium, while some also appear to transport calcium ions (Horie et al., 2011; Kronzucker and Britto, 2011). The biochemical activities of various HKTs have seemingly also been adjusted by how multiple genes in some species are expressed in response to the environment. However, not enough information is yet available about tissue, cell, and subcellular distribution of the proteins to provide sufficient or final clarity. In particular, work on the protein sequences and protein structures of different HKT1 forms must provide information about how the “lifestyle” of a particular species might have fostered changes in amino acid sequence that then shaped the characteristics of this transporter.
Plant salt stress “tolerance,” a qualitative yardstick for a quantity of sodium ions that can be tolerated, is a composite of the combined action of several pathways that are located in different cellular compartments. These include the synthesis of osmolytes and otherwise protective metabolites or proteins (Nelson et al., 1998; Zhu, 2001) and the regulation of Na+ homeostasis, which itself has several components: uptake entities, efflux capacity, cellular and subcellular sequestration, and orchestrated distribution across the plant body. For tolerance to be achieved, the management of sodium ions must be organized against a background protection of potassium ion uptake and distribution. In addition, the reestablishment of general ion balance and nutrient homeostasis are required for a successful salt stress defense (Hasegawa et al., 2000; Tester and Davenport, 2003).
Na+ uptake and distribution are accomplished and influenced by a variety of membrane proteins, antiporters, nonspecific cation channels, cyclic nucleotide gated channels (CNGCs) anion transporters, ATP-binding cassette-type transporters, amino acid permeases, Na+ and K+ transporters, plasma membrane and vacuolar ATPases, and aquaporins (Apse et al., 1999; Shi et al., 2000). Na+ efflux is carried out by the SOS pathway in a reaction chain starting from the sensing of salt stress-induced level of cellular Ca2+. This results in the formation of a complex between a calcium-binding protein, SOS3 (CBL4), and a protein kinase, SOS2. The SOS3/SOS2 complex can phosphorylate and thus activate the Na+/H+ antiporter SOS1, which enhances Na+ export from cells. Export of sodium ions is not restricted to the surface of roots but, most likely even more importantly, for redistribution throughout the plant body (Shi et al., 2000, 2003; Qiu et al., 2002; Oh et al., 2009). Sequestration of sodium ions into vacuoles, which can be a major sink as long as plants can generate new cells and hence new vacuoles, is facilitated by members of the AtNHX family in attempts to mitigate toxic concentrations of cytosolic Na+ (Apse et al., 1999; Pardo et al., 2006). High-affinity K+ transporters in different protein families become engaged in compensatory K+ acquisition. Membrane-intrinsic Na+ and K+ transporters, channels, and cotransporters establish and maintain the intracellular K+/Na+ ratio based on the potential mediated by H+-ATPases (Hasegawa et al., 2000; Zhu, 2003).
In contrast to glycophytic species, a group that includes essentially all crops, species with salinity stress tolerance, termed halophytes, exert exceptional control over Na+ influx combined with export mechanisms, the ability to coordinate distribution to various tissues, and efficient sequestration of Na+ into vacuoles (Oh et al., 2009, 2010b; Flowers et al., 2010). Halophytes accumulate Na+ into vacuoles, and this provides the osmotic potential that supports water influx (Adams et al., 1998; Tester and Davenport, 2003) and accelerates growth (Adams et al., 1998). A distinction among halophytic species for the amount of NaCl required for optimal growth (Flowers et al., 2010) seems to describe a qualitative rather than a fundamental difference between species that may be related more to intrinsic vigor than to a specific set of mechanisms.
Thellungiella salsuginea (or Eutrema salsugineum, previously Thellungiella halophila) provides the model in which the halophytic nature of plants can be studied because it is a close relative of Arabidopsis, with all the advantages of this glycophytic model and with the exceptional ability to grow in seawater-strength concentrations of NaCl (Bressan et al., 2001; Amtmann et al., 2005; Vinocur and Altman, 2005; Oh et al., 2009). Moreover, the genome sequences for T. salsuginea (Q. Xie, personal communication) and the related Thellungiella parvula (Dassanayake et al., 2011a, 2011b) are now known. The two Thellungiella species exemplify the genetic and genomic basis of the extremophile lifestyle of plants and tolerance to salinity (Volkov et al., 2003; Inan et al., 2004; Gong et al., 2005; Vera-Estrella et al., 2005; Volkov and Amtmann, 2006; Amtmann, 2009; Oh et al., 2010a; Orsini et al., 2010). The tolerance, based on several studies in Thellungiella (Wang et al., 2004; Volkov and Amtmann, 2006), appears to be partly due to lower and tightly controlled net Na+ uptake and efficient and selective K+ uptake compared with Arabidopsis.
Here, we report the involvement of a T. salsuginea HKT1-like protein (TsHKT1;2) in contributing to the species’ halophytic behavior. This TsHKT1;2 is induced by salt stress, different from the expression pattern of Arabidopsis HKT1 (AtHKT1). T. salsuginea HKT-RNAi (for RNA interference) lines show sensitivity to NaCl and exhibit K+ deficiency, highlighting a role for this protein in salt tolerance and K+ transport in T. salsuginea. The different function of TsHKT1;2 becomes particularly clear when expressed in Arabidopsis wild-type and hkt1 mutant lines and yeast mutants lacking Na+ or K+ transporters. The results suggest differential Na+ and K+ selectivity for TsHKT1;2 as a salinity stress tolerance-defining difference in this species.
RESULTS
Plant HKT Superfamily Relationships
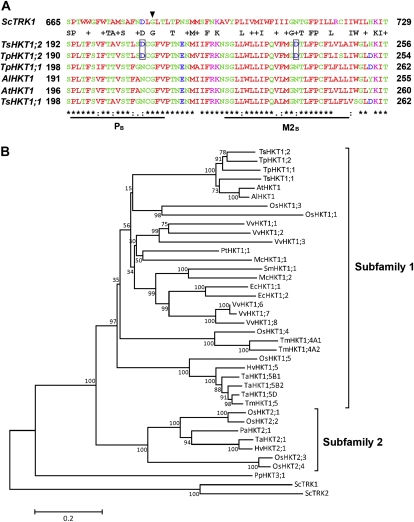
The T. salsuginea genome includes at least two different HKT-like transcripts (F. Quigley and H.J. Bohnert, unpublished data). Similarly, the genome sequence of T. parvula includes two tandemly duplicated HKT genes (Dassanayake et al., 2011a). We focus here on the transcript of the T. salsuginea HKT1;2 gene that shows differences in amino acid sequence that separate its sequence from that of TsHKT1;1 and from Arabidopsis HKT1 (Fig. 1A). TsHKT1;1 and TsHKT1;2 proteins exhibit 79.41% and 79.20% amino acid sequence identity with AtHKT1, respectively. In the phylogenetic tree, Arabidopsis and Thellungiella HKTs are grouped close together in subfamily 1. Within the branch consisting of the single-copy HKT1 sequences from Arabidopsis species, TsHKT1;1 was included, while TsHKT1;2 was separated (Fig. 1B; Supplemental Table S1). The two HKT genes in T. parvula show the same difference in their placement.
Figure 1.
Sequence comparison and phylogenetic analysis of HKT homologs in T. salsuginea. A, Comparison of HKT homologs from Arabidopsis and Thellungiella species. Amino acid sequences in the second pore loop region (PB) and the adjacent transmembrane domain (M2B) were aligned by ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The yeast TRK1 (ScTRK1) sequence is included for comparison. The conserved Gly residues in the PB region (Mäser et al., 2002) are indicated by the arrowhead. The Asp residues specific for TsHKT1;2 and TpHKT1;2 are highlighted with boxes. B, Unrooted minimum-evolution phylogenetic tree of protein sequences of HKT homologs with 10,000 bootstrap replicates. Accession numbers and species for all sequences are listed in Supplemental Table S1. Yeast TRK proteins (ScTRK1 and ScTRK2) are included as an outgroup. Subfamilies are as defined by Platten et al. (2006). The scale bar shows 0.2 substitutions per site. The tree was generated using MEGA5 (http://www.megasoftware.net/). [See online article for color version of this figure.]
The TsHKT1;2 Transcript Is Induced by Salt Stress
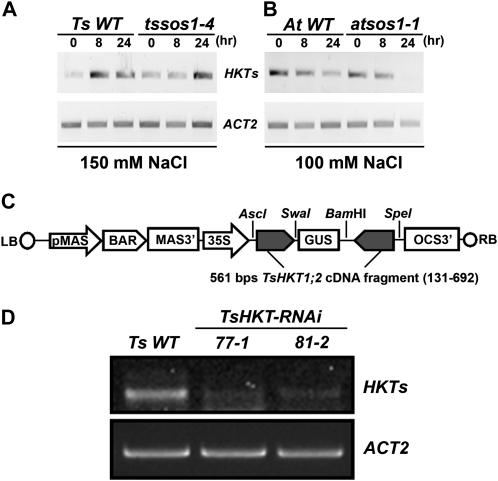
The expression patterns of HKT genes under control and salt stress conditions were determined in Arabidopsis and T. salsuginea plants. In a time-course experiment, HKT expression was induced by NaCl in T. salsuginea (Fig. 2A), whereas HKT1 transcript amounts declined during the same time in Arabidopsis (Fig. 2B). The same patterns were observed in the salt-sensitive T. salsuginea tssos1-4 (Oh et al., 2009) and atsos1-1 (Shi et al., 2000) mutants, suggesting a regulatory pathway independent of SOS1 activity (Fig. 2, A and B). Among the two known HKT1 copies in T. salsuginea, TsHKT1;2 was dramatically induced by salt stress while TsHKT1;1 expression remained at much lower levels (Supplemental Fig. S1). To gauge whether any of the HKT proteins, with HKT1;2 the more likely candidate because of its expression pattern, in T. salsuginea contributed to the halophytic character, lines transformed with a TsHKT-RNAi construct (Fig. 2C) were developed. Several independent TsHKT-RNAi transgenic T. salsuginea lines confirmed reduced expression of TsHKTs compared with the vector control (Fig. 2D).
Figure 2.
Expression of HKT homologs and construction of T. salsuginea TsHKT-RNAi transgenic plants. A and B, HKT expression patterns were analyzed with semiquantitative RT-PCR in T. salsuginea (A) and Arabidopsis (B). Two-week-old T. salsuginea and 10-d-old Arabidopsis seedlings were treated with 150 and 100 mm NaCl, respectively, for the indicated times. Compared are a T. salsuginea Shandong line transformed with the empty RNAi vector (Ts WT) and Arabidopsis Col-gl1 (At WT) and their respective mutant lines with compromised SOS1 expression (tssos1-4 and atsos1-1). Actin (ACT2) is used as a reference transcript. C, Diagrammatic representation of the TsHKT-RNAi construct. BAR, Bialaphos resistance; LB, left border; MAS3′, mannopine synthase transcriptional terminator; OCS-3′, octopine synthase transcriptional terminator; pMAS, mannopine synthase promoter; RB, right border. D, TsHKT expression was inhibited in T. salsuginea lines transformed with the TsHKT-RNAi construct. Shown are two representative TsHKT-RNAi transgenic lines (77-1 and 81-2) in comparison with the vector control (Ts WT).
TsHKT-RNAi Inhibits Growth and K+ Homeostasis in T. salsuginea
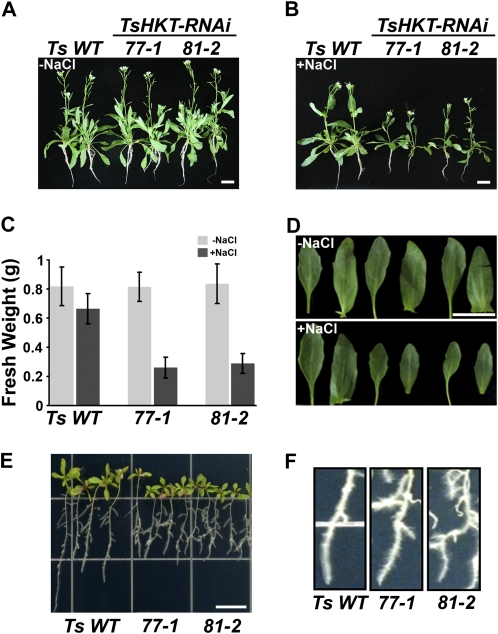
Sodium-specific responses are different in T. salsuginea and Arabidopsis, at least with respect to HKT expression levels (Fig. 2, A and B). To analyze this discrepancy at the activity level, TsHKT-RNAi lines were grown for 1 month on inert artificial soil, as described in “Materials and Methods,” and treated with 300 mm NaCl for 2 weeks. TsHKT-RNAi lines showed no differences in growth under nonstress conditions (Fig. 3A, −NaCl) but showed sensitivity to salt (Fig. 3B, +NaCl) compared with the vector control. The TsHKT-RNAi lines accumulated substantially less fresh weight (Fig. 3C) and produced smaller leaves upon salt stress compared with the wild type (Fig. 3D). In potassium-free medium supplemented with 100 mm NaCl, TsHKT-RNAi line seedlings showed inhibition of primary root growth (Fig. 3E) and developed root hairs at higher density compared with the wild type (Fig. 3F). To confirm a role for K+ accumulation by wild-type TsHKT and TsHKT-RNAi lines, hydroponically grown plants were treated with 250 mm NaCl for 24 h. Analysis of ion contents by inductively coupled plasma optical emission spectroscopy (ICP-OES) showed decreases in K+ and in TsHKT-RNAi lines compared with the wild type (Supplemental Fig. S2, A and C). The K+/Na+ ratio was significantly lower in the shoot tissues while marginally higher in the root tissues of the TsHKT-RNAi lines (Supplemental Fig. S2, B and D). These differences in growth and ion contents suggested a function for TsHKTs in Na+ tolerance and the maintenance of K+ homeostasis.
Figure 3.
Salt-sensitive phenotypes of T. salsuginea TsHKT-RNAi lines. A to D, T. salsuginea plants harboring either the vector control (Ts WT) or the TsHKT-RNAi construct (lines 77-1 and 81-2) were grown on inert artificial soil and treated with no salt (A) or 300 mm NaCl (B) as described in “Materials and Methods.” C, After the salt treatment, the fresh weights were compared, with error bars representing sd values from three independent repeats (n = 30 in each repeat). D, Representative leaf sizes of Ts WT and TsHKT-RNAi lines were also compared. E and F, Root growth of Ts WT and TsHKT-RNAi seedlings under salt stress. Ten-day-old seedlings grown on 1/2 MS were transferred to K+-deficient medium (see “Materials and Methods”) supplemented with 100 mm NaCl. The photograph was taken after an additional 10 d of vertical growth (E). A magnification of the root hair zone is shown in F. Bars = 10 mm in A, B, D, and E. [See online article for color version of this figure.]
TsHKT1;2 Is Different from AtHKT1 in the Na+ Hypersensitivity Response
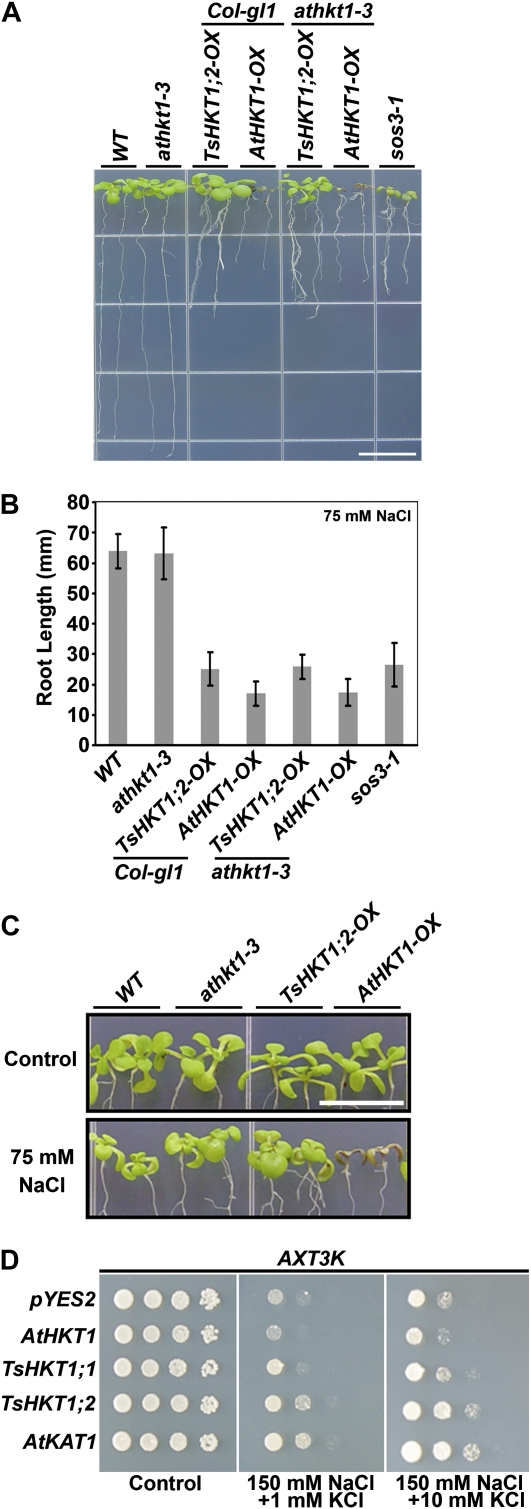
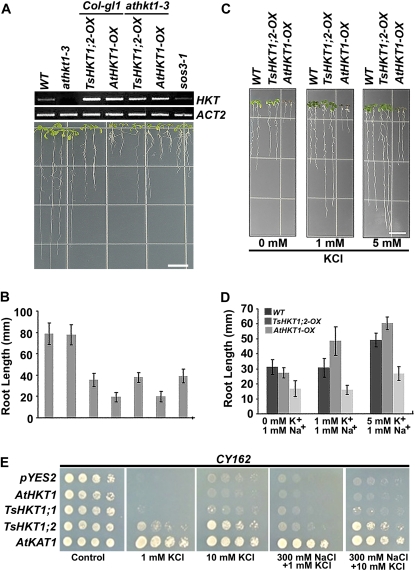
To analyze possible differential sodium-specific responses of T. salsuginea and Arabidopsis HKT1 proteins, seedlings of Arabidopsis lines expressing either AtHKT1 or TsHKT1;2 in the wild type or an AtHKT1 mutant (athkt1-3) background were transferred to medium including 75 mm NaCl. All HKT-expressing lines demonstrated root growth retardation compared with their respective controls. Lines expressing AtHKT1 showed more severe inhibition of primary root growth compared with those expressing TsHKT1;2 (Fig. 4, A and B). Shoot sensitivity responses were observed only in lines expressing AtHKT1 (Fig. 4, A and C).
Figure 4.
Comparison of salt stress responses of Arabidopsis and yeast strains ectopically expressing TsHKT1;2 and AtHKT1. A to C, Four-day-old Arabidopsis seedlings overexpressing TsHKT1;2 (TsHKT1;2-OX) or AtHKT1 (AtHKT1-OX) with a 35S-CaMV promoter, in the wild-type (WT; Col-gl1) or an AtHKT1 knockout mutant (athkt1-3) background, were transferred to salt medium (Supplemental Table S4). The photograph was taken after vertical growth for 7 d (A), and root growth was measured (B) with three independent replicates (n = 30 in each repeat). A magnified image is presented in C to show the shoot phenotype. Bars = 10 mm in A and C. D, TsHKT1;1, TsHKT1;2, and AtHKT1 were ectopically expressed in the S. cerevisiae strain AXT3K (Δena1-4-Δnha1 Δnhx1), which lacks the Na+ efflux system. Cells transformed with a vector control and AtKAT1 were included as negative and positive controls, respectively. Yeast cells of decimal dilution were plated on AP medium containing 1 mm K+ (Control) and 150 mm NaCl with 1 and 10 mm KCl, respectively. [See online article for color version of this figure.]
The affinity for Na+ and K+ ions of AtHKT1 and the TsHKTs was compared in a heterologous system. TsHKTs and AtHKT1 were expressed in yeast strain AXT3K (ena1::HIS3::ena4, nha1::LEU2, nhx1::KanMX4), which lacks Na+ transport activity and is salt sensitive (Quintero et al., 2002). Expression of the plasma membrane K+ channel AtKAT1 served as a control. Salt stress (150 mm NaCl) inhibited the growth of cells expressing AtHKT1, and expression of TsHKT1;1 did not result in a significant difference from the vector control. In contrast, cells expressing TsHKT1;2 showed growth on Arg/phosphate (AP) medium (Rodríguez-Navarro and Ramos, 1984) comparable to cells expressing AtKAT1. The addition of 10 mm K+ further enhanced the growth of cells expressing TsHKT1;2 or AtKAT1 (Fig. 4D). The cells expressing TsHKT1;2 accumulated significantly lower levels of Na+, while AtHKT1-expressing cells showed higher uptake of Na+ than the vector control (Supplemental Fig. S3A). On the contrary, TsHKT1;2-expressing cells accumulated significantly more K+, especially in the presence of 250 mm Na+, than the vector control. AtHKT1-expressing cells showed lower K+ levels than the vector control, suggesting totally opposite activities between AtHKT1 and TsHKT1;2 proteins (Supplemental Fig. S3B).
TsHKT-RNAi Led to Growth Retardation under K+-Limiting Conditions in T. salsuginea
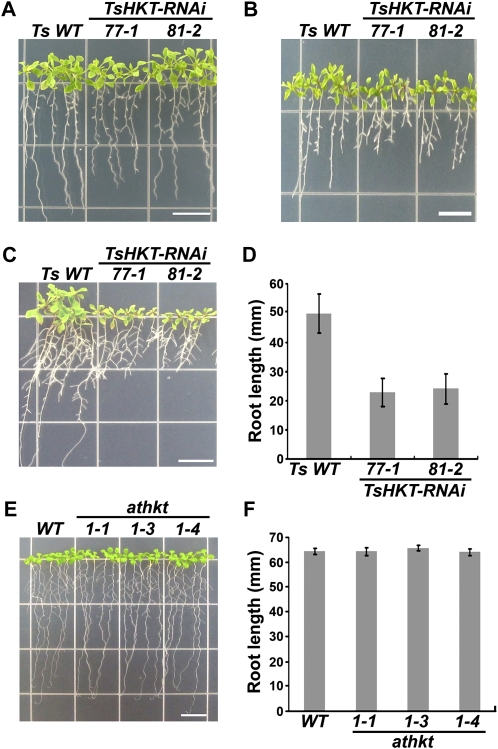
To probe for a possible function for TsHKT1;2 in K+ homeostasis, T3 homozygous TsHKT-RNAi plants grown on one-half-strength Murashige and Skoog medium (1/2 MS) for 10 d were transferred to K+-deficient medium. The K+-deficient medium was made by replacing KNO3 and KH2PO4 in 1/2 MS with NH4NO3 and (NH4)2HPO4, which inhibit K+ transporters (Supplemental Table S2). TsHKT-RNAi lines were marginally smaller when grown in normal medium (Fig. 5A) but were more sensitive to K+ deficiency compared with the wild type (Fig. 5B). The addition of 10 mm K+ was not able to enhance the growth of TsHKT-RNAi lines, unlike the response shown by wild-type seedlings (Fig. 5, C and D). This is in contrast to Arabidopsis, where athkt knockout mutants did not show differences in root growth on K+-deficient medium supplemented with 10 mm K+ (Fig. 5, E and F).
Figure 5.
Compromised growth of T. salsuginea TsHKT-RNAi lines under K+-limiting conditions. A to C, T. salsuginea seedlings harboring either the vector control (Ts WT) or the TsHKT-RNAi construct (lines 77-1 and 81-2) were grown on 1/2 MS for 10 d and transferred to 1/2 MS control medium (A), K+-deficient medium (B), or the same medium supplemented with 10 mm KCl (C). The K+-deficient medium was modified from MS medium containing no KNO3 and with KH2PO4 replaced with (NH4)2HPO4 (see “Materials and Methods”; Supplemental Table S2). Photographs were taken after incubation in a vertical position for 15 d. D, The root growth in C is quantified. E, Arabidopsis wild type (WT) and athkt1 knockout mutants under K+-limiting conditions. Arabidopsis Col-gl1 wild-type plants and Arabidopsis plants harboring different alleles of athkt1 knockout mutants (athkt1-1, -3, and -4) were grown on 1/2 MS for 4 d and transferred to K+-deficient medium supplemented with 10 mm KCl. The photograph was taken after the transferred seedlings had been incubated in a vertical position for 15 d. F, The root growth in E is quantified. Error bars in D and F represent sd values of three repeats (n = 30). Bars = 10 mm in A, B, C, and E. [See online article for color version of this figure.]
TsHKT1;2 Shows Higher Selectivity for K+ over Na+ Than AtHKT1
To analyze the putative K+-associated role of TsHKT1;2, transgenic Arabidopsis lines overexpressing either TsHKT1;2 or AtHKT1 under the control of the cauliflower mosaic virus (CaMV) 35S promoter were selected for the same level of HKT expression (Fig. 6A, top). Seeds from the homozygous T3 lines were germinated and grown for 4 d on 1/2 MS and transferred to K+-deficient medium supplemented with 20 mm NaCl. The growth inhibition symptoms were less severe in TsHKT1;2 transgenic plants compared with AtHKT1-expressing plants (Fig. 6, A and B). To test for ion selectivity of the HKT transporters, the same transgenic lines used in Figure 6A were grown on K+-deficient medium supplemented with 1 mm Na+ and 0, 1, or 5 mm K+ (Fig. 6, C and D). In the absence of K+, the presence of 1 mm Na+ reduced root growth, with a higher degree of inhibition observed in AtHKT1-overexpressing plants. However, TsHKT1;2-overexpressing plants showed root growth comparable or better than the wild type with a gradual increase of K+ concentration in the medium. In contrast, the root growth of AtHKT1-overexpressing plants was not rescued by the addition of K+ to the medium (Fig. 6, C and D).
Figure 6.
Distinct responses to K+-limiting conditions of Arabidopsis and yeast strains ectopically expressing TsHKT1;2 and AtHKT1. A and B, Four-day-old Arabidopsis seedlings overexpressing TsHKT1;2 (TsHKT1;2-OX) or AtHKT1 (AtHKT1-OX), on the wild-type (WT; Col-gl1) or an AtHKT1 knockout mutant (athkt1-3) background, were transferred to K+-deficient medium (Supplemental Table S2) supplemented with 20 mm NaCl. The photograph was taken after 7 d of incubation in a vertical position (A), and the root growth was quantified (B). Semiquantitative RT-PCR was used to select transgenic lines with comparable levels of ectopic HKT expression (A, top). C and D, The root growth of the same transgenic lines is compared with the wild type in the K+-deficient medium (Supplemental Table S2) supplemented with 1 mm Na+ and 0, 1, or 5 mm K+ ions. Photographs were taken as in A (C), and root growth was quantified as in B (D). Bars = 10 mm in A and C. Error bars in B and D represent sd values of three repeats (n = 30). E, Growth of yeast strain CY162 (Mat a, ura3-52, his3D200, his44-15, trkD1, trkD2::pcK64) cells harboring the vector control (pYES2), TsHKT1;1, TsHKT1;2, AtHKT1, and AtKAT1. Cells were plated in decimal dilutions on AP medium containing either 100 mm KCl (Control) or the indicated amounts of KCl and NaCl. Cells transformed with a vector control and AtKAT1 are included as negative and positive controls, respectively. [See online article for color version of this figure.]
The K+ transport activity and preference over Na+ by TsHKT1;2 were further confirmed by ectopic expression of TsHKT1;2 in the K+ transporter-deficient yeast strain CY162 (Mat a, ura3–52, his3D200, his44–15, trkD1, trkD2::pcK64; Ko and Gaber, 1991). On AP medium containing 1 mm K+, cells expressing TsHKT1;2 showed growth comparable to cells expressing AtKAT1, while the expression of AtHKT1 or the vector plasmid failed to restore the growth of the cells (Fig. 6E). Similarly, AtKAT1- and TsHKT1;2-expressing cells grew strongly on 10 mm K+ in the presence of 300 mm NaCl. At this concentration of K+, TsHKT1;1-expressing cells also showed somewhat enhanced growth that was not seen with cells expressing AtHKT1 (Fig. 6E), indicating that a trend toward K+ selectivity, although less pronounced than in TsHKT1;2, is present in TsHKT1;1 as well.
K+ Specificity by TsHKT1;2 Is Based on Subtle Sequence Differences
Among differences in primary amino acid sequence (Supplemental Fig. S4), two amino acids seemed to stand out because of their location in transmembrane and pore domains in a comparison between plant HKT protein sequences (Fig. 1A). In both places, the replacements are Asp (D) residues in TsHKT1;2 (and TpHKT1;2), whereas Asn (N) is found in Arabidopsis and all other known plant sequences (data not shown).
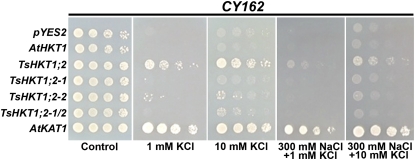
The two D residues, individually and together, were replaced by N residues (D207N, D238N) that are present in AtHKT1 (Fig. 1A). Each single change (lines TsHKT1;2-1 and TsHKT1;2-2, respectively) resulted in reduced growth in the presence of Na+, which was particularly obvious in AP medium with 1 mm KCl and in 1 mm KCl + 300 mm NaCl (Fig. 7). However, in the presence of 10 mm KCl with or without NaCl added, the single mutants and cells harboring the double mutant (line TsHKT1;2-1/2) performed slightly better than AtHKT1, albeit less well than the wild-type TsHKT1;2-expressing cells (Fig. 7).
Figure 7.
Effects of point mutations in positions 207 and 238 of the TsHKT1;2 protein. Yeast strain CY162 (Mat a, ura3-52, his3D200, his44-15, trkD1, trkD2::pcK64) cells harboring TsHKT1;2 were compared with cells expressing point mutants TsHKT1;2-1 (D207N), TsHKT1;2-2 (D238N), and TsHKT1;2-1/2 (D207N and D238N). Cells were spotted in decimal dilution on AP medium containing either 100 mm KCl (Control) or the indicated amounts of KCl and NaCl. Cells transformed with the vector control, AtHKT1, and AtKAT1 are included as controls. [See online article for color version of this figure.]
DISCUSSION
A View of Thellungiella’s Lifestyle
Our study aimed at analyzing the contribution, if any, by a HKT1 isoform of the well-studied AtHKT1 and OsHKT1 proteins to the halophytic character of T. salsuginea. Distinct from glycophytes, the halophytic character has been described as quantitative rather than qualitative (Waisel, 1972), and previous studies have supported this notion (Adams et al., 1998; Inan et al., 2004; Gong et al., 2005; Oh et al., 2009). If so, one might expect that for being salt tolerant or salt requiring, a gradation exists, possibly set up by a number of genes and proteins that might differ not in type but in how responsive the genes or how sensitive the proteins are under this stress condition. By now, the genome sequences of two halophytic close Arabidopsis relatives, T. parvula and T. salsuginea (Dassanayake et al., 2011a; Q. Xie, personal communication), may be consulted for this. While the organization of these genomes is highly conserved in comparison with the Arabidopsis genome, significant differences exist in the number of isoforms for many stress-relevant genes and their expression (Oh et al., 2010a; Dassanayake et al., 2011a, 2011b).
Irrespectively, we cannot yet point precisely to the set of genes that define halophytism, but several pathways and genes have emerged and others, less appreciated or known at present, are most certainly involved. The possible contributions of a large number of monovalent cation transporters and ill-defined other transport proteins in different compartments are examples. There are 197 genes in this category encoded in the Arabidopsis genome (The Arabidopsis Information Resource 9), while 299 genes for proteins in the same category have been identified in the genome sequence of T. parvula (Dassanayake et al., 2011a).
Sodium Transport Systems
Ways by which the influx, efflux, and distribution of sodium ions into and within the plant are controlled have been studied to some degree. Of foremost significance and importance is the SOS pathway. The sequential action of three genes in the pathway allows for sodium excretion and distribution throughout the plant body, to a degree that seems to change during the development of plants, with the Na+/H+ antiporter protein, SOS1, playing an important role (Shi et al., 2000; Qiu et al., 2002). While the dynamic distribution of activated components of the SOS pathway in different tissues and growth phases, under no-stress conditions and during a salt stress episode, is yet unknown, elimination or reduction (RNAi) of genes in the SOS pathway, SOS1 in particular, leads to severe sensitivity to NaCl in glycophytes and halophytes alike (Liu et al., 2000; Shi et al., 2000; Oh et al., 2009).
Another transport process has received attention as well. Wheat TaHKT1 is a Na+/K+ transporter whose conductance depends on the Na+ concentration. At micromolar concentrations, it works as a Na+/K+ symporter, but with Na+ in the millimolar range, HKT1 acts as a Na+ uniporter (Rubio et al., 1995). In contrast, AtHKT1 acts as a high-affinity selective Na+ transporter in heterologous systems such as Xenopus oocytes and yeast (Uozumi et al., 2000; Berthomieu et al., 2003). Rice contains multiple HKT genes (Fig. 1B), with genes in group OsHKT1 behaving as Na+ transporters similar to AtHKT1, while OsHKT2 genes behave as symporters or uniporters, as does TaHKT1 (Hauser and Horie, 2010). Considering that most rice HKT-like transporters have not been studied, it appears possible that this family in rice has taken on crucial roles in monovalent cation homeostasis. K+ deficiency acts as a positive regulator for the expression of both types of OsHKT genes, whereas Na+ leads to negative regulation (Horie et al., 2001). This behavior is reminiscent of fungal TRK K+ transport proteins that are structurally related to HKTs. Fungal TRKs accomplish conditional modulation of Na+/K+ selectivity according to the ionic environment and the K+ status of the cell (Rodríguez-Navarro, 2000).
Early on, the superior ability of T. salsuginea at maintaining a high K+/Na+ ratio under salinity challenges was recognized (Volkov et al., 2003; Vera-Estrella et al., 2005; Volkov and Amtmann, 2006). An extraordinarily strong support for the importance of HKT-like proteins in mediating salinity stress tolerance has been provided by genetic and molecular studies in models (Sunarpi et al., 2005; Møller et al., 2009; Genc et al., 2010; Hauser and Horie, 2010; Plett et al., 2010; Qiu et al., 2011). In several cases, improved salinity tolerance has been associated with regions of genomes that included HKT-type transporters. Here, we provide evidence for the basis of a similar characteristic in T. salsuginea. It is based on the behavior of one of the HKT family transporters present in this true halophyte. TsHKT1;2 is part of the system that shapes salinity tolerance in this species. Our results identify TsHKT1;2 acting in a different function from AtHKT1 and, by sequence signature and expression characteristic, also the TsHKT1;1 protein. This example of neofunctionalization of a duplicated gene with a function in K+ homeostasis in the presence of Na+ ions provides the second example, in addition to gene expression strength, for the halophytic lifestyle that characterizes Thellungiella species.
Evolutionary Implications of HKT Genes in Thellungiella Species
The view of the evolutionary relationships appears to argue for reconsidering the HKT nomenclature, because functional diversity in terms of ion specificity could be more complex than what is implied by the distinction of two subfamilies. HKT proteins in vascular plants can be divided into two subfamilies with putatively distinct ion selectivities (Platten et al., 2006; Hauser and Horie, 2010). It has been suggested that the members of subfamily 2 contain a conserved Gly residue in the first pore loop of the protein, while members of subfamily 1 show a Ser residue in this position (Mäser et al., 2002). Had it previously been assumed that this Gly-to-Ser substitution in the first pore loop might be responsible for K+ or Na+ selectivity (Uozumi et al., 2000), the fact that TsHKT1;2 fails to follow this rule seems to indicate that another site, or sites, must be responsible for the K+ preference over Na+.
Irrespective of the observed difference in ion selectivity, both TsHKT1;2 and AtHKT1 are included in subfamily 1 and grouped together with other crucifer HKT proteins (Fig. 1B). In the tree structure, TsHKT1;1, TsHKT1;2, and AtHKT1 contain the conserved Ser residues in the first pore loop (Supplemental Fig. S4, PA). The positively charged amino acids in the M2D transmembrane domain, suggested as an essential determinant for K+ transport activity (Kato et al., 2007), were also conserved among the crucifer species (Supplemental Fig. S4, arrows in M2D). Amino acid sequence variations distinguishing AtHKT1 and TsHKT1;1 (as well as TpHKT1;1) appeared in the second pore loop region (PB) and the adjacent transmembrane domain (M2B). In the positions where Arabidopsis HKT1 contains conserved Asn residues, both TsHKT1;2 and TpHKT1;2 contained conserved negatively charged Asp residues (Fig. 1A). These Asp residues in the PB region are unique to TsHKT1;2 and TpHKT1;2 and not shared by other plant HKTs known to date (data not shown). However, the yeast potassium transporter ScTrk1 contains an Asp in the same position of the pore loop region, suggesting this amino acid as a possible determinant for potassium-specific transporters (Fig. 1A). It appears that TsHKT1;2 achieves ion selectivity through subtle amino acid sequence variations that are different from the HKT2 proteins included in subfamily 2 (Mäser et al., 2002; Platten et al., 2006). The distinction in two subfamilies seems less a functional characteristic than a reflection of evolutionary distance, with the monocot-specific label HKT2 possibly signifying the retention of, following genome duplications and subsequent amplification, an HKT isoform that became eliminated in dicotyledonous species. Equalizing the HKT2 branch with preferentially K+-transporting proteins may require discussion and possible revision. Based on our results, sequence diversity and functional characteristics appear not connected in a way that can be partitioned into these two HKT subfamilies (Platten et al., 2006).
The Basis of Halophytism?
The inclusion of the TsHKT1;2 protein sequence in alignments provides insights that might not only be useful in understanding how these proteins function. In addition, our results also demonstrate the involvement of TsHKT1;2 in global K+ homeostasis. The ability of Thellungiella species to maintain a low cytosolic Na+/K+ ratio in the presence of salt has been shown before (Orsini et al., 2010). Adding more detail, TsHKT1;2 emerged as part of the system that shapes salinity tolerance in this species. Some HKTs responsible for K+ homeostasis in versions of the protein that act as Na+/K+ symporters were induced by salt or by K+ deficiency, while in other species, expression ceased under salt stress (Rubio et al., 1995; Wang et al., 1998; Rus et al., 2001; Kader et al., 2006). Salt stress positively regulates TsHKT1;2, whereas the expression of its isoform TsHKT1;1 was only insignificantly altered by salinity (Supplemental Fig. S1), while AtHKT1 was down-regulated by salt (Fig. 2B). Our analysis of this salt-specific induction, documented by RNAi lines and overexpression, suggests a major role for TsHKT1;2 in the acquisition of K+ in the presence of salt. High Na+ content in the cell results in K+ deficiency (Maathius and Amtmann, 1999), alters the Na+/K+ ratio, and induces K+ transporters (Horie et al., 2001). Mutations in SOS1 or its expression level reduce salt tolerance in Arabidopsis (Shi et al., 2000) and in its halophytic relative T. salsuginea (Oh et al., 2009). T. salsuginea tssos1-4 (RNAi line of TsSOS1 sensitive to salt) showed strong salt-dependent up-regulation of TsHKT1;2 (Fig. 2A; Supplemental Fig. S1), while strong repression of AtHKT1 expression was seen in atsos1-1 plants under salt stress (Fig. 2B). These responses highlight the fundamental divergence of expression and activity between the two TsHKT proteins. AtHKT1 is a Na+ uniporter with no affinity for K+ (Uozumi et al., 2000). Similarly HKT1-disrupted Arabidopsis lines (athkt1-1, athkt1-3, and athkt1-4) did not demonstrate sensitivity to K+ deficiency (Fig. 5E). Conversely, TsHKT down-regulation by RNAi demonstrated reduced fresh weight, the formation of small leaves, as well as root growth retardation upon salt stress on K+-deficient medium (Fig. 3). These results highlight an essential role for TsHKT1;2 in K+ homeostasis and salt tolerance. The juxtapositioning of Arabidopsis and T. salsuginea seems to point toward copy number variation, in gene number and expression, as well as biochemical differences in activities that maintain a high K+/Na+ ratio as the major determinants in their behavior to salt stress that decide growth and reproductive success.
Halophytes take advantage of an excess of Na+ for maintaining cellular osmotic potential to support water uptake and growth, by maintaining low cytosolic [Na+], if K+ uptake in the presence of salt stress can be maintained (Adams et al., 1998; Tester and Davenport, 2003; Orsini et al., 2010). The low affinity of TsHKT1;2 for Na+ supports this physiological mechanism. The same rationale demands that AtHKT1, lacking any specificity for K+, be down-regulated when external sodium ions increase (Fig. 2B). AtHKT1 also fails to complement CY162 (a yeast strain deficient in the K+ transport system) cells (Uozumi et al., 2000; Fig. 6E) in contrast to TsHKT1;2 (Fig. 6E). Our results demonstrate a critical involvement of TsHKT1;2 in the halophytic behavior of T. salsuginea and strongly support findings in other species about the importance of HKT in salinity stress tolerance (Kader et al., 2006; Møller et al., 2009). Earlier results indicating strong induction and activity after salt stress of TsSOS1 (Oh et al., 2009, 2010b) might imply that the coevolution of these two ion transport systems provides a crucial component in shaping halophytic, extremophile lifestyles.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) seeds of ecotype Columbia (Col)-gl1 (wild type) as well as athkt1-1, athkt1-3, athkt1-4, and sos3-1 in the Col-gl1 background were provided by Prof. P.M. Hasegawa (Purdue University). Thellungiella salsuginea v162, a line lacking the vernalization requirement, was used for development of the transgenic plants.
Generation of TsHKT-RNAi Thellungiella and Arabidopsis Plants
The TsHKT-RNAi construct was made by inserting a 561-bp (131–692 bp) cDNA TsHKT fragment back and forth at both ends of GUS (used as intron) through restriction/ligation of a PCR-amplified product in the pGSA1285 vector. Primers used for the amplification of forward and reverse fragments of the 561-bp cDNA are given in Supplemental Table S3. This construct should silence all HKT copies in T. salsuginea. T. salsuginea line v162 plants were transformed by floral dip using Agrobacterium tumefaciens suspension cells harboring the TsHKT-RNAi construct. The transformed plants were selected by BASTA and reconfirmed by PCR using the primers listed in Supplemental Table S3. Four independent homozygous T3 lines were used in the experiments.
The cDNAs of TsHKT1;2 and AtHKT1 were amplified by reverse transcription (RT)-PCR with the primers listed in Supplemental Table S3. The cloned HKT cDNAs were inserted between BamHI and XbaI sites downstream of a 35S-CaMV promoter in the 1300PT (Multi) binary vector, and wild-type Col-gl1 and athkt1-3 plants were transformed by floral dip. Transgenic plants were selected based on hygromycin resistance and confirmed with the primers listed in Supplemental Table S3. Lines showing 3:1 segregation with resistance to hygromycin were selected, and homozygous T3 plants showing comparable levels of TsHKT1;2 or AtHKT1 expression were used for comparative experiments.
Growth Responses to Different Ion Compositions
To test growth responses to NaCl of the mature plants, 10-d-old T. salsuginea transgenic plants were transferred to inert soil (Isolite CG-1; Isolite Insulating Products), grown for 1 month with irrigation with one-eighth-strength MS medium solution every other day, and treated with 300 mm NaCl for 2 weeks. For all other in vitro experiments, Arabidopsis and T. salsuginea plants were germinated, incubated on 1/2 MS salts, 30 g L−1 Suc, and 12 g L−1 agar plates for 4 and 10 d, respectively, and transferred to medium containing Na+ or deficient in K+ accordingly. Detailed medium recipes are included in Supplemental Tables S2 and S4. For HKT expression analyses, Arabidopsis and T. salsuginea plants were grown, treated with salt, and harvested as described by Oh et al. (2009).
RNA Extraction and Quantitative RT-PCR
RNAs from Thellungiella and Arabidopsis both were extracted with the RNeasy Plant Mini Kit (Qiagen). RT-PCR was carried out with 3 μg of total RNA using the ThermoScript RT-PCR System (Invitrogen) with the primers listed in Supplemental Table S3. Quantitative RT-PCR (Supplemental Fig. S1) was done essentially as described by Gong et al. (2005).
Yeast Expression and Growth
Yeast strains AXT3K (ena1::HIS3::ena4, _nha1::LEU2, _nhx1::KanMX4; Quintero et al., 2002) and CY162 (Mat a, ura3-52, his3D200, his44-15, trkD1, trkD2::pcK64; Ko and Gaber, 1991) were used. The cDNAs, amplified with the primers listed in Supplemental Table S3, were cloned into the BamHI and NotI sites of the pYES2 vector (Invitrogen). Yeast cells were transformed with LiAc methods, selected on −Ura synthetic dropout medium, and subjected to growth on AP medium (Rodríguez-Navarro and Ramos, 1984) with K+ and Na+ added as indicated.
Analysis of Ion Content
Ionic content analyses in plants were carried out as described (Rus et al., 2001), except that plants were grown hydroponically for 4 weeks. Samples were dried at 65°C for 2 d, and 100 mg of ground tissue was extracted with 10 mL of 0.1 n HNO3 for 30 min. Samples were filtered, and K+ ion content analysis was carried out with ICP-OES (Optima 4300DV/5300DV; Perkin-Elmer). Analyses of ion uptake in yeast followed a protocol by Takahashi et al. (2007) with minor modifications. Briefly, yeast (Saccharomyces cerevisiae strain AXT3K) transformed with pYES2 (vector control), AtHKT1, TsHKT1;1, and TsHKT1;2 were grown in synthetic dropout medium (dropout medium without uracil) to an optical density at 600 nm of 0.7 (1.0 × 108 cells). Cells were suspended in 10 mL of AP medium with 2.0% Glc, 10 mm MES, pH 6.0 adjusted with Tris base, and supplemented with the indicated concentrations of KCl and NaCl. Cells were incubated for 2 h, collected at 1,250 rpm for 5 min, and acid extracted overnight in 10 mL of 0.1 m HCl. Samples were centrifuged at 5,000g for 5 min to remove debris, and ion contents of the supernatant were determined by ICP-OES (Optima 4300DV/5300DV; Perkin-Elmer).
Site-Directed Mutagenesis of TsHKT1;2
In T. salsuginea HKT1;2, two conserved amino acids (Asp) in the second pore loop region (Fig. 1A, PB) and the adjacent transmembrane domain were mutated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). The changes introduced the amino acid Asn that is found in Arabidopsis HKT1 in these positions (Fig. 1A; Supplemental Fig. S4). Individual amino acid mutations (D207N [TsHKT1;2-1] and D238N [TsHKT1;2-2]) were made to the template of pYES2-TsHKT1;2 cDNA with their respective primers (Supplemental Table S3). To make a mutation of both (D207N and D238N; double mutant TsHKT1;2-1/2), the mutated pYES2-TsHKT1;2-1 was used as a template with the primers designed for TsHKT1;2-2.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers BAJ34563.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression pattern of TsHKT1;1 and TsHKT1;2.
Supplemental Figure S2. K+ and Na+ content of TsHKT-RNAi lines.
Supplemental Figure S3. Ion content in yeast cells expressing AtHKT1, TsHKT1;1, and TsHKT1;2.
Supplemental Figure S4. Alignment of HKT1 protein sequences of T. salsuginea, T. parvula, Arabidopsis, and Arabidopsis lyrata.
Supplemental Table S1. HKT-like protein sequences used for phylogenetic analysis.
Supplemental Table S2. K+-deficient media recipe.
Supplemental Table S3. List of primers.
Supplemental Table S4. Minimal Media for salt stress treatment.
Acknowledgments
We are grateful to Dr. Nobuyuki Uozumi (Tohoku University) for providing cDNAs and yeast strain CY162 with permission from Dr. R.F. Gaber (Northwestern University). We also thank Prof. Paul M. Hasegawa (Purdue University) and Dr. Ana Rus (California State University) for providing materials and Dr. Qi Xie (Chinese Academy of Genetics) for providing unpublished information.
References
- Adams P, Nelson DE, Yamada S, Chmara W, Jensen RG, Bohnert HJ, Griffiths H. (1998) Growth and development of Mesembryanthemum crystallinum. New Phytol 138: 171–190 [DOI] [PubMed] [Google Scholar]
- Amtmann A. (2009) Learning from evolution: Thellungiella generates new knowledge on essential and critical components of abiotic stress tolerance in plants. Mol Plant 2: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amtmann A, Bohnert HJ, Bressan RA. (2005) Abiotic stress and plant genome evolution: search for new models. Plant Physiol 138: 127–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apse MP, Aharon GS, Snedden WA, Blumwald E. (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285: 1256–1258 [DOI] [PubMed] [Google Scholar]
- Berthomieu P, Conéjéro G, Nublat A, Brackenbury WJ, Lambert C, Savio C, Uozumi N, Oiki S, Yamada K, Cellier F, et al. (2003) Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J 22: 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan RA, Zhang C, Zhang H, Hasegawa PM, Bohnert HJ, Zhu JK. (2001) Learning from the Arabidopsis experience: the next gene search paradigm. Plant Physiol 127: 1354–1360 [PMC free article] [PubMed] [Google Scholar]
- Dassanayake M, Oh DH, Haas JS, Hernandez A, Hong H, Ali S, Yun DJ, Bressan RA, Zhu JK, Bohnert HJ, Cheeseman JM. (2011a) The genome of the extremophile crucifer Thellungiella parvula. Nat Genet 43: 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassanayake M, Oh DH, Hong H, Bohnert HJ, Cheeseman JM. (2011b) Transcription strength and halophytic lifestyle. Trends Plant Sci 16: 1–3 [DOI] [PubMed] [Google Scholar]
- Flowers TJ, Galal HK, Bromham L. (2010) Evolution of halophytes: multiple origins of salt tolerance in land plants. Funct Plant Biol 37: 604–612 [Google Scholar]
- Genc Y, Oldach K, Verbyla AP, Lott G, Hassan M, Tester M, Wallwork H, McDonald GK. (2010) Sodium exclusion QTL associated with improved seedling growth in bread wheat under salinity stress. Theor Appl Genet 121: 877–894 [DOI] [PubMed] [Google Scholar]
- Gong Q, Li P, Ma S, Indu Rupassara S, Bohnert HJ. (2005) Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J 44: 826–839 [DOI] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51: 463–499 [DOI] [PubMed] [Google Scholar]
- Hauser F, Horie T. (2010) A conserved primary salt tolerance mechanism mediated by HKT transporters: a mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant Cell Environ 33: 552–565 [DOI] [PubMed] [Google Scholar]
- Horie T, Brodsky DE, Costa A, Kaneko T, Lo Schiavo F, Katsuhara M, Schroeder JI. (2011) K+ transport by the OsHKT2;4 transporter from rice with atypical Na+ transport properties and competition in permeation of K+ over Mg2+ and Ca2+ ions. Plant Physiol 156: 1493–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Yoshida K, Nakayama H, Yamada K, Oiki S, Shinmyo A. (2001) Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J 27: 129–138 [DOI] [PubMed] [Google Scholar]
- Inan G, Zhang Q, Li P, Wang Z, Cao Z, Zhang H, Zhang C, Quist TM, Goodwin SM, Zhu J, et al. (2004) Salt cress: a halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles. Plant Physiol 135: 1718–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader MA, Seidel T, Golldack D, Lindberg S. (2006) Expressions of OsHKT1, OsHKT2, and OsVHA are differentially regulated under NaCl stress in salt-sensitive and salt-tolerant rice (Oryza sativa L.) cultivars. J Exp Bot 57: 4257–4268 [DOI] [PubMed] [Google Scholar]
- Kato N, Akai M, Zulkifli L, Matsuda N, Kato Y, Goshima S, Hazama A, Yamagami M, Guy HR, Uozumi N. (2007) Role of positively charged amino acids in the M2D transmembrane helix of Ktr/Trk/HKT type cation transporters. Channels (Austin) 1: 161–171 [DOI] [PubMed] [Google Scholar]
- Ko CH, Gaber RF. (1991) TRK1 and TRK2 encode structurally related K+ transporters in Saccharomyces cerevisiae. Mol Cell Biol 11: 4266–4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronzucker HJ, Britto DT. (2011) Sodium transport in plants: a critical review. New Phytol 189: 54–81 [DOI] [PubMed] [Google Scholar]
- Liu JP, Ishitani M, Halfter U, Kim CS, Zhu J-K. (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA 97: 3730–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis FJM, Amtmann A. (1999) K+ nutrition and Na+ toxicity: the basis of cellular K+/Na+ ratios. Ann Bot (Lond) 84: 123–133 [Google Scholar]
- Mäser P, Eckelman B, Vaidyanathan R, Horie T, Fairbairn DJ, Kubo M, Yamagami M, Yamaguchi K, Nishimura M, Uozumi N, et al. (2002) Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett 531: 157–161 [DOI] [PubMed] [Google Scholar]
- Møller IS, Gilliham M, Jha D, Mayo GM, Roy SJ, Coates JC, Haseloff J, Tester M. (2009) Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 21: 2163–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Shen B, Bohnert HJ. (1998) Salinity tolerance: mechanisms, models and the metabolic engineering of complex traits. Genet Eng (NY) 20: 153–176 [DOI] [PubMed] [Google Scholar]
- Oh DH, Dassanayake M, Haas JS, Kropornika A, Wright C, d’Urzo MP, Hong H, Ali S, Hernandez A, Lambert GM, et al. (2010a) Genome structures and halophyte-specific gene expression of the extremophile Thellungiella parvula in comparison with Thellungiella salsuginea (Thellungiella halophila) and Arabidopsis. Plant Physiol 154: 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DH, Lee SY, Bressan RA, Yun DJ, Bohnert HJ. (2010b) Intracellular consequences of SOS1 deficiency during salt stress. J Exp Bot 61: 1205–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DH, Leidi E, Zhang Q, Hwang SM, Li Y, Quintero FJ, Jiang X, D’Urzo MP, Lee SY, Zhao Y, et al. (2009) Loss of halophytism by interference with SOS1 expression. Plant Physiol 151: 210–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsini F, D’Urzo MP, Inan G, Serra S, Oh DH, Mickelbart MV, Consiglio F, Li X, Jeong JC, Yun DJ, et al. (2010) A comparative study of salt tolerance parameters in 11 wild relatives of Arabidopsis thaliana. J Exp Bot 61: 3787–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo JM, Cubero B, Leidi EO, Quintero FJ. (2006) Alkali cation exchangers: roles in cellular homeostasis and stress tolerance. J Exp Bot 57: 1181–1199 [DOI] [PubMed] [Google Scholar]
- Platten JD, Cotsaftis O, Berthomieu P, Bohnert HJ, Davenport RJ, Fairbairn DJ, Horie T, Leigh RA, Lin HX, Luan S, et al. (2006) Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci 11: 372–374 [DOI] [PubMed] [Google Scholar]
- Plett D, Safwat G, Gilliham M, Skrumsager Møller I, Roy S, Shirley N, Jacobs A, Johnson A, Tester M. (2010) Improved salinity tolerance of rice through cell type-specific expression of AtHKT1;1. PLoS ONE 5: e12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L, Wu D, Ali S, Cai S, Dai F, Jin X, Wu F, Zhang G. (2011) Evaluation of salinity tolerance and analysis of allelic function of HvHKT1 and HvHKT2 in Tibetan wild barley. Theor Appl Genet 122: 695–703 [DOI] [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK. (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99: 8436–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Ohta M, Shi H, Zhu JK, Pardo JM. (2002) Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci USA 99: 9061–9066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Navarro A. (2000) Potassium transport in fungi and plants. Biochim Biophys Acta 1469: 1–30 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Navarro A, Ramos J. (1984) Dual system for potassium transport in Saccharomyces cerevisiae. J Bacteriol 159: 940–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio F, Gassmann W, Schroeder JI. (1995) Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270: 1660–1663 [DOI] [PubMed] [Google Scholar]
- Rus A, Baxter I, Muthukumar B, Gustin J, Lahner B, Yakubova E, Salt DE. (2006) Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet 2: e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus A, Yokoi S, Sharkhuu A, Reddy M, Lee BH, Matsumoto TK, Koiwa H, Zhu JK, Bressan RA, Hasegawa PM. (2001) AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. Proc Natl Acad Sci USA 98: 14150–14155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman DP, Schroeder JI. (1994) Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature 370: 655–658 [DOI] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim CS, Zhu JK. (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97: 6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Lee B-H, Wu S-J, Zhu JK. (2003) Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat Biotechnol 21: 81–85 [DOI] [PubMed] [Google Scholar]
- Sunarpi HT, Horie T, Motoda J, Kubo M, Yang H, Yoda K, Horie R, Chan WY, Leung HY, Hattori K, et al. (2005) Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J 44: 928–938 [DOI] [PubMed] [Google Scholar]
- Takahashi R, Liu S, Takano T. (2007) Cloning and functional comparison of a high-affinity K+ transporter gene PhaHKT1 of salt-tolerant and salt-sensitive reed plants. J Exp Bot 58: 4387–4395 [DOI] [PubMed] [Google Scholar]
- Tester M, Davenport R. (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot (Lond) 91: 503–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uozumi N, Kim EJ, Rubio F, Yamaguchi T, Muto S, Tsuboi A, Bakker EP, Nakamura T, Schroeder JI. (2000) The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol 122: 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera-Estrella R, Barkla BJ, García-Ramírez L, Pantoja O. (2005) Salt stress in Thellungiella halophila activates Na+ transport mechanisms required for salinity tolerance. Plant Physiol 139: 1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinocur B, Altman A. (2005) Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Biotechnol 16: 123–132 [DOI] [PubMed] [Google Scholar]
- Volkov V, Amtmann A. (2006) Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana, has specific root ion-channel features supporting K+/Na+ homeostasis under salinity stress. Plant J 48: 342–353 [DOI] [PubMed] [Google Scholar]
- Volkov V, Wang B, Dominy PJ, Fricke W, Amtmann A. (2003) Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana, possesses effective mechanisms to discriminate between potassium and sodium. Plant Cell Environ 27: 1–14 [Google Scholar]
- Waisel Y. (1972) Biology of Halophytes. Academic Press, New York [Google Scholar]
- Wang TB, Gassmann W, Rubio F, Schroeder JI, Glass ADM. (1998) Rapid up-regulation of HKT1, a high-affinity potassium transporter gene, in roots of barley and wheat following withdrawal of potassium. Plant Physiol 118: 651–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZI, Li P, Fredricksen M, Gong ZH, Kim CS, Zhang CQ, Bohnert HJ, Zhu J-K, Bressan RA, Hasegawa PM, et al. (2004) Expressed sequence tags from Thellungiella halophila, a new model to study plant salt-tolerance. Plant Sci 166: 609–616 [Google Scholar]
- Zhu J-K. (2001) Plant salt tolerance. Trends Plant Sci 6: 66–71 [DOI] [PubMed] [Google Scholar]
- Zhu J-K. (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6: 441–445 [DOI] [PubMed] [Google Scholar]