Abstract
In legumes, the symbiotic nodules are formed as a result of dedifferentiation and reactivation of cortical root cells. A shoot-acting receptor complex, similar to the Arabidopsis (Arabidopsis thaliana) CLAVATA1 (CLV1)/CLV2 receptor, regulating development of the shoot apical meristem, is involved in autoregulation of nodulation (AON), a mechanism that systemically controls nodule number. The targets of CLV1/CLV2 in the shoot apical meristem, the WUSCHEL (WUS)-RELATED HOMEOBOX (WOX) family transcription factors, have been proposed to be important regulators of apical meristem maintenance and to be expressed in apical meristem “organizers.” Here, we focus on the role of the WOX5 transcription factor upon nodulation in Medicago truncatula and pea (Pisum sativum) that form indeterminate nodules. Analysis of temporal WOX5 expression during nodulation with quantitative reverse transcription-polymerase chain reaction and promoter-reporter fusion revealed that the WOX5 gene was expressed during nodule organogenesis, suggesting that WOX genes are common regulators of cell proliferation in different systems. Furthermore, in nodules of supernodulating mutants, defective in AON, WOX5 expression was higher than that in wild-type nodules. Hence, a conserved WUS/WOX-CLV regulatory system might control cell proliferation and differentiation not only in the root and shoot apical meristems but also in nodule meristems. In addition, the link between nodule-derived CLE peptides activating AON in different legumes and components of the AON system was investigated. We demonstrate that the identified AON component, NODULATION3 of pea, might act downstream from or beside the CLE peptides during AON.
Legume plants can thrive on nitrogen-poor soils because they interact symbiotically with soil-resident bacteria, the rhizobia. After a complex signal exchange between both partners, new root organs are formed: the nodules, in which the bacteria fix atmospheric nitrogen for the plant in return for a protective niche and carbon sources. Nodulation requires well-controlled bacterial invasion and initiation of cortical cell division after perception of the bacterially produced Nodulation (Nod) factors. Studies in several legumes have elucidated many elements of the signaling cascade (Catoira et al., 2001; Ben Amor et al., 2003; Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003; Lévy et al., 2004; Mitra et al., 2004; Kaló et al., 2005; Smit et al., 2005, 2007; Heckmann et al., 2006; Kanamori et al., 2006; Middleton et al., 2007). Mature nodules can be of the indeterminate or determinate type depending on whether an apical meristem is sustained through development or not. Which type grows relies on the host: typical models for indeterminate nodule development are Medicago truncatula and pea (Pisum sativum).
Nodule number and nitrogen fixation are strictly regulated by the legume plants to balance the nitrogen demand with nutrient supply and environmental conditions. As a result, the plant regulates the number of nodules and the size of the nodulation zone locally (Gonzalez-Rizzo et al., 2006; Tirichine et al., 2007), but also by systemic mechanisms (Bauer, 1981; Pierce and Bauer, 1983; Kosslak and Bohlool, 1984; Caetano-Anollés and Gresshoff, 1991). The best studied systemic control mechanism is called autoregulation of nodulation (AON). During this AON process, the first nodules that are formed inhibit further nodule development on the entire root system via currently unknown long-distance signals (Kosslak and Bohlool, 1984). AON is activated early after the perception of the Nod factors, at the onset of cell division for primordium formation, and its strength increases as nodule development proceeds (Pierce and Bauer, 1983; Caetano-Anollés and Gresshoff, 1991; Li et al., 2009).
AON-defective mutants in Lotus japonicus, Medicago truncatula, soybean (Glycine max), and pea (Carroll et al., 1985; Sagan and Duc, 1996; Szczyglowski et al., 1998; Krusell et al., 2002; Nishimura et al., 2002; Penmetsa et al., 2003; Searle et al., 2003; Schnabel et al., 2005, 2010) developed many more nodules than the corresponding wild-type plants. Grafting studies and analysis of mutants have shown that AON involves a root-shoot signal communication (Delves et al., 1986; Olsson et al., 1989; Sheng and Harper, 1997; Wopereis et al., 2000; Penmetsa et al., 2003). Later, a shoot-acting receptor complex, similar to the receptors that control the balance between cell division and differentiation in the shoot apical meristem (SAM), were found to play a central role (Krusell et al., 2002; Nishimura et al., 2002; Searle et al., 2003; Schnabel et al., 2005). Indeed, individual mutants were affected in a gene encoding a CLAVATA1-like leucine-rich repeat receptor-like kinase (CLV1-like LRR-RLK; har1/sunn/sym29/NARK; Krusell et al., 2002; Nishimura et al., 2002; Searle et al., 2003; Schnabel et al., 2005), whereas other mutants were impaired in CLV2-like LRR-RLK (clv2/sym28; Krusell et al., 2011). In addition, the supernodulating mutant klavier isolated from L. japonicus was altered in a gene encoding an LRR-RLK structurally unrelated to CLV1-like RLKs but closely homologous to the Arabidopsis (Arabidopsis thaliana) TOADSTOOL2/RECEPTOR-LIKE PROTEIN KINASE2 (TOAD2/RPK2; Oka-Kira et al., 2005; Miyazawa et al., 2010). All the Arabidopsis homologs transmitted the signal of the stem cell-specific peptide hormone CLV3 in the Arabidopsis SAM to control stem cell homeostasis by forming distinct homodimers or heterodimers (Oka-Kira and Kawaguchi, 2006; Bleckmann et al., 2010; Guo et al., 2010; Kinoshita et al., 2010; Miyazawa et al., 2010).
The elucidation of this shoot-acting receptor complex has led to the hypothesis that AON is initiated by a nodulation-dependent root-derived signal Q that is perceived in the shoot by the receptor, after which a shoot-determined inhibitor migrates from shoot to root to suppress further nodule formation (Gresshoff, 2003; Ferguson et al., 2010). Although both signals are currently unknown, the nature of the receptors (CLV1, CLV2, and TOAD2/RPK2-like components) might hint at a CLV3-like peptide as the Q signal. CLV3 belongs to the large group of CLAVATA3/EMBRYO-SURROUNDING REGION (CLE) peptides, which are small secreted peptides consisting of 12 to 13 conserved amino acids that are cleaved from the C-terminal end of CLE preproproteins (for review, see Wang and Fiers, 2010). Interestingly, in L. japonicus, M. truncatula, and soybean, when ectopically expressed, CLE genes that encode structurally similar peptides block or reduce nodulation systemically and depending on HYPERNODULATION ABERRANT ROOT1 (HAR1)/SUPERNUMEROUS NODULES (SUNN)/NODULE AUTOREGULATION RECEPTOR KINASE (NARK; Okamoto et al., 2009; Mortier et al., 2010; Reid et al., 2011; Saur et al., 2011). Albeit without ultimate proof, these nodulation-related CLE peptides have been hypothesized to act as the Q signal to activate the CLV-like receptor complex in the shoot for AON. In addition to the CLE peptides, several mutants have been identified that might be affected in genes involved in the control of AON in the root, such as nod3, rdn1, rdh1, tml, and plenty (Postma et al., 1988; Ishikawa et al., 2008; Magori et al., 2009; Novák, 2010; Yoshida et al., 2010; Novák et al., 2011; Schnabel et al., 2011). These mutants might be defective in the genes that control the root-derived signals or the root-to-shoot processing/transducing signals or vice versa (Li et al., 2009; Novák, 2010). Recently, rdn1 and nod3 have been found to encode unknown proteins that act in the vascular system, suggesting that these proteins might be involved in the vascular transport of a mobile signal acting between roots and shoots (Schnabel et al., 2011).
The downstream processes activated via AON are still unknown, but typical cell proliferation regulators might be the targets of the shoot-derived signals to further restrict the nodule number. The phytohormones auxin and cytokinin are central in the control of cell division and differentiation, and both, but especially the cytokinins, are essential for nodule formation (Schnabel and Frugoli, 2004; Gonzalez-Rizzo et al., 2006; Tirichine et al., 2007; Crespi and Frugier, 2008; Frugier et al., 2008; Plet et al., 2011). Based on expression analysis, the nodulation-related MtCLE12 peptide has been proposed to control nodulation by negatively influencing cytokinin signaling (Saur et al., 2011). Additionally, the auxin marker GH3:GUS was up-regulated in roots ectopically expressing the same nodulation-related CLE peptide (Saur et al., 2011).
The similarities with the CLV3 signaling pathway might also hint at putative targets of the AON pathway. In the SAM, a cellular, nonautonomous feedback loop between CLV3 signaling and the homeodomain transcription factor WUSCHEL (WUS) regulates stem cell homeostasis (Schoof et al., 2000). WUS acts in the organizing center of the SAM and is essential for the specification and maintenance of stem cell proliferation in the central zone of the meristem (Mayer et al., 1998; Schoof et al., 2000). The CLV3 signaling pathway, including the CLV1/CLV2 receptor kinases and the CLV3 regulatory peptide, negatively controls WUS expression, thereby restricting the size of the stem cell population (Brand et al., 2000; Schoof et al., 2000). In the root apical meristem (RAM), a similar signaling system involving the WUSCHEL-RELATED HOMEOBOX5 (WOX5) functions in the quiescent center (QC) to regulate the balance between cell division and differentiation (Kamiya et al., 2003; Haecker et al., 2004). Complementation experiments proved that WUS and WOX5 are functionally equivalent (Sarkar et al., 2007) and, therefore, could be involved in common regulatory pathways that control meristem maintenance and development in roots and shoots. In addition, a CLE peptide (CLE40) might control the WOX5 expression domain in the RAM through the interaction with the RLK ARABIDOPSIS CRINKLY4 (Stahl and Simon, 2009).
In view of the similarities between the AON-controlling RLKs in legumes and the SAM and RAM stem cell homeostasis, we hypothesized that a WOX transcription factor might also be a target during AON. Previously, by means of gene expression studies, a few members of the WOX family in M. truncatula had been identified, including WOX5, which is expressed in root tissues upon somatic embryogenesis (Chen et al., 2009). We analyzed whether its expression is modified upon nodulation and is regulated by AON and CLE peptide signaling. Furthermore, we addressed the question of whether the M. truncatula CLE peptide, MtCLE13, which inhibits nodulation in a SUNN-dependent manner in gain-of-function analyses (Mortier et al., 2010), could also influence the nodulation of other known AON mutants. To this end, we tested whether the MtCLE13 gain-of-function effect on nodulation also occurred in pea and studied its effect on the pea mutants sym28, sym29, and nod3. We present a comparative analysis of two legume plants that might be useful to understand the role of WOX5 upon nodule development in indeterminate nodule-forming legumes.
RESULTS
Quantitative PCR Analysis of WOX5 Expression upon Nodulation in M. truncatula and Pea
To identify a WOX gene with expression modulated upon nodulation, the Mt2.1 genomic sequence was searched with BLASTX followed by an analysis of the expression patterns of each hit via the M. truncatula Gene Expression Atlas (Benedito et al., 2008). WOX5 (CU326389) was the only WOX transcription factor induced upon nodulation that already had been studied previously during somatic embryogenesis (Chen et al., 2009). The WOX5 gene and mRNA were identified in pea (GenBank accession nos. JN603579 and JN603580) with degenerate primers designed based on the M. truncatula WOX5 nucleotide sequences and subsequent RACE analyses (see “Materials and Methods”). The coding sequence of the WOX sequence of pea was 92.7% similar within and 85.5% similar outside the homeobox region of the M. truncatula WOX5 (CU326389) and, therefore, was designated WOX5 (Supplemental Fig. S1).
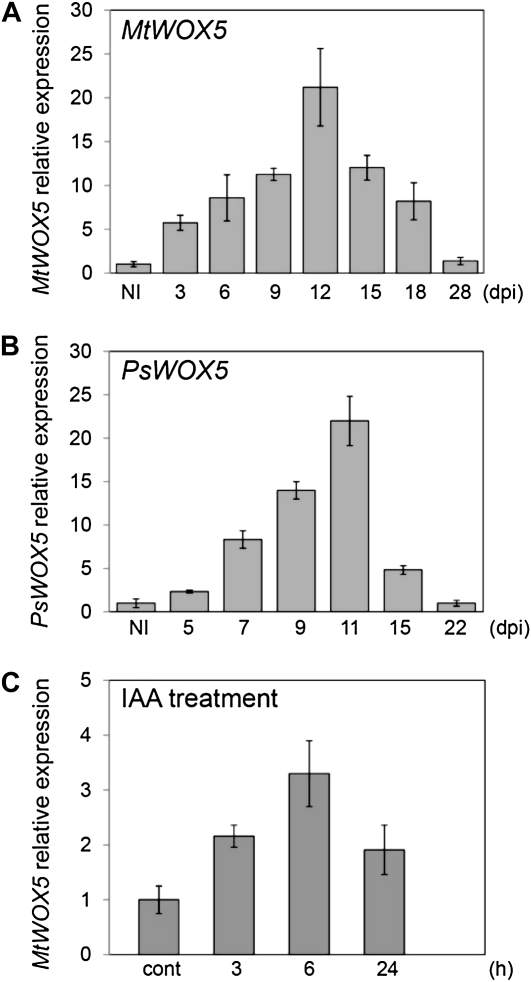
To study the temporal expression during nodule development, the relative transcript level of WOX5 was analyzed at different stages after inoculation in both M. truncatula and pea. For M. truncatula (A17) and pea (cv Frisson), expression was tested from 3 to 28 d post inoculation (dpi; Fig. 1A) and from 5 to 22 dpi (Fig. 1B), respectively, and compared with the expression levels in uninoculated roots. The expression levels of WOX5 transiently increased during nodule development to reach a maximum at approximately 11 to 12 dpi, whereafter they decreased again both in M. truncatula and pea (Fig. 1, A and B).
Figure 1.
Temporal expression of WOX5 during M. truncatula and pea nodulation and upon auxin treatment. A, qRT-PCR analysis of WOX5 in noninoculated M. truncatula roots (NI) and at 3, 6, 9, 12, 15, 18, and 28 dpi. Expression is shown relative to the expression found in NI. B, qRT-PCR analysis of WOX5 in noninoculated pea roots (NI) and at 5, 7, 9, 11, 15, and 22 dpi. Expression is shown relative to the expression found in NI. C, Effect of treatment with 10−6 m indole-3-acetic acid (IAA) on WOX5 gene expression level at different days after treatment. Expression is shown relative to the expression found in untreated roots (cont). The error bars represent sd of three technical repeats. The graphs show the results of one biological repeat, representative for two additional independent experiments.
A correct auxin/cytokinin balance is a prerequisite for nodule formation (Oldroyd and Downie, 2008; Ding and Oldroyd, 2009). To determine whether auxins and/or cytokinins affected WOX5 expression, 10−6 m indole-3-acetic acid or 10−7 m 6-benzylaminopurine was supplemented to the growth medium of 5-d-old M. truncatula seedlings. The hormone-treated roots were harvested under each condition after 3, 6, and 24 h. Roots from plates without hormone addition were used as a negative control. WOX5 expression was temporarily induced after auxin treatment (Fig. 1C), whereas 6-benzylaminopurine treatment had no clear effect (data not shown).
Tissue-Specific Expression Pattern of WOX5 upon Nodulation in Pea and M. truncatula by Promoter-GUS Analysis
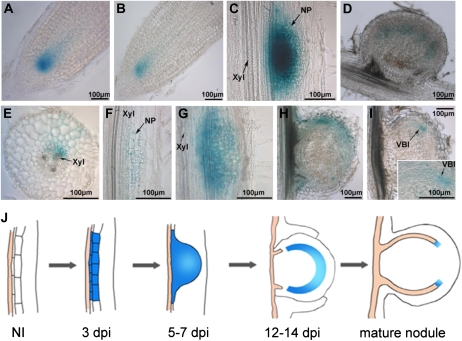
To localize WOX5 expression during nodule development, a 2,310-bp M. truncatula WOX5 promoter region was isolated and used to construct pMtWOX5:GUS (see “Materials and Methods”). This construct was introduced into both M. truncatula and pea roots. In noninoculated transgenic roots of both legumes, GUS staining was observed in the meristematic zones of the root tips (RAM), corresponding to the QC area (Fig. 2, A and B). WOX5 was also expressed in primordia of lateral roots (Supplemental Fig. S2).
Figure 2.
Analysis of the tissue-specific expression pattern of WOX5 by promoter-reporter analysis in M. truncatula and pea. A, WOX5 expression in root tips of pea. B, WOX5 expression in root tips of M. truncatula. C and D, WOX5 expression at different stages of nodule development in pea at 5 dpi (C) and 21 dpi (D). E to I, WOX5 expression at different stages of nodule development in M. truncatula at 3 dpi (E and F), 5 dpi (G), 14 dpi (H), and 21 dpi (I). The inset in I shows an enlargement of the provascular strand. NP, Nodule primordium; VBI, vascular bundle initials; Xyl, xylem. Bars = 100 μm. J, Schematic representation of the WOX5 expression domain at different stages of nodule development. On average, for each stage, approximately 30 to 50 transgenic roots were analyzed with similar results. NI, Noninoculated M. truncatula root.
Next, expression was analyzed at various stages of nodule development both in M. truncatula and pea. At 3 to 4 dpi, GUS staining in M. truncatula was visible in some dividing cells of the pericycle, endodermis, and cortex located opposite the xylem poles, corresponding to the sites of nodule primordium initiation (Fig. 2, E and F). Also, a low expression level was seen in the outer cortical cells. At slightly more advanced stages (5–7 dpi), GUS staining was observed in the developing nodule primordium (Fig. 2G), and as nodule development progressed, WOX5 expression gradually decreased and remained in the periphery of the developing nodules (12–14 dpi; Fig. 2H). Finally, in mature nodules (21–28 dpi), WOX5 expression was limited to some cells at the tip of the vascular bundles (Fig. 2I).
The expression pattern was similar in transgenic roots of pea carrying the pMtWOX5:GUS construct. Before inoculation, GUS staining was visible in a local area in the RAM (Fig. 2B). WOX5 expression was activated very early upon inoculation in the proliferation zone and afterward within developing primordia (Fig. 2C). The area of WOX5 expression in mature nodules of pea was restricted to the tips of the vascular bundles, although it was slightly more expanded than that in M. truncatula (Fig. 2D). Thus, the tissue-specific expression pattern of WOX5 upon nodulation was relatively similar in the two indeterminate nodule-forming legumes. The results of the WOX5 expression analysis with the pMtWOX5:GUS fusion were consistent with the quantitative reverse transcription (qRT)-PCR data. The enhanced level of WOX5 expression at early nodulation stages (7–12 dpi) corresponded to the stage when most of the nodule primordia developed. WOX5 expression reached a maximum preceding the nodule meristem formation, whereas its decrease at later stages matched the restricted WOX5 expression zone in developing nodules.
qRT-PCR Analysis of WOX5 Expression in Mutants Affected in the CLV-Like Receptor Complex Involved in AON
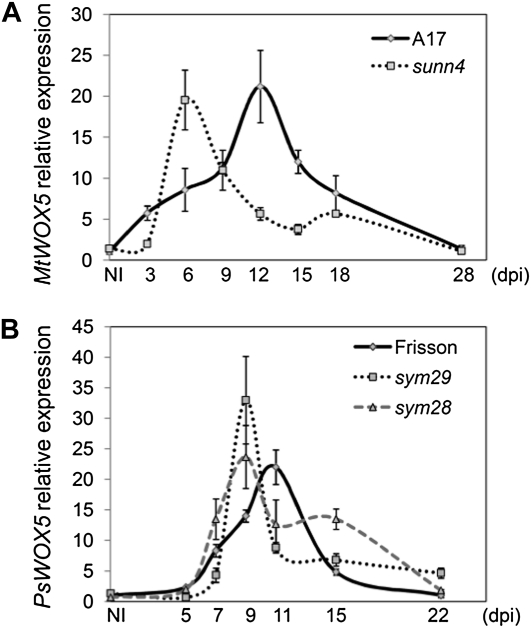
To study the possible interaction of WOX5 with the CLV system during AON, we analyzed WOX5 expression in various supernodulating mutants. If any interaction between WOX5 and the CLV system existed, the defects in the components of the CLV system should be connected with changeable levels of WOX5 expression. In M. truncatula, WOX5 expression was tested upon inoculation of the sunn-4 mutant (Fig. 3A), and in pea, it was tested upon inoculation of sym29 (P88) and sym28 (P64; Fig. 3B). For each mutant, the temporal expression pattern was compared with the respective wild-type controls (A17 for sunn-4 and Frisson for sym28 and sym29).
Figure 3.
qRT-PCR analysis of WOX5 gene expression upon nodule development in supernodulating mutants of M. truncatula and pea. A, Relative WOX5 expression in noninoculated M. truncatula roots (NI) and at different dpi. Expression is shown relative to that in NI. B, Relative WOX5 expression in noninoculated pea roots (NI) and at different dpi. Expression is shown relative to the expression in NI. The graphs show the results of one biological repeat, representative for one and two additional independent experiments in M. truncatula and pea, respectively. The error bars represent sd of three technical repeats.
The WOX5 temporal expression pattern upon nodule development differed between wild-type plants and the supernodulating mutants (Fig. 3). In both M. truncatula and pea, the highest WOX5 expression occurred earlier in the supernodulating mutant than in the wild-type plants. Upon inoculation of the sunn-4 mutant, the maximal WOX5 expression level was reached at 6 dpi, but only at 12 dpi in wild-type plants. Similarly, in pea sym28 and sym29, the WOX5 expression was maximal at 9 dpi, but only at 11 dpi in wild-type pea. At later nodulation stages in pea, WOX5 expression was slightly higher in the supernodulating mutants than in the wild type. These results hint at an interaction between the AON shoot receptor complex and WOX5 expression.
Tissue-Specific Expression Pattern of WOX5 in Supernodulating Mutants of M. truncatula and Pea
To control whether WOX5 expression was altered locally within developing nodules of supernodulating mutants, we studied WOX5 expression in the M. truncatula sunn-4 and sunn-3 mutants as well as in pea mutants sym28 and sym29. For this purpose, the pMtWOX5:GUS construct was introduced into the mutant roots, and the expression patterns were compared with those of wild-type plants. For the comparison of wild-type and mutant alleles, GUS staining was done in an identical manner for all samples (see “Materials and Methods”).
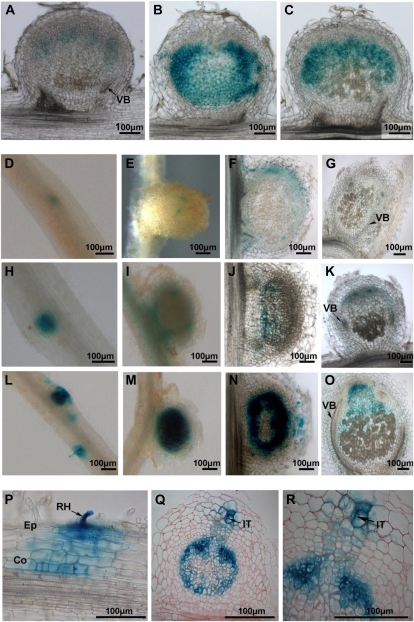
The GUS activity was significantly higher in nodules of the supernodulating pea mutants sym29 and sym28 than in those of the wild-type pea cv Frisson (Fig. 4, A–C). The expression in wild-type nodules was hardly visible in pea cv Frisson in the apical region, whereas in the AON mutants, GUS staining was seen all over the meristem and infection zone (Fig. 4, A–C). Consequently, WOX5 expression was altered locally, at the level of a particular nodule, in supernodulating mutants of pea.
Figure 4.
Analysis of the tissue-specific expression pattern of WOX5 using promoter-reporter fusion in supernodulating mutants of M. truncatula and pea. A to C, WOX5 expression in the wild type (A) and in sym28 (B) and sym29 (C) mutants of pea. GUS staining through a 21-dpi nodule is shown. D to R, WOX5 expression at different stages of nodule development in the wild type (D–G) and in sunn-4 (H–K) and sunn-3 (L–R) mutants of M. truncatula A17. D, H, and L, WOX5:GUS staining during an incipient nodulation event. E, I, and M, WOX5:GUS staining in a young developing nodule at 14 dpi. F, J, and N, Young WOX5:GUS-stained nodule (14 dpi). G, K, and O, Mature elongated nodule (21 dpi). Stereoscopic images (D, E, H, I, L, and M) and light micrographs of agarose sections (50 μm; A–C, F, G, J, K, N, and O) are shown. P to R, Technovit sections (5 μm) stained with ruthenium red through WOX5:GUS developing nodules at 3 dpi (P) and 12 dpi (Q and R). Co, Cortex; Ep, epidermis; It, infection thread; RH, root hair; VB, vascular bundle. Bars = 100 μm.
Next, WOX5 expression was analyzed in two M. truncatula sunn mutants (sunn-3 and sunn-4 alleles), both carrying a nonsense mutation (Schnabel et al., 2005). At 5 to 7 dpi, GUS staining was more intense in sunn-3 and sunn-4 than in wild-type plants, although the expression pattern did not change (Fig. 4, D, H, and L). At later stages in nodule development, at 14 dpi (Fig. 4, E, I, and M [stereoscopic images] and Fig. 4, F, J, and N [agarose sections]) and at 21 dpi (Fig. 4, G, K, and O), the expression was still higher in sunn-3 (Fig. 4, N and O) and sunn-4 (Fig. 4, J and K) mutants than in wild-type nodules (Fig. 4, F and G). Interestingly, GUS staining was always more intense in sunn-3 than in sunn-4 nodules. Sectioning revealed that GUS staining was restricted to the vasculature tips in wild-type nodules, whereas this expression pattern extended over the meristem and infection zone and, to a lesser extent, in the fixation zone in sunn-3 nodules (Fig. 4, N, and O).
In developing nodules of sunn-3, an additional expression pattern was seen that was clearly associated with the infection process when compared with the wild-type and sunn-4 nodules (Fig. 4, P–R). GUS staining was observed in the epidermal and cortical cells that contained infection threads and was strong in the apical region, outside the meristem, and in mature nodules, corresponding to the initially infected, old root cortex (Fig. 4, P–R) during nodule primordium development. As a consequence, two WOX5 expression domains were clearly visible in the sunn-3 mutants: one associated with proliferating cells of developing nodules and one with infection threads penetrating the outer cortical cells. At later stages of nodule development (21 dpi), WOX5 expression was still higher in sunn-3 than in wild-type plants. In other words, the sunn, sym28 and sym29 mutations affect WOX5 expression.
Effect of the Ectopic Expression of 35S:MtWOX5 on Nodule and Root Development
To investigate the role of WOX5 in nodule organogenesis, transgenic roots overexpressing 35S:MtWOX5 were constructed and analyzed by microscopy. qPCR analysis confirmed the ectopic expression of M. truncatula WOX5 in individual transgenic roots (Supplemental Fig. S3). Analysis of root length and nodule number revealed no significant changes in these parameters between 35S:MtWOX5 and 35S:GUS control roots (Supplemental Fig. S3). However, the 35S:MtWOX5 roots often displayed slightly thickened root tips and roots with arrested growth (Supplemental Fig. S3). These roots contained more small dividing cells than those of the wild type, corresponding to the prolonged stimulation of cell proliferation. No visible changes in the organization of the 35S:MtWOX5 nodules were found (Supplemental Fig. S4).
Influence of Nodulation-Specific CLE Peptides on the Supernodulating Phenotype of Pea Mutants (sym28, sym29, and nod3)
Previously, ectopic expression of structurally related CLE peptide genes that were induced upon nodulation had been shown to inhibit or significantly reduce nodulation in L. japonicus, soybean, and M. truncatula. In addition, this gain-of-function effect depended on HAR1, NARK, and SUNN, respectively (Okamoto et al., 2009; Mortier et al., 2010; Saur et al., 2011). To analyze this effect, we controlled whether an M. truncatula peptide, MtCLE13, could modulate nodulation in pea, and if so, we wanted to test the effect of 35S:MtCLE13 on the different AON mutants in pea, including the root-determined nod3.
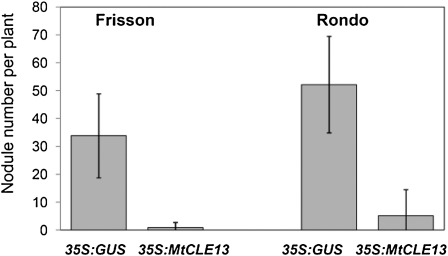
Four-day-old pea seedlings were transformed with the Agrobacterium rhizogenes strain Arqua, containing either a 35S:MtCLE13 or a 35S:GUS construct as a control. Plants were analyzed at 21 dpi, and only nodules on transgenic roots were counted (Fig. 5). In wild-type pea (Frisson and Rondo lines), ectopic expression of MtCLE13 resulted in a severe reduction in nodulation. Whereas, on average, 35 and 50 nodules were obtained on Frisson and Rondo roots carrying the control construct, roots carrying the 35S:MtCLE13 construct only carried one to two and five nodules, respectively. Thus, 35S:MtCLE13 had a similar effect on M. truncatula and pea nodulation, indicating that MtCLE13 is functional in pea (Fig. 5).
Figure 5.
Influence of 35S:MtCLE13 overexpression on the nodule number in wild-type pea plants Frisson and Rondo lines). 35S:MtCLE13, MtCLE13-overexpressing roots; 35S:GUS, GUS-overexpressing roots (control). Error bars indicate sd on three technical repeats. The difference between 35S:GUS control and 35S:MtCLE13 plants in nodule number is statistically significant (Mann-Whitney U test, P < 0.001).
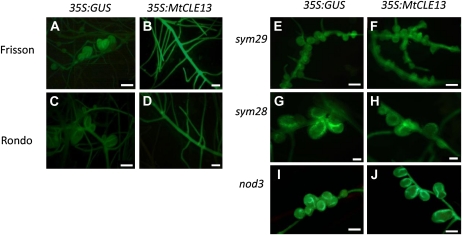
We also analyzed the effect of MtCLE13 overexpression on the pea AON mutants sym29, sym28, and nod3 (Fig. 6). In contrast to the wild type, in which a clear reduction in nodulation was seen by stereomicroscopy (Fig. 6, A–D), roots of mutants carrying a 35S:MtCLE13 construct developed as many nodules as control roots (Fig. 6, E–J). Hence, the effect of 35S:MtCLE13 expression on nodulation is abolished in these mutants.
Figure 6.
Effects of 35S:MtCLE13 on nodulation in the supernodulating pea mutants sym29, sym28, and nod3. A to D, Examples of the nodulation phenotype of GFP-positive transgenic roots carrying the 35S:GUS (control) or 35S:MtCLE13 construct on Frisson (A and B) and Rondo (C and D) wild-type plants. E to J, Examples of the nodulation phenotype of GFP-positive transgenic roots carrying the 35S:GUS (control) or 35S:MtCLE13 construct on the sym29 (E and F), sym28 (G and H), and nod3 (I and J) mutants. Approximately 18 to 25 plants were analyzed with similar results. Bars = 1,000 μm.
DISCUSSION
We investigated the role of the WOX5 transcription factor upon nodule development in two legume plants, M. truncatula (a model legume) and pea. Analysis of the WOX5 expression profile upon nodule development showed that WOX5 expression increases transiently early after rhizobium inoculation and is initially expressed in the pericycle, endodermis, and inner cortex of the infected root. At a later stage of nodule development, WOX5 is expressed in cells of the developing nodule primordia and, during maturation of the primordia, gradually decreases and is restricted to the apical region of fully grown nodules, especially in cells of the provascular bundles.
The WOX5 expression pattern observed at the onset of nodulation coincides with the site of nodule primordium formation (Mathesius et al., 1998a, 1998b; Timmers et al., 1999; Mathesius, 2008). In plant species forming indeterminate nodules, the first divisions are initiated in the pericycle, endodermis, and inner cortical cells, the sites where WOX5 expression occurs. Accumulating data show that changes in cytokinin and auxin signaling underlie nodule development (Hirsch and Fang, 1994; Mathesius et al., 1998a, 1998b; Boot et al., 1999; Pacios-Bras et al., 2003; Huo et al., 2006; Wasson et al., 2006, 2009; Frugier et al., 2008; Mathesius, 2008; Grunewald et al., 2009; Plet et al., 2011). Upon inoculation, at the vasculature and inner cortical cells, cytokinin signaling has been proposed to perturb the expression and accumulation of PIN-FORMED auxin efflux carriers, temporarily inhibiting polar auxin transport to allow local auxin accumulation in the inner cortex for nodule primordium development (Plet et al., 2011). In agreement, a gene encoding an auxin influx carrier, AUXIN RESISTANT1-LIKE, is expressed in cells of the nodule primordia (de Billy et al., 2001; Schnabel and Frugoli, 2004). Moreover, in legumes forming indeterminate nodules, the auxin-responsive reporters GH3:GUS and DR5:GUS were expressed in developing primordia and later in nodule vasculature (Mathesius et al., 1998a; Huo et al., 2006). Because WOX5 is activated by auxin (Chen et al., 2009; this study), the transcription factor might function downstream of cytokinin signaling and polar auxin transport inhibition to form nodule primordia. Expression in the developing nodule primordia is consistent with the function of other members of the WOX family that stimulate cells to proliferate while suppressing their differentiation (van der Graaff et al., 2009).
Interestingly, the WOX5 expression pattern at the onset of the nodule development is similar to that of cytokinin-signaling and cytokinin-responsive genes (Plet et al., 2011). This colocalization raises the question of whether cytokinin signaling and WOX5 expression might be linked similarly as in SAM. In Arabidopsis, WUS is known to be involved in the local regulation of the cytokinin response by repressing cytokinin A-type response regulators, which negatively control cytokinin action (Skylar and Wu, 2011), and, as a result, locally stimulates cytokinin action and cell proliferation. If a similar interaction existed for nodulation and because cytokinin is known to trigger nodule development, a higher nodule number would have been expected in the WOX5-overexpressing roots. However, no increase in nodule number was observed on 35S:MtWOX5 roots, indicating that WOX5 might not be involved in the amplification of cytokinin signaling. As the interaction might be more subtle, it would still be interesting to compare the expression of WOX5 and the different cytokinin markers available for nodulation. In addition, analysis of WOX5 in a cre1 mutant background might give important information, as would analysis of the expression of cytokinin response regulators in 35S:WOX5 roots and nodules. In mature nodules of M. truncatula and pea, the WOX5 gene is preferentially expressed near the tips of vascular bundles, which consist of small dividing cells that give rise to the vascular bundles. Also, in the tips of M. truncatula roots, WOX5 was expressed at the site of vascular bundle initiation in cells that might correspond to the QC of the root meristem (Chen et al., 2009). Based on this expression pattern, the WOX5-expressing nodule cells might represent the organizing centers of the nodule meristems, in analogy to the RAM and SAM. In agreement, the transcription factor NOOT that suppresses root meristem identity in nodule meristems is also expressed at these sites (P. Ratet, personal communication). The origin of nodules has been debated in the literature, because they are unique structures with both shoot and root properties (Hirsch and LaRue, 1997; Markmann and Parniske, 2009). The central nodular tissue is surrounded by several vascular bundles that originate from independent provascular strands at the apical meristem. In an individual nodule of M. truncatula and pea, mostly two vascular bundles are found. Based on the similarity between the WOX5 expression pattern in root tips and mature nodules, it is tempting to speculate that nodule development might involve the fusion of several (lateral) root-like structures. However, because nodules are unique structures, nodule meristem development will surely be more complex than a simple fusion between root meristems. Analysis of more QC markers together with mutant analysis and cell-specific transcript profiling will undoubtedly shed light on this interesting and undiscovered aspect of nodule ontogenesis.
The ectopic expression of WOX5 did not result in changes in nodule number or in root length. Microscopic analyses revealed no modification in nodule morphology and only a minor effect on root morphology. In Arabidopsis, expression of 35S:WOX5 did not produce any morphological changes (Gonzali et al., 2005), whereas the direct activation of WOX5 by a dexamethasone-inducible method resulted in altered differentiation of columella cells (Sarkar et al., 2007). The observed differences between the two methods used in Arabidopsis might be explained by a tight control on WOX5 expression by posttranscriptional and posttranslational mechanisms. Hence, the lack or small effect of 35S:MtWOX5 expression on nodulation and root growth might also be due to posttranscriptional and posttranslational regulations that control WOX5 protein levels. Hence, the analysis of WOX5 levels would provide more information.
In legume plants, nodule number and SAM homeostasis are controlled by a similar shoot-localized CLV signaling complex consisting of different receptors that perceive CLE peptides (Krusell et al., 2002, 2011; Nishimura et al., 2002; Searle et al., 2003). In Arabidopsis, CLV3 suppresses WUS expression to provide the fine balance between cell proliferation and differentiation at the SAM. We addressed the question of whether the CLV signaling complex that controls AON in the shoot also exerts its effect on the expression of WOX5 in the nodules by long-distance regulation. Indeed, qRT-PCR as well as pMtWOX5:GUS analyses demonstrated that WOX5 expression in the nodules changed in sunn and in sym29 and sym28 mutants. Interestingly, in these mutants, the level and area of WOX5 expression had significantly increased in individual nodules. Instead of a restricted expression in the provascular strands as seen in the wild-type nodules, WOX5 expression occurred in the meristem, the infection zone, and the young fixation zone. Hence, signaling via CLV1-like and CLV2-like proteins repressed WOX5 expression in developing nodules. sunn mutants did not show the inhibition of shoot-to-root auxin transport that is usually seen at the onset of nodulation (van Noorden et al., 2006). As WOX5 is auxin inducible, changes in auxin levels might mediate the influence of the CLV signaling complex on WOX5 expression. Thus, a WUS/WOX-CLV regulatory pathway seems to act in the nodule meristem similarly to that of RAM and SAM in plants. A characteristic property of RAM and SAM homeostasis is that both the WUS/WOX proteins and the CLV signaling complex act at short distances in neighboring cells to influence each other’s expression. Although the legume CLV1-like RLK has been shown to be produced in the vasculature of roots and shoots, grafting has indicated that its activity in the shoot controls AON (Krusell et al., 2002; Nishimura et al., 2002; Searle et al., 2003; Schnabel et al., 2005). Whether the shoot or root activity regulates WOX5 expression during nodulation is currently unknown, but tests via grafting experiments would be interesting.
Microscopic analysis of sunn, sym29, and sym28 mutants did not reveal differences in nodule organization, indicating that high WOX5 expression does not disorganize the central nodule tissue. Accordingly, no essential structural changes in the 35S:WOX5 nodules were found. Interestingly, in sunn-3, but not in the other AON mutants, the WOX5 promoter was active also in the outer cortical cells through which infection threads grow toward the nodule primordium. This expression pattern is still visible in remnants of this region at the apex of mature nodules. Outer cortical cells preparing for infection thread passage also dedifferentiate, enter the cell cycle, but, in contrast to the inner cortical cells, get arrested in G2 to make the preinfection threads through which the infection threads will pass (Yang et al., 1994). In agreement with the suppressive effect of the CLV signaling complex on WOX5 expression, a low level of WOX5 might exist in these wild-type cells, and it might be needed transiently to organize this dedifferentiation step. Why this expression pattern was not observed in the other AON mutants is currently unknown. As both sunn-3 and sunn-4 carry nonsense mutations, they might represent strong alleles (Schnabel et al., 2005, 2010; Mortier et al., 2012a), but additional functional analysis, including of sunn-1 and sunn-2 mutants, is undoubtedly needed to assess this issue.
Nodule-derived CLE peptides have been shown to activate AON in different legumes. As a result, ectopic expression of these CLE peptides systemically inhibited nodulation that depended on a functional CLV signaling complex (Okamoto et al., 2009; Mortier et al., 2010; Reid et al., 2011). However, an unsettled question is whether these CLE peptides are able to migrate to the shoot and interact with the CLV receptor complex during AON (Mortier et al., 2012b). To answer this question, the effect of ectopic expression of the MtCLE13 on the wild type and AON mutants of pea was assessed. The M. truncatula CLE13 peptide was active in pea roots, because the ectopic expression of 35S:MtCLE13 strongly reduced pea nodulation, similar to that in M. truncatula (Mortier et al., 2010), indicating that the perception of the nodule-derived AON CLE peptide is conserved between legume species. This observation is also supported by the conserved amino acid sequence of the AON-related CLE peptides of various legumes (Mortier et al., 2011). The effect was absent in the sym28 and sym29 mutants, indicating that PsSYM29 (CLV1 kinase) and PsSYM28 (CLV2 receptor) act downstream of the CLE peptide, consistent with previously published data for L. japonicus, M. truncatula, and soybean (Okamoto et al., 2009; Mortier et al., 2010). Hence, these data confirm the existence of a conserved mechanism of AON in different species of legumeе plants.
Similar to sym28 and sym29 mutants, the ectopic expression of the CLE peptide did not affect nodulation in the pea nod3 mutant that, defective in a gene orthologous to the M. truncatula ROOT DETERMINED NODULATION1 (RDN1), is disturbed in a root-determined component of AON (Schnabel et al., 2011). Experiments have shown that it might control the production/transportation of a root-derived signal that is sent to the shoot at the onset of AON rather than be involved in the perception of the return signal from the shoot (Li et al., 2009; Novák, 2010; Schnabel et al., 2011). In view of this hypothesis, our results on the nod3 mutant demonstrate that AON CLE peptides might act upstream or at the level of the RDN1/NOD3 protein. Although we cannot rule out that RDN1/NOD3, synthesized in the vasculature, might aid the CLE peptide transport toward the shoot, our data might also hint at an indirect interaction between nodule-related CLE peptides and the CLV receptor complex in the shoot. As a result, different CLV signaling systems in the root and shoot might act on a common component, such as auxin homeostasis, to control the nodule number.
The relation between WOX5 and MtCLE13 expression is currently unresolved. At the onset of nodulation, the expression of WOX5 overlaps with that of MtCLE13. In mature nodules, MtCLE13 is expressed predominantly in the apical region of the nodules (Mortier et al., 2010), whereas WOX5 is more restricted to the provascular strands. Interestingly, infrequently, the MtCLE13 expression level is enhanced in these provascular strands (Mortier et al., 2010). Hence, short-distance communication between the CLE peptides and WOX5 is very plausible. The expression of WOX5 and MtCLE13 remained unchanged in 35S:CLE13 and 35S:MtWOX5 roots, respectively (data not shown). More sensitive investigations, involving stable marker lines and careful microscopy, will be needed to unravel these interactions.
In summary, we have shown that WOX5 expression in nodules is suppressed by the CLV signaling complex that controls AON. Thereby, we provide another component of the AON mechanism in addition to the CLV signaling mechanism in the shoot and the CLE peptides in the root. Hence, a conserved WUS/WOX-CLV regulatory system might regulate cell proliferation and differentiation not only in RAM and SAM (regular plant apical meristems) but also in nodule meristems. The identified AON component, NOD3, might act downstream or alongside the CLE peptides during AON. The long-distance model in which root-derived CLE peptides would migrate to the shoot to be perceived and activate AON would be more complex than anticipated previously. Thus, although the exact interaction between the different AON components remains unsolved, we have found two more pieces of the AON puzzle that can be used in the future to gain an integrated insight into how AON works.
MATERIALS AND METHODS
Plant Material, Bacterial Strains, and Growth Conditions
Medicago truncatula Jemalong plants (wild-type A17 and supernodulating mutants sunn-3 and sunn-4) and pea (Pisum sativum) cv Frisson (wild type and supernodulating mutants P64 [sym28] and P88 [sym29]) and cv Rondo (wild type and supernodulating mutant P79 [nod3]) were grown in growth chambers (16-h/8-h day/night regime, 21°C, and 75% relative humidity). M. truncatula plants were inoculated with Sinorhizobium meliloti strains Sm2011 pBHR-mRFP and Sm2011 pHC60-GFP and pea plants with Rhizhobium leguminosarum CIAM1026 from the All-Russia Research Institute for Agricultural Microbiology collection (http://www.arriam.spb.ru/eng/lab10/WDCM 966). Seeds were surface sterilized with concentrated sulfuric acid for 10 min and washed five to six times with sterile water. For temporal expression during nodulation, M. truncatula plants inoculated with Sm1021 pHC60-GFP were grown in aeroponic systems with nitrogen-poor SOLi medium (Blondon, 1964). Nodules were obtained at different stages after inoculation from the infected sites of roots by visualizing the green fluorescent bacteria with an MZFLII stereomicroscope (Leica Microsystems) equipped with a blue light source and a GFP Plus filter set (λex = 480/40, λem = 510-nm long pass barrier filter; Leica Microsystems). Pea plants were grown in vermiculate-containing pots, and infected root tissue was harvested at different stages after inoculation with R. leguminosarum CIAM1026. Only segments between emerged lateral roots were collected to avoid harvesting of lateral root primordia.
Auxin Treatment
Five-day-old in vitro-grown plants were treated with 10−6 m indole-3-acetic acid supplemented to the medium. As a control, plants were grown without supplemented hormones. The growth conditions of the seedlings were the same as above. After 0, 3, 6, and 24 h of incubation, the roots of five to seven plants under each condition were harvested and analyzed by qRT-PCR. The experiment was repeated four times with comparable results.
Molecular Cloning
The WOX5 gene fragment of pea was amplified with degenerate primers, cloned in the pAL-TA vector (Evrogen), and subsequently sequenced. The Mint RACE cDNA amplification kit (Evrogen) was used according to the manufacturer’s protocol. As a result, an 872-bp sequence of the WOX5 gene was identified. The WOX5 promoter (1,965 bp) of M. truncatula was first cloned into the pENTR-D/TOPO vector (Invitrogen) and then in pBGFWS7.0 with the LR Clonase enzyme (Invitrogen). The primers used were 5′-GGGGACAACTTTGTATAGAAAAGTTGCAATTTTTGGCGACCAGATT-3′ and 5′-GGGGACTGCTTTTTTGTACAAACTTGCATGCTCTCTTCCATATTTCAATTC-3′. The coding sequence of WOX5 was cloned into the pENTR-D/TOPO vector (Invitrogen) and then in pB7WG2D with the LR Clonase enzyme (Invitrogen).
qRT-PCR Analysis
Total RNA was isolated with the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. After a DNase treatment, the samples were purified through NH4Ac precipitation, quality controlled, and quantified with a Nanodrop spectrophotometer (Isogen). RNA (1 μg) was used for cDNA synthesis with the iScript cDNA synthesis kit (Bio-Rad). The qRT-PCR experiments were done on a Lightcycler 480 (Roche Diagnostics), and SYBR Green was used for detection. All reactions were done in triplicate and averaged. Cycle threshold values were obtained with the accompanying software, and data were analyzed with the 2−ΔΔCt method (Livak and Schmittgen, 2001). The relative expression was normalized against the constitutively expressed 40S ribosomal S8 protein (TC100533; Medicago Gene Index) for M. truncatula gene expression analysis and the ubiquitin gene in the experiments with pea. The following primers were used for qRT-PCR analysis: MtWOX5_forward, 5′-GCCTGATAGAGTGATTGAGAC-3′; MtWOX5_reverse, 5′-GGGTGTTCCATTTGTTCTCC-3′; PsWOX5_forward, 5′-GGTTTCAAAATCATAAGGCTAGGGA-3′; and PsWOX5_reverse, 5′-TCAACCGTAAGTCTAATGGTGGATG-3′. Each experiment was repeated at least three times with independent biological tissues.
Agrobacterium rhizogenes-Mediated Plant Transformation
A. rhizogenes-mediated M. truncatula plants were transformed as described (Mortier et al., 2010). For pea, the M. truncatula protocol was used and 3- to 4-d-old seedlings were infected by stabbing the hypocotyls with a fine needle containing an A. rhizogenes culture. After 7 to 10 d, calli appeared with emerged transgenic roots at the infection site. Plants were transferred into pots with vermiculite, watered with nitrogen-poor SOLi medium, grown for approximately 10 d at 21°C with a 16-h photoperiod and light at 70 μE m−2 s−1, and subsequently inoculated with rhizobia.
Histochemical Localization of GUS Activity
GUS activity in cotransformed roots and nodules was analyzed with 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid as substrate (Van den Eede et al., 1992). Roots and nodules were vacuum infiltrated during 20 min and subsequently incubated in GUS buffer at 37°C. After staining, root nodules were fixed, embedded with agarose (3%), and sectioned (50 μm) with a microtome with a vibrating blade (НМ 650V; Thermo Scientific). For thin sections (3–5 μm), plant tissues were embedded with the Technovit 7100 kit (Heraeus Kulzer) according to the manufacturer’s instructions and sectioned with a microtome (Leica Microsystems). For tissue-specific staining, thin sections were submerged in a 0.05% (w/v) ruthenium red solution (Sigma-Aldrich), washed in distilled water, and dried. Finally, sections were mounted with Depex (BHD Chemicals). Photographs were taken with a Diaplan microscope equipped with bright-field and dark-field optics (Leitz) or a laser confocal microscope (LSM 510 META NLO; Carl Zeiss).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers JN603579 and JN603580.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of WOX5 coding sequences of pea and M. truncatula.
Supplemental Figure S2. WOX5 expression in lateral root primordia analyzed by promoter-reporter fusion in M. truncatula and pea.
Supplemental Figure S3. Phenotype of WOX5-overexpressing roots of M. truncatula.
Supplemental Figure S4. Phenotypes of WOX5-overexpressing and control nodules of M. truncatula.
Acknowledgments
We thank Dr. E. Andronov and A. Pinaev (All-Russian Research Institute of Agricultural Microbiology) for assistance with the use of the Core Facility; Dr. Julia Frugoli (Department of Genetics and Biochemistry, Clemson University) for providing seeds of the Medicago mutant lines; Annick De Keyser and Christa Verplancke (VIB-Ghent University) for technical assistance; and Martine De Cock (Ghent University) for help in preparing the manuscript.
References
- Bauer WD. (1981) Infection of legumes by rhizobia. Annu Rev Plant Physiol 32: 407–449 [Google Scholar]
- Ben Amor B, Shaw SL, Oldroyd GED, Maillet F, Penmetsa RV, Cook D, Long SR, Dénarié J, Gough C. (2003) The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J 34: 495–506 [DOI] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al. (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55: 504–513 [DOI] [PubMed] [Google Scholar]
- Bleckmann A, Weidtkamp-Peters S, Seidel CAM, Simon R. (2010) Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol 152: 166–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondon F. (1964) Contribution à l’étude du développement de graminées fourragères: ray-grass et dactyle. Rev Gen Bot 71: 293–381 [Google Scholar]
- Boot KJM, van Brussel AAN, Tak T, Spaink HP, Kijne JW. (1999) Lipochitin oligosaccharides from Rhizobium leguminosarum bv. viciae reduce auxin transport capacity in Vicia sativa subsp. nigra roots. Mol Plant Microbe Interact 12: 839–844 [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Caetano-Anollés G, Gresshoff PM. (1991) Plant genetic control of nodulation. Annu Rev Microbiol 45: 345–382 [DOI] [PubMed] [Google Scholar]
- Carroll BJ, McNeil DL, Gresshoff PM. (1985) Isolation and properties of soybean [Glycine max (L.) Merr.] mutants that nodulate in the presence of high nitrate concentrations. Proc Natl Acad Sci USA 82: 4162–4166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R, Timmers ACJ, Maillet F, Galera C, Penmetsa RV, Cook D, Dénarié J, Gough C. (2001) The HCL gene of Medicago truncatula controls Rhizobium-induced root hair curling. Development 128: 1507–1518 [DOI] [PubMed] [Google Scholar]
- Chen S-K, Kurdyukov S, Kereszt A, Wang X-D, Gresshoff PM, Rose RJ. (2009) The association of homeobox gene expression with stem cell formation and morphogenesis in cultured Medicago truncatula. Planta 230: 827–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi M, Frugier F. (2008) De novo organ formation from differentiated cells: root nodule organogenesis. Sci Signal 1: re11; erratum Crespi M, Frugier F (2009) Sci Signal 2: er1 [DOI] [PubMed] [Google Scholar]
- de Billy F, Grosjean C, May S, Bennett M, Cullimore JV. (2001) Expression studies on AUX1-like genes in Medicago truncatula suggest that auxin is required at two steps in early nodule development. Mol Plant Microbe Interact 14: 267–277 [DOI] [PubMed] [Google Scholar]
- Delves AC, Mathews A, Day DA, Carter AS, Carroll BJ, Gresshoff PM. (1986) Regulation of the soybean-Rhizobium nodule symbiosis by shoot and root factors. Plant Physiol 82: 588–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Oldroyd GED. (2009) Positioning the nodule, the hormone dictum. Plant Signal Behav 4: 89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson BJ, Indrasumunar A, Hayashi S, Lin M-H, Lin Y-H, Reid DE, Gresshoff PM. (2010) Molecular analysis of legume nodule development and autoregulation. J Integr Plant Biol 52: 61–76 [DOI] [PubMed] [Google Scholar]
- Frugier F, Kosuta S, Murray JD, Crespi M, Szczyglowski K. (2008) Cytokinin: secret agent of symbiosis. Trends Plant Sci 13: 115–120 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S, Crespi M, Frugier F. (2006) The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18: 2680–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzali S, Novi G, Loreti E, Paolicchi F, Poggi A, Alpi A, Perata P. (2005) A turanose-insensitive mutant suggests a role for WOX5 in auxin homeostasis in Arabidopsis thaliana. Plant J 44: 633–645 [DOI] [PubMed] [Google Scholar]
- Gresshoff PM. (2003) Post-genomic insights into plant nodulation symbioses. Genome Biol 4: 201.1–201.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W, van Noorden G, Van Isterdael G, Beeckman T, Gheysen G, Mathesius U. (2009) Manipulation of auxin transport in plant roots during Rhizobium symbiosis and nematode parasitism. Plant Cell 21: 2553–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Han L, Hymes M, Denver R, Clark SE. (2010) CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J 63: 889–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T. (2004) Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131: 657–668 [DOI] [PubMed] [Google Scholar]
- Heckmann AB, Lombardo F, Miwa H, Perry JA, Bunnewell S, Parniske M, Wang TL, Downie JA. (2006) Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol 142: 1739–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch AM, Fang Y. (1994) Plant hormones and nodulation: what’s the connection? Plant Mol Biol 26: 5–9 [DOI] [PubMed] [Google Scholar]
- Hirsch AM, LaRue TA. (1997) Is the legume nodule a modified root or stem or an organ sui generis? Crit Rev Plant Sci 16: 361–392 [Google Scholar]
- Huo X, Schnabel E, Hughes K, Frugoli J. (2006) RNAi phenotypes and the localization of a protein::GUS fusion imply a role for Medicago truncatula PIN genes in nodulation. J Plant Growth Regul 25: 156–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K, Yokota K, Li YY, Wang Y, Liu C-T, Suzuki S, Aono T, Oyaizu H. (2008) Isolation of a novel root-determined hypernodulation mutant rdh1 of Lotus japonicus. Soil Sci Plant Nutr 54: 259–263 [Google Scholar]
- Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al. (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kamiya N, Nagasaki H, Morikami A, Sato Y, Matsuoka M. (2003) Isolation and characterization of a rice WUSCHEL-type homeobox gene that is specifically expressed in the central cells of a quiescent center in the root apical meristem. Plant J 35: 429–441 [DOI] [PubMed] [Google Scholar]
- Kanamori N, Madsen LH, Radutoiu S, Frantescu M, Quistgaard EMH, Miwa H, Downie JA, James EK, Felle HH, Haaning LL, et al. (2006) A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc Natl Acad Sci USA 103: 359–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A, Betsuyaku S, Osakabe Y, Mizuno S, Nagawa S, Stahl Y, Simon R, Yamaguchi-Shinozaki K, Fukuda H, Sawa S. (2010) RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137: 3911–3920; erratum Kinoshita A, Betsuyaku S, Osakabe Y, Mizuno S, Nagawa S, Stahl Y, Simon R, Yamaguchi-Shinozaki K, Fukuda H, Sawa S (2010) 137: 4327 [DOI] [PubMed] [Google Scholar]
- Kosslak RM, Bohlool BB. (1984) Suppression of nodule development of one side of a split-root system of soybeans caused by prior inoculation of the other side. Plant Physiol 75: 125–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusell L, Madsen LH, Sato S, Aubert G, Genua A, Szczyglowski K, Duc G, Kaneko T, Tabata S, de Bruijn F, et al. (2002) Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420: 422–426 [DOI] [PubMed] [Google Scholar]
- Krusell L, Sato N, Fukuhara I, Koch BEV, Grossmann C, Okamoto S, Oka-Kira E, Otsubo Y, Aubert G, Nakagawa T, et al. (2011) The Clavata2 genes of pea and Lotus japonicus affect autoregulation of nodulation. Plant J 65: 861–871 [DOI] [PubMed] [Google Scholar]
- Lévy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet E-P, Ané J-M, Lauber E, Bisseling T, et al. (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364 [DOI] [PubMed] [Google Scholar]
- Li D, Kinkema M, Gresshoff PM. (2009) Autoregulation of nodulation (AON) in Pisum sativum (pea) involves signalling events associated with both nodule primordia development and nitrogen fixation. J Plant Physiol 166: 955–967 [DOI] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R. (2003) LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302: 630–633 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al. (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- Magori S, Oka-Kira E, Shibata S, Umehara Y, Kouchi H, Hase Y, Tanaka A, Sato S, Tabata S, Kawaguchi M. (2009) TOO MUCH LOVE, a root regulator associated with the long-distance control of nodulation in Lotus japonicus. Mol Plant Microbe Interact 22: 259–268 [DOI] [PubMed] [Google Scholar]
- Markmann K, Parniske M. (2009) Evolution of root endosymbiosis with bacteria: how novel are nodules? Trends Plant Sci 14: 77–86 [DOI] [PubMed] [Google Scholar]
- Mathesius U. (2008) Auxin: at the root of nodule development? Funct Plant Biol 35: 651–668 [DOI] [PubMed] [Google Scholar]
- Mathesius U, Bayliss C, Weinman JJ, Schlaman HRM, Spaink HP, Rolfe BG, McCully ME, Djordjevic MA. (1998a) Flavonoids synthesized in cortical cells during nodule initiation are early developmental markers in white clover. Mol Plant Microbe Interact 12: 1223–1232 [Google Scholar]
- Mathesius U, Schlaman HRM, Spaink HP, Sautter C, Rolfe BG, Djordjevic MA. (1998b) Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J 14: 23–34 [DOI] [PubMed] [Google Scholar]
- Mayer KFX, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Middleton PH, Jakab J, Penmetsa RV, Starker CG, Doll J, Kaló P, Prabhu R, Marsh JF, Mitra RM, Kereszt A, et al. (2007) An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19: 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra RM, Gleason CA, Edwards A, Hadfield J, Downie JA, Oldroyd GE, Long SR. (2004) A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: gene identification by transcript-based cloning. Proc Natl Acad Sci USA 101: 4701–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa H, Oka-Kira E, Sato N, Takahashi H, Wu G-J, Sato S, Hayashi M, Betsuyaku S, Nakazono M, Tabata S, et al. (2010) The receptor-like kinase KLAVIER mediates systemic regulation of nodulation and non-symbiotic shoot development in Lotus japonicus. Development 137: 4317–4325 [DOI] [PubMed] [Google Scholar]
- Mortier V, Den Herder G, Whitford R, Van de Velde W, Rombauts S, D’Haeseleer K, Holsters M, Goormachtig S. (2010) CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol 153: 222–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortier V, De Wever E, Vuylsteke M, Holsters M, Goormachtig S. (2012a) Nodule numbers are governed by interaction between CLE peptides and cytokinin signaling. Plant J (in press) [DOI] [PubMed] [Google Scholar]
- Mortier V, Fenta BA, Martens C, Rombauts S, Holsters M, Kunert K, Goormachtig S. (2011) Search for nodulation-related CLE genes in the genome of Glycine max. J Exp Bot 62: 2571–2583 [DOI] [PubMed] [Google Scholar]
- Mortier V, Holsters M, Goormachtig S. (2012b) Never too many? How legumes control nodule numbers. Plant Cell Environ 35: 245–258 [DOI] [PubMed] [Google Scholar]
- Nishimura R, Hayashi M, Wu G-J, Kouchi H, Imaizumi-Anraku H, Murakami Y, Kawasaki S, Akao S, Ohmori M, Nagasawa M, et al. (2002) HAR1 mediates systemic regulation of symbiotic organ development. Nature 420: 426–429 [DOI] [PubMed] [Google Scholar]
- Novák K. (2010) Early action of pea symbiotic gene NOD3 is confirmed by adventitious root phenotype. Plant Sci 179: 472–478 [DOI] [PubMed] [Google Scholar]
- Novák K, Lisá L, Škrdleta V. (2011) Pleiotropy of pea RisfixC supernodulation mutation is symbiosis-independent. Plant Soil 342: 173–182 [Google Scholar]
- Oka-Kira E, Kawaguchi M. (2006) Long-distance signaling to control root nodule number. Curr Opin Plant Biol 9: 496–502 [DOI] [PubMed] [Google Scholar]
- Oka-Kira E, Tateno K, Miura K-i, Haga T, Hayashi M, Harada K, Sato S, Tabata S, Shikazono N, Tanaka A, et al. (2005) klavier (klv), a novel hypernodulation mutant of Lotus japonicus affected in vascular tissue organization and floral induction. Plant J 44: 505–515 [DOI] [PubMed] [Google Scholar]
- Okamoto S, Ohnishi E, Sato S, Takahashi H, Nakazono M, Tabata S, Kawaguchi M. (2009) Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol 50: 67–77 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JA. (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59: 519–546 [DOI] [PubMed] [Google Scholar]
- Olsson JE, Nakao P, Bohlool BB, Gresshoff PM. (1989) Lack of systemic suppression of nodulation in split root systems of supernodulating soybean (Glycine max [L.] Merr.) mutants. Plant Physiol 90: 1347–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacios-Bras C, Schlaman HRM, Boot K, Admiraal P, Langerak JM, Stougaard J, Spaink HP. (2003) Auxin distribution in Lotus japonicus during root nodule development. Plant Mol Biol 52: 1169–1180 [DOI] [PubMed] [Google Scholar]
- Penmetsa RV, Frugoli JA, Smith LS, Long SR, Cook DR. (2003) Dual genetic pathways controlling nodule number in Medicago truncatula. Plant Physiol 131: 998–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce M, Bauer WD. (1983) A rapid regulatory response governing nodulation in soybean. Plant Physiol 73: 286–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plet J, Wasson A, Ariel F, Le Signor C, Baker D, Mathesius U, Crespi M, Frugier F. (2011) MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J 65: 622–633 [DOI] [PubMed] [Google Scholar]
- Postma JG, Jacobsen E, Feenstra W. (1988) Three pea mutants with an altered nodulation studied by genetic analysis and grafting. J Plant Physiol 132: 424–430 [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al. (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Reid DE, Ferguson BJ, Gresshoff PM. (2011) Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Mol Plant Microbe Interact 24: 606–618 [DOI] [PubMed] [Google Scholar]
- Sagan M, Duc G. (1996) Sym28 and Sym29, two new genes involved in regulation of nodulation in pea (Pisum sativum L.). Symbiosis 20: 229–245 [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T. (2007) Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814 [DOI] [PubMed] [Google Scholar]
- Saur IML, Oakes M, Djordjevic MA, Imin N. (2011) Crosstalk between the nodulation signaling pathway and the autoregulation of nodulation in Medicago truncatula. New Phytol 190: 865–874 [DOI] [PubMed] [Google Scholar]
- Schnabel E, Journet E-P, de Carvalho-Niebel F, Duc G, Frugoli J. (2005) The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol Biol 58: 809–822 [DOI] [PubMed] [Google Scholar]
- Schnabel E, Mukherjee A, Smith L, Kassaw T, Long S, Frugoli J. (2010) The lss supernodulation mutant of Medicago truncatula reduces expression of the SUNN gene. Plant Physiol 154: 1390–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel EL, Frugoli J. (2004) The PIN and LAX families of auxin transport genes in Medicago truncatula. Mol Genet Genomics 272: 420–432 [DOI] [PubMed] [Google Scholar]
- Schnabel EL, Kassaw TK, Smith LS, Marsh JF, Oldroyd GE, Long SR, Frugoli JA. (2011) The ROOT DETERMINED NODULATION1 gene regulates nodule number in roots of Medicago truncatula and defines a highly conserved, uncharacterized plant gene family. Plant Physiol 157: 328–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KFX, Jürgens G, Laux T. (2000) The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Searle IR, Men AE, Laniya TS, Buzas DM, Iturbe-Ormaetxe I, Carroll BJ, Gresshoff PM. (2003) Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299: 109–112 [DOI] [PubMed] [Google Scholar]
- Sheng C, Harper JE. (1997) Shoot versus root signal involvement in nodulation and vegetative growth in wild-type and hypernodulating soybean genotypes. Plant Physiol 113: 825–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skylar A, Wu X. (2011) Regulation of meristem size by cytokinin signaling. J Integr Plant Biol 53: 446–454 [DOI] [PubMed] [Google Scholar]
- Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, Bisseling T. (2007) Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol 145: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R. (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Stahl Y, Simon R. (2009) Is the Arabidopsis root niche protected by sequestration of the CLE40 signal by its putative receptor ACR4? Plant Signal Behav 4: 634–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczyglowski K, Shaw RS, Wopereis J, Copeland S, Hamburger D, Kasiborski B, Dazzo FB, de Bruijn FJ. (1998) Nodule organogenesis and symbiotic mutants of the model legume Lotus japonicus. Mol Plant Microbe Interact 11: 684–697 [Google Scholar]
- Timmers ACJ, Auriac M-C, Truchet G. (1999) Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126: 3617–3628 [DOI] [PubMed] [Google Scholar]
- Tirichine L, Sandal N, Madsen LH, Radutoiu S, Albrektsen AS, Sato S, Asamizu E, Tabata S, Stougaard J. (2007) A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315: 104–107 [DOI] [PubMed] [Google Scholar]
- Van den Eede G, Deblaere R, Goethals K, Van Montagu M, Holsters M. (1992) Broad host range and promoter selection vectors for bacteria that interact with plants. Mol Plant Microbe Interact 5: 228–234 [DOI] [PubMed] [Google Scholar]
- van der Graaff E, Laux T, Rensing SA. (2009) The WUS homeobox-containing (WOX) protein family. Genome Biol 10: 248.1–248.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Noorden GE, Ross JJ, Reid JB, Rolfe BG, Mathesius U. (2006) Defective long-distance auxin transport regulation in the Medicago truncatula super numeric nodules mutant. Plant Physiol 140: 1494–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Fiers M. (2010) CLE peptide signaling during plant development. Protoplasma 240: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson AP, Pellerone FI, Mathesius U. (2006) Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell 18: 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson AP, Ramsay K, Jones MGK, Mathesius U. (2009) Differing requirements for flavonoids during the formation of lateral roots, nodules and root knot nematode galls in Medicago truncatula. New Phytol 183: 167–179 [DOI] [PubMed] [Google Scholar]
- Wopereis J, Pajuelo E, Dazzo FB, Jiang Q, Gresshoff PM, De Bruijn FJ, Stougaard J, Szczyglowski K. (2000) Short root mutant of Lotus japonicus with a dramatically altered symbiotic phenotype. Plant J 23: 97–114 [DOI] [PubMed] [Google Scholar]
- Yang W-C, de Blank C, Meskiene I, Hirt H, Bakker J, van Kammen A, Franssen H, Bisseling T. (1994) Rhizobium Nod factors reactivate the cell cycle during infection and nodule primordium formation, but the cycle is only completed in primordium formation. Plant Cell 6: 1415–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida C, Funayama-Noguchi S, Kawaguchi M. (2010) plenty, a novel hypernodulation mutant in Lotus japonicus. Plant Cell Physiol 51: 1425–1435 [DOI] [PubMed] [Google Scholar]