Abstract
The rolB (for rooting locus of Agrobacterium rhizogenes) oncogene has previously been identified as a key player in the formation of hairy roots during the plant-A. rhizogenes interaction. In this study, using single-cell assays based on confocal microscopy, we demonstrated reduced levels of reactive oxygen species (ROS) in rolB-expressing Rubia cordifolia, Panax ginseng, and Arabidopsis (Arabidopsis thaliana) cells. The expression of rolB was sufficient to inhibit excessive elevations of ROS induced by paraquat, menadione, and light stress and prevent cell death induced by chronic oxidative stress. In rolB-expressing cells, we detected the enhanced expression of antioxidant genes encoding cytosolic ascorbate peroxidase, catalase, and superoxide dismutase. We conclude that, similar to pathogenic determinants in other pathogenic bacteria, rolB suppresses ROS and plays a role not only in cell differentiation but also in ROS metabolism.
During agrobacterial infection, the rolA, rolB, and rolC genes of the plant pathogen Agrobacterium rhizogenes are transferred into the plant genome, causing tumor formation and hairy root disease (for review, see Nilsson and Olsson, 1997). The expression of the rol genes and, most importantly, the rolB (for rooting locus of Agrobacterium rhizogenes) gene, is critical for hairy root production (Nilsson and Olsson, 1997). The function of rolB is not restricted to root formation; the gene promotes the de novo formation of floral and shoot meristems (Altamura et al., 1994; Koltunow et al., 2001), induces parthenocarpy (Carmi et al., 2003), causes a delay in pistil and anther development (Cecchetti et al., 2004), and modifies the balance between the proliferation of procambial cells and xylem differentiation during stamen development (Cecchetti et al., 2007). The mechanism by which the RolB oncoprotein exerts such different morphological alterations remains unknown. RolB was shown to exhibit Tyr phosphatase activity (Filippini et al., 1996) and interact with 14-3-3 proteins (Moriuchi et al., 2004). RolB has no homology to any prokaryotic or eukaryotic proteins except the RolB (PLAST) family of oncoproteins in Agrobacterium species (Levesque et al., 1988; Otten and Schmidt, 1998). These RolB-related oncoproteins have been proposed to alter the developmental plasticity of transformed plants (Levesque et al., 1988; Moriuchi et al., 2004).
A new function for the rol genes in plant-Agrobacterium interactions was revealed with the discovery that these genes are potential activators of secondary metabolism in transformed cells from different plant families (Bulgakov, 2008). An investigation of the rolA-, rolB-, rolC,- rolABC-, and pRiA4-transformed cells (wild-type A. rhizogenes, strain A4) of Rubia cordifolia revealed that each of the rol genes appears to have its own individual mechanism of secondary metabolism activation (Shkryl et al., 2008).
Recently, we performed experiments to understand the relationship between the activation of secondary metabolism and the production of reactive oxygen species (ROS) in R. cordifolia cells transformed with A. rhizogenes pRiA4 and the rolC gene (Bulgakov et al., 2008; Shkryl et al., 2010). Single-cell assays based on confocal microscopy showed that rolC significantly lowers intracellular ROS levels, thus acting as a powerful suppressor of ROS. The transformation of R. cordifolia calli with the wild-type A. rhizogenes A4 strain resulted in the decrease of ROS levels in pRiA4-transformed cells. However, this effect was weaker than that observed with the expression of the single rolC gene (Shkryl et al., 2010). The suppression of ROS in pRiA4-cells was accompanied by the enhanced expression of several genes encoding ROS-detoxifying enzymes (Shkryl et al., 2010).
The effect of the rolB gene on ROS metabolism in transformed cells has not, to our knowledge, been studied. As far as the rolB and rolC genes act together in the process of neoplastic transformation, it is reasonable to expect that rolB would act in concert with rolC to decrease ROS levels. However, the participation of rolB in the induction of cellular death (necrosis) in the callus and leaves of transformed plants (Schmülling et al., 1988) and activation of secondary metabolism (Bulgakov, 2008), i.e. the processes that are often associated with the increased production of ROS, would indicate increased ROS levels in transformed tissues. The aim of this investigation was to discriminate between these possibilities.
RESULTS
Steady-State ROS Levels in rolB-Transformed Cells
Three cell lines, RB-L (low rolB expression), RB-M (moderate rolB expression), and RB-H (high rolB expression), were established several years ago and recently reexamined in terms of their gene expression, growth, and anthraquinone production (Shkryl et al., 2008). In these cell lines, rolB is expressed at a ratio of 1:4:10, respectively. The stability of gene expression was controlled during this work. The RB-L, RB-M, and RB-H cultures consisted of cell aggregates with yellow, deep-yellow, and orange-red colors, respectively. The deep-colored RB-H culture occasionally forms small black zones of necrotic cells and represents a culture with the maximum possible rolB transcript abundance; the increased expression of rolB in these cells induces cell death.
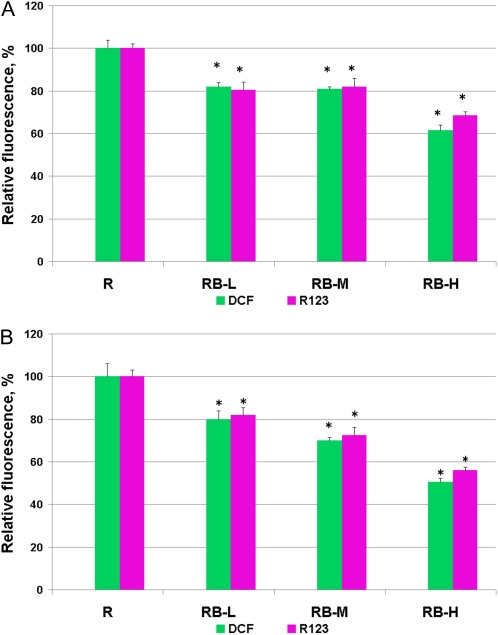
2,7-Dichlorodihydrofluorescein diacetate (H2DCF-DA) is currently the most widely used fluorogenic probe for real-time ROS imaging in plants (Swanson et al., 2011). Subsequent to the cleavage of the diacetate ester by intracellular esterase, this dye reacts with ROS, such as hydrogen peroxide (H2O2), peroxyl radicals, and peroxynitrite (Crow, 1997). When H2DCF-DA was used as a fluorogenic dye, the rolB-expressing lines showed a highly reproducible decrease of the steady-state levels of ROS (Fig. 1).
Figure 1.
Steady-state ROS levels in control (R) and rolB-expressing (RB-L, RB-M, and RC-H) R. cordifolia cell suspension (A) and callus (B) cultures. The cells cultivated for 5 (A) or 21 (B) d were loaded with H2DCF-DA or H2R123, and fluorescence of dichlorofluorescein (DCF) or rhodamine 123 (R123) was visualized by laser-scanning confocal microscopy. Fluorescence of DCF and R123 reflects intracellular ROS abundance. The results are presented as mean ± se from six independent experiments. * P < 0.05 versus values of the R culture, ANOVA followed by Fisher's Plsd test. [See online article for color version of this figure.]
These results were confirmed using another fluorescent probe, dihydrorhodamine 123 (H2R123). The specificity of H2R123 and H2DCF-DA for ROS is similar (Crow, 1997; Abele et al., 2002). H2R123 has less molar fluorescence than H2DCF-DA, but the former penetrates the mitochondrial membrane and thereby reflects the total cytosolic and mitochondrial ROS levels (Hempel et al., 1999). The level of ROS in the RB-L and RB-M cultures was 81% to 83% of that in the R culture, similar to the measurements obtained with H2DCF-DA (80%–83%; Fig. 1A). Low levels of ROS were detected in the RB-H line (Fig. 1). The cells were analyzed during the exponential growth phase (4–5 d of cultivation). An analogous result was obtained when we analyzed the cells during the linear growth phase (d 7) and in the stationary growth phase (11–12 d). These results were confirmed using callus cultures (Fig. 1B).
To test whether this effect was species specific, we included other model systems in our investigation, i.e. the long-standing test system based on Panax ginseng-transformed cell cultures (Bulgakov et al., 1998) and the recently transformed Arabidopsis (Arabidopsis thaliana) cells. Measurements of ROS in these cultures revealed that ROS inhibition by rolB was also observed in these systems (Table I).
Table I. Intracellular ROS levels in P. ginseng and Arabidopsis control and rolB-expressing cell cultures measured by laser confocal microscopy.
P. ginseng and Arabidopsis calli or cell suspensions were cultivated for 21 or 5 d, respectively, and analyzed by confocal microscopy. ROS levels are presented as mean ± se from a single experiment. *P < 0.05 versus values of the corresponding controls, Student's t test.
| Cell Line | No. of Cells Analyzed | DCF Fluorescence, Pixels |
| P. ginseng calli (vector control) | 58 | 65.4 ± 3.9 |
| P. ginseng rolB-expressing calli | 79 | 48.2 ± 2.4* |
| P. ginseng cell suspension (vector control) | 80 | 60.0 ± 4.5 |
| P. ginseng rolB-expressing cell suspension | 80 | 48.5 ± 2.0* |
| Arabidopsis calli (vector control) | 100 | 75 ± 3.5 |
| Arabidopsis low-rolB-expressing calli AtB-1 | 100 | 60 ± 2.5* |
| Arabidopsis high-rolB-expressing calli AtB-2 | 100 | 47.5 ± 1.9* |
| Arabidopsis cell suspension (vector control) | 80 | 86.1 ± 5.0 |
| Arabidopsis low-rolB-expressing cell suspension AtB-1 | 80 | 66.4 ± 4.5* |
| Arabidopsis high-rolB-expressing cell suspension AtB-2 | 80 | 56.5 ± 4.2* |
ROS measurements were also performed on the callus extracts of all species studied using an independent method, luminol-based luminometric determination. In this assay, the ROS were determined as a sum of H2O2, superoxide anion, and hydroxyl radicals (Komrskova et al., 2006). The values obtained from these measurements were consistent with confocal microscopy data and revealed that the ROS concentrations in rolB-transformed calli from all species were 10% to 30% less than in the control calli (Table II).
Table II. ROS levels in extracts of rolB-expressing callus cultures measured by the luminol-based luminometric determination.
The data are presented as mean from three independent experiments. * P < 0.05 versus values of the control cultures, ANOVA followed by Fisher's Plsd test.
| Callus Line | H2O2 Content |
|
| 21 d of Cultivation (Linear Phase of Growth) | 28 d of Cultivation (Stationary Phase of Growth) | |
| % of the Control | ||
| R. cordifolia, R (control) | 100 | 100 |
| R. cordifolia, RB-L | 92 | 90 |
| R. cordifolia, RB-M | 80* | 83* |
| R. cordifolia, RB-H | 68* | 70* |
| P. ginseng (vector control) | 100 | 100 |
| P. ginseng rolB-expressing | 74* | 85* |
| Arabidopsis (vector control) | 100 | 100 |
| Arabidopsis low-rolB-expressing AtB-1 | 83* | 86* |
| Arabidopsis high-rolB-expressing AtB-2 | 70* | 72* |
Taken together, our results indicate that rolB expression reproducibly decreases the steady-state ROS level in transformed plant cells.
ROS Accumulation in Stressed Cells
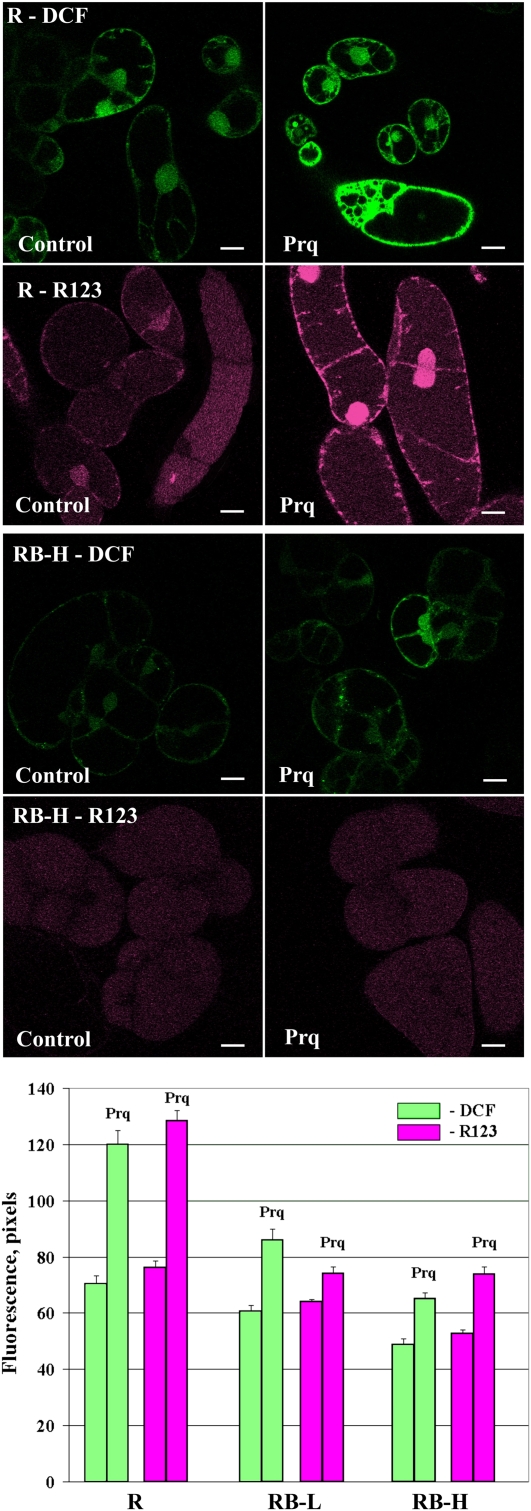
Different treatments were applied to trigger ROS production in rolB-transformed cell cultures of R. cordifolia. For this purpose, we used paraquat, menadione, and light stress. To study the effect of paraquat on ROS levels, moderate treatment conditions (10 μm paraquat and 1-h light incubation) were used, in which paraquat did not cause cell death. As shown in Figure 2, the measurements performed with H2DCF-DA and H2R123 revealed lower ROS levels in the paraquat-treated rolB-transformed cells than in the control cells. From these results, we concluded that rolB effectively prevented the excessive increase in ROS levels induced by paraquat. In the R culture, R123 fluorescence revealed ROS localization at the periphery of the cells, around the nuclear envelope, and in the cytosol (Fig. 2). In the RB-H culture, R123 fluorescence showed that ROS localized inside the cells, with almost no ROS in the plasma membrane or nucleus (Fig. 2). This result indicates that rolB mainly suppresses intracellular ROS related to the plasma membrane and nuclear regions.
Figure 2.
Effects caused by paraquat (Prq). Intracellular ROS abundance in control (R) and rolB-expressing R. cordifolia cell suspension cultures was determined by laser-scanning confocal microscopy. The cultures were cultivated in the dark for 4 d and treated with paraquat (10 μm) for 1 h under continuous light exposure. A diagram at the bottom represents ROS levels obtained from two independent experiments (mean ± se). Scale bars, 20 μm.
The experiments using menadione and H2DCF-DA as a probe showed similar results. Although rolB-expressing cells responded to paraquat treatment with a slight but noticeable ROS induction, these cells were almost insensitive to menadione, showing no ROS elevation compared with the control cells (Table III). Menadione produces superoxide radicals and H2O2 at the plasma membrane by the single-electron reduction of O2 in a reaction catalyzed by NAD(P)H:quinone-acceptor oxidoreductase (Schopfer et al., 2008). Paraquat has another mechanism of ROS generation: It acts as a terminal oxidant of PSI. In light, it reduces oxygen to a superoxide radical, which subsequently dismutates to H2O2 (Mehler, 1951). In our experiments, paraquat caused the rapid elevation of ROS production after 1 h of treatment; menadione caused a gradual elevation of ROS levels over a more prolonged period (20 h).
Table III. Suppression of ROS elevations induced by menadione and light stress in R. cordifolia rolB-expressing cells.
| Treatment | DCF Fluorescence |
||
| R (Control) | RB-L | RB-H | |
| Pixels | |||
| Menadione, μma | |||
| 0 | 82.0 ± 4.3 | 70.9 ± 3.9 | 48.0 ± 2.4 |
| 100 | 123.5 ± 5.0 | 74.0 ± 4.6 | 53.4 ± 4.1 |
| Light (time of illumination, s)b | |||
| 2 | 77.0 ± 4.0 | 59.8 ± 5.3 | 62.5 ± 3.2 |
| 600 | 113.9 ± 5.3 | 83.5 ± 3.8 | 63.8 ± 5.4 |
| 1,000 | 130.7 ± 6.4 | 87.4 ± 5.2 | 64.9 ± 6.1 |
Menadione was added to 4-d suspension cultures. After 20 h of incubation, cells were analyses by laser confocal microscopy using H2DCF-DA as a probe.
The data were obtained by confocal imaging of ROS in cells subjected to illumination from an Ar laser (excitation at 488 nm, intensity of the laser 5.9% of maximal). Images of single cells were captured and video files of the images were analyzed.
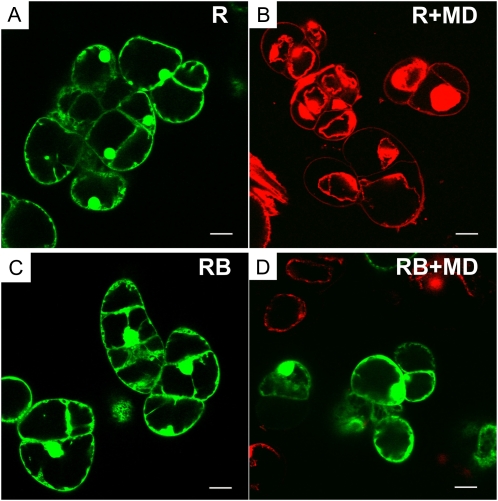
To test the viability of the cells during prolonged cultivation, the cells were stained with propidium iodide after 24 and 48 h of cultivation with 0 to 500 μm of menadione. Propidium iodide can enter only cells with damaged membranes, whereupon it intercalates into double-stranded nucleic acids, resulting in a bright-red fluorescence in nonviable cells, particularly in the nucleus (Fig. 3). From these experiments, the IC50 values (the concentration of menadione that decreases cell viability by 50%) were calculated. At 24 h of cultivation, the IC50 values were 100 and 250 μm for the R and RB-H cultures, respectively. At 48 h of cultivation, the same level of resistance in the rolB-transformed cells to menadione was observed (a 2.5-fold difference); the IC50 values were 70 and 180 μm, respectively. The rolB-transformed cells were viable even in the presence of 500 μm menadione (Fig. 3). This is probably the highest level of resistance to the inhibitor reported for plant cells.
Figure 3.
Viability of R. cordifolia cells (R and RB-H lines) in the presence of a high concentration of menadione. Confocal imaging of intact cells (A and C) and cells treated with menadione (500 μm; B and D) for 24 h. Living cells (green) were visualized by fluorescein diacetate, and dead cells (red) with collapsed nuclei were visualized with propidium iodide. Scale bars, 50 μm.
The results of these experiments suggest that rolB-expressing cells could sustain a permanently active mechanism suitable for ROS detoxification.
In the subsequent experiment, the R and RB cells were subjected to light stress by argon (Ar) laser illumination (488 nm) for 16 min. In the control R culture, there was a 1.7-fold increase in the ROS level during this time (Table III). In the low rolB-expressing cells, the increase in ROS levels was less significant (1.45-fold). The RB-H culture showed no elevation of ROS levels. Because the rolB-transformed cells initially contained less ROS, the light-induced ROS levels were similar to those observed in the control cells before light treatment (Table III). Thus, rolB prevented excessive ROS accumulation during light-induced stress.
The rolB Gene Prevents Cell Death during Long-Term Application of Paraquat
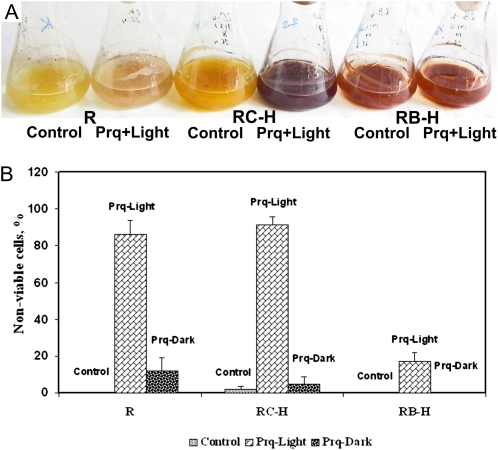
In the experiments described above, the cell cultures were subjected to the acute action of ROS-inducing stimuli. We were interested in examining the effect of rolB on long-term ROS stress. For this experiment, we added paraquat (100 μm) to actively growing 4-d-old R. cordifolia cell suspension cultures. These cultures were subsequently cultivated for 4 d. To assess the effect on ROS, one-half of the culture vessels was incubated in the dark and the other one-half in the light (paraquat induces ROS only under light conditions). Subsequently, the cells were stained with propidium iodide to determine the percentage of nonviable cells. In the presence of paraquat, 11% and 85% of the cells in the R culture were nonviable under dark and light conditions, respectively (Fig. 4). In contrast, no dead cells were detected for the rolB culture under dark conditions, and only 16% of the cells were damaged under light conditions. This result indicates that rolB expression strongly protects cells against ROS-induced cell death.
Figure 4.
The rolB gene prevents cell death induced by paraquat. A, Phenotypes of the cultures; B, cell count. The control cells (R line), high-rolB-expressing cells (RB-H line), and high-rolC-expressing cells (RC-H line) were cultivated in the presence of 100 μm paraquat for 8 d in the dark or in the light (200 μmol m−2 s−1 radiation). Then, the cells were stained with propidium iodide and analyzed by confocal microscopy. Forty to 50 cells were counted in each variant, and the percentage of nonviable cells was calculated. The data are presented as mean ± se from a single experiment with three replicates.
For comparison, we included rolC-expressing cells in this experiment. Interestingly, rolC did not prevent cell death because 92% of the rolC cells were damaged. This damage resulted in distinct phenotypical effects: The R and RC-H cultures demonstrated a dying phenotype, whereas the RB-H culture was viable (Fig. 4).
In an additional experiment, we applied H2O2 exogenously to suspension cultures of R. cordifolia and measured their growth for 6 d. At the 2 mm concentration, H2O2 inhibited the growth of the R- and rolC-transformed cultures but not that of the RB-H culture. This culture was viable even under treatment with H2O2 at concentrations as high as 10 mm (data not shown).
Expression of Genes Participating in ROS Detoxification in rolB Cells
It is known that the mechanism of ROS detoxification in plants involves the enhanced expression of genes encoding antioxidant enzymes, such as superoxide dismutase, ascorbate peroxidase, catalase, glutathione peroxidase, and other enzymes (for review, see Mittler et al., 2004).
We studied whether the expression of genes encoding antioxidant enzymes was changed in rolB-transformed cells as compared with the control cells. The expression of Arabidopsis genes encoding ascorbate peroxidase (EC 1.11.1.11), superoxide dismutase (EC 1.15.1.1), and catalase (EC 1.11.1.6) and the corresponding R. cordifolia genes, described previously (Shkryl et al., 2010), was studied using quantitative real-time reverse transcriptase (qRT)-PCR (Table IV). In this table, we also show the expression of the rolB gene measured in parallel with that of the antioxidant genes (Table IV, top).
Table IV.
Expression of genes encoding antioxidant enzymes in nontransformed and rolB-transformed cellsa
| Genes | Cell Cultures |
||||||
| Arabidopsis |
R. cordifolia |
||||||
| At | AtB-1 | AtB-2 | R | RB-L | RB-M | RB-H | |
| rolB | –b | 0.054± 0.013 | 0.191 ± 0.018 | – | 0.062 ± 0.004 | 0.266 ± 0.018 | 0.706 ± 0.059 |
| Catalase | AtCat1 | RcCat1 | |||||
| 0.278 ± 0.014 | 0.651 ± 0.027* | 0.939 ± 0.061* | 0.379 ± 0.012 | 0.397 ± 0.048 | 0.485 ± 0.015* | 0.738 ± 0.027* | |
| Ascorbate peroxidase | AtApx1 | RcApx1 | |||||
| 0.288 ± 0.019 | 0.511 ± 0.004* | 0.991 ± 0.009* | 0.292 ± 0.009 | 0.581 ± 0.011* | 0.978 ± 0.021* | 0.301 ± 0.012 | |
| AtApx2 | RcApx2 | ||||||
| 0.324 ± 0.003 | 0.646 ± 0.006* | 1.016 ± 0.016* | 0.164 ± 0.013 | 0.324 ± 0.042* | 0.404 ± 0.015* | 0.155 ± 0.008 | |
| AtApx3 | RcApx3 | ||||||
| 0.341 ± 0.041 | 0.553 ± 0.061* | 0.901 ± 0.099* | 0.342 ± 0.005 | 0.514 ± 0.003* | 0.696 ± 0.028* | 0.332 ± 0.011 | |
| Cu/Zn superoxide dismutase | AtCSD1 | RcCSD1 | |||||
| 0.498 ± 0.025 | 0.496 ± 0.038 | 0.966 ± 0.033* | 0.601 ± 0.005 | 0.611 ± 0.009 | 0.761 ± 0.006* | 0.873 ± 0.009* | |
| AtCSD2 | RcCSD2 | ||||||
| 0.882 ± 0.013 | 0.911 ± 0.051 | 0.915 ± 0.086 | 0.571 ± 0.054 | 0.611 ± 0.006 | 0.535 ± 0.011 | 0.598 ± 0.038 | |
| AtCSD3 | RcCSD3 | ||||||
| 0.802 ± 0.043 | 0.779 ± 0.014 | 0.918 ± 0.083 | 0.597 ± 0.017 | 0.577 ± 0.027 | 0.621 ± 0.023 | 0.517 ± 0.002 | |
RNA samples were isolated from callus cultures during the linear phase of growth (20–22 d). Two RNA samples were analyzed with three analytical repetitions.
Dashes indicate the absence of expression. The data are presented as mean ± se. * P < 0.05 versus values of the control cultures, ANOVA followed by Fisher's Plsd test.
The expression of the AtCat1 gene (GenBank accession no. NP_564121.1) and the orthologous R. cordifolia catalase RcCat1 gene (GQ380493) showed a 2- to 3-fold increase in rolB-expressing cells. This effect was dependent on the strength of rolB expression. These genes were previously shown to be the main catalase genes participating in ROS detoxification in Arabidopsis (Frugoli et al., 1996) and R. cordifolia (Shkryl et al., 2010).
The expression of Arabidopsis ascorbate peroxidase genes AtApx1 (GenBank accession no. AT1G07890), AtApx2 (AT3G09640), and AtApx3 (NP_195226) was compared with that of the orthologous genes RcApx1 (GQ380494), RcApx2 (GU949549), and RcApx3 (GU949550). According to the literature, AtApx1 and AtApx2 of Arabidopsis (Panchuk et al., 2002; Davletova et al., 2005) and RcApx1 and RcApx2 of R. cordifolia (Shkryl et al., 2010) are cytosolic isoforms of ascorbate peroxidases that play a pivotal role in ROS degradation. AtApx3 and RcApx3 are peroxisomal membrane-bound isoforms. We found that all these forms were up-regulated in rolB-transformed cells as compared with normal cells, but this up-regulation was observed only in cultures with a low and moderate expression of rolB (Table IV). In cells with high rolB expression (RB-H line), the expression of the Apx genes was similar to that in the control. Expression of the rolB gene in this line was 11 times higher than that of the RB-L line, i.e. the rolB gene was strongly overexpressed (Table IV). We could not select Arabidopsis cells with an analogous high expression of rolB because such cells were not viable. Thus, in our test systems, we observed the rolB dose-dependent process of Apx gene regulation. The behavior of Apx genes can be explained by a phenomenon known as the mystery of APX silencing during excessive stress (Foyer and Shigeoka, 2011), where the inactivation of APX is associated with the increased expression of catalase.
Three Arabidopsis copper (Cu)/zinc (Zn) superoxide dismutase genes and the corresponding genes of R. cordifolia were also studied. Arabidopsis contains three Cu/Zn superoxide dismutases: cytosolic AtCSD1 (AT1G08830), chloroplastic AtCSD2 (AT2G28190), and peroxisomal AtCSD3 (AT5G18100; Kliebenstein et al., 1998). The AtCSD2 and AtCSD3 genes, like the orthologous genes of R. cordifolia (RcCSD2, GU949547, and RcCSD3, GU949548), showed no up-regulation in rolB-expressing cells. In contrast, AtCSD1 and RcCSD1 (GQ380492) were up-regulated.
Effect of rolB on Reduced Glutathione/Oxidized Glutathione Ratio
Permanent transcriptional activation of antioxidant genes in rolB-expressing cells is expected to cause perturbations of redox homeostasis in cells. Here, we provide a short overview of the redox balance to estimate the degree to which rolB expression and antioxidant activation modify the redox balance of transformed cells.
The balance between the reduced glutathione (GSH) and oxidized glutathione (GSSG) is a central factor in maintaining the cellular redox state (Foyer and Noctor, 2005). It has been reported that when the intensity of a stress increases, GSH concentrations decline and the redox state becomes more oxidized, leading to the deterioration of the system. An elevated GSH concentration is correlated with the ability of plants to withstand induced oxidative stress.
The contents of GSH and GSSG in plant and callus tissues of R. cordifolia were measured by mass spectrometry, and the data are presented in Table V. The concentration ranges of GSH and GSSG in leaves of R. cordifolia were 156 nmol g−1 fresh weight (FW; GSH) and 44 nmol g−1 FW (GSSG), which are consistent with the values reported for other plant species (Rellán-Alvarez et al., 2006). The GSH/GSSG ratio was 3.6. In the control R calli, we detected the decreased concentration of GSH and a corresponding decreased GSH/GSSG ratio (2.2). In the rolB-transformed cells, the total pool of glutathione (GSH + GSSG) and the GSH/GSSG ratio was slightly higher than the corresponding values in the normal cells.
Table V. Content of GSH and GSSG and the GSH/GSSG ratio in leaves and callus cultures of R. cordifolia and Arabidopsis.
Measurements were performed in three independent samples. Callus cultures were analyzed at the linear (20–22 d) phase of growth with three biological replicates. Averaged data (mean ± se) are presented. * P < 0.05 versus values of control calli, ANOVA followed by Fisher's Plsd test.
| Plant Samples and Callus Cultures | GSH | GSSG | GSH to GSSG |
| nmol g−1 FW | |||
| R. cordifolia leaves | 156 ± 31 | 44 ± 15 | 3.6 ± 1.1 |
| R | 118 ± 21 | 53 ± 0.3 | 2.2 ± 0.3 |
| RB-L | 114 ± 10 | 53 ± 1 | 2.2 ± 0.3 |
| RB-H | 128 ± 12 | 51 ± 2 | 2.5 ± 0.3 |
| Arabidopsis (vector control) | 78 ± 4.5 | 15 ± 3 | 5.2 ± 1.1 |
| Arabidopsis low-rolB-expressing AtB-1 | 80 ± 12 | 13 ± 5 | 6.2 ± 1.6 |
| Arabidopsis high-rolB-expressing AtB-2 | 97 ± 10* | 11 ± 3 | 8.8 ± 2.1* |
According to the literature data, Arabidopsis leaves in normal physiological conditions contain 152 to 263 nmol g−1 FW GSH and 21–75 nmol g−1 FW GSSG, thus maintaining the GSH/GSSG ratio in the range of 2.0 to 12.5 (Vanhoudt et al., 2011). In transformed Arabidopsis cells, rolB caused a moderate increase of the GSH content and GSH/GSSG ratio (compared with vector control cells), but these values remained within the normal physiological parameters (Table V). Thus, nonstressed rolB-transformed cells maintain the normal redox balance.
Tolerance to Salt
ROS contributes to stress damage, as evidenced by observations that transgenic plants overexpressing ROS scavengers show increased tolerance to environmental stresses (Xiong et al., 2002). An example of this effect is the increased resistance of rolC-expressing cells to salt stress (Bulgakov et al., 2008). The R, RB-L, and RB-H suspension cultures were grown in the presence of varying NaCl concentrations. The IC50 of the R culture was 16 mm. The RB-L and RB-H cells were more tolerant to NaCl than the control culture, with the IC50 values of 21 and 25 mm, respectively (the difference is statistically significant; P < 0.05 versus value of the R culture, Student's t test.). Notably, the rolC gene alone was capable of increasing the IC50 to 70 mm (Bulgakov et al., 2008). The IC50 values for the RABC and RA4 cultures were 45 and 41 mm, respectively (Shkryl et al., 2010). Therefore, the effects of the rol genes on salt tolerance were not additive.
DISCUSSION
The rolB Gene as a ROS Suppressor
We have previously reported that rolB-transformed cells of R. cordifolia contain a large amount of anthraquinones, especially in those cells where the gene is highly transcribed (Shkryl et al., 2008). This effect is combined with the necrotizing effect of rolB. Because these features are associated with a high ROS level (Bulgakov et al., 2011; Shkryl et al., 2011), one would expect that the gene could induce ROS production. Our results, however, contradict this hypothesis. The rolB gene suppresses ROS in resting plant cells (Fig. 1; Tables I and II) and prevents or attenuates the elevation of intracellular ROS levels caused by external stimuli (Figs. 2–4; Table III). For this reason, rolB cells have enhanced resistance to salt, paraquat, menadione, light stress, and external H2O2.
The mechanism by which RolB permanently supports an active antioxidative status of transformed cells is probably the up-regulation of antioxidant genes. The majority of the antioxidant genes studied, including those encoding the Cu/Zn superoxide dismutases, catalases, and ascorbate peroxidases, were up-regulated in rolB-expressing cells (Table IV). The activation was dependent on the strength of rolB expression and, in particular, on the cell line and type of antioxidant gene. For example, low doses of rolB failed to activate the expression of the Cu/Zn superoxide dismutase genes AtCSD1, AtCSD2, AtCSD3, RcCSD1, RcCSD2, and RcCSD3. High doses of rolB failed to activate the expression of the ascorbate peroxidase genes AtApx1, AtApx2, AtApx3, RcApx1, RcApx2, and RcApx3. RolB activated the complete set of the ascorbate peroxidase genes but did not activate all of the Cu/Zn superoxide dismutase genes (Table IV). Such patterns of antioxidant gene expression in transformed cells may reflect a nonspecific (secondary) effect of rolB on ROS metabolism.
Therefore, we suggest that the activation of antioxidant genes is not a consequence of a direct action of rolB. Alternatively, one can propose a scenario in which transformed cells receive an unknown deleterious signal from the RolB protein. The cells try to compensate for this effect by cellular compensatory mechanisms, adjusting available antioxidant systems at the right place and time. In many cases, the compensation is successful, and cells maintain almost normal redox balance (Table V). In cases where it is not possible because of excessive rolB expression, cells die by necrosis.
From the physiological point of view, the effect of rolB is similar to the phenomenon known as stress acclimation or, more specifically, systemic acquired acclimation (Mullineaux et al., 2000; Gechev et al., 2006). During ROS-induced stress acclimation, plants produce catalases, ascorbate peroxidases, and other ROS-detoxifying enzymes to protect their cells against new stresses (Gechev et al., 2006). This leads to sustained antioxidant defenses and the protection of the plants from subsequent stresses.
Similarity and Dissimilarity between RolB- and Pseudomonas syringae HopAO1-Mediated Effects
An interesting analogy between the effects mediated by RolB of A. rhizogenes and some type III proteins of P. syringae emerges from our results. The effector HopAO1 (HopPtoD2) protein of P. syringae is injected from bacterial to plant cells to promote bacterial growth through suppression of the innate immunity of host cells. It was shown that HopAO1 possesses protein Tyr phosphatase activity (Bretz et al., 2003; Espinosa et al., 2003) and suppresses induced ROS in plants (Bretz et al., 2003). The observation that the rolB gene causes ROS inhibition in plant cells indicates a functional analogy between the RolB and HopAO1 Tyr phosphatases. Pseudomonas and Agrobacterium use different mechanisms to deliver pathogenic determinants to plant cells, using type III and IV secretion systems, respectively. However, a strategy aimed at the suppression of plant defense responses seems to be logical for both pathogens.
One could propose that HopAO1 and RolB are related proteins that originate from lateral gene transfer between Agrobacterium and Pseudomonas, because it is known that both microorganisms are amenable to such genetic innovation (Kado, 2009). However, our comparison of the HopAO1 and RolB amino acid sequences showed only limited amino acid similarity (24% amino acid identity). The localization of these proteins in plant cells is also different. HopAO1 is localized to the soluble fraction of protein extracts (Underwood et al., 2007), whereas RolB is localized in the plasma membrane (Filippini et al., 1996) or in the nucleus (Moriuchi et al., 2004). Therefore, these proteins have probably evolved independently.
Combined Effect of the rolB and rolC Genes
Although the rolB and C genes promote root formation synergistically, an antagonistic effect of the rol genes has been demonstrated at different levels. The stimulatory effect of the rolB gene on anthraquinone formation was weaker when this gene was combined with rolC (Shkryl et al., 2008). Constitutive rolB expression suppressed the growth of tobacco (Nicotiana tabacum) cells, and the rolC gene was able to attenuate this growth inhibition (Schmülling et al., 1988). Likewise, rolC diminished the rolB-induced high sensitivity to auxin in transformed cells (Maurel et al., 1991) and the severity of rolB-induced phenotypes (Capone et al., 1989; Vanaltvorst et al., 1992). Recently, a contrasting difference between the action of rolB and rolC on class III peroxidase gene expression has been demonstrated (Veremeichik et al., 2012). Although overexpression of a single rolB gene caused dramatic up-regulation of R. cordifolia class III peroxidase genes, the effect of the rolC gene on peroxidase transcript abundance was minimal. Interestingly, the effect of the rolB gene was almost totally suppressed in pRiA4 calli, where rolB and rolC were expressed simultaneously.
It has been shown that the combined actions of the rolB and C genes do not cause significant ROS suppression (Shkryl et al., 2010). If it were otherwise, the combined effect of the rol genes could cause totally disturbed ROS homeostasis and cell death. However, the strategy of the phytopathogen A. rhizogenes is not to kill cells. Instead, the bacteria, acting via the transferred genes, render cells to be more tolerant of environmental stresses and increase their defense potential. In many cases, the rol genes ensure a high growth rate of transformed cells and their hormonal independence. In this context, the actions of each of the rol genes seem to be in tune with the actions of the other, providing physiological conditions for better cell fitness in the face of changing environmental conditions. Perhaps this is the main effect of the rol genes as members of the RolB (plast) gene family. The rol-induced perturbations are beneficial to transformed cells but not to the whole organism, as in the case with animal tumors. A. rhizogenes-infected plants have abnormal metabolism and produce large amounts of opines, which are necessary for bacterial growth but cannot be utilized by plants. An interesting question arises: What kind of cells can be active producers of opines? It is logical to propose that transformed plant cells with increased growth and viability fit this criterion.
The Interplay between ROS Production and Morphogenetic Responses
Although some biochemical perturbations caused by the rolC and rolB genes can be explained, the root-forming activity of these genes and the phenotypical abnormalities caused by them are more difficult to understand. It is known that ROS control cell expansion and root elongation (for review, see Gapper and Dolan, 2006). Recently published data suggest a complex and dynamic role for ROS in stress-induced morphogenetic responses, indicating the involvement of ROS in cell developmental programs (Tsukagoshi et al., 2010; Blomster et al., 2011). Transgenic plants with reduced ROS levels showed a reduced apical dominance, enhanced branching, decreased chlorophyll content, abnormal flower development (abnormal petal number, fasciated styles and ovaries), parthenocarpy, reduced leaf lobing, and curled leaflets (Sagi et al., 2004).
Most of these traits, excluding the epinastic curling in the leaf margin, are similar to those described previously for pRiA4-, rolABC-transformed plants, or plants transformed with single rol genes (for review, see Nilsson and Olsson, 1997). The most typical effects of rolB on plant development are heterostyly, altered leaf and flower morphology, and the increased formation of adventitious roots on stems (Schmülling et al., 1988). RolB promotes de novo meristem formation in cultured tissues (Altamura et al., 1994) and plants (Koltunow et al., 2001). The type of organ that is formed from these meristems (roots, shoots, vegetative rosettes, or capitula) depends on the developmental and hormonal context. Furthermore, RolB perturbs the growth of plant reproductive organs by altering the developmental potential and reproductive fate of the ovule and affecting the processes of anther dehiscence and style elongation (Koltunow et al., 2001; Carmi et al., 2003; Cecchetti et al., 2004). RolB is thought to be involved in auxin signal perception/transduction pathways (Cecchetti et al., 2004). Because recent data indicate a clear interplay between auxin and ROS level in altering the leaf developmental program (Blomster et al., 2011), the interaction between auxin signaling and ROS metabolism in rolB-induced morphological responses is especially interesting. There may be a link between the morphological responses and ROS level perturbations induced by rolB.
MATERIALS AND METHODS
Plant Cell Cultures
Rubia cordifolia (Rubiaceae) cell cultures described in this work were established in 2000 using clonally cultivated plantlets (Bulgakov et al., 2002). The plantlets were transformed with Agrobacterium tumefaciens strain GV3101 harboring the pPCV002-CaMVBT construct (rolB under the 35S Cauliflower mosaic virus promoter; Spena et al., 1987). The control nontransformed culture (R) was established from the same plantlets and cultivated under the same conditions as the transformed ones. Independently transformed lines with low, moderate, and high expression of the rolB gene (RB- l, RB-M, and RB-H callus cultures, respectively) were obtained by selection of homogenous yellow, deep-yellow, and orange-red colors, respectively. These lines have been previously characterized to have stable morphology, growth, biosynthetic parameters, and levels of rolB expression (Shkryl et al., 2008). Cell suspensions were cultivated using WB/A liquid medium (Bulgakov et al., 1998) supplemented with 0.5 mg L−1 6-benzylaminopurine and 2.0 mg L−1 α-naphthaleneacetic acid in the dark (excluding experiments with paraquat, where cells were cultivated in the light; see below) at 24°C with 12-d subculture intervals.
The Panax ginseng GV (vector control) and rolB-transformed callus cultures were cultivated using W4CPA medium supplemented with 0.4 mg L−1 4-chlorophenoxyacetic acid in the dark as described previously (Bulgakov et al., 1998). The suspension variants of these cultures were grown in W4CPA liquid medium.
The Arabidopsis (Arabidopsis thaliana) vector control and rolB-transgenic callus cultures were obtained from Columbia seedlings using the pPCV002-CaMVBT construct as described previously (Bulgakov et al., 2010). The calli were cultivated using W2,4-d medium (Bulgakov et al., 1998) supplemented with 0.4 mg L−1 2,4-dichlorophenoxyacetic acid in the dark at 24°C with 30-d subculture intervals. Two rolB-expressing callus lines (AtB-1 and AtB-2) were selected from these primary calli as described previously (Bulgakov et al., 2010) to obtain lines with different strengths of rolB expression. The AtB-2 line showed a 4-fold higher expression of rolB in comparison with AtB-1. These cultures were 1 year of age. The cell suspension cultures AtB-1 and AtB-2 were cultivated using W2,4-d liquid medium (Bulgakov et al., 1998) in the dark at 24°C with 14-d subculture intervals.
Laser Confocal Imaging of Intracellular ROS
Measurements of intracellular ROS were performed as described previously (Bulgakov et al., 2008). The experiments were based on the ability of plant cells to oxidize fluorogenic dyes to their corresponding fluorescent analogs that allowed ROS determination in living cells. Suspensions of plant cells were grown in liquid nutrient medium for 4 to 12 d and filtrated through 100 μm of mesh nylon to separate cell clusters. Single cells and 10 to 20 cell aggregates were gently centrifuged and resuspended in liquid WB/A medium containing 50 μm H2DCF-DA (Molecular Probes) or 10 μm H2R123 (Molecular Probes) and incubated at 25°C ± 1°C in the dark. Cells were incubated with H2DCF-DA and H2R123 for 10 min. Slices from calli were prepared by using a vibratome HM650V (Microm). Dye-loaded cells were washed in the medium and resuspended. Intracellular oxidation of H2DCF-DA and H2R123 yielded DCF and R123 that were detected by microscopy. Examination of fluorescence in single living cells was performed with an LSM 510 META confocal laser scanning microscope (Carl Zeiss) equipped with an Ar laser with an effective power of 30 mW. The intensity of the Ar laser was 5.9% of the maximal value for H2DCF-DA and 10% for H2R123. All confocal images were recorded as 40-s time series at intervals of 0.5 ms. Video files of the captured images were recorded using the above described settings and analyzed with LSM 510 Release 3.5 software (Carl Zeiss). Data were presented as the mean from several separate experiments (at least 30–40 cells were analyzed in each experiment).
Luminometric Determination of ROS
The production of ROS in callus cultures was measured by the luminol-based luminometric method according to Piedras et al. (1998). The control and rolB-transformed calli were harvested at 21 and 28 d of cultivation and analyzed using a RF-1501 instrument (Shimadzu EUROPA GmbH). The following settings were used: excitation, 355 nm; emission, 420 nm; response, auto; number of iterations, three; reaction, 8 s; and analysis, 5 s. The calibration curve used was linear in concentrations from 50 to 800 μmol L−1 H2O2. Luminol (5-amino-2,3-dihydro-1,4-phthalazinedione) was obtained from ICN Pharmaceuticals.
Cell Viability
The viability of cells was tested by the addition of propidium iodide (Sigma, 0.3 mg mL−1, final concentration in water) to cell suspension cultures. Confocal images were obtained after excitation at 536 nm and emission at 617 nm (laser wave, 543; intensity, 20%; and filter LP, 560).
Paraquat, Menadione, and Light Treatments
Suspension-cultivated R. cordifolia cells were grown for 4 d in the dark and treated with paraquat (Aldrich, 10 μm final concentration) for 1 h under continuous light exposure (200 μmol m−2 s−1 radiation). Menadione (Sigma, 100 μm final concentration) was added to 4-d suspension cultures, which were subsequently cultivated for 20 h in the dark. Light stress was caused by continuous illumination of cells with the LSM 510 META Ar laser (effective power, 30 mW; 5.9% maximal laser intensity) at 488 nm.
Tolerance to Salt
Resistance of the R. cordifolia-transformed cell cultures to salt stress was determined as described previously (Bulgakov et al., 2008). Cell suspensions were cultivated in the presence of NaCl (0, 30, 60, and 120 mm) for 10 d. Data were obtained from two separate experiments consisting of 10 replicates each.
qRT-PCR
qRT-PCR was performed as described previously (Shkryl et al., 2010). RNA concentration and 28S-18S ratios were determined using an RNA StdSens LabChip kit and Experion Automated Electrophoresis Station (Bio-Rad Laboratories) with Experion Software System Operation and Data Analysis Tools (version 3.0) following the manufacturer’s protocol and recommendations.
The qRT-PCR analysis was performed using the Bio-Rad CFX96 Real-Time System (Bio-Rad Laboratories) with a 2.5× SYBR green PCR master mix containing ROX as a passive reference dye (Syntol). Two biological replicates, resulting from two different RNA extractions, were used for analysis, and three technical replicates were analyzed for each biological replicate. No-template controls and RNA-RT controls were included in the analysis to verify the absence of contamination. The absence of nonspecific products or primer-dimer artifacts in the samples was confirmed by melting curve analysis at the end of each run and by product visualization using electrophoresis. Primer efficiency of less than 95% was confirmed with a standard curve spanning 7 orders of magnitude. Data were analyzed using CFX Manager Software (Version 1.5; Bio-Rad Laboratories).
Analysis of GSH and GSSG by Mass Spectrometry
GSH and GSSG were extracted from R. cordifolia cells quantitatively as described by Rellán-Alvarez et al. (2006) and analyzed according to the recommendations of these authors in the Instrumental Centre for Biotechnology and Gene Engineering at the Institute of Biology and Soil Science using a HCTultra PTM Discovery System (Bruker Daltonik). The HCTultra is equipped with a high-capacity ion trap that enables the acquisition of tandem mass spectrometry data on low-abundance precursor ions and is designed to determine low-weight peptides. Cell extracts or solutions of commercial GSH or GSSG (Sigma) were directly injected in the spectrometer, outfitted with an electrospray ion source, at a flow rate of 120 μL/h (mass range mode, ultra scan; ion polarity, positive or negative; ion source type, ESI; scan mode, standard-normal). The identity of GSH was confirmed by analysis of the masses of the deprotonated molecules of GSH [M-H]− with a mass-to-charge ratio (m/z) of 306.0 and GSH [M+H]+ with m/z 308.0 as well as product ions with m/z 161.9, 179.0, 233.0, and 290.1 specific for GSH (Supplemental Fig. S1). The identity of GSSG was confirmed by analysis of the masses of GSSG [M-H]− with m/z 611.1 and [M+H]+ with m/z 613.1 as well as product ions with m/z 355.1 and 484.1 (Supplemental Fig. S1). The analysis parameters (in mass spectrometry and tandem mass spectrometry modes) were optimized for the production of characteristic precursor and product ions in the positive ionization mode. GSH and GSSG levels were determined based on a comparison of the averaged peak heights of [M+H]+ ions in concentrations of 0.1 to 5 μm (GSH) and 0.05 to 0.5 μm (GSSG). GSH and GSSG solutions and extracts were measured using identical conditions.
Statistical Analysis
In statistical evaluation, the Student’s t test was used for the comparison between two independent groups. For comparison among multiple data, ANOVA followed by a multiple comparison procedure was employed. Fisher’s protected lsd (Plsd) posthoc test was employed for the intergroup comparison. A difference of P < 0.05 was considered significant.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Mass spectrometry of the reduced (GSH) and oxidized (GSSG) forms of glutathione.
References
- Abele D, Heise K, Pörtner HO, Puntarulo S. (2002) Temperature-dependence of mitochondrial function and production of reactive oxygen species in the intertidal mud clam Mya arenaria. J Exp Biol 205: 1831–1841 [DOI] [PubMed] [Google Scholar]
- Altamura MM, Capitani F, Gazza L, Capone I, Costantino P. (1994) The plant oncogene rolB stimulates the formation of flowers and root meristemoids in tobacco thin cell layers. New Phytol 126: 283–293 [Google Scholar]
- Blomster T, Salojärvi J, Sipari N, Brosché M, Ahlfors R, Keinänen M, Overmyer K, Kangasjärvi J. (2011) Apoplastic reactive oxygen species transiently decrease auxin signaling and cause stress-induced morphogenic response in Arabidopsis. Plant Physiol 157: 1866–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretz JR, Mock NM, Charity JC, Zeyad S, Baker CJ, Hutcheson SW. (2003) A translocated protein tyrosine phosphatase of Pseudomonas syringae pv. tomato DC3000 modulates plant defence response to infection. Mol Microbiol 49: 389–400 [DOI] [PubMed] [Google Scholar]
- Bulgakov VP. (2008) Functions of rol genes in plant secondary metabolism. Biotechnol Adv 26: 318–324 [DOI] [PubMed] [Google Scholar]
- Bulgakov VP, Aminin DL, Shkryl YN, Gorpenchenko TY, Veremeichik GN, Dmitrenok PS, Zhuravlev YN. (2008) Suppression of reactive oxygen species and enhanced stress tolerance in Rubia cordifolia cells expressing the rolC oncogene. Mol Plant Microbe Interact 21: 1561–1570 [DOI] [PubMed] [Google Scholar]
- Bulgakov VP, Gorpenchenko TY, Shkryl YN, Veremeichik GN, Mischenko NP, Avramenko TV, Fedoreyev SA, Zhuravlev YN. (2011) CDPK-driven changes in the intracellular ROS level and plant secondary metabolism. Bioeng Bugs 2: 327–330 [DOI] [PubMed] [Google Scholar]
- Bulgakov VP, Khodakovskaya MV, Labetskaya NV, Tchernoded GK, Zhuravlev YN. (1998) The impact of plant rolC oncogene on ginsenoside production by ginseng hairy root cultures. Phytochemistry 49: 1929–1934 [Google Scholar]
- Bulgakov VP, Shkryl YN, Veremeichik GN. (2010) Engineering high yields of secondary metabolites in Rubia cell cultures through transformation with rol genes. Methods Mol Biol 643: 229–242 [DOI] [PubMed] [Google Scholar]
- Bulgakov VP, Tchernoded GK, Mischenko NP, Khodakovskaya MV, Glazunov VP, Radchenko SV, Zvereva EV, Fedoreyev SA, Zhuravlev YN. (2002) Effect of salicylic acid, methyl jasmonate, ethephon and cantharidin on anthraquinone production by Rubia cordifolia callus cultures transformed with the rolB and rolC genes. J Biotechnol 97: 213–221 [DOI] [PubMed] [Google Scholar]
- Capone I, Spanò L, Cardarelli M, Bellincampi D, Petit A, Costantino P. (1989) Induction and growth properties of carrot roots with different complements of Agrobacterium rhizogenes T-DNA. Plant Mol Biol 13: 43–52 [DOI] [PubMed] [Google Scholar]
- Carmi N, Salts Y, Dedicova B, Shabtai S, Barg R. (2003) Induction of parthenocarpy in tomato via specific expression of the rolB gene in the ovary. Planta 217: 726–735 [DOI] [PubMed] [Google Scholar]
- Cecchetti V, Altamura MM, Serino G, Pomponi M, Falasca G, Costantino P, Cardarelli M. (2007) ROX1, a gene induced by rolB, is involved in procambial cell proliferation and xylem differentiation in tobacco stamen. Plant J 49: 27–37 [DOI] [PubMed] [Google Scholar]
- Cecchetti V, Pomponi M, Altamura MM, Pezzotti M, Marsilio S, D’Angeli S, Tornielli GB, Costantino P, Cardarelli M. (2004) Expression of rolB in tobacco flowers affects the coordinated processes of anther dehiscence and style elongation. Plant J 38: 512–525 [DOI] [PubMed] [Google Scholar]
- Crow JP. (1997) Dichlorodihydrofluorescein and dihydrorhodamine 123 are sensitive indicators of peroxynitrite in vitro: implications for intracellular measurement of reactive nitrogen and oxygen species. Nitric Oxide 1: 145–157 [DOI] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R. (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17: 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa A, Guo M, Tam VC, Fu ZQ, Alfano JR. (2003) The Pseudomonas syringae type III-secreted protein HopPtoD2 possesses protein tyrosine phosphatase activity and suppresses programmed cell death in plants. Mol Microbiol 49: 377–387 [DOI] [PubMed] [Google Scholar]
- Filippini F, Rossi V, Marin O, Trovato M, Costantino P, Downey PM, Lo Schiavo F, Terzi M. (1996) A plant oncogene as a phosphatase. Nature 379: 499–500 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. (2005) Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17: 1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Shigeoka S. (2011) Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol 155: 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frugoli JA, Zhong HH, Nuccio ML, McCourt P, McPeek MA, Thomas TL, McClung CR. (1996) Catalase is encoded by a multigene family in Arabidopsis thaliana (L.) Heynh. Plant Physiol 112: 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel SL, Buettner GR, O’Malley YQ, Wessels DA, Flaherty DM. (1999) Dihydrofluorescein diacetate is superior for detecting intracellular oxidants: comparison with 2′,7′-dichlorodihydrofluorescein diacetate, 5(and 6)-carboxy-2′,7′-dichlorodihydrofluorescein diacetate, and dihydrorhodamine 123. Free Radic Biol Med 27: 146–159 [DOI] [PubMed] [Google Scholar]
- Gapper C, Dolan L. (2006) Control of plant development by reactive oxygen species. Plant Physiol 141: 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev TS, Van Breusegem F, Stone JM, Denev I, Laloi C. (2006) Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays 28: 1091–1101 [DOI] [PubMed] [Google Scholar]
- Kado CI. (2009) Horizontal gene transfer: sustaining pathogenicity and optimizing host-pathogen interactions. Mol Plant Pathol 10: 143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Monde RA, Last RL. (1998) Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol 118: 637–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow AM, Johnson SD, Lynch M, Yoshihara T, Costantino P. (2001) Expression of rolB in apomictic Hieracium piloselloides Vill. causes ectopic meristems in planta and changes in ovule formation, where apomixis initiates at higher frequency. Planta 214: 196–205 [DOI] [PubMed] [Google Scholar]
- Komrskova D, Lojek A, Hrbac J, Ciz M. (2006) A comparison of chemical systems for luminometric determination of antioxidant capacity towards individual reactive oxygen species. Luminescence 21: 239–244 [DOI] [PubMed] [Google Scholar]
- Levesque H, Delepelaire P, Rousé P, Slightom J, Tepfer D. (1988) Common evolutionary origin of the central portions of the Ri TL-DNA of Agrobacterium rhizogenes and the Ti T-DNAs of Agrobacterium tumefaciens. Plant Mol Biol 11: 731–744 [DOI] [PubMed] [Google Scholar]
- Maurel C, Barbier-Brygoo H, Spena A, Tempé J, Guern J. (1991) Single rol genes from the Agrobacterium rhizogenes T(L)-DNA alter some of the cellular responses to auxin in Nicotiana tabacum. Plant Physiol 97: 212–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehler AH. (1951) Studies on reactions of illuminated chloroplasts. II. Stimulation and inhibition of the reaction with molecular oxygen. Arch Biochem Biophys 34: 339–351 [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9: 490–498 [DOI] [PubMed] [Google Scholar]
- Moriuchi H, Okamoto C, Nishihama R, Yamashita I, Machida Y, Tanaka N. (2004) Nuclear localization and interaction of RolB with plant 14-3-3 proteins correlates with induction of adventitious roots by the oncogene rolB. Plant J 38: 260–275 [DOI] [PubMed] [Google Scholar]
- Mullineaux P, Ball L, Escobar C, Karpinska B, Creissen G, Karpinski S. (2000) Are diverse signalling pathways integrated in the regulation of Arabidopsis antioxidant defence gene expression in response to excess excitation energy? Philos Trans R Soc Lond B Biol Sci 355: 1531–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson O, Olsson O. (1997) Getting to the root: the role of the Agrobacterium rhizogenes rol genes in the formation of hairy roots. Physiol Plant 100: 463–473 [Google Scholar]
- Otten L, Schmidt J. (1998) A T-DNA from the Agrobacterium tumefaciens limited-host-range strain AB2/73 contains a single oncogene. Mol Plant Microbe Interact 11: 335–342 [DOI] [PubMed] [Google Scholar]
- Panchuk II, Volkov RA, Schöffl F. (2002) Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol 129: 838–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piedras P, Hammond-Kosack KE, Harrison K, Jones JDG. (1998) Rapid, Cf-9- and Avr9-dependent production of active oxygen species in tobacco suspension cultures. Mol Plant Microbe Interact 11: 1155–1166 [Google Scholar]
- Rellán-Alvarez R, Hernández LE, Abadía J, Alvarez-Fernández A. (2006) Direct and simultaneous determination of reduced and oxidized glutathione and homoglutathione by liquid chromatography-electrospray/mass spectrometry in plant tissue extracts. Anal Biochem 356: 254–264 [DOI] [PubMed] [Google Scholar]
- Sagi M, Davydov O, Orazova S, Yesbergenova Z, Ophir R, Stratmann JW, Fluhr R. (2004) Plant respiratory burst oxidase homologs impinge on wound responsiveness and development in Lycopersicon esculentum. Plant Cell 16: 616–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmülling T, Schell J, Spena A. (1988) Single genes from Agrobacterium rhizogenes influence plant development. EMBO J 7: 2621–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer P, Heyno E, Drepper F, Krieger-Liszkay A. (2008) Naphthoquinone-dependent generation of superoxide radicals by quinone reductase isolated from the plasma membrane of soybean. Plant Physiol 147: 864–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkryl YN, Veremeichik GN, Bulgakov VP, Gorpenchenko TY, Aminin DL, Zhuravlev YN. (2010) Decreased ROS level and activation of antioxidant gene expression in Agrobacterium rhizogenes pRiA4-transformed calli of Rubia cordifolia. Planta 232: 1023–1032 [DOI] [PubMed] [Google Scholar]
- Shkryl YN, Veremeichik GN, Bulgakov VP, Tchernoded GK, Mischenko NP, Fedoreyev SA, Zhuravlev YN. (2008) Individual and combined effects of the rolA, B, and C genes on anthraquinone production in Rubia cordifolia transformed calli. Biotechnol Bioeng 100: 118–125 [DOI] [PubMed] [Google Scholar]
- Shkryl YN, Veremeichik GN, Bulgakov VP, Zhuravlev YN. (2011) Induction of anthraquinone biosynthesis in Rubia cordifolia cells by heterologous expression of a calcium-dependent protein kinase gene. Biotechnol Bioeng 108: 1734–1738 [DOI] [PubMed] [Google Scholar]
- Spena A, Schmülling T, Koncz C, Schell JS. (1987) Independent and synergistic activity of rol A, B and C loci in stimulating abnormal growth in plants. EMBO J 6: 3891–3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson SJ, Choi WG, Chanoca A, Gilroy S. (2011) In vivo imaging of Ca2+, pH, and reactive oxygen species using fluorescent probes in plants. Annu Rev Plant Biol 62: 273–297 [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN. (2010) Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616 [DOI] [PubMed] [Google Scholar]
- Underwood W, Zhang S, He SY. (2007) The Pseudomonas syringae type III effector tyrosine phosphatase HopAO1 suppresses innate immunity in Arabidopsis thaliana. Plant J 52: 658–672 [DOI] [PubMed] [Google Scholar]
- Vanaltvorst AC, Bino RJ, Vandijk AJ, Lamers AMJ, Lindhout WH, van der Mark F, Dons JJM. (1992) Effects of the introduction of Agrobacterium rhizogenes rol genes on tomato plant and flower development. Plant Sci 83: 77–85 [Google Scholar]
- Vanhoudt N, Cuypers A, Horemans N, Remans T, Opdenakker K, Smeets K, Bello DM, Havaux M, Wannijn J, Van Hees M, et al. (2011) Unraveling uranium induced oxidative stress related responses in Arabidopsis thaliana seedlings: II. Responses in the leaves and general conclusions. J Environ Radioact 102: 638–645 [DOI] [PubMed] [Google Scholar]
- Veremeichik GN, Shkryl YN, Bulgakov VP, Avramenko TV, Zhuravlev YN. (January 12, 2012) Molecular cloning and characterization of seven class III peroxidases induced by overexpression of the agrobacterial rolB gene in Rubia cordifolia transgenic callus cultures. Plant Cell Rep http://dx.doi.org/10.1007/s00299-011-1219-3 [DOI] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK. (2002) Cell signaling during cold, drought, and salt stress. Plant Cell (Suppl) 14: S165–S183 [DOI] [PMC free article] [PubMed] [Google Scholar]