Abstract
Leaves between species vary in their size, serration, complexity, and shape. However, phylogeny is not the only predictor of leaf morphology. The shape of a leaf is the result of intricate developmental processes, including heteroblastic progression (changes in leaf size and shape at different nodes) and the developmental stage of an organ. The leaflets that arise from complex leaves are additionally modified by their positioning along the proximal-distal axis of a leaf and whether they fall on the left or right side of leaves. Even further, leaves are environmentally responsive, and their final shape is influenced by environmental inputs. Here, we comprehensively describe differences in leaflet shape between wild tomato (Solanum section Lycopersicon) species using a principal component analysis on elliptical Fourier descriptors arising from >11,000 sampled leaflets. We leverage differences in developmental rate to approximate a developmental series, which allows us to resolve the confounding differences in intrinsic leaflet form and developmental stage along positions of the heteroblastic leaf series and proximal-distal axis of leaves. We find that the resulting developmental trajectory of organs at different positions along these axes are useful for describing the changes in leaflet shape that occur during the shade avoidance response in tomato. We argue that it is the developmental trajectory, the changes in shape that occur over developmental time in organs reiterated at multiple positions, that is the relevant phenotype for discerning differences between populations and species, and to understand the underlying developmental processes that change during evolution.
The diversity of leaf forms among species is staggering. Leaves can either be complex or simple, serrated or entire, or a variety of different morphs that vary by aspect ratio, the shape of their tips, and the distribution of laminar outgrowth along the proximal-distal axis (Nicotra et al., 2011). Great progress has been made identifying the genetic basis for a variety of leaf forms (Champagne and Sinha, 2004). Principal to understanding leaf form is auxin, which not only patterns the initiation of leaves during phyllotaxis (Reinhardt et al., 2003), but also foretells the positioning of leaflets and serrations (Barkoulas et al., 2008; Koenig et al., 2009). In addition, leaf serration is controlled by the activity of members of the CUP-SHAPED COTYLEDONS family and miR164 activity (Blein et al., 2008). The length and width of leaves is regulated by ANGUSTIFOLIA and ROTUNDIFOLIA3 (Tsuge et al., 1996). How rugose (wrinkled) a leaf is and the duration of cell proliferation depend on TCP (TB1/CYC/PCF) activity regulated by miR319 (Nath et al., 2003; Palatnik et al., 2003; Ori et al., 2007), and the complexity of leaves is controlled, at least in part, by a network of genes regulated through members of the KNOTTED1-LIKE HOMEOBOX (KNOX) family (Bharathan et al., 2002).
Most of our knowledge concerning the regulation of leaf forms derives from mutagenesis strategies, in which leaf phenotypes arising from mutations at single loci are selected based on their severity. However, the genetic basis for the vast majority of natural variation in leaf form remains unknown. Only recently have a few examples of genes regulating differences in leaf shape between species or populations been uncovered. One such study used linkage mapping to identify PETROSELINUM (PTS), a KNOX gene lacking a homeodomain, as the cause of increased leaf complexity in a Galápagan tomato (Solanum galapagense) species. The polymorphism consisted of a single nucleotide deletion in the PTS promoter, leading to increased gene expression (Kimura et al., 2008). As sequencing technologies advance, newer methods for identifying the genetic basis of leaf variants have arisen. One recent study, for example, leveraged nested-association mapping in a large, controlled crossing scheme to identify liguleless genes as regulators of leaf angle in maize (Zea mays; Tian et al., 2011).
As the study of leaf form progresses from investigations of mutations within a defined genotype to studies of natural variation between populations and species, methods to quantify leaf shape variation will have to be developed. Fortunately, a number of morphometric techniques, based on landmarks (Zimmerman et al., 2000; Klingenberg et al., 2001) or outline analysis methods such as elliptic Fourier descriptors (EFD; Kuhl and Giardina, 1982; Iwata et al., 1998; Iwata and Ukai, 2002), have been developed to quantify shape. Importantly, the measures resulting from a principal component analysis (PCA) on the output of these methods can be treated as traits, and have been used to find quantitative trait loci regulating the variance in shape that they explain (Langlade et al., 2005). In Antirrhinum, particularly, a modified landmark analysis has been used to study biological questions ranging from allometry, heterosis, and combinatorial gene control in leaves and flowers (Feng et al., 2009; Cui et al., 2010; Rosas et al., 2010).
When we quantify differences in organs between species and populations, or even mutants, it is important to consider what these differences represent with respect to the underlying development of the organism. A leaf exists as a continuous range of morphs during its developmental progression postinception, eventually stabilizing into a mature, final form. We might measure differences in leaf form at a single time point and describe these as essential characters defining a species. However, these differences might radically change if the leaves are measured at alternative points during development. When two species are being compared the situation becomes even more complicated if development between them is at such incongruous rates that the differences we observe are unintelligible, creating differences in the developmental stage of leaves at different nodes.
However, differences in developmental rate and heteroblasty, which mask intrinsic differences in leaf shape, can also be leveraged to statistically account for these effects. There are not only genetic effects (species and populations) that change developmental rate and the heteroblastic progression, but also environmental effects. For example, foliar shade is known to accelerate the progression to flowering and presumably the juvenile-to-adult phase transition as well (Willmann and Poethig, 2011; Chitwood et al., 2012). Surprisingly few studies have measured changes in leaf shape at different nodes or as modified by genetic by environmental effects (Iwata et al., 2002; Weight et al., 2008; Pryer and Hearn, 2009). In each of these cases, the variance in shape is partitioned by an explanatory factor. Still, to our knowledge, no current study has looked at a time series of leaves from each node, nor how heteroblasty is modified by the environment to properly resolve these factors quantitatively. Except for a few studies (Pryer and Hearn, 2008; Piazza et al., 2010), compound leaves and the unique developmental context of leaflets on such leaves have not been analyzed either.
What is required to accurately document the changes in leaf shape between species, without undue biases in our understanding of the underlying biology, is a time series of changes in leaf shape at each node. We term the changes in shape that occur through time developmental trajectory, and it is this measure that should be studied with respect to the heteroblastic series and, ideally, under multiple environmental conditions that modulate the shape of leaves. One way to approximate a time series is to leverage natural variation in developmental rate, such that leaves at different nodes are sampled from multiple developmental time points from young and old plants.
Here, we present a comprehensive analysis of leaves, under a variety of developmental and environmental contexts, among wild tomato species using EFD. We look at six different factors that influence the morphology of leaflets borne on leaves, including the species from which leaves are sampled, the heteroblastic series, the proximal-distal and left-right axes of leaves, simulated sun versus foliar shade conditions, and developmental stage (Fig. 1). The complex leaves of tomato present an opportunity to study a multitude of developmental axes in the leaf, and we describe the effects of the proximal-distal and left-right axes of the leaf on leaflet form. Such effects cannot be documented in simple-leafed species. We find that the developmental stage of a plant has a profound effect on the shape of leaves arising from different nodes and is necessary to explain the differences in morphology that arise as a consequence of the shade avoidance response. Key to this insight is the leveraging of inherent differences in developmental rate to approximate a developmental series. We argue that the relevant differences between species and the response to foliar shade are alterations in the developmental trajectory of leaf morphogenesis. As studies of natural variation progress to discern subtle differences in leaf form between populations and species, we propose the importance of considering developmental trajectory as the underlying phenotype representative of morphological evolution.
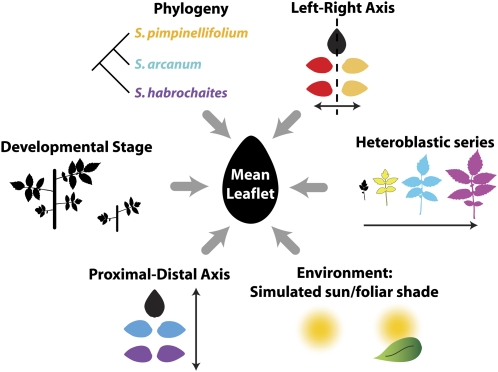
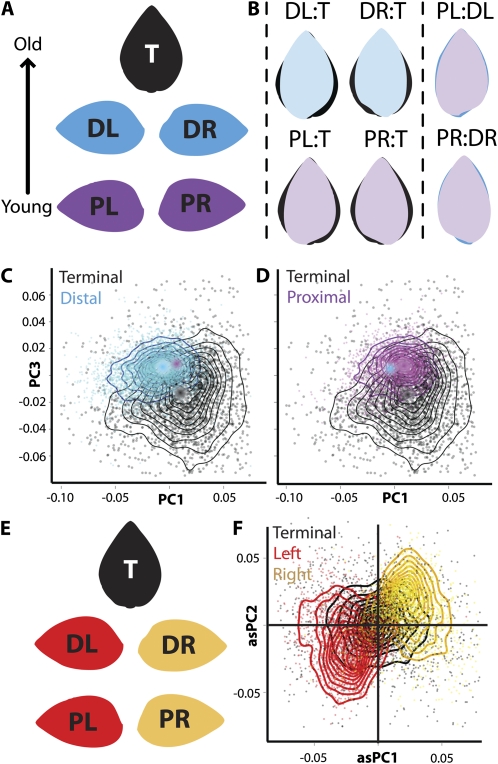
Figure 1.
Factors contributing to variation in leaflet shape. Phylogeny, the heteroblastic leaf series, the proximal-distal and left-right axes of leaves, environment, and developmental stage all contribute to leaflet shape. Shown is the mean leaflet resulting from the analysis of >11,000 leaflets in this study. Colors correspond to those used in other figures to depict factor levels.
RESULTS
Analysis of Leaflet Shape in Wild Tomato Species
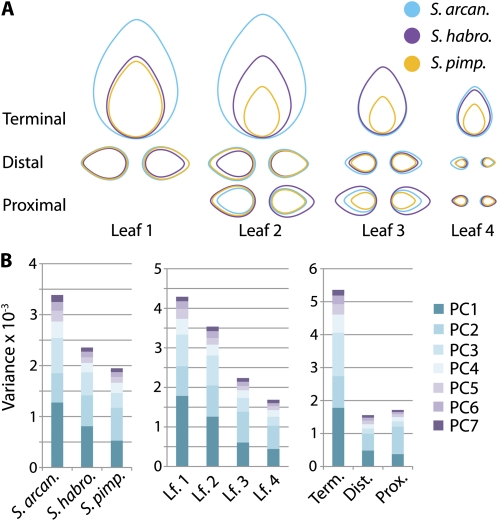
In a previous study, we analyzed the correlations between leaf dimensions and the environment in accessions of wild tomato species originating from Peru and Ecuador (Chitwood et al., 2012). Unlike the previous study that focused on leaf size, here we analyze the morphological attributes of leaflet shape. The accessions of wild tomato selected represent three different species—Solanum arcanum, Solanum habrochaites, and Solanum pimpinellifolium. Eight accessions of each species were chosen such that a diverse range of longitude, latitude, altitude, and vegetation density levels from their point of origin are represented. Importantly, the accessions chosen show a stunning array of leaflet morphs (Fig. 2) and natural variation is apparent at both inter- and intraspecific levels, as discussed in the next section.
Figure 2.
Variation in leaflet morphology across wild tomato accessions. Representative terminal leaflets from leaf 3 are shown for accessions used in this study. Species include S. arcanum (cyan), S. habrochaites (purple), and S. pimpinellifolium (gold). Note the extensive variation in leaflet shape, serration, and size. Both inter- and intraspecies variation in leaflet shape is apparent. [See online article for color version of this figure.]
To quantitatively analyze leaflet shape, we used an EFD-based method (Iwata et al., 1998; Iwata and Ukai, 2002). The Fourier analysis of shape decomposes outline information into a weighted sum of wave functions differing by frequency. The Fourier coefficients determine the contribution of each waveform to the analyzed shape, and lower-order harmonics describe overall shape whereas higher-order harmonics describe local variation in outlines. The coefficients therefore describe global and local features of outlines and are a quantitative means to analyze variance of shape in populations. An important advantage of EFD-based methods is that symmetric variance can be separated and analyzed independently from asymmetric variance in shape (Iwata et al., 1998). Additionally, the shape variation can be analyzed independent of size (Iwata and Ukai, 2002), as is done in this study.
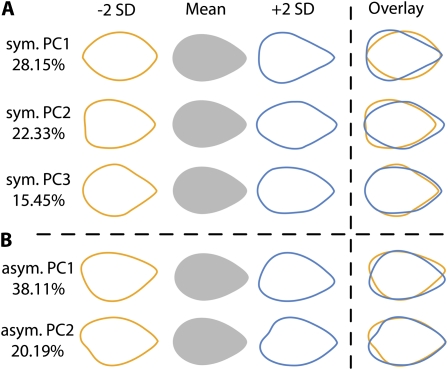
PCA is traditionally used to simplify and make comprehensible the variation in shape resulting from outline analyses. We chose to focus on only the first three symmetric principal components (PCs) and first two asymmetric PCs (Fig. 3) from the resulting EFDs as (1) the first PCs describe greater amounts of shape variance and therefore represent important attributes of shape that vary in the leaflets analyzed and (2) the PCs representing smaller amounts of shape variance become subject to noise, and are therefore difficult to interpret with respect to their underlying relevance to biology. Together, the first three symmetric PCs explain approximately two-thirds of all symmetric variance in shape, and each individual PC explains at least 15% of the variance (Supplemental Fig. S1). For the asymmetric PCs, more asymmetric shape variance is explained in the first PCs compared to the symmetric analysis. We only analyze the first two asymmetric PCs (explaining approximately 60% of asymmetric shape variance) to demonstrate the mirrored relationship between left and right leaflets.
Figure 3.
PCs explaining variance in leaflet shape. A, PCs 1 to 3 and the percent of symmetric shape variance in leaflets they explain. PCs are derived from EFDs that decompose and quantify shape variance in leaflet outlines. Shown are theoretical eigenleaves at values ±2 sds for each PC and their overlay. B, Similar to PCs explaining symmetric shape variance, shown are PCs 1 and 2 explaining asymmetric variance in leaflet shape.
We analyzed variance in shape with respect to six different factors, including species, the heteroblastic leaf series, the proximal-distal and left-right axes of leaves, developmental stage, and the shade avoidance response (Fig. 1). We noticed a skew in the distribution of some PCs (Supplemental Fig. S2), and variance in shape differed widely between leaflets by species, leaf node, and position on the proximal-distal axis of leaves, therefore violating assumptions of homoscedasticity. This prompted us to analyze our data nonparametrically. By a Kruskal-Wallis test, all factors except light treatment are significant predictors of PCs 1 to 3 (Table I). We detail at least two PCs for most factors, choosing which PCs to analyze based on their patterns of variance with respect to factor levels (Supplemental Figs. S3–S5).
Table I. P values for PCs varying by factors analyzed in this study.
Table of Kruskal-Wallis P values for PC1, PC2, and PC3 differing by factors analyzed in this study. Note the extremely significant P values for PCs by all factors. Despite relatively less significant P values compared to other factors, differences in shape with respect to light treatment are detected, especially for PC2.
| Factor | PC1 | PC2 | PC3 |
| Species | <2.2 × 10−16 | <2.2 × 10−16 | <2.2 × 10−16 |
| Leaf series | <2.2 × 10−16 | <2.2 × 10−16 | <2.2 × 10−16 |
| Proximal-distal axis | <2.2 × 10−16 | <2.2 × 10−16 | <2.2 × 10−16 |
| Leaf no. | <2.2 × 10−16 | <2.2 × 10−16 | <2.2 × 10−16 |
| Light treatment | 1.5 × 10−7 | 3.9 × 10−11 | 5.8 × 10−3 |
Leaflet Shape as a Function of Species
Of all the factors by which the leaflets we analyzed can be grouped, perhaps the most obvious is the species from which they are derived. Indeed, by simply working with different wild tomato accessions, distinct leaflet morphologies are evident. S. pimpinellifolium has been noted as having cordate (heart-shaped) leaves, with unique bulges on the proximal ends of its leaflet (Peralta et al., 2008) and a deltoid tip. S. habrochaites accessions generally possess obtuse leaflets, with relatively blunt tips and have an elliptic shape. The leaflets of S. arcanum accessions, on the other hand, tend to be relatively acuminate (with a tapering point) and thinner, sometimes to the point of being lanceolate or even acicular (lance or needle like). These observations, like many botanical descriptions, are qualitative and determined by individual researchers. Additionally, such species-level descriptions are, by their nature, generalized, and do not account for variation in shape among intraspecies populations and individuals, which is evident in the selected accessions (Fig. 2).
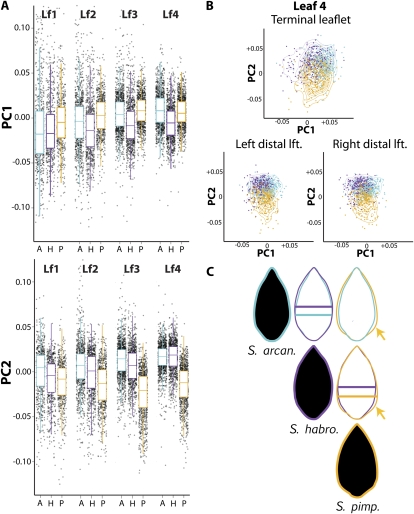
It is therefore worth commenting that, despite considerable variation within species, our analysis discriminates shape differences at a species level that quantify defining morphological characteristics that we and others have observed when working with wild tomato species. S. habrochaites, for example, has lower PC1 values than either S. pimpinellifolium or S. arcanum (Fig. 4A; Supplemental Fig. S3). Low PC1 values correspond to an obtuse, elliptic/ovate morph (Fig. 3A) whereas high PC1 values represent a more deltoid shape. If the mean outlines of leaflets from different species are compared it is apparent that the distribution of area in S. habrochaites leaflets is shifted distally relative to the other species (Fig. 4C, indicated by bars), accounting for its unique shape. S. pimpinellifolium, contrastingly, is defined by lower PC2 values (Fig. 4A). Low PC2 values correspond to a cordate morph with proximal leaflet bulges (Fig. 3A). Again, these features can be observed when comparing mean leaflet outlines, in which the proximal, cordate bulges of S. pimpinellifolium are evident compared to accessions from other species (Fig. 4C, indicated by arrows).
Figure 4.
Leaflet shape differences between wild tomato species. A, Box plots overlaid upon jitter plots for PC1 and PC2 in different species and leaves. Defining features for species include lower PC1 values associated with S. habrochaites leaflets and lower PC2 values in S. pimpinellifolium. A, S. arcanum; H, S. habrochaites; P, S. pimpinellifolium. B, Scatterplot of PCs 1 and 2 overlaid with contours to aid visualization showing the relative distribution of PC values by species separated by leaf 4 terminal and distal lateral leaflets. As for the per-leaf data shown in A, note that morphological distinctions between species are well preserved across the leaf series and the proximal-distal axis of leaves. C, Mean leaflet shapes for each species. Shape differences between species are evident in overlays. For example, the low PC2 values of S. pimpinellifolium predict a cordate morph, evidenced by a proximal bulge of laminar outgrowth (gold arrow). Similarly, PC values for S. habrochaites predict the distally shifted mass in its mean leaflets, indicated by bars spanning the widest point of leaflets for each species. Cyan, S. arcanum; purple, S. habrochaites; gold, S. pimpinellifolium.
In addition to the species-level shape differences apparent in these wild tomato accessions, it is also clear that they exhibit heteroblasty; that is, leaves emerging from different nodes have intrinsically different shapes (Poethig, 2003). Because of this, we wanted to determine if the differences we observe are particular to a node or leaflet type. This does not seem to be the case, and comparing PC values across the leaf series, the general shape contrasts between species are preserved (Fig. 4A). Additionally, analyzing leaflets along the proximal-distal axis of a single leaf (leaf 4, Fig. 4B), the general patterns between species remain constant. That is not to say that there are no differences in shape by node or leaflet position, just that the shape differences between species remain in a wide range of developmental contexts, which we discuss next.
Leaflet Shape as a Function of the Leaf Series
We next analyzed shape changes that occur in leaflets that arise on leaves at different nodes. In our analysis, we terminally harvest plants at a single time point. We later show how variation in developmental rate can be used as a surrogate for a time series, but first analyze difference in shape relating to node position. PC1 to 3 trend higher in leaves emerging from later nodes (Fig. 5, A and B). The trend of increasing values in PCs 1 to 3 almost universally holds for the different species, except notably for PC2 in S. pimpinellifolium (Fig. 4A), where instead of increasing in value PC2 decreases. This reflects the defining cordate morph of this species that evidently becomes more prominent in later emerging leaves. The increasing PC values in PCs 1 to 3 through the leaf series correspond to either thinner, more defined leaflet tips (PC1) or a more even distribution of weight throughout the leaflet (PCs 2 and 3; Fig. 3). Collectively, these shape changes correspond to a thinner leaflet morph relative to more orbicular leaflets that arise from the first leaves in the series (Fig. 5C).
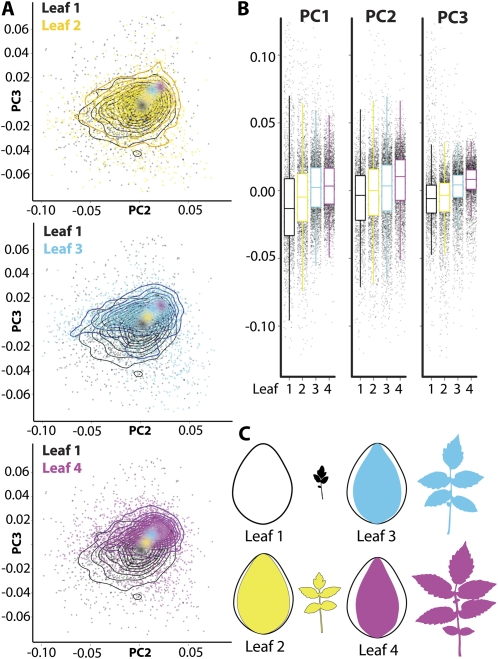
Figure 5.
Leaflet shape differences across the heteroblastic series. A, Scatterplots overlaid with contours to aid visualization. Shown are PC2 and PC3, which increase in value across the leaf series. Data are highly variable, but increasing PC values can be seen by colored dots indicating mean position of each leaf in PC space. Shown are plots of each leaf in turn plotted against leaf 1. B, Box plots overlaid on jitter plots for PC 1 to 3 values by leaf. Note increasing PC values across the leaf series. C, Mean leaflet shapes across the leaf series juxtaposed upon leaf 1. Note the increasing thinness of leaflets progressing through the leaf series. Black, leaf 1; yellow, leaf 2; cyan, leaf 3; magenta, leaf 4.
Leaflet Shape as a Function of the Proximal-Distal Axis and Left-Right Asymmetries
With respect to leaf morphology, the compound leaves of tomato present a developmental situation more complex relative to simple leaves. The rachis of the complex leaf is a developmental axis in itself, similar to the heteroblastic series just described. Like the leaf series, the proximal-distal axis of each compound leaf from which leaflets arise can be confounded for (1) intrinsic differences between leaflet shape at varying positions and (2) the developmental stage of the leaflets, and we discuss later how variance in developmental rate can resolve these contributing factors. In the case of tomato species, the leaf develops basipetally (that is, the distal tip of the leaf is developmentally older than the proximal base). The most proximal pair of lateral leaflets is always younger than the distal pair, and in turn the distal pair of lateral leaflets is younger than the terminal leaflet. Additionally, the terminal leaflet is (on average) symmetrical, whereas the lateral leaflets exhibit asymmetries (Fig. 6).
Figure 6.
Leaflet shape differences across the proximal-distal and left-right axes of leaves. A, Mean leaflet shapes of terminal (T, black), distal lateral (D, blue), and proximal lateral (P, purple) leaflets showing their arrangement in leaves. L and R indicate left and right, respectively. Arrow indicates the basipetal development of tomato leaves. B, Mean leaflet shapes of terminal, distal, and proximal lateral leaflets juxtaposed upon each other. Note that, like the heteroblastic series, younger leaflets are thinner relative to the older terminal leaflet. C and D, Scatterplots overlaid with contours to aid visualization of PC1 and PC3. Shape differences between distal and proximal lateral leaflets are small, but high variance and low PC3 values are unique to terminal leaflets. Proximal (C) and distal (D) leaflet points plotted against terminal leaflets. Dots indicate mean values of leaflets, as indicated by color. E, Mean leaflet shapes of terminal (T, black), left (L, red), and right (R, gold) leaflets showing their arrangement in leaves. F, Scatterplot overlaid with contours to aid visualization of asymmetric PC1 and PC2. Note that left and right leaflets are easily separated by asymmetric PCs, and that on average terminal leaflets are symmetric, centered at the origin.
Like the leaf series, there are symmetrical differences in leaflet shape between the older, first-emerged leaflets (beginning with the terminal leaflet) and younger, later-emerged leaflets (for our analysis, ending with the most proximal pair of lateral leaflets; Supplemental Fig. S4). These shape differences are predominated by extremely high shape variance in the terminal leaflet relative to the lateral leaflets (Fig. 6, C and D; and discussed in the next section). However, there are intrinsic shape differences between leaflet types separate from variance. Most notably, terminal leaflets have lower PC3 values relative to lateral leaflets, which correspond to bulges in the blade near the midpoint of the terminal leaflet (Fig. 3A). We interpret this quantitative difference to represent the lobing present in terminal relative to lateral leaflets. This lobing in its severest form can manifest as a trifoliate-like morph. Differences between the distal and proximal pairs of lateral leaflets are slight (Fig. 6, B–D). Nonetheless, juxtaposing leaflet shape means upon each other, it is interesting that lateral leaflets become less round and more acicular and lanceolate relative to the terminal leaflet (Fig. 6B). This is reminiscent of the less-round leaves traversing the leaf series (Fig. 5C), and for both the leaf series and the leaf proximal-distal axis, the rounder/orbicular morph is associated with the older, first-emerged organ.
The most obvious difference between the terminal and lateral leaflets is their symmetry: Terminal leaflets are symmetrical, whereas the left and right lateral leaflets are asymmetrical and are mirror images of each other (Fig. 6E). The asymmetry in lateral leaflets is such that they bulge at their base, on the side facing the proximal end of the leaf (Fig. 6). Although speculative, the source of the asymmetry may be auxin based, as the net flow of auxin in the leaf primordium during leaflet formation is in a basipetal direction, and the asymmetric bulges may be an effect of auxin drag. To eliminate noise from our analysis and focus on true morphological differences, the previous mentioned PCA focuses only on symmetrical variance in the data. However, if only asymmetric aspects of the variance are analyzed, the resulting PCs easily separate right and left lateral leaflets from each other as having, on average, opposite signed PC values from each other (Figs. 3B and 6F). Contrastingly, the terminal leaflet is centered on the origin of asymmetric PCs. Asymmetries in leaves remain an understudied aspect of leaf development. Analyses such as EFD, which can quantitatively isolate variance relating to asymmetry, promise to be a powerful tool in the future to study the basis of leaf asymmetry.
Variance and Canalization
While the simplest analysis is to look at differences in leaflet morphs in terms of organ position and species, other factors might be more relevant to underlying developmental mechanisms. Two such factors are variance and temporal developmental trajectories.
Although shape differences in leaflets by species (Fig. 4), the leaf series (Fig. 5), and the proximal-distal axis (Fig. 6) are evident, an obvious trait separating the leaflets at a population level for these factors is variance. By eye, it is obvious that terminal leaflets, leaflets arising off leaves from the first nodes, and the leaflets of S. arcanum are all more variable than their counterparts. The first two leaves of all the accessions examined were noticeably amorphous compared to leaves later in the series. Looking at the variances in PCs for different leaflet types by species, these trends are evident (Fig. 7A; Supplemental Table S1). That the trends in variance are apparent in all PCs, each of which explains a distinct aspect of shape, suggests that the variance reflects inherent randomness in the system (Fig. 7B). The randomness in shape, like nonrandom patterns in shape, exhibits similar trends in both the heteroblastic series and proximal-distal axis, with variance decreasing through the leaf series and contrasting degrees of variance between the terminal and lateral leaflets (Fig. 7B). At least for the heteroblastic series, the decreasing variance through the series might be a symptom of canalization. Phyllotactic patterning is known to be irregular in the first initiated leaves of a shoot as a result of instabilities in auxin transport, which also affects the patterning of leaves along their proximal-distal axis (Reinhardt et al., 2003; Smith et al., 2006; Barkoulas et al., 2008; Koenig et al., 2009).
Figure 7.
Variance by species, the leaf series, and leaflet position along the proximal-distal axis of leaves. A, Diagrammatic leaflet area is proportional to variance in PC1 values for each leaflet shown. Note the increased variance in S. arcanum leaflets and leaflets arising from leaves in the beginning of the series. Also, terminal leaflets have large variance relative to others. Cyan, S. arcanum; purple, S. habrochaites; gold, S. pimpinellifolium. B, Variance in PCs 1 to 7, representing >85% of all symmetric shape variance, showing (left to right) trends in variance by species, node, and the placement of leaflets along the proximal-distal axis of leaves.
Resolving Developmental Stage and Series: Developmental Trajectory
The trends in variance across the leaf series and proximal-distal axis of leaves bring up again the confounding of developmental processes in these axes. In an analysis of any trait for these axes at a single time point, heteroblasty and developmental stage will be confounded. The resolution of these two processes requires sampling leaves at each node for multiple time points. The design of such an experiment reflects the underlying developmental processes that give rise to variation in leaf morphology between populations and species: the differences between reiterated organs throughout their development. We term the resulting spatiotemporal phenotype the developmental trajectory of a trait.
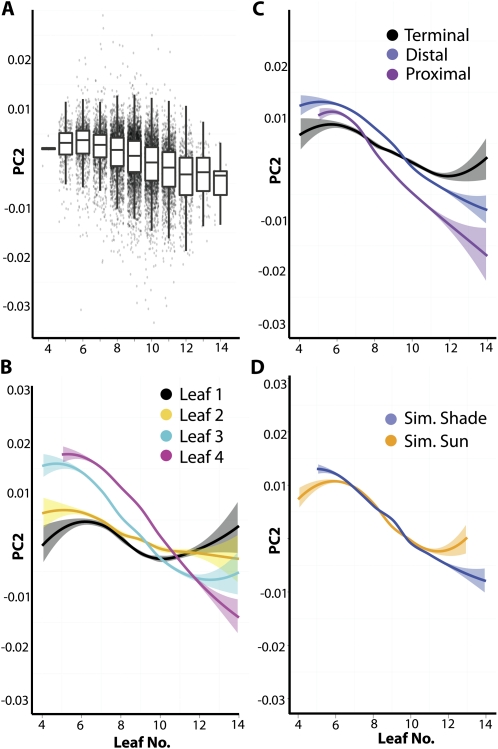
Developmental stage can be defined by any number of traits, but the proxy that we use for this study is leaf number, as measured by all observable primordia (including millimeter-length leaves, corresponding to approximately P4 and 5). If PCs are analyzed with respect to leaf number, PC2 shows an interesting pattern that the other PCs do not (Fig. 8A; Supplemental Fig. S5). In plants with few leaves (slow growers, early in developmental time), PC2 values are relatively high. As one progresses to plants with more leaves (faster growers, relatively farther in developmental time), PC2 values decrease in a sigmoidal fashion reminiscent of growth rate (Fig. 8A). As plants with fewer leaves are earlier in their development than those with many leaves, the changes in the overall shape of leaves across leaf number (developmental time) reflect changes in shape during development: a developmental trajectory.
Figure 8.
Developmental trajectory in leaflet shape across the heteroblastic series, the proximal-distal axis of leaves, and the shade avoidance response. A, PC2 values show a unique pattern of variance across plants early or late in their development, as indicated by leaf number. Shown are box plots overlaid on jitter plots, showing a sigmoidal decrease in PC2 values in plants with higher leaf numbers. B, Loess regression of PC2 values versus leaf number by leaf. Note that mature leaves, such as leaf 1 and 2, have stable PC2 values, whereas leaves 3 and 4 exhibit more dynamic PC2 values, indicating growth across the developmental stages sampled. Light coloring indicates se. Black, leaf 1; yellow, leaf 2; cyan, leaf 3; magenta, leaf 4. C, Loess regression for PC2 values versus leaf number by leaflet position along the proximal-distal axis. Terminal leaflets, older relative to distal and proximal lateral leaflets, exhibit relatively stable PC2 values. Black, terminal leaflet; blue, distal lateral leaflet; purple, proximal lateral leaflet. D, Loess regression of simulated foliar shade and sun leaves. Shade avoidance is characterized by two different aspects indicated by the developmental trajectory. Like the younger leaves in the leaf series and proximal-distal axis, shade avoided leaves have dynamic PC2 values across the developmental stages sampled. Additionally, shade leaves have lower terminal PC2 values, indicating an accelerated transition to adult leaflet forms, consistent with shade-induced acceleration toward flowering. Blue, simulated foliar shade; orange, simulated sun.
This approximation of developmental time is one way to begin separating the confounding of differences between organs in a series and their developmental stage. For instance, separating PC2 values by leaf and leaf number (Fig. 8B), a number of predictable trends are observed. The oldest organs (in this case, leaves 1 and 2) are relatively static in their shape across the developmental stages sampled. This is because, as the first-emerged organs, leaves 1 and 2 were sampled at a relatively mature stage, probably during the plateau stage of their growth. Contrastingly, leaves 3 and 4, as newly emerged organs, would either be extremely young (from plants with fewer leaves) or older (from plants with many leaves). As expected if PC2 reflects a shape component that changes as leaves develop, leaves 3 and 4 show a greater range of PC2 values across plants with different leaf numbers. These trends are exhibited in another developmental axis, the proximal-distal axis of leaves (Fig. 8C). Note that the terminal leaflets (the oldest leaflets on this axis) show relatively stagnant PC2 values across developmental time, just like leaflets from leaves 1 or 2. As one progresses to younger leaflets (to the distal lateral leaflets, and then to the proximal lateral leaflets), the changes in PC2 values across developmental time become more severe, reflecting the exponential phase of growth these organs are traversing through during the sampled developmental times relative to the older, terminal leaflets.
The slopes of these curves and their movement through a Cartesian plane defined by developmental stage and shape is the developmental trajectory of the organ. However, if we wanted to isolate the effects of the heteroblastic series from developmental trajectory, we would look to the shape differences between organs in plants at the most mature stage of development. Just as leaves 1 and 2 show relatively stable shape across developmental time, presumably the shape of leaves 3 and 4 in plants with the highest leaf numbers reflects intrinsic shape differences between different leaves, as all leaves are compared at a mature stage. This is also true for the proximal-distal axis. In this regard, it is interesting that as one traverses the leaf series or the proximal-distal axis from the first-emerged leaves to the last, that the defining shape component is decreasing PC2 values (Fig. 8, B and C). However, the opposite pattern is observed earlier in developmental time, reflecting the increased PC2 values characteristic of young leaves.
A quantitative framework for the changes in leaf shape that occur during normal development is a prerequisite for understanding the changes in shape induced by genetic or environmental changes. The differences in shape between leaves in simulated sun or simulated foliar shade conditions, although highly significant, were the least statistically supported of factors analyzed, demonstrating the subtlety of these effects on shape (Table I). It is interesting that of all the PCs, PC2 is the most significant for the shade avoidance response.
There is a long-standing debate as to whether differences between sun and shade leaves reflect changes in the types of leaves produced (heteroblastic differences, such as a prolonged production of juvenile leaves) or changes in the developmental progression of leaf forms (either differences in developmental rate or plasticity in leaf form in response to the environment). Beginning with Goebel, the first interpretations of shade response hypothesized that leaf shape changes resulted from prolonged juvenility, and that shade induces the production of juvenile forms at later shoot positions from the inception of the leaf (Goebel, 1908; Ashby, 1948; Njoku, 1956; Montaldi et al., 1963; Cameron, 1970). Contrastingly, subsequent analyses hinted at a developmental component, that leaves initiate a normal heteroblastic progression in the shade, but through subsequent development and plasticity, appear juvenile in their mature forms (Jones, 1995). It should be noted that in the majority of these studies, it is light intensity, rather than changes in light quality that were tested. Many of the hypotheses concerning prolonged juvenility focus on nutrient availability and the ability to produce adult forms. This is not necessarily a concern under our light conditions, as photosynthetically active radiation is equal under the two light treatments (only the red to far-red ratios of light change). Nonetheless, these classic studies are the most relevant insights we have into plasticity of leaf form in response to light.
Looking at developmental trajectory, which reflects both effects of the heteroblastic series and developmental rate, it seems that both processes may be at work in the leaves of wild tomato species during the shade avoidance response (Fig. 8D). The overall developmental trajectory of shade leaves resembles that of younger, newly emerged adult leaves. With respect to shape, it is as if the development of leaves was prolonged, as if they defer the mature leaf form and are proceeding through an exponential phase of growth relative to sun leaves. This is reflected in the wider range, and more severe changes in PC2 values across developmental stages in shade leaves (Fig. 8D). Looking at the terminal, mature PC2 values of shade leaves that arise from older plants, shade leaves appear shifted to leaf forms that usually appear later in the leaf series relative to sun leaves. That is, rather than prolonged juvenility, it seems that simulated foliar shade instead stimulates the production of adult leaf forms, progressing through the heteroblastic series more rapidly. Especially in tomato, in which reproductive development is connected with transitions to sympodial growth, we might expect that the early progression to flowering during shade avoidance would be associated with rapid transitions from juvenile to adult leaf forms, exactly as we observe (Fig. 8D). Indeed, genetic or environmental regulation of flowering time is known to modulate vegetative phase change (Willmann and Poethig, 2011).
DISCUSSION
Our results demonstrate the importance of accounting for both (1) developmental time and (2) the heteroblastic series when defining morphological differences between environmental conditions and/or genotypes. The response in leaf shape to foliar shade is exemplary of the developmental mechanisms that differ between populations and species that can mask intrinsic shape variation. Not only is developmental rate accelerated under simulated foliar shade (Chitwood et al., 2012), but there are heteroblastic differences as well. This leads to the observation that shade leaves appear to be under extended growth and have shape characteristics of leaves that under normal conditions emerge later in the heteroblastic series.
Especially with regard to the study of natural variation, inherent differences in developmental rate can be leveraged to approximate a developmental series. The incorporation of developmental stage into models of traits can be important to correct for differences in developmental rate. In a previous study, we found that correcting for slow- versus fast-growing tomato accessions was vital to uncover correlations between the dimensions of leaves and the environment (Chitwood et al., 2012). Here, we use a similar strategy to enhance our understanding of how shape changes in leaflets occur through developmental time, the leaf series, and the proximal-distal axis. The sampling of all leaflets from the first four leaves of 24 accessions of tomato (representing three different species) under two light treatments in high replication would be near impossible to achieve across different time points. In this scheme of a replicated single time point, >11,000 leaflets from approximately 726 plants were analyzed in total. We reason that large-scale natural variation experiments will always suffer from the drawback of terminal harvest points, but usually such experiments have a large degree of variance in developmental rate. Instead of masking relevant developmental mechanisms, this variation should be leveraged to maximize the information gathered from such experiments.
Although intrinsic shape differences between species are apparent (Fig. 4), unless these changes occur ubiquitously across all developmental contexts, sampling single developmental instances will undoubtedly lead to biases in the perception of actual underlying variation between species. Differences between species may show varying patterns across the heteroblastic leaf series (Fig. 5). In the case of complex leafed species such as tomato, leaflet shape between populations may also manifest differentially with respect to positioning along the proximal-distal axis of leaves (Fig. 6). Additionally, we document less-studied aspects of leaflet shape that nonetheless vary by species and developmental context, including leaf asymmetry and variance (Figs. 6 and 7). It is not unreasonable to suspect that the differences in size, shape, and complexity of leaves between species and populations exhibit similar underlying complexities, and that it is developmental trajectory and other elemental mechanisms that we should aim to study when describing natural variation.
MATERIALS AND METHODS
Growth Conditions
Accessions were obtained from the C.M. Rick Tomato Genetics Resource Center (University of California, Davis), which maintains accessions as an outcrossing population of 10 individuals. Plants were grown in a (Conviron) walk-in chamber in which the left and right sides were divided into low and high red to far-red light treatments, which we refer to as simulated foliar shade and simulated sun. Each side consisted of six shelves fitting five 11 × 22 inch trays each. Temperature was adjusted to 22°C and photoperiod to a 16:8 h light-dark cycle. Lighting consisted of alternating fluorescent (F48T12CWHO) and far-red (F48T12FRHO, peak emission 750 nm, Interlectric) bulbs. High red to far-red wavelength ratios were achieved by blocking far-red irradiance with sleeves whereas all bulbs (both normal fluorescent and far red) transmitted light in the low-red to far-red treatment. Shade cover was placed perpendicularly over bulbs in the low-red to far-red treatment (simulated foliar shade) to adjust overall light intensity to match that of the high-red to far-red treatment (simulated sun).
Wild tomato (Solanum section Lycopersicon) seed was sterilized for 10 min in 50% bleach, washed in autoclaved water, and subsequently plated on one-half Murashige and Skoog plates. Plates were placed in darkness covered in foil at ambient room temperature for 3 d and then placed into the simulated sun treatment for 9 d, after which seedlings were transferred one per approximately 5 × 5 inch pots placed eight per 11 × 22 inch trays in Sunshine (SunGro) soil mix. Plants were watered by filling trays with just enough water to cover the bottoms of pots and waiting for water to evaporate before watering again. Plants were harvested for phenotypic analysis starting 28 d postplating.
Two experiments were performed to replicate results, and the sides of the chamber used for simulated sun and simulated foliar shade treatments were switched between each experiment. Up to 10 seedlings per accession per treatment per experiment were transplanted for analysis. The mean and median numbers of plants analyzed under the simulated sun treatment overall for each accession was 15.3 and 16, respectively; for the simulated foliar shade treatment these statistics were 14.9 and 15, respectively. A total of 726 plants were analyzed in total.
Photography
Leaf number was counted on all measured plants and included young primordia that could be observed by eye (approximately 2–3 mm in overall length) that likely corresponded to P4 and P5 (that is, the fourth and fifth oldest leaf primordia). At the time of harvest, only the first four leaves of each plant had expanded sufficiently for analysis. Leaves were dissected, placed under nonreflective glass, and their terminal and primary leaflets removed at the base. Olympus SP-500 UZ cameras were mounted on copy stands (Adorama, 36-inch deluxe copy stand) and controlled remotely by computer using Cam2Com software (Sabsik).
EFD Analysis
For shape analysis, photographs were first converted to binary form using ImageJ (Abramoff et al., 2004) and individual leaflets extracted from the leaf series and named appropriately as separate files. Over 11,000 leaflets were analyzed in total.
The analysis of leaflet shape was conducted using EFD followed by PCA using the program SHAPE (Iwata and Ukai, 2002). Object contours were extracted as chain code. Chain code was subsequently used to calculate normalized EFDs. Normalization was based upon manual orientation with respect to the proximal-distal axis of the leaflet. PCA was performed on the EFDs resulting from the first 20 harmonics of Fourier coefficients. For the analysis of symmetrical shape, a and d coefficients were analyzed, while for analysis of asymmetrical shape, b and c coefficients were analyzed (Iwata et al., 1998). Coefficients of EFD were calculated at −2 and +2 sds for each PC and the respective contour shapes reconstructed from an inverse Fourier transformation. PCs were then analyzed for statistical differences between various factors.
All basic statistic functions were performed in R (R Development Core Team, 2011) and visualized in the package ggplot2 (Wickham, 2009). PCs arising from EFD for tomato leaflets were not normal, and statistical significance was determined individually for each explanatory variable using nonparametric tests.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Symmetric and asymmetric variance explained by each PC.
Supplemental Figure S2. Distribution of PC values.
Supplemental Figure S3. PC values by species.
Supplemental Figure S4. PC values across the proximal-distal axis of leaves.
Supplemental Figure S5. PC values by developmental stage.
Supplemental Table S1. Variance in PC values by leaflet type and species.
References
- Abramoff MD, Magalhaes PJ, Ram SJ. (2004) Image processing with ImageJ. Biophotonics International 11: 36–42 [Google Scholar]
- Ashby E. (1948) Studies in the morphogenesis of leaves. I. An essay on leaf shape. New Phytol 47: 152–176 [Google Scholar]
- Barkoulas M, Hay A, Kougioumoutzi E, Tsiantis M. (2008) A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat Genet 40: 1136–1141 [DOI] [PubMed] [Google Scholar]
- Bharathan G, Goliber TE, Moore C, Kessler S, Pham T, Sinha NR. (2002) Homologies in leaf form inferred from KNOXI gene expression during development. Science 296: 1858–1860 [DOI] [PubMed] [Google Scholar]
- Blein T, Pulido A, Vialette-Guiraud A, Nikovics K, Morin H, Hay A, Johansen IE, Tsiantis M, Laufs P. (2008) A conserved molecular framework for compound leaf development. Science 322: 1835–1839 [DOI] [PubMed] [Google Scholar]
- Cameron RJ. (1970) Light intensity and the growth of Eucalyptus seedlings. I. Ontogenic variation in E. fastigata. Aust J Bot 18: 29–43 [Google Scholar]
- Champagne C, Sinha N. (2004) Compound leaves: equal to the sum of their parts? Development 131: 4401–4412 [DOI] [PubMed] [Google Scholar]
- Chitwood DH, Headland LR, Filiault DL, Kumar R, Jimenez-Gomez J, Schrager AV, Park DS, Peng J, Sinha NR, Maloof JN. (2012) Native environment modulates leaf size and response to simulated foliar shade across wild tomato species. PLoS ONE 7: e29570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui ML, Copsey L, Green AA, Bangham JA, Coen E. (2010) Quantitative control of organ shape by combinatorial gene activity. PLoS Biol 8: e1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Wilson Y, Bowers J, Kennaway R, Bangham A, Hannah A, Coen E, Hudson A. (2009) Evolution of allometry in antirrhinum. Plant Cell 21: 2999–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel K. (1908) Einleitung in die Experimentelle Morphologie der Pflanzen. B.G. Teubner, Leipzig, Germany [Google Scholar]
- Iwata H, Nesumi H, Ninomiya S, Takano Y, Ukai Y. (2002) The evaluation of genotype x environment interaction of citrus leaf morphology using image analysis and Elliptic Fourier Descriptors. Breed Sci 52: 243–251 [Google Scholar]
- Iwata H, Niikura S, Matsuura S, Takano Y, Ukai Y. (1998) Evaluation of variation of root shape of Japanese radish (Raphanus sativus L.) based on image analysis using Elliptic Fourier Descriptors. Euphytica 102: 143–149 [Google Scholar]
- Iwata H, Ukai Y. (2002) SHAPE: a computer program package for quantitative evaluation of biological shapes based on elliptic Fourier descriptors. J Hered 93: 384–385 [DOI] [PubMed] [Google Scholar]
- Jones CS. (1995) Does shade prolong juvenile development? A morphological analysis of leaf shape changes in Cucurbita argyrosperma subsp. Sororia (Cucurbitaceae). Am J Bot 82: 346–359 [Google Scholar]
- Kimura S, Koenig D, Kang J, Yoong FY, Sinha N. (2008) Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr Biol 18: 672–677 [DOI] [PubMed] [Google Scholar]
- Klingenberg CP, Leamy LJ, Routman EJ, Cheverud JM. (2001) Genetic architecture of mandible shape in mice: effects of quantitative trait loci analyzed by geometric morphometrics. Genetics 157: 785–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig D, Bayer E, Kang J, Kuhlemeier C, Sinha N. (2009) Auxin patterns Solanum lycopersicum leaf morphogenesis. Development 136: 2997–3006 [DOI] [PubMed] [Google Scholar]
- Kuhl FP, Giardina CR. (1982) Elliptic Fourier features of a closed contour. Computer Graphics and Image Processing 18: 236–258 [Google Scholar]
- Langlade NB, Feng X, Dransfield T, Copsey L, Hanna AI, Thébaud C, Bangham A, Hudson A, Coen E. (2005) Evolution through genetically controlled allometry space. Proc Natl Acad Sci USA 102: 10221–10226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montaldi ER, Caso OH, Lewin IJ. (1963) Algunos factores que afectan la morfologia de las hojas en una planta de desarrollo heteroblastico. Revisita de Investigaciones Agricoles (Buenos Aires) 17: 321–340 [Google Scholar]
- Nath U, Crawford BC, Carpenter R, Coen E. (2003) Genetic control of surface curvature. Science 299: 1404–1407 [DOI] [PubMed] [Google Scholar]
- Nicotra AB, Leigh A, Boyce CK, Jones CS, Niklas KJ, Royer DL, Tsukaya H. (2011) The evolution and functional significance of leaf shape in the angiosperms. Funct Plant Biol 38: 535–552 [DOI] [PubMed] [Google Scholar]
- Njoku E. (1956) Studies in the morphogenesis of leaves. XI. The effect of light intensity on leaf shape in Ipomoea caerulea. New Phytol 55: 91–110 [Google Scholar]
- Ori N, Cohen AR, Etzioni A, Brand A, Yanai O, Shleizer S, Menda N, Amsellem Z, Efroni I, Pekker I, et al. (2007) Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet 39: 787–791 [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Peralta IE, Spooner DM, Knapp S. (2008) Taxonomy of wild tomatoes and their relatives (Solanum sect. Lycopersicoides, sect. Juglandifolia, sect. Lycopersicon Solanaceae). Syst Bot Monogr 84: 1–186 [Google Scholar]
- Piazza P, Bailey CD, Cartolano M, Krieger J, Cao J, Ossowski S, Schneeberger K, He F, de Meaux J, Hall N, et al. (2010) Arabidopsis thaliana leaf form evolved via loss of KNOX expression in leaves in association with a selective sweep. Curr Biol 20: 2223–2228 [DOI] [PubMed] [Google Scholar]
- Poethig RS. (2003) Phase change and the regulation of developmental timing in plants. Science 301: 334–336 [DOI] [PubMed] [Google Scholar]
- Pryer KM, Hearn DJ. (2009) Evolution of leaf form in marsileaceous ferns: evidence for heterochrony. Evolution 63: 498–513 [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2011) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C. (2003) Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Rosas U, Barton NH, Copsey L, Barbier de Reuille P, Coen E. (2010) Cryptic variation between species and the basis of hybrid performance. PLoS Biol 8: e1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RS, Guyomarc’h S, Mandel T, Reinhardt D, Kuhlemeier C, Prusinkiewicz P. (2006) A plausible model of phyllotaxis. Proc Natl Acad Sci USA 103: 1301–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F, Bradbury PJ, Brown PJ, Hung H, Sun Q, Flint-Garcia S, Rocheford TR, McMullen MD, Holland JB, Buckler ES. (2011) Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat Genet 43: 159–162 [DOI] [PubMed] [Google Scholar]
- Tsuge T, Tsukaya H, Uchimiya H. (1996) Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 122: 1589–1600 [DOI] [PubMed] [Google Scholar]
- Weight C, Parnham D, Waites R. (2008) LeafAnalyser: a computational method for rapid and large-scale analyses of leaf shape variation. Plant J 53: 578–586 [DOI] [PubMed] [Google Scholar]
- Wickham H. (2009) ggplot2: Elegant Graphics for Data Analysis. Springer, New York
- Willmann MR, Poethig RS. (2011) The effect of the floral repressor FLC on the timing and progression of vegetative phase change in Arabidopsis. Development 138: 677–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman E, Palsson A, Gibson G. (2000) Quantitative trait loci affecting components of wing shape in Drosophila melanogaster. Genetics 155: 671–683 [DOI] [PMC free article] [PubMed] [Google Scholar]