Abstract
All higher plants contain an ent-kaurene oxidase (KO), as such a cytochrome P450 (CYP) 701 family member is required for gibberellin (GA) phytohormone biosynthesis. While gene expansion and functional diversification of GA-biosynthesis-derived diterpene synthases into more specialized metabolism has been demonstrated, no functionally divergent KO/CYP701 homologs have been previously identified. Rice (Oryza sativa) contains five CYP701A subfamily members in its genome, despite the fact that only one (OsKO2/CYP701A6) is required for GA biosynthesis. Here we demonstrate that one of the other rice CYP701A subfamily members, OsKOL4/CYP701A8, does not catalyze the prototypical conversion of the ent-kaurene C4α-methyl to a carboxylic acid, but instead carries out hydroxylation at the nearby C3α position in a number of related diterpenes. In particular, under conditions where OsKO2 catalyzes the expected conversion of ent-kaurene to ent-kaurenoic acid required for GA biosynthesis, OsKOL4 instead efficiently reacts with ent-sandaracopimaradiene and ent-cassadiene to produce the corresponding C3α-hydroxylated diterpenoids. These compounds are expected intermediates in biosynthesis of the oryzalexin and phytocassane families of rice antifungal phytoalexins, respectively, and can be detected in rice plants under the appropriate conditions. Thus, it appears that OsKOL4 plays a role in the more specialized diterpenoid metabolism of rice, and our results provide evidence for divergence of a KO/CYP701 family member from GA biosynthesis. This further expands the range of enzymes recruited from the ancestral GA primary pathway to the more complex and specialized labdane-related diterpenoid metabolic network found in rice.
The are phytohormones required for normal plant growth and development in all higher plants (Yamaguchi, 2008). Accordingly, the genes encoding the relevant biosynthetic enzymes must be present in all plant genomes. These have provided a genetic reservoir from which other, more specialized diterpenoid metabolism has evolved (Peters, 2010). Indeed, such derivation of more specialized metabolism from hormone biosynthesis appears to be an emerging theme in plant metabolism (Chu et al., 2011).
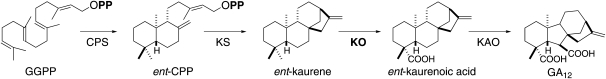
GA biosynthesis is initiated by cyclization of the general diterpenoid precursor (E,E,E)-geranylgeranyl diphosphate to ent-labdadienyl/copalyl diphosphate (ent-CPP), leading to designation of the derived natural products as labdane-related diterpenoids (Peters, 2010). The GAs are more precisely defined by their tetracyclic 6-5-6-5 ring structure (MacMillan and Takahashi, 1968). However, their biosynthesis proceeds through the 6-6-6-5 tetracycle ent-kaur-16-ene, which is formed from the bicyclic ent-CPP by ent-kaurene synthases (KSs; Yamaguchi, 2008). The ring contraction catalyzed by the cytochrome P450 (CYP) ent-kaurenoic acid oxidase (KAO) is then the committed step in GA-specific biosynthesis (Fig. 1). Hence, the upstream enzymes, ent-CPP synthase (CPS), KS, and kaurene oxidase (KO)/CYP701, can be diverted to alternative labdane-related diterpenoid biosynthesis. Many examples of CPS and KS-like (KSL) diterpene synthases that operate in more specialized metabolism are known (Cho et al., 2004; Nemoto et al., 2004; Otomo et al., 2004a, 2004b; Prisic et al., 2004; Wilderman et al., 2004; Xu et al., 2004, 2007; Harris et al., 2005; Kanno et al., 2006; Morrone et al., 2006; Gao et al., 2009; Toyomasu et al., 2009; Falara et al., 2010). By contrast, no KO/CYP701 family member has been shown to act as anything other than a KO—i.e. catalyze conversion of C19 to a carboxylic acid in ent-kaurene.
Figure 1.
GA biosynthesis. Shown are the three pre-GA steps catalyzed by the two relevant diterpene synthases, CPS and KS, and CYP KO/CYP701, as well as formation of the characteristic 6-5-6-5 tetracyclic ring structure by the subsequently acting KAO.
Rice (Oryza sativa) produces more than 20 labdane-related diterpenoids that act as antimicrobial phytoalexins (Peters, 2006; Toyomasu, 2008). Sequencing of the rice genome demonstrated the presence of multiple CPS, KS(L), and KO family members (IRGSP, 2005), although only one of each is involved in GA biosynthesis (Sakamoto et al., 2004). Biochemical characterization of the rice CPS and KS(L) families has revealed the expected functional divergence—i.e. as necessary for the known production of alternative labdane-related diterpenoids (Peters, 2006; Toyomasu, 2008). However, these diterpene synthases produce olefins that require further elaboration, particularly the insertion of oxygen by CYP (Kato et al., 1995), to form bioactive phytoalexins (Peters, 2006).
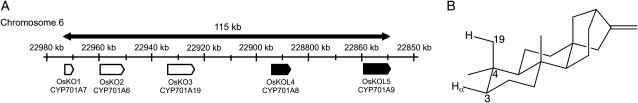
Intriguingly, the rice genome contains multiple paralogs of KO/CYP701 as a five-gene tandem array on chromosome 6 (Fig. 2A), despite the fact that only one (CYP701A6, termed OsKO2, as enumerated by chromosomal order) is apparently involved in GA biosynthesis (Sakamoto et al., 2004). Notably, almost all of the rice labdane-related diterpenoid phytoalexins contain oxygen at C3, and in many cases this can be traced back to insertion/hydroxylation at the C3α (pro-R) position (Peters, 2006), which is very close to the C4α-methyl (i.e. C19) that KO acts upon (Fig. 2B). In addition, the two more phylogenetically distant rice KO paralogs (thus termed KO like [KOL], hence OsKOL4/CYP701A8 and OsKOL5/CYP701A9) exhibit inducible transcription in response to elicitation with the fungal cell wall component chitin oligosaccharide, and OsKOL4 is not able to complement the GA biosynthetic deficiency of an OsKO2 mutant, although the more closely related OsKO1/CYP701A7 can (Itoh et al., 2004). Together, this suggests that the OsKOL might be involved in more specialized metabolism (Peters, 2006).
Figure 2.
Rice KO and KOL, with potential targeting of neighboring hydrocarbon bonds—i.e. C3α instead of C19 by KOL. A, Tandem array of KO paralogs in the rice genome. Phylogenetically related and coregulated genes, as discussed in the text, share common gene symbols (noninducible KO in white, inducible KOL in black). B, Three-dimensional rendering of ent-kaurene showing position of the hydrocarbon bond targeted by KO (C19) and its proximity to the C3α (pro-R) proton typically targeted for hydroxylation in rice labdane-related diterpenoid phytoalexin biosynthesis.
Here, we demonstrate that under conditions where OsKO2 exhibits KO activity, OsKOL4 does not, instead catalyzing C3α hydroxylation of ent-sandaracopimaradiene and ent-cassadiene, as well as ent-kaurene. We further report that in planta OsKOL4 transcripts accumulate in response to methyl jasmonate, followed by accumulation of 3α-hydroxy-ent-sandaracopimaradiene and 3α-hydroxy-ent-cassadiene, which are expected intermediates in the similarly inducible production of oryzalexins A to F and phytocassanes A to E, consistent with a role for this CYP in early oxidative steps of the biosynthetic pathways for these phytoalexins.
RESULTS
Recombinant Expression
Based on previously reported evidence suggesting that OsKOL4 and OsKOL5 might be involved in rice labdane-related diterpenoid phytoalexin biosynthesis (Itoh et al., 2004), we were interested in characterizing their biochemical activity. This was originally attempted using the native genes, obtained from the KOME rice cDNA database (Kikuchi et al., 2003). Due to our previous successful expression of plant CYP in Escherichia coli (Swaminathan et al., 2009; Morrone et al., 2010a; Wang et al., 2011; Wu et al., 2011), this was carried out using our bacterial modular metabolic engineering system (Cyr et al., 2007). In particular, by coexpression with the requisite CYP reductase (CPR) and functional pairings of upstream CPS and KS(L) to produce all of the labdane-related diterpenes found in rice (see Supplemental Fig. S1). Unfortunately, this did not lead to any further transformation, even when these native genes were modified at their N termini to optimize functional bacterial expression.
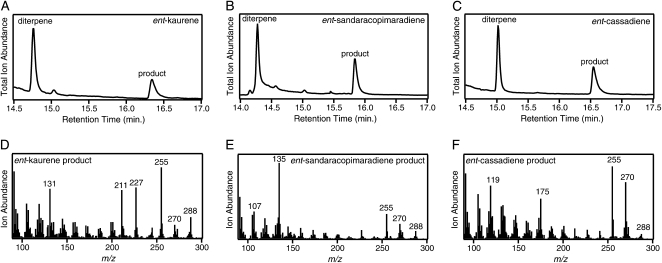
Codon optimization has been reported to increase expression efficiency of plant CYP (Chang et al., 2007), and we have found that complete gene recoding to optimize codon usage for expression in E. coli can lead to activity when none was observed with the native gene sequence (Wang et al., 2011; Wu et al., 2011). Thus, we had such gene constructs synthesized for OsKOL4 and OsKOL5. For comparative purposes, we also had such a gene construct synthesized for OsKO2 as well. Each of these was further N-terminally modified for bacterial expression, and both full-length and modified constructs were again incorporated into our metabolic engineering system as described above. The N-terminally modified synthetic OsKO2 construct exhibited the KO activity expected from the previously reported genetic studies (Itoh et al., 2004; Sakamoto et al., 2004), as well as biochemical characterization of the native gene recombinantly expressed in Pichia pastoris (Ko et al., 2008), selectively reacting with ent-kaurene to produce ent-kauren-19-oic acid via sequential oxidation to ent-kauren-19-ol and ent-kauren-19-al intermediates (Supplemental Fig. S2). By contrast, while no activity was observed for either OsKOL5 construct, the N-terminally modified synthetic OsKOL4 construct reacted with three diterpenes (ent-kaurene, ent-sandaracopimaradiene, and ent-cassadiene), with reasonable conversion of these diterpene olefins to apparently hydroxylated diterpenoids (molecular mass = 288 D) in each case (Fig. 3). Furthermore, the ent-kaurene product was not ent-kauren-19-ol, demonstrating a change in regiochemistry relative to KO.
Figure 3.
OsKOL4 catalyzes diterpene hydroxylation. A, GC-MS chromatogram of extract from E. coli engineered for production of ent-kaurene with coexpression of the synthetic and N-terminally modified OsKOL4 and OsCPR1. B, GC-MS chromatogram of extract from E. coli engineered for production of ent-sandaracopimaradiene and coexpression of the synthetic and N-terminally modified OsKOL4 and OsCPR1. C, GC-MS chromatogram of extract from E. coli engineered for production of ent-cassadiene and coexpression of the synthetic and N-terminally modified OsKOL4 and OsCPR1. D, Mass spectra for ent-kaurene-derived product (3α-hydroxy-ent-kaurene). E, Mass spectra for ent-sandaracopimaradiene-derived product (3α-hydroxy-ent-sandaracopimaradiene). F, Mass spectra for peak 2 (3α-hydroxy-ent-cassadiene).
Product Identification
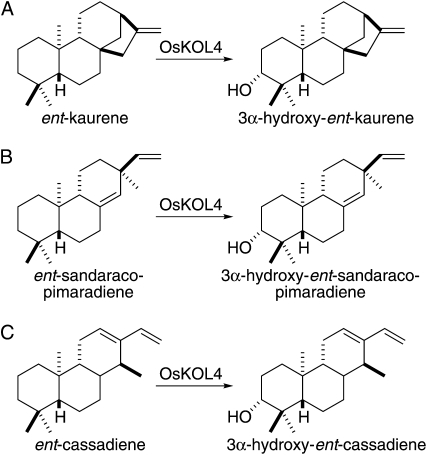
To produce enough of the OsKOL4 products for structural characterization by NMR, we increased the yield of our metabolic engineering system by incorporation of the bottom half of the yeast (Saccharomyces cerevisiae) mevalonate-dependent pathway, enabling production of isoprenoid precursors from fed mevalonate, which significantly increases accumulation of the diterpenoid end product (Morrone et al., 2010b). It was then possible to produce and purify several milligrams of each product by extraction from reasonable quantities of these recombinant cultures (3-L each). From the subsequent NMR analysis (Supplemental Fig. S3; Supplemental Tables S1–S3), it was found that each product contained a C3α-hydroxyl [i.e. (R)-3-OH] group (Fig. 4).
Figure 4.
OsKOL4 catalyzed reactions. A, Hydroxylation of ent-kaurene to 3α-hydroxy-ent-kaurene. B, Hydroxylation of ent-sandaracopimaradiene to 3α-hydroxy-ent-sandaracopimaradiene. C, Hydroxylation of ent-cassadiene to 3α-hydroxy-ent-cassadiene.
Enzymatic Characterization
The hydroxylase activity of OsKOL4 was further characterized by in vitro enzymatic analysis. This was accomplished via coexpression with a rice CPR (OsCPR1), with determination of the level of functional CYP present by measurement of the carbon monoxide (CO) difference binding spectra from the resulting clarified lysates (Supplemental Fig. S4). However, it should be noted that such spectra are rather inconsistent, with some preparations exhibiting activity in the absence of the characteristic peak at 450 nm. Thus, the resulting catalytic rates must be viewed with some caution. Nevertheless, steady-state kinetic analysis of enzymatic activity was carried out with preparations for which CO difference binding was apparent, which indicated that OsKOL4 exhibits similar catalytic efficiency with ent-sandaracopimaradiene and ent-cassadiene, but an approximately 3-fold reduction in affinity for ent-kaurene (Table I).
Physiological Relevance
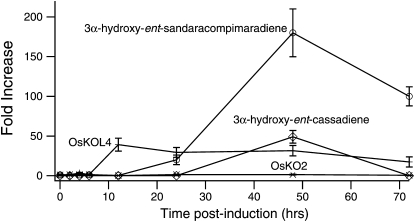
C3α-hydroxy-ent-sandaracopimaradiene and C3α-hydroxy-ent-cassadiene already have been suggested as potential intermediates in the production of rice oryzalexin and phytocassane phytoalexins (Peters, 2006). Transcription of the genes encoding the relevant diterpene synthases, as well as production of the diterpene precursors themselves, is inducible by methyl jasmonate (Prisic et al., 2004; Morrone et al., 2011). Thus, we investigated the effect of such induction on transcription of OsKOL4 and accumulation of its enzymatic products. Similar to the upstream diterpene synthases, OsKOL4 mRNA levels were dramatically increased in rice leaves by induction with methyl jasmonate, accumulating to more than 10-fold-higher levels within 12 h (Fig. 5; Supplemental Fig. S5). By contrast, OsKO2 mRNA levels were relatively unchanged. In addition, it was possible to detect increased accumulation of both C3α-hydroxy-ent-sandaracopimaradiene and C3α-hydroxy-ent-cassadiene following induction, although C3α-hydroxy-ent-kaurene was not detected at any point (Fig. 5; Supplemental Fig. S6). These results are consistent with both the availability of the precursor diterpenes (i.e. ent-kaurene is produced at much lower levels under these conditions; Morrone et al., 2011), and the expression pattern of OsKOL4 (Fig. 5), indicating a causative relationship.
Figure 5.
Accumulation of OsKOL4 (+) and OsKO2 (x) mRNA, along with that of 3α-hydroxy-ent-sandaracopimaradiene (white circles) and 3α-hydroxy-ent-cassadiene (white diamonds) in response to methyl jasmonate induction (as indicated). Data points shown are the average values from duplicate samples, each measured twice, with error bars representing the corresponding sd. 3α-Hydroxy-ent-casssadiene was only detected at the 48-h time point, thus fold-induction was calculated assuming a detection limit of 50 pg/g tissue fresh weight.
DISCUSSION
The mutually exclusive activity reported here for OsKOL4 relative to OsKO2 demonstrates biochemical divergence between these rice KO paralogs. Specifically, while the OsKO2 recombinant construct used here exhibited the expected KO activity (Itoh et al., 2004; Sakamoto et al., 2004; Ko et al., 2008), the analogous OsKOL4 construct was found to only catalyze C3α hydroxylation, even with their common substrate ent-kaurene (Fig. 4). Accordingly, although much more distantly related CYP701 family members retain KO activity—e.g. PpKO/CYP701B1, which falls into a separate subfamily and shares less than 42% amino acid sequence identity with any member of the CYP701A subfamily (Miyazaki et al., 2011)—OsKOL4, which shares 71% amino acid sequence identity with OsKO2, has evolved novel enzymatic function.
While we were unable to identify any activity for OsKOL5, it should be noted that, although its transcription is similarly inducible, OsKOL5 mRNA accumulates at a much lower level than does that of OsKOL4 (Itoh et al., 2004). Thus, it is possible that OsKOL5 may be an inactive pseudogene. However, OsKOL5 is rather divergent, sharing only 79% amino acid sequence identity with OsKOL4, its closest relative. Accordingly, OsKOL5 may function at some later step in rice diterpenoid, or in some other type of natural products biosynthesis.
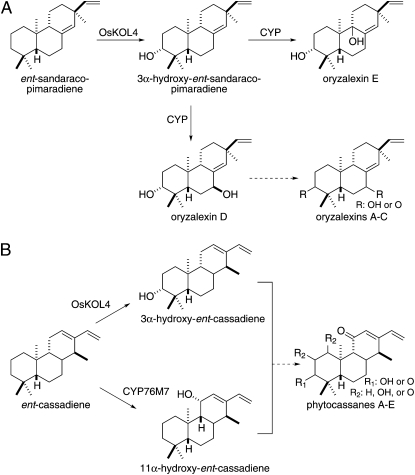
In any case, our results further indicate that OsKOL4 acts in rice diterpenoid phytoalexin biosynthesis. In particular, the C3α hydroxylation of ent-sandaracopimaradiene and ent-cassadiene catalyzed by OsKOL4 is correlated with the presence of C3(α)-oxy moieties in the respectively derived oryzalexins and phytocassanes (Fig. 6), and induction with the plant defense signaling molecule methyl jasmonate elicits both increased levels OsKOL4 mRNA and subsequent accumulation of its enzymatic products (Fig. 5). Moreover, UV irradiation similarly increases OsKOL4 mRNA levels (Itoh et al., 2004), which further has been shown to lead to accumulation of at least C3α-hydroxy-ent-sandaracopimaradiene (Kato et al., 1995). Perhaps more critically, consistent with suggestions that the oryzalexins and phytoalexins serve as phytoalexins against the blast pathogen Magnaporthe oryzae (Peters, 2006), increased mRNA levels of not only OsKOL4, but also the relevant upstream diterpene synthases OsCPS2, OsKSL7, and OsKSL10 are observed upon infection with this fungus (Marcel et al., 2010).
Figure 6.
OsKOL4 plays a putative role in rice diterpenoid phytoalexin biosynthesis. A, Hypothetical role for OsKOL4 in orzyalexin biosynthesis. B, Hypothetical role for OsKOL4 in phytocassane biosynthesis, with potential bifurcation as shown.
While the role of OsKOL4 in oryzalexin biosynthesis seems straightforward, its exact role in phytocassane production is not as clear (Fig. 6). These phytoalexins all contain C3(α)-oxy and C11-keto moieties, and we have previously demonstrated that CYP76M7 catalyzes C11α hydroxylation of ent-cassadiene, with C11α-hydroxy-ent-cassadiene also being detected in induced rice leaf extracts (Swaminathan et al., 2009). Hence, both C3α- and C11α-hydroxylated ent-cassadienes are present in rice, which would seem to suggest the possibility of a bifurcated biosynthetic network (Fig. 6B). However, at least in vitro, OsKOL4 does not react with C11α-hydroxy-ent-cassadiene, and nor does CYP76M7 react with C3α-hydroxy-ent-cassadiene, leaving the exact roles of these CYP in phytocassane biosynthesis unclear at this time.
The ability of OsKOL4 to react with ent-kaurene offers potential competition to the use of this intermediate in GA biosynthesis by OsKO2. However, C3α-hydroxy-ent-kaurene is not observed in planta, and OsKOL4 exhibits a lower affinity for ent-kaurene than its other substrates (Table I). Moreover, while ent-kaurene is constitutively produced in rice plants, it is a minor component of the diterpenes produced following elicitation (Wickham and West, 1992; Mohan et al., 1996; Morrone et al., 2011). Given that OsKO2 is constitutively expressed throughout rice plants, while OsKOL4 exhibits a very limited expression pattern, with significant expression levels only observed upon elicitation (Itoh et al., 2004), the ability of OsKOL4 to react with ent-kaurene does not seem to be physiologically relevant.
Table I. OsKOL4 steady-state kinetic constants.
| Substrate | kcat | KM | kcat/KM |
| s−1 | μm | ||
| ent-cassadiene | 0.017 ± 0.001 | 2.0 ± 0.4 | 9 × 103 |
| ent-sandaracopimaradiene | 0.013 ± 0.001 | 2.3 ± 0.8 | 6 × 103 |
| ent-kaurene | 0.022 ± 0.001 | 7 ± 2 | 3 × 103 |
Regardless of exact role, the biochemical divergence of OsKOL4 provides a clear example of a KO/CYP701 family member associated with more specialized, rather than GA metabolism. While stevioside biosynthesis seems likely to proceed via ent-kaurenoic acid presumably produced by one of the two distinct KO family members found in Stevia rebaudiana, it remains unclear which, or even if one or the other is dedicated to stevioside versus GA biosynthesis, and these both catalyze the prototypical KO reaction in any case (Humphrey et al., 2006). By contrast, the novel enzymatic activity exhibited by OsKOL4 precludes any function in GA biosynthesis (compare Figs. 1 and 4), and the transcriptional regulation of this gene also is more clearly associated with more specialized (i.e. phytoalexin) metabolism (Fig. 5; Itoh et al., 2004).
The organization of the rice KO/CYP701 family members as a five-gene tandem array with distinct division between phylogenetically related and coregulated KO and KOL has suggested a plausible evolutionary history (Itoh et al., 2004). In particular, this array presumably originated with tandem gene duplication of the ancestral KO required for GA metabolism. This enabled neofunctionalization, including at least divergence in transcriptional regulation of one of these KO genes (i.e. to form an inducible KOL), with subsequent gene duplication/expansion of this ancestral KO and derived KOL pair to yield the currently observed five-gene tandem array. The data presented here indicates divergence of the enzymatic activity of at least one of the KOL (i.e. the targeting of C3α instead of C19 by OsKOL4). Notably, although the roles of the rice KO(L) family members other than OsKO2 and OsKOL4 remain unknown, at least one of these presumably then provided a selective advantage—i.e. in order for this later gene expansion to sweep through the rice population. For example, while no activity was found here for OsKOL5, it still remains possible that this acts later in rice diterpenoid, or in other natural products biosynthesis.
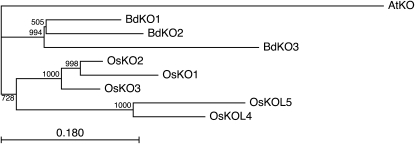
Intriguingly, examination of the recently sequenced Brachypodium distachyon genome (International Brachypodium Initiative, 2010) demonstrates the existence of a similar tandem array of KO/CYP701A subfamily members (BdKO1–3). However, molecular phylogenetic analysis further indicates that this arose separately from the rice KO cluster (Fig. 7). Thus, it seems likely that similar functional diversification as that we report here for OsKOL4 may have occurred multiple times, at least in the grass plant family where diversion of GA biosynthesis to the production of more specialized metabolism seems to be widespread (Peters, 2006). This latter point perhaps also indicates why it has been possible to divert OsKOL4 to more specialized metabolism with relatively few changes (i.e. this is still a member of the CYP701A subfamily, sharing 65% to 79% amino acid sequence identity with the other OsKO). By contrast, diversion of other CYP from primary to secondary metabolism has been associated with the establishment of novel subfamilies, which by definition share less than 55% identity with other family members—e.g. more specialized triterpenoid biosynthesis has been shown to variously utilize a novel KAO/CYP88D subfamily member diverted from GA metabolism (Seki et al., 2008), or a novel CYP51H subfamily member derived from the C14-demethylase operating in sterol metabolism (Qi et al., 2006). Accordingly, the close relationship between GA and other labdane-related diterpenoid biosynthesis, in particular the use of structurally similar multicyclic diterpene intermediates, presumably enabled the relatively facile diversion of OsKOL4 to more specialized metabolism observed here.
Figure 7.
Molecular phylogenetic analysis of rice and B. distachyon KO/CYP701A subfamily members. Based on amino acid sequence alignment, with inclusion of a verified KO from the dicot Arabidopsis (Arabidopsis thaliana; AtKO/CYP701A3; Helliwell et al., 1998) as the outgroup sequence.
CONCLUSION
In summary, our results demonstrate that rice contains at least one functionally divergent KO/CYP701 family member. The distinct enzymatic activity of OsKOL4 identified here precludes its action in GA metabolism, and it instead appears to play a role in rice diterpenoid phytoalexin biosynthesis. This then further expands the range of enzymes recruited from GA/primary metabolism to play functionally distinct roles in more specialized labdane-related diterpenoid metabolism beyond the diterpene synthases, and supports the emerging paradigm that the requirement for phytohormone production provides a biosynthetic reservoir that often is tapped in the evolution of secondary metabolism.
MATERIALS AND METHODS
General Procedure
Unless otherwise noted, chemicals were purchased from Fisher Scientific, and molecular biology reagents from Invitrogen. Gene mapping was based on the annotated rice (Oryza sativa) genome sequence at GenBank, along with the previously assigned OsKO(L) nomenclature (Itoh et al., 2004; Sakamoto et al., 2004). CYP nomenclature was determined via BLAST searches at the CYP homepage maintained by Dr. David Nelson (http://drnelson.uthsc.edu/CytochromeP450.html). Gas chromatography (GC) was performed with a Varian 3900 GC with Saturn 2100 ion trap mass spectrometer (MS) in electron ionization (70 eV) mode. Samples (1 μL) were injected in splitless mode at 50°C and, after holding for 3 min at 50°C, the oven temperature was raised at a rate of 14°C/min to 300°C, where it was held for an additional 3 min. MS data from 90 to 600 mass-to-charge ratio (m/z) were collected starting 12 min after injection until the end of the run.
Recombinant Constructs
The OsKOL4 and OsKOL5 native cDNA were obtained from the KOME rice cDNA databank (GenBank accessions AY579214 and AY660664). Synthetic constructs for OsKO2, OsKOL4, and OsKOL5, codon optimized for expression in Escherichia coli, were obtained from Genscript (see Supplemental Information S1 for the corresponding nucleotide sequences). All of these were subcloned into the Gateway vector pENTR/SD/D-TOPO by directional topoisomerization. N-terminal modification of these genes for improved functional bacterial expression was performed in a two-stage PCR process, first removing the N-terminal transmembrane helix (OsKO2, 39 amino acid; OsKOL4, 42 amino acid; and OsKOL5, 36 amino acid) from the 5′ end of the open reading frame and then adding 10 new codons (encoding the amino acid sequence MAKKTSSKGK), based on the modifications used for bacterial expression of the mammalian CYP2B subfamily (Scott et al., 2001). All constructs were verified by complete gene sequencing and then transferred via directional recombination into a modified pCDF-Duet vector (Novagen), which contains a DEST cassette in the first multiple cloning site and a rice CPR (OsCPR1) in the second multiple cloning site, as previously described (Swaminathan et al., 2009).
Recombinant Expression
All CYP were expressed in the C41 overexpress strain of E. coli (Lucigen) using our previously described modular diterpene metabolic engineering system (Cyr et al., 2007). Briefly, these CYP were coexpressed with not only OsCPR1 (i.e. using the constructs described above), but also with a geranylgeranyl diphosphate synthase and CPS carried on cocompatible pGGxC vectors, as well as OsKSL expressed from the additionally cocompatible pET-based pDEST14 or pDEST15 (i.e. for expression as a fusion to glutathione-S-transferase). The resulting diterpenoids were extracted from liquid cultures (media and cells), typically 50-mL volumes grown for 72 h at 16°C after induction, with an equal volume of hexane, and analyzed by GC-MS, including samples that were methylated to examine potential acid formation. In every case the expected diterpene olefin product (i.e. given the coexpressed diterpene synthases) was observed, with hydroxylated diterpenoids detected as described above (Fig. 3), such that the overall yields were consistent.
Diterpenoid Production
The novel enzymatic products were obtained in sufficient amounts for NMR analysis by both increasing flux into isoprenoid metabolism and scaling up the culture volumes. Flux toward isoprenoid biosynthesis was increased by incorporation of the bottom half of the mevalonate-dependent isoprenoid precursor pathway from yeast (Saccharomyces cerevisiae), using the previously described pMBI (Martin et al., 2003). This enables production of the isoprenoid precursors isopentenyl diphosphate and dimethylallyl diphosphate from mevalonate, such that feeding of 20 mm mevalonolactone significantly increases diterpenoid production, as previously described (Morrone et al., 2010b). The resulting diterpenoids were extracted from 3 L of culture (media and cells) with an equal volume of a 1:1 mixture of ethyl acetate and hexanes, and the organic extract then dried by rotary evaporation. The resulting residue was dissolved in 5 mL 45% methanol/45% acetonitrile/10% distilled water, and the hydroxylated diterpenoids purified by HPLC. This was carried out using an Agilent 1100 series instrument equipped with autosampler, fraction collector, and diode array UV detection, over a ZORBAX Eclipse XDB-C8 column (4.6 × 150 mm, 5 μm) at a 0.5 mL/min flow rate. The column was preequilibrated with 20% acetonitrile/distilled water, sample loaded, then the column washed with 20% acetonitrile/distilled water (0–2 min), and eluted with 20% to 100% acetonitrile (2–7 min), followed by a 100% acetonitrile wash (7–27 min). Following purification, each compound was dried under a gentle stream of N2, and then dissolved in 0.5 mL deuterated methanol (CD3OD; Sigma-Aldrich), with this evaporation-resuspension process repeated two more times to completely remove the protonated acetonitrile solvent, resulting in a final estimated approximately 5 to 10 mg of each novel diterpenoid.
Chemical Structure Identification
NMR spectra for the diterpenoids were recorded at 25°C on a Bruker Avance 500 spectrometer equipped with a cryogenic probe for 1H and 13C. Structural analysis was performed using 1 D 1H, 1 D DQF-COSY, HSQC, HMQC, HMBC, and NOESY experiment spectra acquired at 500 MHz and 13C (125.5 MHz) and DEPT135 spectra using standard experiments from the Bruker TopSpin v1.3 software. All samples were placed in NMR tubes purged with nitrogen gas for analyses, and chemical shifts were referenced using known methanol [13C 49.15(7), 1H 3.31(5) ppm (m)] signals offset from tetramethylsilane (Supplemental Tables S1–S3). Correlations from the HMBC spectra were used to propose a partial structure, while COSY correlations between protonated carbons and HSQC spectra were used to complete the partial structure and assign proton chemical shifts. The configuration of the A and B rings (C1–C10) is predetermined by the configuration of the CPP intermediate, since chemical bonds in that portion of the molecule are not altered. Thus, nuclear Overhauser effect dipole-dipole signals observed between the C5 proton and the C3 alcohol methine proton (Supplemental Fig. S4) could be used to assign the α (R) configuration of the C3 hydroxyl group (Supplemental Table S1–S3).
Kinetic Analysis
Kinetic analysis was carried out using the synthetic and N-terminally modified OsKOL4 construct expressed in E. coli using the OsCPR1 coexpression construct described above. Expression cultures in TB medium were supplemented with 1 mm thiamine, 5 mg/L riboflavin, and 75 mg/L 5-aminolevulinic acid, along with induction with 1 mm isopropylthio-β-galactoside at A600 of 0.8 to 1.0. After 72 h at 16°C, the cells were harvested and clarified lysates prepared for in vitro kinetic assays with quantification of CYP by reduced CO-binding difference spectra using an extinction coefficient of 91 mm−1 cm−1 (Omura and Sato, 1964). Kinetic assays were performed using 220 pm active OsKOL4 in each 1-mL reaction. The three diterpenes were used as substrates with varying concentrations (0.8–196 μm ent-sandaracopimaradiene, 1–200 μm ent-cassadiene, and 1–240 μm ent-kaurene), but otherwise these assays were carried out as previously described (Swaminathan et al., 2009). After 30 min, 50 μL 1 m HCL was added to stop the reaction, enzymatic products extracted by ethyl acetate, confirmed by GC-MS, and quantified by GC with flame ionization detection, using an external standard curve of ent-kauren-19-ol.
Plant Analyses
The rice plants (ssp. Nipponbare) and subsequent mRNA and metabolite analyses were largely carried out as previously described (Wang et al., 2011). Briefly, the plants were cultivated in growth chambers under 12-h light (28°C) and 12-h dark (24°C) cycles to the sixth leaf stage. Induced plants were treated with 1-mL 0.2% (v/v) methyl jasmonate per plant, while control/uninduced plants were only treated with the 0.1% Tween 20 carrier solution, and all plants then incubated (separately) and duplicate samples harvested at 0, 2, 4, 6, 12, 24, 48, and 72 h after induction. OsKOL4 mRNA levels were analyzed by reverse transcription-PCR using the gene-specific primers (forward: TCGTAAAAGTATGGTCGCAATC; reverse: ATACATGGTCCAT-TCGGTGGT), with PrimeSTAR HS DNA polymerase (Takara Bio) and the following program: 2 min at 94°C followed by 35 cycles of 30 s at 94°C, 30 s at 55°C, and 90 s at 72°C—with normalization of the resulting yield against that from similar amplification of the rice actin gene. Metabolite analysis was carried out with approximately 2-g leaf samples, which were frozen and ground to powder in liquid nitrogen, then extracted with 50-mL ethyl acetate. These rice extracts were fractioned by HPLC and analyzed by GC-MS in single ion monitoring mode. To detect the hydroxylated diterpenoids of interest here, the MS was operated in chemical ionization mode with methanol as the reaction solvent, selecting the base peak (m/z = 271, MH+-water) for fragmentation (0.6 eV), with quantification of the fragment at m/z = 215. Each of the biological duplicate samples was independently analyzed twice, for both mRNA and metabolites (Fig. 5).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Functional map of rice diterpene synthases.
Supplemental Figure S2. KO activity of OsKO2.
Supplemental Figure S3. NMR correlations.
Supplemental Figure S4. OsKOL4 CO difference binding spectra.
Supplemental Figure S5. Reverse transcription-PCR expression data.
Supplemental Figure S6. Rice metabolite analysis (liquid chromatography with mass selective mass spectrometry detection).
Supplemental Table S1. 3a-Hydroxy-ent-kaurene chemical shifts.
Supplemental Table S2. 3a-Hydroxy-ent-sandaracopimaradiene chemical shifts.
Supplemental Table S3. 3a-Hydroxy-ent-cassadiene chemical shifts.
References
- Chang MCY, Eachus RA, Trieu W, Ro D-K, Keasling JD. (2007) Engineering Escherichia coli for production of functionalized terpenoids using plant P450s. Nat Chem Biol 3: 274–277 [DOI] [PubMed] [Google Scholar]
- Cho E-M, Okada A, Kenmoku H, Otomo K, Toyomasu T, Mitsuhashi W, Sassa T, Yajima A, Yabuta G, Mori K, et al. (2004) Molecular cloning and characterization of a cDNA encoding ent-cassa-12,15-diene synthase, a putative diterpenoid phytoalexin biosynthetic enzyme, from suspension-cultured rice cells treated with a chitin elicitor. Plant J 37: 1–8 [DOI] [PubMed] [Google Scholar]
- Chu HY, Wegel E, Osbourn A. (2011) From hormones to secondary metabolism: the emergence of metabolic gene clusters in plants. Plant J 66: 66–79 [DOI] [PubMed] [Google Scholar]
- Cyr A, Wilderman PR, Determan M, Peters RJ. (2007) A modular approach for facile biosynthesis of labdane-related diterpenes. J Am Chem Soc 129: 6684–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falara V, Pichersky E, Kanellis AK. (2010) A copal-8-ol diphosphate synthase from the angiosperm Cistus creticus subsp. creticus is a putative key enzyme for the formation of pharmacologically active, oxygen-containing labdane-type diterpenes. Plant Physiol 154: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Hillwig ML, Huang L, Cui G, Wang X, Kong J, Yang B, Peters RJ. (2009) A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org Lett 11: 5170–5173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris LJ, Saparno A, Johnston A, Prisic S, Xu M, Allard S, Kathiresan A, Ouellet T, Peters RJ. (2005) The maize An2 gene is induced by Fusarium attack and encodes an ent-copalyl diphosphate synthase. Plant Mol Biol 59: 881–894 [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Sheldon CC, Olive MR, Walker AR, Zeevaart JA, Peacock WJ, Dennis ES. (1998) Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc Natl Acad Sci USA 95: 9019–9024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey TV, Richman AS, Menassa R, Brandle JE. (2006) Spatial organisation of four enzymes from Stevia rebaudiana that are involved in steviol glycoside synthesis. Plant Mol Biol 61: 47–62 [DOI] [PubMed] [Google Scholar]
- International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Itoh H, Tatsumi T, Sakamoto T, Otomo K, Toyomasu T, Kitano H, Ashikari M, Ichihara S, Matsuoka M. (2004) A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol Biol 54: 533–547 [DOI] [PubMed] [Google Scholar]
- Kanno Y, Otomo K, Kenmoku H, Mitsuhashi W, Yamane H, Oikawa H, Toshima H, Matsuoka M, Sassa T, Toyomasu T. (2006) Characterization of a rice gene family encoding type-A diterpene cyclases. Biosci Biotechnol Biochem 70: 1702–1710 [DOI] [PubMed] [Google Scholar]
- Kato H, Kodama O, Akatsuka T. (1995) Characterization of an inducible P450 hydroxylase involved in the rice diterpene phytoalexin biosynthetic pathway. Arch Biochem Biophys 316: 707–712 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, et al. (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301: 376–379 [DOI] [PubMed] [Google Scholar]
- Ko KW, Lin F, Katsumata T, Sugai Y, Miyazaki S, Kawaide H, Okada K, Nojiri H, Yamane H. (2008) Functional identification of a rice ent-kaurene oxidase, OsKO2, using the Pichia pastoris expression system. Biosci Biotechnol Biochem 72: 3285–3288 [DOI] [PubMed] [Google Scholar]
- MacMillan J, Takahashi N. (1968) Proposed procedure for the allocation of trivial names to the gibberellins. Nature 217: 170–171 [DOI] [PubMed] [Google Scholar]
- Marcel S, Sawers R, Oakeley E, Angliker H, Paszkowski U. (2010) Tissue-adapted invasion strategies of the rice blast fungus Magnaporthe oryzae. Plant Cell 22: 3177–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin VJJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. (2003) Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol 21: 796–802 [DOI] [PubMed] [Google Scholar]
- Miyazaki S, Katsumata T, Natsume M, Kawaide H. (2011) The CYP701B1 of Physcomitrella patens is an ent-kaurene oxidase that resists inhibition by uniconazole-P. FEBS Lett 585: 1879–1883 [DOI] [PubMed] [Google Scholar]
- Mohan RS, Yee NKN, Coates RM, Ren YY, Stamenkovic P, Mendez I, West CA. (1996) Biosynthesis of cyclic diterpene hydrocarbons in rice cell suspensions: conversion of 9,10-syn-labda-8(17),13-dienyl diphosphate to 9β-pimara-7,15-diene and stemar-13-ene. Arch Biochem Biophys 330: 33–47 [DOI] [PubMed] [Google Scholar]
- Morrone D, Chen X, Coates RM, Peters RJ. (2010a) Characterization of the kaurene oxidase CYP701A3, a multifunctional cytochrome P450 from gibberellin biosynthesis. Biochem J 431: 337–344 [DOI] [PubMed] [Google Scholar]
- Morrone D, Hillwig ML, Mead ME, Lowry L, Fulton DB, Peters RJ. (2011) Evident and latent plasticity across the rice diterpene synthase family with potential implications for the evolution of diterpenoid metabolism in the cereals. Biochem J 435: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrone D, Jin Y, Xu M, Choi S-Y, Coates RM, Peters RJ. (2006) An unexpected diterpene cyclase from rice: functional identification of a stemodene synthase. Arch Biochem Biophys 448: 133–140 [DOI] [PubMed] [Google Scholar]
- Morrone D, Lowry L, Determan MK, Hershey DM, Xu M, Peters RJ. (2010b) Increasing diterpene yield with a modular metabolic engineering system in E. coli: comparison of MEV and MEP isoprenoid precursor pathway engineering. Appl Microbiol Biotechnol 85: 1893–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto T, Cho E-M, Okada A, Okada K, Otomo K, Kanno Y, Toyomasu T, Mitsuhashi W, Sassa T, Minami E, et al. (2004) Stemar-13-ene synthase, a diterpene cyclase involved in the biosynthesis of the phytoalexin oryzalexin S in rice. FEBS Lett 571: 182–186 [DOI] [PubMed] [Google Scholar]
- Omura T, Sato R. (1964) The carbon monoxide-binding pigment of liver microsome. II. Solubilization, purification, and properties. J Biol Chem 239: 2379–2385 [PubMed] [Google Scholar]
- Otomo K, Kanno Y, Motegi A, Kenmoku H, Yamane H, Mitsuhashi W, Oikawa H, Toshima H, Itoh H, Matsuoka M, et al. (2004a) Diterpene cyclases responsible for the biosynthesis of phytoalexins, momilactones A, B, and oryzalexins A-F in rice. Biosci Biotechnol Biochem 68: 2001–2006 [DOI] [PubMed] [Google Scholar]
- Otomo K, Kenmoku H, Oikawa H, König WA, Toshima H, Mitsuhashi W, Yamane H, Sassa T, Toyomasu T. (2004b) Biological functions of ent- and syn-copalyl diphosphate synthases in rice: key enzymes for the branch point of gibberellin and phytoalexin biosynthesis. Plant J 39: 886–893 [DOI] [PubMed] [Google Scholar]
- Peters RJ. (2006) Uncovering the complex metabolic network underlying diterpenoid phytoalexin biosynthesis in rice and other cereal crop plants. Phytochemistry 67: 2307–2317 [DOI] [PubMed] [Google Scholar]
- Peters RJ. (2010) Two rings in them all: the labdane-related diterpenoids. Nat Prod Rep 27: 1521–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prisic S, Xu M, Wilderman PR, Peters RJ. (2004) Rice contains two disparate ent-copalyl diphosphate synthases with distinct metabolic functions. Plant Physiol 136: 4228–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Bakht S, Qin B, Leggett M, Hemmings A, Mellon F, Eagles J, Werck-Reichhart D, Schaller H, Lesot A, et al. (2006) A different function for a member of an ancient and highly conserved cytochrome P450 family: from essential sterols to plant defense. Proc Natl Acad Sci USA 103: 18848–18853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, et al. (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134: 1642–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EE, Spatzenegger M, Halpert JR. (2001) A truncation of 2B subfamily cytochromes P450 yields increased expression levels, increased solubility, and decreased aggregation while retaining function. Arch Biochem Biophys 395: 57–68 [DOI] [PubMed] [Google Scholar]
- Seki H, Ohyama K, Sawai S, Mizutani M, Ohnishi T, Sudo H, Akashi T, Aoki T, Saito K, Muranaka T. (2008) Licorice beta-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proc Natl Acad Sci USA 105: 14204–14209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S, Morrone D, Wang Q, Fulton DB, Peters RJ. (2009) CYP76M7 is an ent-cassadiene C11α-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell 21: 3315–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyomasu T. (2008) Recent advances regarding diterpene cyclase genes in higher plants and fungi. Biosci Biotechnol Biochem 72: 1168–1175 [DOI] [PubMed] [Google Scholar]
- Toyomasu T, Kagahara T, Hirose Y, Usui M, Abe S, Okada K, Koga J, Mitsuhashi W, Yamane H. (2009) Cloning and characterization of cDNAs encoding ent-copalyl diphosphate synthases in wheat: insight into the evolution of rice phytoalexin biosynthetic genes. Biosci Biotechnol Biochem 73: 772–775 [DOI] [PubMed] [Google Scholar]
- Wang Q, Hillwig ML, Peters RJ. (2011) CYP99A3: functional identification of a diterpene oxidase from the momilactone biosynthetic gene cluster in rice. Plant J 65: 87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham KA, West CA. (1992) Biosynthesis of rice phytoalexins: identification of putative diterpene hydrocarbon precursors. Arch Biochem Biophys 293: 320–332 [DOI] [PubMed] [Google Scholar]
- Wilderman PR, Xu M, Jin Y, Coates RM, Peters RJ. (2004) Identification of syn-pimara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol 135: 2098–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Hillwig ML, Wang Q, Peters RJ. (2011) Parsing a multifunctional biosynthetic gene cluster from rice: biochemical characterization of CYP71Z6 & 7. FEBS Lett 585: 3446–3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Hillwig ML, Prisic S, Coates RM, Peters RJ. (2004) Functional identification of rice syn-copalyl diphosphate synthase and its role in initiating biosynthesis of diterpenoid phytoalexin/allelopathic natural products. Plant J 39: 309–318 [DOI] [PubMed] [Google Scholar]
- Xu M, Wilderman PR, Morrone D, Xu J, Roy A, Margis-Pinheiro M, Upadhyaya NM, Coates RM, Peters RJ. (2007) Functional characterization of the rice kaurene synthase-like gene family. Phytochemistry 68: 312–326 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251 [DOI] [PubMed] [Google Scholar]