Abstract
Potato virus X (PVX) requires three virally encoded proteins, the triple gene block (TGB), for movement between cells. TGB1 is a multifunctional protein that suppresses host gene silencing and moves from cell to cell through plasmodesmata, while TGB2 and TGB3 are membrane-spanning proteins associated with endoplasmic reticulum-derived granular vesicles. Here, we show that TGB1 organizes the PVX “X-body,” a virally induced inclusion structure, by remodeling host actin and endomembranes (endoplasmic reticulum and Golgi). Within the X-body, TGB1 forms helically arranged aggregates surrounded by a reservoir of the recruited host endomembranes. The TGB2/3 proteins reside in granular vesicles within this reservoir, in the same region as nonencapsidated viral RNA, while encapsidated virions accumulate at the outer (cytoplasmic) face of the X-body, which comprises a highly organized virus “factory.” TGB1 is both necessary and sufficient to remodel host actin and endomembranes and to recruit TGB2/3 to the X-body, thus emerging as the central orchestrator of the X-body. Our results indicate that the actin/endomembrane-reorganizing properties of TGB1 function to compartmentalize the viral gene products of PVX infection.
Plant RNA viruses, like their animal counterparts, replicate on the cytoplasmic face of host endomembranes, which are remodeled in the process. The reorganized membranes and the viral replication machinery that they support (virus-encoded RNA-dependent RNA polymerase with host- and virus-encoded cofactors) are collectively referred to as a viral replication complex (VRC). VRCs are thought to facilitate efficient replication of the viral genome in a protected environment. Additionally, they may also be intimately associated with virion assembly and budding (Sanfaçon, 2005; Miller and Krijnse-Locker, 2008; den Boon et al., 2010; Laliberté and Sanfaçon, 2010). Plant viruses differ from animal viruses in that they are mostly nonenveloped particles. Furthermore, viral cell-to-cell transport occurs through intercellular channels (plasmodesmata) rather than via plasma membrane budding (Lucas, 2006). Nevertheless, virion assembly can be closely associated with membranous VRCs for nonenveloped viruses (den Boon et al., 2010).
At late infection stages, many plant viruses induce the formation of a single large inclusion body, which is often localized next to the nucleus and has historically been termed an “X-body” because of its unclear function (Goldstein, 1924). Although such plant viral inclusion bodies have been observed for nearly a century and are important diagnostic features (Martelli and Russo, 1977), the cell biological aspects of their assembly and function have not been studied for most viruses. This is the case for Potato virus X (PVX), the type member of the potexviruses, a widespread group of economically important plant RNA viruses (Adams et al., 2004). PVX has been instrumental in the discovery and experimental use of RNA silencing (Batten et al., 2003; Verchot-Lubicz et al., 2007) and also serves as a model system for viruses encoding three “movement proteins” (MPs) in overlapping open reading frames, the “triple gene block” (TGB; Morozov and Solovyev, 2003; Verchot-Lubicz et al., 2007, 2010). MPs are nonstructural, virus-encoded proteins required for viral cell-to-cell transport through plasmodesmata (Lucas, 2006).
Two of the PVX MPs, TGB2 and TGB3, are transmembrane proteins that reside in the endoplasmic reticulum (ER). TGB2 induces the formation of ER-derived granular vesicles (Boevink et al., 1996; Ju et al., 2005), to which TGB3 is recruited during infection (Schepetilnikov et al., 2005; Samuels et al., 2007; Bamunusinghe et al., 2009). The granular vesicles are also associated with ribosomes, virions, and viral RNA-dependent RNA polymerase (“replicase”; Ju et al., 2005; Bamunusinghe et al., 2009) and probably constitute VRCs (Verchot-Lubicz et al., 2010).
The largest PVX MP, TGB1, modifies plasmodesmata and moves between cells (Angell et al., 1996; Yang et al., 2000; Howard et al., 2004). It also functions as an ATPase/RNA helicase (Kalinina et al., 2002), a translational activator (Atabekov et al., 2000; Rodionova et al., 2003), and a suppressor of RNA silencing (Voinnet et al., 2000; Bayne et al., 2005; Chiu et al., 2010). In uninfected tissue, fluorescent protein (FP) fusions to the TGB1 N terminus result in cytoplasmic fluorescence, whereas C-terminal fusions form aggregates (Yang et al., 2000; Howard et al., 2004; Samuels et al., 2007; Tilsner et al., 2009). In infected tissue, N-terminal GFP-TGB1 has been observed in plasmodesmata and aggregates (Samuels et al., 2007). In electron microscopy (EM), large proteinaceous aggregates in the form of “beaded sheets” are typical for PVX infections (Kozar and Sheludko, 1969; Stols et al., 1970; Shalla and Shepard, 1972), and these are heavily decorated with antibodies against TGB1 (Davies et al., 1993; Santa Cruz et al., 1998). The protein also has a strong tendency to oligomerize in vitro (Leshchiner et al., 2008), indicating that the aggregates observed with FP fusions to TGB1 may not be fusion-dependent artifacts.
We previously described an in vivo protocol for imaging viral (v)RNA using the sequence-specific RNA-binding protein, Pumilio, coupled to bimolecular fluorescence complementation (PUM-BiFC; Ozawa et al., 2007; Tilsner et al., 2009). In mature PVX infections, the bulk of the vRNA was localized in circular “whorls” within the perinuclear X-body. The X-body also harbors the beaded sheets that contain TGB1 (Kozar and Sheludko, 1969; Stols et al., 1970; Shalla and Shepard, 1972; Davies et al., 1993; Santa Cruz et al., 1998). A fluorescent TGB1-mCherry fusion was localized precisely to the center of the RNA whorls (Tilsner et al., 2009), prompting us to investigate the role of TGB1 in the organization of the PVX X-body in more detail.
Recently, it was shown that PVX requires an intact actin cytoskeleton for successful cell-to-cell movement (Harries et al., 2009). Here, we demonstrate that TGB1 alone is responsible for organizing the X-body through extensive remodeling of the actin/ER network of infected cells. Our results indicate that TGB1-mediated actin/ER-remodeling functions in compartmentalizing viral gene products during PVX infection.
RESULTS
TGB1 Aggregates Form the Core of the Layered X-Body
PVX-infected tissue contains characteristic proteinaceous aggregates (beaded sheets) within the X-body that are heavily labeled by anti-TGB1 antibody (Fig. 1, A and B; Kozar and Sheludko, 1969; Stols et al., 1970; Shalla and Shepard, 1972). Recently, we found that a C-terminal TGB1-mCherry fusion, expressed ectopically from a 35S promoter within infected tissue, is localized in aggregates inside the X-body, morphologically similar to the beaded sheets (Tilsner et al., 2009). To verify that this was not due to ectopic expression, we engineered PVX to endogenously express the TGB1-mCherry fusion instead of the native protein (PVX.TGB1-mCherry) and infected the model host plant Nicotiana benthamiana with this modified virus. PVX.TGB1-mCherry produced systemic symptoms on inoculated plants, but mCherry fluorescence was detected only in the initially inoculated area, indicating that the virus lost the FP insertion during infection. In the initially infected cells, TGB1-mCherry was found in similar aggregates to those shown within the X-body by EM (Fig. 1, C and D).
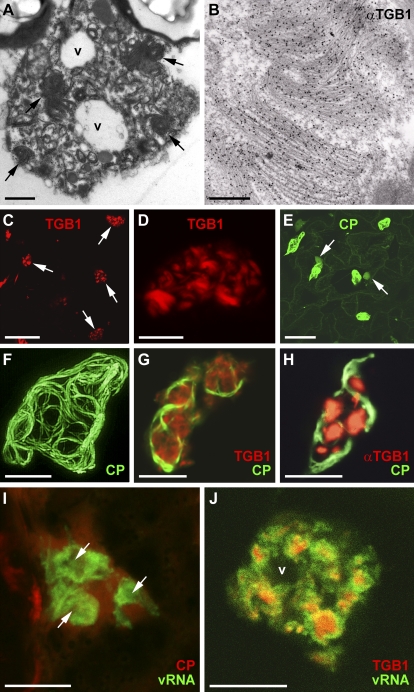
Figure 1.
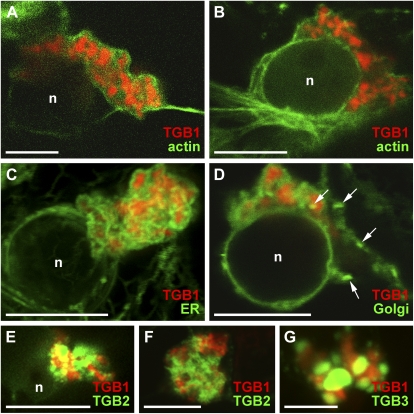
Distribution of viral components in the PVX X-body. A, EM of an X-body with proteinaceous beaded sheet aggregates (arrows) and small vacuoles (v). B, Immuno-EM with TGB1-specific antibody, showing heavy gold labeling of the beaded sheets within the X-body. C and D, TGB1-mCherry expressed from the viral genome (35S::PVX.TGB1-mCherry) decorates the X-bodies in infected cells (arrows in C) and is arranged in curved or semicircular aggregates morphologically similar to the beaded sheets. (Note that panel E, which shows cell outlines, is almost identical in magnification to C). E and F, GFP-CP expressed from a green overcoat virus construct (Santa Cruz et al., 1996) labels virions that form cages around the X-body. In E, weak nucleocytoplasmic GFP-CP fluorescence also highlights the nuclei next to X-bodies (arrows) and outlines the peripheral cytoplasm. G, Aggregates of TGB1-mCherry (expressed from a bombardment construct) localize within the cage formed by GFP-CP-decorated virions in green overcoat PVX-infected tissue. H, The same localization is observed when native TGB1 expressed from the virus is labeled by immunofluorescence. I, PUM-BiFC (Tilsner et al., 2009)-labeled PVX RNA forms circular whorls with dark centers (arrows) that are surrounded by, but do not colocalize with, an mCherry-CP fusion expressed from a red overcoat virus. J, TGB1-mCherry (expressed from a bombardment construct) is located within the centers of the PUM-BiFC-labeled vRNA whorls. All confocal images are maximum projections of entire z-stacks except for G and J, which are individual z-sections. Bars = 10 μm except for A (1 μm), B (250 nm), and C and E (50 μm).
For most experiments, we used PVX green and red “overcoat” constructs modified to express a fluorescent GFP- or mCherry-coat protein (CP) fusion (Santa Cruz et al., 1996, 1998; Tilsner et al., 2009). These overcoat viruses encapsidate and move locally and systemically (Santa Cruz et al., 1996), enabling the identification of infected cells and the imaging of encapsidated virions. GFP-CP virions accumulate in “cages” around the X-body, similar to wild-type virions observed by immuno-EM (Fig. 1, E–H; Oparka et al., 1996; Santa Cruz et al., 1998; Tilsner et al., 2009). TGB1-mCherry aggregates were localized within the spaces enclosed by these virion cages (Fig. 1G). The same localization was observed with a fluorescent antibody to TGB1 (Fig. 1H). As shown previously (Tilsner et al., 2009), mCherry-CP-encapsidated virions surround whorls of nonencapsidated vRNA visualized by PUM-BiFC (Fig. 1I), but the two signals do not overlap, with TGB1-mCherry localizing to the center of the whorls (Fig. 1J). Thus, the X-body has a layered structure, with TGB1 aggregates at the center, nonencapsidated vRNA in the middle, and encapsidated virions at the cytoplasmic periphery.
Host Endomembranes Are Recruited into the X-Body
Because PVX is known to replicate on ER membranes and induce their proliferation (Doronin and Hemenway, 1996; Bamunusinghe et al., 2009) and the presence of nonencapsidated vRNA suggested that the X-body might be a site of virus replication, we analyzed the contribution of endomembranes to the X-body using FPs targeted to the ER lumen (via an N-terminal signal peptide and a C-terminal HDEL ER-retention signal; Haseloff et al., 1997) or the Golgi membrane (membrane-tethered by a rat sialyl transferase transmembrane domain; Boevink et al., 1998). ER was heavily recruited into the X-body but remained excluded from some areas within it (Fig. 2, A–C). Within the X-body, the ER did not retain its usual structure as a polygonal tubular network (Sparkes et al., 2009) but was instead modified into small granules (Fig. 2, B and C) and/or diffuse membrane reservoirs within the X-body (Fig. 2D). These different structures likely represent progressive accumulation and remodeling of ER membranes. Colocalization with TGB1 showed that the ER membranes surrounded the TGB1 aggregates (Fig. 2E), similar to vRNA (Fig. 1J). Exclusion of ER from irregularly shaped areas with diffuse boundaries (Fig. 2, A–C) thus corresponds to the position of large TGB1 aggregates (Fig. 1A), whereas rounded unlabeled areas with clear boundaries are vacuoles (Figs. 1A and 2, A, B, and D).
Figure 2.
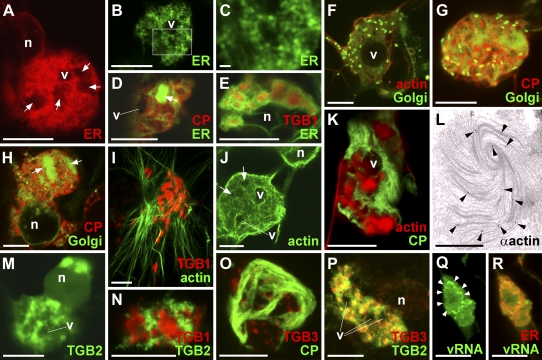
Reorganization of host components in the X-body. A, The X-body contains a large amount of remodeled ER (labeled by lumenally targeted FPs). Irregular areas from which the ER is excluded are indicated (arrows), as well as a vacuole (v; compare with Fig. 1A). n, Nucleus. B to D, Detailed examination of ER within the X-body shows rearrangement into granules (B and C) and diffuse membrane reservoirs (arrow; D). The box in B indicates the area enlarged in C. E, ER membranes within the X-body are wrapped around TGB1-mCherry aggregates, which explains the exclusion of ER from some nonvacuolar areas of the X-body in A to D (compare with Fig. 1A). F to H, Recruitment of GFP-labeled Golgi membranes into the X-body. Individual Golgi stacks cluster in a vacuolated X-body surrounded and traversed by a network of actin filaments (F). The X-body also contains diffuse Golgi membrane labeling. With increasing resorption of Golgi membranes into the X-body, this diffuse labeling becomes stronger (G). Eventually, the X-body no longer contains intact Golgi stacks but only disassembled Golgi membranes (H), which accumulate in diffuse membrane reservoirs, similar to ER (arrows). I, Aggregates of TGB1-mCherry (expressed from the viral genome) within the X-body reside in cages of FABD2-GFP-labeled actin filaments. J, Actin cables encircle the X-body, but finer filaments also criss-cross it. Dark areas containing TGB1 aggregates (arrows) and small vacuoles are visible. K, Texas Red-phalloidin labeling also stains more amorphous actin assemblies localized within the cages of GFP-CP overcoat virions similar to the localization of TGB1 (compare with Fig. 1, G and H). L, Anti-actin immuno-EM with gold-conjugated secondary antibody shows labeling of a beaded sheet TGB1 aggregate (arrowheads). An enlarged view of L is provided in Supplemental Figure S1. M and N, GFP-TGB2 within the X-body in infected cells localizes to sheets and granular vesicles that surround TGB1-mCherry aggregates (N), similar to ER membranes. TGB constructs were bombarded into infected tissue. O, Agroinfiltrated TGB3-TagRFP localizes to granular vesicles within the virion cage surrounding the X-body in a green overcoat PVX.GFP-CP infection. P, The X-body consists of TGB2/3-containing granular vesicles labeled by agroinfiltrated GFP-TGB2 and TGB3-TagRFP fusions. Note that the GFP-TGB2 fluorescence is more dispersed over the ER network than TGB3-TagRFP. Q and R, PUM-BiFC-labeled vRNA whorls (arrowheads) consist of granular hot spots (Q) that colocalize with ER (R). All confocal images are individual z-sections except for A to C, O, and P, which are maximum projections of entire z-stacks. Bars = 10 μm except for C (1 μm) and L (500 nm).
Surprisingly, Golgi bodies were also recruited into the X-body (Fig. 2, F–H). This was unexpected, because no association of the PVX TGB proteins or replicase with Golgi membranes is known (Ju et al., 2005; Bamunusinghe et al., 2009) and TGB3 does not traffic in the secretory pathway (Schepetilnikov et al., 2005). Individual Golgi bodies initially became clustered around the X-body (Fig. 2F). Later, the Golgi marker became more dispersed and was eventually found in the same diffuse membrane reservoirs as observed for the ER (Fig. 2, G and H). The presence of both lumenal and membrane-tethered endomembrane markers in the same compartment suggests that they are incorporated within densely stacked membranes that are heavily reorganized within the X-body.
F-Actin Is Recruited into the X-Body
In plants, the cortical ER and Golgi bodies are highly motile and attached to an underlying actin cytoskeleton (Boevink et al., 1998; Sparkes et al., 2009). As it was shown recently that PVX requires an intact actin cytoskeleton for cell-to-cell movement (Harries et al., 2009), we used in vivo imaging of F-actin with the Lifeact and Fimbrin Actin-Binding Domain2 (FABD2) markers (Sheahan et al., 2004; Riedl et al., 2008) to examine the influence of PVX infection on actin organization.
X-bodies in cells infected with PVX.TGB1-mCherry were associated with a dense meshwork of microfilaments and encircled by large actin cables (Fig. 2, I and J). When actin was labeled with the drug Texas Red-phalloidin, which both stains and stabilizes actin filaments (Cooper, 1987), large accumulations of amorphous actin were found within the X-body (Fig. 2K). These “balls” of phalloidin-stabilized actin were surrounded by aggregated virions, similar to the localization of the TGB1 protein (Fig. 1, G and H). To analyze this at higher resolution, we obtained immunogold-EM images using an actin-specific antibody. Gold particles were associated with the beaded sheets that harbor TGB1 (Fig. 2L; Supplemental Fig. S1), indicating that actin filaments may be directly linked to the inclusions.
In contrast to F-actin, when we tested the effects of PVX infection on transgenic plants expressing a GFP fusion to α-tubulin (Ueda et al., 1999), no intact microtubules were recruited to the X-body and only a diffuse pool of unpolymerized tubulin was apparent around it (Supplemental Fig. S2A).
The X-Body Consists of Aggregated Granular Vesicles Containing TGB2 and TGB3
Because both the TGB2/3-decorated ER-derived granular vesicles and the X-body appear to be possible sites of PVX replication, we also investigated the spatial relationship of TGB2/3 to the TGB1 aggregates within the X-body. TGB2 and -3 had previously been localized to the X-body by live-cell imaging, but due to high fluorescence intensity, no substructure was resolved (Ju et al., 2005; Samuels et al., 2007). Expression of GFP-TGB2 from a 35S promoter in infected tissue showed that the protein was localized in granules accumulated around the TGB1 aggregates within the X-body, very similar to the ER (Fig. 2, M and N). Similarly, TGB3-TagRFP (for red fluorescent protein) ectopically expressed in tissue infected with GFP-CP overcoat virus was localized in granules in the X-body, encased by virions (Fig. 2O). GFP-TGB2 and TGB3-TagRFP coexpressed in infected tissue showed clearly that the X-body consists of accumulated TGB2/3 granular vesicles (Figs. 2P and 3B). Because the TGB2/3 granular vesicles associate with ribosomes and viral replicase (Ju et al., 2005; Bamunusinghe et al., 2009) and localize in the same area as the replication-supporting ER (Doronin and Hemenway, 1996; Bamunusinghe et al., 2009) and nonencapsidated vRNA (Figs. 2, Q and R, and 3B), they are probably VRCs. Indeed, the vRNA whorls consisted of granular “hot spots” that colocalized with ER within the X-body, as shown by PUM-BiFC images with sufficient signal-to-noise ratio (Fig. 2, Q and R). When TGB3-labeled granular vesicles were colocalized with vRNA, they encircled the vRNA whorls (Fig. 3A). Thus, the nonencapsidated RNA is found between the granular vesicles comprising the putative replication sites and the aggregates of the TGB1 RNA helicase (Fig. 3B).
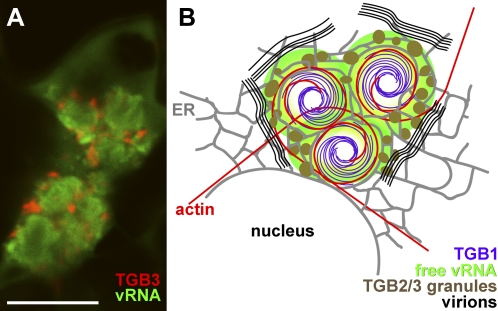
Figure 3.
Structure of the PVX X-body. A, TGB3-TagRFP-labeled granular vesicles surround the PUM-BiFC-labeled vRNA whorls. B, Schematic model of the X-body structure. Endomembranes (gray; ER) and actin microfilaments (red) are wrapped around the central TGB1 beaded sheet aggregates (purple) accumulating in the perinuclear region. Replicative granular vesicles containing TGB2 and -3 (brown) accumulate on the ER within the X-body. Nonencapsidated progeny vRNA (green) surrounds the TGB1 aggregates, whereas encapsidated virions (black) accumulate at the X-body periphery. The confocal image is an individual z-section. Bar = 10 μm.
To analyze the link between the X-body and the TGB2/3 granular vesicles, we observed the formation of the X-body during PVX infection in more detail. For this purpose, we focused on the leading edges of expanding fluorescent PVX overcoat lesions, which contain the most recently infected cells (i.e. are representative of early infection events). Toward the lesion center, the infection stages get progressively older. Using TGB3, TGB2, and PUM-BiFC reporter constructs, we observed various intermediate stages between TGB2/3 granular vesicles and the X-body from earlier to later infection stages (Fig. 4). At the leading edge, no single, perinuclear X-bodies were observed. TGB3-GFP, which labels the ER network in uninfected cells (Krishnamurthy et al., 2003), was recruited to the TGB2-induced granular vesicles in infected cells (Fig. 4, A and B), as described previously (Schepetilnikov et al., 2005; Samuels et al., 2007; Bamunusinghe et al., 2009). Approximately one to two cells behind the infection front, slightly larger TGB3 granular vesicles were associated with fluorescent overcoat virions (Fig. 4C). Within two to three cells into the lesion, the granular vesicles began to accumulate around larger virus packets. However, these aggregates were not perinuclear and still moved with the ER surface (Sparkes et al., 2009) or cytoplasmic streaming (Fig. 4D), similar to the granular vesicles (Boevink et al., 1996; Ju et al., 2005; Verchot-Lubicz et al., 2010). Similar small VRCs were also observed with TagRFP-TGB2 and PUM-BiFC reporters (Fig. 4, E and F). Eventually, these peripheral VRCs showed the vRNA whorls (Tilsner et al., 2009) and vacuolization typical of the X-body (Fig. 4G), before finally accumulating in the perinuclear region (Fig. 4H). Collectively, these data demonstrate continuity between the TGB2/3 granular vesicles and the X-body. Thus, the X-body constitutes a large virus “factory” comprising accumulated granular VRCs.
Figure 4.
Continuity between TGB2/3 granular vesicles and the X-body. A and B, At the leading edge of a PVX.mCherry-CP infection, TGB3-GFP fluorescence is ER associated in uninfected cells (top right cell; circular structures are chloroplasts, visible due to chlorophyll autofluorescence). In infected cells (red mCherry-CP labeling), TGB3-GFP is recruited to TGB2-induced, ER-associated granular vesicles (Ju et al., 2005; Samuels et al., 2007; Bamunusinghe et al., 2009). C, Farther into the infection site, TGB3-TagRFP-labeled granular vesicles are associated with GFP-CP-coated virion aggregates. D, Several TGB3-TagRFP granules have accumulated around a large virion raft. The insets show individual optical sections, whereas the main image is a maximum projection, showing motility of the VRC (time points from the first frame are indicated). E, With a TagRFP-TGB2 fusion, both large and small (boxed) VRCs with associated virions are visible in cells approximately two to three intercellular boundaries behind the infection front. n, Nucleus. F, PUM-BiFC vRNA imaging at the leading edge of an infection shows accumulation of the RNA reporter in medium-sized VRCs. G, Farther into the infection site, VRCs with intense PUM-BiFC signal are visible before a perinuclear X-body has accumulated. These smaller VRCs already show the vacuolization (arrowhead) and vRNA whorls typical of the X-body (inset). v, Vacuole. H, At a similar infection stage as in G, a group of accumulated VRCs showing typical vRNA whorls accumulates near the nucleus. Confocal images (C, D, D insets, G, G inset, and H) are individual z-sections. The other images (A, B, and d–F) are maximum projections of entire z-stacks, which encompassed the upper cortex to the median region of cells in A, B, and F and the median region only in D and E. Bars = 50 μm except for C, D, D insets, G inset, and H (10 μm).
TGB1 Is the Central Organizer of the X-Body
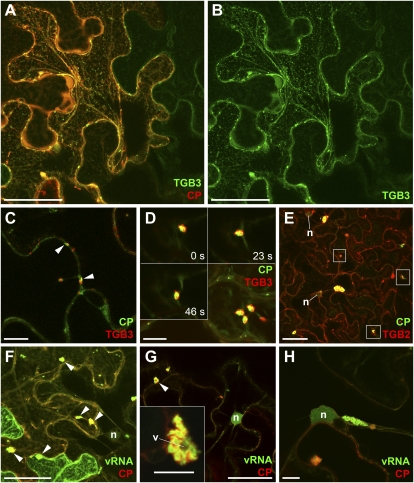
We next tested whether TGB1 aggregates could recruit actin, endomembranes, and TGB2/3 in the absence of other viral factors. When 35S-driven TGB1-mCherry was expressed in uninfected tissue, aggregates similar in shape and structure to those within the X-body were formed (Fig. 5). Using the same actin, ER, and Golgi markers as before, we found that all of these components were recruited to the TGB1 aggregates (Fig. 5, A–D). Importantly, the transmembrane Golgi marker did not remain restricted to individual Golgi stacks but formed sheets wrapped around the inclusion bodies, as occurs during infection (Fig. 5D). Similar to infected tissue, no intact microtubules were recruited by TGB1 (Supplemental Fig. S2B). When GFP-TGB2 or TGB3-GFP fusions were coexpressed with TGB1-mCherry, they were also recruited to the TGB1 aggregates (Fig. 5, E–G). Notably, the largest TGB1 aggregates, and their associated recruited organelles and TGB2/3, were found in the perinuclear region. Thus, TGB1 induces the formation of a “pseudo-X-body” in the absence of virus infection (Fig. 5, A–E).
Figure 5.
Generation of X-body-like structures by TGB1 alone. A and B, Ectopically expressed TGB1-mCherry in uninfected plants. TGB1 forms perinuclear assemblies of aggregates similar to infected tissue, which recruit FABD2-GFP-labeled actin filaments. C, Recruitment of ER by perinuclear TGB1-mCherry aggregates in uninfected tissue. The ER is wrapped tightly around the aggregates. D, Recruitment of Golgi membranes and disassembly of Golgi stacks by TGB1-mCherry. Similar to the ER, Golgi membranes are wrapped tightly around the perinuclear aggregates. A few individual Golgi stacks are visible (arrows). E and F, Recruitment of GFP-TGB2-labeled granular vesicles by perinuclear TGB1-mCherry aggregates. G, Recruitment of TGB3-GFP-labeled membranes. All confocal images are individual z-sections except for C, which is a maximum projection of an entire z-stack. n, Nucleus. Bars = 10 μm, except for G (5 μm).
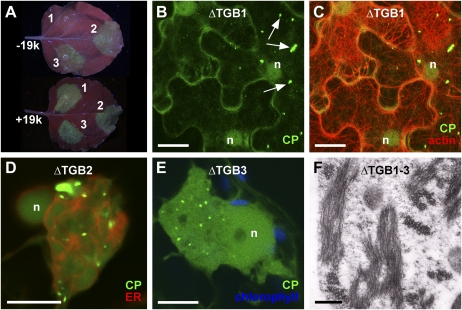
To further confirm the role of TGB1 in organizing the X-body, we individually knocked out all three TGB genes in a GFP-CP overcoat construct. To facilitate colocalizations with these movement-deficient (Beck et al., 1991; Verchot et al., 1998; Morozov and Solovyev, 2003; Verchot-Lubicz et al., 2010) viruses, their expression was driven from a 35S promoter (35S::PVX.ΔTGB1/2/3.GFP-CP). Agroinfiltration of the ΔTGB1 construct resulted in much weaker fluorescence than the ΔTGB2 and -3 mutants (Fig. 6A). Because TGB1 also functions as a suppressor of RNA silencing (Voinnet et al., 2000), we coexpressed the tomato bushy stunt virus 19k silencing suppressor to functionally complement for the lack of TGB1 silencing suppression activity (Voinnet et al., 2003), which restored the fluorescence of the ΔTGB1 construct to that of the other mutants (Fig. 6A). However, while the ΔTGB1 virus produced aggregates of encapsidated virions (Fig. 6B), no X-body and no rearrangement of actin microfilaments (Fig. 6C) or ER (Supplemental Fig. S3B) were observed. Without 19k, similar results were obtained but GFP-CP fluorescence was fainter (Supplemental Fig. S3). By contrast, the ΔTGB2 and ΔTGB3 viruses produced X-bodies (Fig. 6, D and E). Interestingly, the GFP-CP virions often formed dispersed punctae rather than strings or cages around the X-body (Fig. 6, D and E).
Figure 6.
TGB1, but not TGB2 or TGB3, is required for the formation of the X-body. A, Modified PVX genomes with one TGB gene knocked out and encoding the GFP-CP green overcoat fusion, expressed from a 35S promoter after agroinfiltration (1, PVX.ΔTGB1.GFP-CP; 2, PVX.ΔTGB2.GFP-CP; 3, PVX.ΔTGB3.GFP-CP). Without the tomato bushy stunt virus 19k silencing suppressor (−19k), the absence of TGB1 results in strongly attenuated GFP-CP fluorescence, whereas coexpression of 19k (+19k) restores fluorescence to the level of the TGB2 and -3 knockouts. B and C, In the absence of TGB1, no perinuclear X-body is formed, even though green overcoat virions accumulate (arrows). Lifeact-TagRFP-labeled F-actin shows no actin cage in the perinuclear region. D and E, In the absence of TGB2 (D) or TGB3 (E), a perinuclear X-body is visible, which shows rearranged but not granular ER (D). Note the accumulation of virions in punctae at the X-body periphery, rather than strings or cages. In E, chlorophyll autofluorescence is shown for contrast. F, In N. benthamiana protoplasts infected with PVX lacking all three TGB genes, no X-body is formed, but virions still accumulate. Confocal images (B and C) are maximum projections of entire z-stacks, whereas D and E are individual z-sections. n, Nucleus. Bars = 10 μm except for F (500 nm).
The X-Body Is Not Required for Virus Accumulation and Encapsidation
The TGB proteins are not required for PVX replication (Beck et al., 1991; Verchot et al., 1998; Batten et al., 2003; Morozov and Solovyev, 2003; this study). A TGB1-organized X-body was also not necessary for virus accumulation if an alternative silencing suppressor was supplied. Furthermore, in the absence of any of the individual TGB proteins, virion assembly still occurred, as aggregates of fluorescent virions were visible (Fig. 6, B, D, and E). To confirm that the TGB proteins are not required for PVX encapsidation, we also expressed a virus with a deletion of the entire triple gene block (PVX.ΔTGB.GFP-CP; Santa Cruz et al., 1998) in protoplasts. Assembled virus particles were visible in the EM images of these protoplasts (Fig. 6F). However, in the individual TGB mutants, packets of assembled fluorescent virions were less abundant than in overcoat constructs with an intact TGB (compare Fig. 1, E and F, with Fig. 6, B, D, and E). This indicates that virus assembly was less efficient in the mutants, possibly due to the changes in VRC compartmentation.
DISCUSSION
What Is the Role of the X-Body?
RNA viruses induce the formation of viral replication complexes on reorganized host membranes, which serve as scaffolds and protective compartments for virus replication and can be involved in virus assembly (Sanfaçon, 2005; Miller and Krijnse-Locker, 2008; den Boon et al., 2010; Laliberté and Sanfaçon, 2010). Several observations make it highly likely that the intricately structured PVX X-body (Fig. 3B) is a large replication site. These include the incorporation of TGB2/3-granular vesicles associated with replicase and ribosomes (Ju et al., 2005; Bamunusinghe et al., 2009), the localization of nonencapsidated vRNA within the X-body (Tilsner et al., 2009; this study), the accumulation of progeny virions at its periphery (Oparka et al., 1996; Santa Cruz et al., 1998; Tilsner et al., 2009; this study), and the dependence of the arrangement of virions on the presence of the granule-forming TGB2/3 proteins. Here, we have shown that the TGB1 protein is the viral gene product that organizes the X-body, adding another dimension to this highly multifunctional protein. The extensive rearrangements of actin and endomembranes induced by TGB1, and the resorption of the Golgi and ER into the same subcellular compartment, are reminiscent of the effects of the fungal toxin brefeldin A on plant cells (Boevink et al., 1998), although it remains unclear whether TGB1 exerts these effects through direct or indirect membrane interactions.
None of the TGB proteins are required for replication, as demonstrated by individual and combined knockout mutants (Beck et al., 1991; Verchot et al., 1998; Batten et al., 2003; Morozov and Solovyev, 2003; this study), and limited viral replication and assembly can proceed without an X-body. However, the intricate and organized structure of the X-body indicates that it fulfills a function during infection. One possibility is that it compartmentalizes TGB1 at late infection stages. Potexviruses are mechanically transmitted (Adams et al., 2004), and maximizing the production of progeny virions might be the main “objective” once cell-to-cell movement has been accomplished. TGB1 functions as a MP but also as a translational activator that destabilizes virus particles by binding CP subunits at their 5′ end (Atabekov et al., 2000; Kiselyova et al., 2003; Rodionova et al., 2003; Karpova et al., 2006). This destabilization would be problematic for mechanical virion transmission. Indeed, PVX virions isolated from infected tissue are nontranslatable, indicating that they are not bound to TGB1 (Karpova et al., 1997; Atabekov et al., 2000). Additionally, TGB1 and CP coaggregate in vitro, and Karpova et al. (2006) have speculated that a “direct interaction of unassembled CP and TGB1 does not occur in vivo.” Removal of excess TGB1 protein into aggregates may thus be necessary to enable or maximize virion production during later stages of infection. Interestingly, the TGB1 proteins of other potexviruses also form aggregates (Rouleau et al., 1994; Chang et al., 1997) that associate with viral RNA (Lin et al., 1993), and many potexviruses produce protein aggregates surrounded by accumulating virions (Zettler et al., 1968; Diaz-Ruiz and Feldman, 1990; Lin et al., 1993; Rouleau et al., 1994; Chang et al., 1997), indicating a possible conserved regulatory mechanism.
In the absence of any or all three of the TGB proteins, virus assembly can still proceed, but at reduced efficiency. Encapsidation of nonenveloped RNA viruses can be functionally coupled to membrane-associated replication even though membranes are not directly required for virion formation, possibly to facilitate packaging specificity or efficiency (Annamalai and Rao, 2005; Venter et al., 2005; Annamalai and Rao, 2006; Shin et al., 2010). In vitro, full-length filamentous PVX particles cannot be assembled from vRNA and CP (Karpova et al., 2006). Thus, the organization of PVX VRCs into the X-body, while not absolutely required for encapsidation, may function to increase the efficiency of vRNA packaging in a natural infection. Additionally, potexviral CP mutants defective for interaction with TGB1 can encapsidate virus particles in vitro and in protoplasts but fail to do so in intact tissue (Fedorkin et al., 2000; Zayakina et al., 2008; J. Tilsner and K.J. Oparka, unpublished data). Hsu et al. (2004) have shown that aggregates of the TGB1 protein of the related Bamboo mosaic potexvirus are a pool of active protein and suggested that the inclusions were sites of vRNA processing. In replication-coupled packaging of Turnip yellow mosaic virus, the protease/helicase domain of the viral replicase has been implicated in virion assembly (Shin et al., 2010). It is tempting to speculate that expression of the CP-interacting TGB1 movement protein, which has an RNA helicase domain related to viral replicases (Morozov and Solovyev, 2003), continues during later infection stages because TGB1 plays some role in linking PVX replication and encapsidation.
TGB1-Mediated Actin/Endomembrane Remodeling
Besides VRC compartmentation, it is possible that the actin/endomembrane-remodeling activities of TGB1 are also related to its functions as a suppressor of RNA silencing and/or its role as a MP. The current model for silencing suppression by TGB1 is that it targets Argonaute1 for degradation (Chiu et al., 2010). While this implicates no obvious requirement for actin/ER remodeling, miRNA-induced silencing complexes containing Argonaute2, another TGB1 interactor (Chiu et al., 2010), are associated with endosomal membranes (Gibbings et al., 2009), raising the possibility that other silencing pathways might also be endomembrane associated. On the other hand, an intact actin cytoskeleton is required for PVX cell-to-cell movement (Harries et al., 2009), and both actin and the ER are components of plasmodesmata (Tilsner et al., 2011). Recently, we have shown that TGB1 is required for directing PVX CP into plasmodesmata (J. Tilsner and K.J. Oparka, unpublished data). It is thus very likely that actin/ER remodeling by TGB1 also plays a role in dilating plasmodesmata (Angell et al., 1996; Yang et al., 2000; Howard et al., 2004; Tilsner et al., 2011) and inserting PVX into the intercellular channels.
In summary, the long-familiar X-body emerges as a structure that is induced specifically by the remarkably versatile PVX TGB1, a protein that appears to play multiple roles in the organization of virus compartmentation throughout infection. To understand its multiple roles, it will now be important to characterize in detail how TGB1 interacts with endomembranes and actin during infection.
MATERIALS AND METHODS
Plasmid Constructions
See Supplemental Table S1 for primers. The virus constructs PVX.GFP-CP (Santa Cruz et al., 1996), PVX.pum-mCherry-CP (Tilsner et al., 2009), and PVX.ΔTGB.GFP-CP (Santa Cruz et al., 1998), carrying modified cDNAs of the PVX genome under the control of a T7 promoter, and the binary vector 35S::PVX.GFP (pGR106.GFP; Jones et al., 1999) have been described previously. pGR106.TGB1-mCherry was generated by assembling two PCR products in the cloning vector pGEM-T Easy (Promega): a NotI-TGB1-mCherry-PmeI-SacI fragment was generated by overlap PCR with primers Not-TGB1for, Cher-TGB1rev, TGB1-Cherfor, and Sac-Cherrev and ligated into NotI/SacI-treated pGEM-T Easy; a PmeI-TGB2/3-SacI fragment was amplified with primers Pme-TGB2for/Sac-TGB3rev and ligated into the PmeI/SacI sites of the previous construct. The complete TGB1-mCherry.TGB2/3 cassette was then subcloned into the ApaI/AscI sites of pGR106.GFP, and finally, the GFP gene was removed by AscI/NotI digestion, blunting, and religation. To generate TGB deletion mutants in pGR106.GFP, the entire triple gene block was amplified with Not-TGB1for/Sac-TGB3rev primers and ligated into NotI/SacI-treated pGEM-T Easy. Frame-shift mutations in the TGB2 and TGB3 genes were generated by digestion with XbaI or EcoNI, respectively, blunting, and religation. The ΔTGB2 and ΔTGB3 triple gene bock cassettes were reinserted into pGR106.GFP as ApaI/AscI fragments. The ΔTGB1 construct was created by treating pGR106.GFP with ApaI, blunting, and religating. The GFP open reading frame of the mutant constructs was replaced with the GFP-CP fusion (Santa Cruz et al., 1996) inserted as an EagI (blunted)/SpeI fragment into the AscI (blunted)/SpeI sites.
The bombardment constructs pRTL2.GFP-TGB2, pRTL2.TGB3-GFP (Ju et al., 2005), and pRTL2.TGB1-mCherry (Tilsner et al., 2009) were described elsewhere. Expression vectors for agroinfiltration of TGB constructs were obtained by amplifying individual open reading frames with or without stop codons as appropriate and with Gateway adaptors. The PCR products were recombined into the pDONR207 vector using Gateway recombinase (Invitrogen). Sequenced inserts were recombined into the destination vectors pGWB402Ω (unfused expression), pGWB405 (C-terminal GFP fusion), pGWB406 (N-terminal GFP fusion), pGWB460 (C-terminal TagRFP fusion), and pGWB461 (N-terminal TagRFP fusion; Nakagawa et al., 2007) as required.
The construction of the PUM-BiFC RNA-imaging system was described by Tilsner et al. (2009). For agroinfiltration, the PUMHD-split-mCitrine fusions were subcloned into pENTR1A (Invitrogen) and then recombined into pGWB402Ω (Nakagawa et al., 2007).
The generation of the Lifeact-TagRFP actin marker was described by Berepiki et al. (2010). Binary vectors for transient expression of Lifeact-TagRFP and ER-TagRFP markers were generated using Gateway technology. The ER-TagRFP construct was generated by adding an N-terminal signal peptide and a C-terminal HDEL ER retention signal (Boevink et al., 1996) through PCR extension. Lifeact-TagRFP and ER-TagRFP constructs were amplified with Gateway adapters, and the respective PCR products were recombined into pDONR207, sequenced, and recombined into pGWB402Ω. The plasmid pBIN.19k was described by Voinnet et al. (2003).
Plant Material, and Transgenic and Transient Reporter Expression
Nicotiana benthamiana was used as the experimental host. Plants were grown at 20°C with a 16-h/8-h light/dark cycle. T7 polymerase in vitro transcription of PVX cDNA clones and leaf inoculation were as described by Santa Cruz et al. (1998). For coexpression of FP reporters in infected tissue, transgenic plants and transient expression were used. Transgenic N. benthamiana lines expressing ER-GFP (Haseloff et al., 1997), FABD2-GFP (Sheahan et al., 2004), Golgi-GFP (Boevink et al., 1998), and α-tubulin-GFP (Ueda et al., 1999) were described previously. Microprojectile bombardment and agroinfiltration of 35S expression constructs were done as described by Tilsner et al. (2009) and Voinnet et al. (2003), respectively. Agrobacteria were infiltrated at an optical density at 600 nm of 0.5 for individuals, 0.25 each for combinations of two, and 0.2 each for three constructs. The Agrobacterium tumefaciens strain AGL1 was used with pSoup helper plasmid for pGR106 derivatives (Jones et al., 1999; Hellens et al., 2000). The isolation and PVX infection of N. benthamiana protoplasts were done as described by Chapman et al. (1992).
Confocal Imaging
Infected, agroinfiltrated, or bombarded leaves were removed from plants and mounted, whole, onto glass microscope slides using double-sided sticky tape (Banner). Virus-expressed constructs were imaged after 2 to 5 d for inoculated leaves and 5 to 16 d for systemic leaves, agroinfiltrated constructs after 2 to 4 d, and bombarded constructs after 1 to 2 d. All imaging was performed on a Leica SP2 microscope equipped with water-dipping lenses (Leica) with the following excitation wavelengths: mCitrine (PUM-BiFC), 514 nm; TagRFP, AlexaFluor568, and Texas Red, 561 nm; GFP, 488 nm; mCherry, 594 nm. Detection ranges were optimized for each fluorophore combination to minimize bleed-through, and, where necessary, sequential scanning was used. Microscope power settings were adjusted to optimize contrast for each individual image. Images were collected using Leica LCS software and imported into Adobe Photoshop (Adobe Systems) for the preparation of figures. Staining of actin with Texas Red-phalloidin (Invitrogen/Molecular Probes) was carried out as described by Goodbody and Lloyd (1990).
Immunofluorescence Detection of TGB1
Infected leaf patches were excised and mounted on slides, and the upper epidermis was abraded. The exposed tissue was fixed with 4% paraformaldehyde in 50 mm PIPES, 5 mm EGTA, and 5 mm MgSO4 (PEM). After washing in PEM, the tissue was digested with 0.2% cellulase, 0.005% Triton X-100, and 0.01% bovine serum albumin (BSA) in PEM. The buffer was then changed to phosphate-buffered saline (PBS; 137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, and 2 mm KH2PO4), and the tissue was permeabilized with 1% Triton X-100 and 0.05% Nonidet in PBS. After washing with 0.05% Tween 20 in PBS, samples were blocked with 2.5% BSA and 0.05% Tween 20 in PBS. Incubation with anti-TGB1 primary antibody (Santa Cruz et al., 1998), 1:100 in PBS with 2.5% BSA, was done overnight at 4°C. Samples were washed for at least 2 h in 1% BSA and 0.05% Tween 20 in PBS before incubation with secondary antibody (AlexaFluor568-conjugated goat anti-rabbit [Invitrogen/Molecular Probes], 1:200 in PBS with 1% BSA) for 1 h at room temperature. Before imaging, the tissue was again washed in 1% BSA in PBS.
EM
Infected leaves were fixed in glutaraldehyde and postfixed in osmium tetroxide as described previously (Roberts, 1994). For immunogold labeling of actin and PVX TGB1, the procedures and TGB1 antibody described by Oparka et al. (1996) and Santa Cruz et al. (1998) were used. Rabbit anti-actin antibody was obtained from Invitrogen/Molecular Probes. Conventional and immunogold electron micrographs were obtained on a JEOL 1200EXII electron microscope.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Actin-specific immuno-electron micrograph of a TGB1 aggregate.
Supplemental Figure S2. No recruitment of microtubules to the X-body by TGB1.
Supplemental Figure S3. Absence of X-body in tissue infected by a PVX ΔTGB1 mutant.
Supplemental Table S1. Oligonucleotide primers used in this study.
Acknowledgments
We thank David Baulcombe for the pGR106.GFP plasmid and Jeanmarie Verchot-Lubicz for the pRTL2.GFP-TGB2 and pRTL2.TGB3-GFP constructs. We thank David Baulcombe for the pGR106.GFP plasmid, Jeanmarie Verchot-Lubicz for the pRTL2.GFP-TGB2 and pRTL2.TGB3-GFP constructs, and Tsuyoshi Nakagawa for the pGWB vectors.
References
- Adams MJ, Antoniw JF, Bar-Joseph M, Brunt AA, Candresse T, Foster GD, Martelli GP, Milne RG, Zavriev SK, Fauquet CM. (2004) The new plant virus family Flexiviridae and assessment of molecular criteria for species demarcation. Arch Virol 149: 1045–1060 [DOI] [PubMed] [Google Scholar]
- Angell SM, Davies C, Baulcombe DC. (1996) Cell-to-cell movement of potato virus X is associated with a change in the size-exclusion limit of plasmodesmata in trichome cells of Nicotiana clevelandii. Virology 216: 197–201 [DOI] [PubMed] [Google Scholar]
- Annamalai P, Rao ALN. (2005) Replication-independent expression of genome components and capsid protein of brome mosaic virus in planta: a functional role for viral replicase in RNA packaging. Virology 338: 96–111 [DOI] [PubMed] [Google Scholar]
- Annamalai P, Rao ALN. (2006) Packaging of brome mosaic virus subgenomic RNA is functionally coupled to replication-dependent transcription and translation of coat protein. J Virol 80: 10096–10108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atabekov JG, Rodionova NP, Karpova OV, Kozlovsky SV, Poljakov VY. (2000) The movement protein-triggered in situ conversion of potato virus X virion RNA from a nontranslatable into a translatable form. Virology 271: 259–263 [DOI] [PubMed] [Google Scholar]
- Bamunusinghe D, Hemenway CL, Nelson RS, Sanderfoot AA, Ye CM, Silva MAT, Payton M, Verchot-Lubicz J. (2009) Analysis of potato virus X replicase and TGBp3 subcellular locations. Virology 393: 272–285 [DOI] [PubMed] [Google Scholar]
- Batten JS, Yoshinari S, Hemenway C. (2003) Potato virus X: a model system for virus replication, movement and gene expression. Mol Plant Pathol 4: 125–131 [DOI] [PubMed] [Google Scholar]
- Bayne EH, Rakitina DV, Morozov SY, Baulcombe DC. (2005) Cell-to-cell movement of potato potexvirus X is dependent on suppression of RNA silencing. Plant J 44: 471–482 [DOI] [PubMed] [Google Scholar]
- Beck DL, Guilford PJ, Voot DM, Andersen MT, Forster RLS. (1991) Triple gene block proteins of white clover mosaic potexvirus are required for transport. Virology 183: 695–702 [DOI] [PubMed] [Google Scholar]
- Berepiki A, Lichius A, Shoji JY, Tilsner J, Read ND. (2010) F-actin dynamics in Neurospora crassa. Eukaryot Cell 9: 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink P, Oparka KJ, Santa Cruz S, Martin B, Betteridge A, Hawes C. (1998) Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J 15: 441–447 [DOI] [PubMed] [Google Scholar]
- Boevink P, Santa Cruz S, Hawes C, Harris N, Oparka KJ. (1996) Virus-mediated delivery of the green fluorescent protein to the endoplasmic reticulum of plant cells. Plant J 10: 935–941 [Google Scholar]
- Chang BY, Lin NS, Liou DY, Chen JP, Liou GG, Hsu YH. (1997) Subcellular localization of the 28 kDa protein of the triple-gene-block of bamboo mosaic potexvirus. J Gen Virol 78: 1175–1179 [DOI] [PubMed] [Google Scholar]
- Chapman S, Kavanagh T, Baulcombe D. (1992) Potato virus X as a vector for gene expression in plants. Plant J 2: 549–557 [DOI] [PubMed] [Google Scholar]
- Chiu MH, Chen IH, Baulcombe DC, Tsai CH. (2010) The silencing suppressor P25 of Potato virus X interacts with Argonaute1 and mediates its degradation through the proteasome pathway. Mol Plant Pathol 11: 641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA. (1987) Effects of cytochalasin and phalloidin on actin. J Cell Biol 105: 1473–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies C, Hills G, Baulcombe DC. (1993) Sub-cellular localization of the 25-kDa protein encoded in the triple gene block of potato virus X. Virology 197: 166–175 [DOI] [PubMed] [Google Scholar]
- den Boon JA, Diaz A, Ahlquist P. (2010) Cytoplasmic viral replication complexes. Cell Host Microbe 8: 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Ruiz JR, Feldman JM. (1990) Crystalline structures associated with the infection of the potexvirus argentine plantago virus (AplaV). J Phytopathol 129: 187–192 [Google Scholar]
- Doronin SV, Hemenway C. (1996) Synthesis of potato virus X RNAs by membrane-containing extracts. J Virol 70: 4795–4799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorkin ON, Merits A, Lucchesi J, Solovyev AG, Saarma M, Morozov SY, Mäkinen K. (2000) Complementation of the movement-deficient mutations in potato virus X: potyvirus coat protein mediates cell-to-cell trafficking of C-terminal truncation but not deletion mutant of potexvirus coat protein. Virology 270: 31–42 [DOI] [PubMed] [Google Scholar]
- Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. (2009) Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol 11: 1143–1149 [DOI] [PubMed] [Google Scholar]
- Goldstein B. (1924) Cytological study of living cells of tobacco plants affected with mosaic disease. Bull Torrey Bot Club 51: 261–272 [Google Scholar]
- Goodbody KC, Lloyd CW. (1990) Actin filaments line up across Tradescantia epidermal cells, anticipating wound-induced division planes. Protoplasma 157: 92–101 [Google Scholar]
- Harries PA, Park JW, Sasaki N, Ballard KD, Maule AJ, Nelson RS. (2009) Differing requirements for actin and myosin by plant viruses for sustained intercellular movement. Proc Natl Acad Sci USA 106: 17594–17599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S. (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94: 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Howard AR, Heppler ML, Ju HJ, Krishnamurthy K, Payton ME, Verchot-Lubicz J. (2004) Potato virus X TGBp1 induces plasmodesmata gating and moves between cells in several host species whereas CP moves only in N. benthamiana leaves. Virology 328: 185–197 [DOI] [PubMed] [Google Scholar]
- Hsu H-T, Hsu Y-H, Bi I-P, Lin N-S, Chang B-Y. (2004) Biological functions of the cytoplasmic TGBp1 inclusions of bamboo mosaic potexvirus. Arch Virol 149: 1027–1035 [DOI] [PubMed] [Google Scholar]
- Jones L, Hamilton AJ, Voinnet O, Thomas CL, Maule AJ, Baulcombe DC. (1999) RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell 11: 2291–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju H-J, Samuels TD, Wang Y-S, Blancaflor E, Payton M, Mitra R, Krishnamurthy K, Nelson RS, Verchot-Lubicz J. (2005) The potato virus X TGBp2 movement protein associates with endoplasmic reticulum-derived vesicles during virus infection. Plant Physiol 138: 1877–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinina NO, Rakitina DV, Solovyev AG, Schiemann J, Morozov SY. (2002) RNA helicase activity of the plant virus movement proteins encoded by the first gene of the triple gene block. Virology 296: 321–329 [DOI] [PubMed] [Google Scholar]
- Karpova OV, Ivanov KI, Rodionova NP, Dorokhov YuL, Atabekov JG. (1997) Nontranslatability and dissimilar behavior in plants and protoplasts of viral RNA and movement protein complexes formed in vitro. Virology 230: 11–21 [DOI] [PubMed] [Google Scholar]
- Karpova OV, Zayakina OV, Arkhipenko MV, Sheval EV, Kiselyova OI, Poljakov VY, Yaminsky IV, Rodionova NP, Atabekov JG. (2006) Potato virus X RNA-mediated assembly of single-tailed ternary ‘coat protein-RNA-movement protein’ complexes. J Gen Virol 87: 2731–2740 [DOI] [PubMed] [Google Scholar]
- Kiselyova OI, Yaminsky IV, Karpova OV, Rodionova NP, Kozlovsky SV, Arkhipenko MV, Atabekov JG. (2003) AFM study of potato virus X disassembly induced by movement protein. J Mol Biol 332: 321–325 [DOI] [PubMed] [Google Scholar]
- Kozar FE, Sheludko YM. (1969) Ultrastructure of potato and Datura stramonium plant cells infected with potato virus X. Virology 38: 220–229 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy K, Heppler M, Mitra R, Blancaflor E, Payton M, Nelson RS, Verchot-Lubicz J. (2003) The potato virus X TGBp3 protein associates with the ER network for virus cell-to-cell movement. Virology 309: 135–151 [DOI] [PubMed] [Google Scholar]
- Laliberté J-F, Sanfaçon H. (2010) Cellular remodeling during plant virus infection. Annu Rev Phytopathol 48: 69–91 [DOI] [PubMed] [Google Scholar]
- Leshchiner AD, Minina EA, Rakitina DV, Vishnichenko VK, Solovyev AG, Morozov SY, Kalinina NO. (2008) Oligomerization of the potato virus X 25-kD movement protein. Biochemistry (Mosc) 73: 50–55 [DOI] [PubMed] [Google Scholar]
- Lin NS, Chen CC, Hsu YH. (1993) Post-embedding in situ hybridization for localization of viral nucleic acid in ultra-thin sections. J Histochem Cytochem 41: 1513–1519 [DOI] [PubMed] [Google Scholar]
- Lucas WJ. (2006) Plant viral movement proteins: agents for cell-to-cell trafficking of viral genomes. Virology 344: 169–184 [DOI] [PubMed] [Google Scholar]
- Martelli GP, Russo M. (1977) Plant virus inclusion bodies. Adv Virus Res 21: 175–266 [DOI] [PubMed] [Google Scholar]
- Miller S, Krijnse-Locker J. (2008) Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol 6: 363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov SY, Solovyev AG. (2003) Triple gene block: modular design of a multifunctional machine for plant virus movement. J Gen Virol 84: 1351–1366 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. (2007) Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Oparka KJ, Roberts AG, Roberts IM, Prior DAM, Santa Cruz S. (1996) Viral coat protein is targeted to, but does not gate, plasmodesmata during cell-to-cell movement of potato virus X. Plant J 10: 805–813 [Google Scholar]
- Ozawa T, Natori Y, Sato M, Umezawa Y. (2007) Imaging dynamics of endogenous mitochondrial RNA in single living cells. Nat Methods 4: 413–419 [DOI] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. (2008) Lifeact: a versatile marker to visualize F-actin. Nat Methods 5: 605–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts IM. (1994) Factors affecting the efficiency of immunogold labelling of plant virus antigens in thin sections. J Virol Methods 50: 155–166 [DOI] [PubMed] [Google Scholar]
- Rodionova NP, Karpova OV, Kozlovsky SV, Zayakina OV, Arkhipenko MV, Atabekov JG. (2003) Linear remodeling of helical virus by movement protein binding. J Mol Biol 333: 565–572 [DOI] [PubMed] [Google Scholar]
- Rouleau M, Smith RJ, Bancroft JB, Mackie GA. (1994) Purification, properties, and subcellular localization of foxtail mosaic potexvirus 26-kDa protein. Virology 204: 254–265 [DOI] [PubMed] [Google Scholar]
- Samuels TD, Ju H-J, Ye C-M, Motes CM, Blancaflor EB, Verchot-Lubicz J. (2007) Subcellular targeting and interactions among the Potato virus X TGB proteins. Virology 367: 375–389 [DOI] [PubMed] [Google Scholar]
- Sanfaçon H. (2005) Replication of positive-strand RNA viruses in plants: contact points between plant and virus components. Can J Bot 83: 1529–1549 [Google Scholar]
- Santa Cruz S, Chapman S, Roberts AG, Roberts IM, Prior DAM, Oparka KJ. (1996) Assembly and movement of a plant virus carrying a green fluorescent protein overcoat. Proc Natl Acad Sci USA 93: 6286–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa Cruz S, Roberts AG, Prior DAM, Chapman S, Oparka KJ. (1998) Cell-to-cell and phloem-mediated transport of potato virus X: the role of virions. Plant Cell 10: 495–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov MV, Manske U, Solovyev AG, Zamyatnin AA, Jr, Schiemann J, Morozov SY. (2005) The hydrophobic segment of Potato virus X TGBp3 is a major determinant of the protein intracellular trafficking. J Gen Virol 86: 2379–2391 [DOI] [PubMed] [Google Scholar]
- Shalla TA, Shepard JF. (1972) The structure and antigenic analysis of amorphous inclusion bodies induced by potato virus X. Virology 49: 654–667 [DOI] [PubMed] [Google Scholar]
- Sheahan MB, Staiger CJ, Rose RJ, McCurdy DW. (2004) A green fluorescent protein fusion to actin-binding domain 2 of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol 136: 3968–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin HI, Kim HY, Cho TJ. (2010) The Pro/Hel region is indispensable for packaging non-replicating turnip yellow mosaic virus RNA, but not replicating viral RNA. Mol Cells 29: 463–469 [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Frigerio L, Tolley N, Hawes C. (2009) The plant endoplasmic reticulum: a cell-wide web. Biochem J 423: 145–155 [DOI] [PubMed] [Google Scholar]
- Stols ALH, Hill-van der Meulen GW, Toen MKI. (1970) Electron microscopy of Nicotiana glutinosa leaf cells infected with potato virus X. Virology 40: 168–170 [DOI] [PubMed] [Google Scholar]
- Tilsner J, Amari K, Torrance L. (2011) Plasmodesmata viewed as specialised membrane adhesion sites. Protoplasma 248: 39–60 [DOI] [PubMed] [Google Scholar]
- Tilsner J, Linnik O, Christensen NM, Bell K, Roberts IM, Lacomme C, Oparka KJ. (2009) Live-cell imaging of viral RNA genomes using a Pumilio-based reporter. Plant J 57: 758–770 [DOI] [PubMed] [Google Scholar]
- Ueda K, Matsuyama T, Hashimoto T. (1999) Visualization of microtubules in living cells of transgenic Arabidopsis thaliana. Protoplasma 206: 201–206 [Google Scholar]
- Venter PA, Krishna NK, Schneemann A. (2005) Capsid protein synthesis from replicating RNA directs specific packaging of the genome of a multipartite, positive-strand RNA virus. J Virol 79: 6239–6248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot J, Angell SM, Baulcombe DC. (1998) In vivo translation of the triple gene block of potato virus X requires two subgenomic mRNAs. J Virol 72: 8316–8320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot-Lubicz J, Torrance L, Solovyev AG, Morozov SY, Jackson AO, Gilmer D. (2010) Varied movement strategies employed by triple gene block-encoding viruses. Mol Plant Microbe Interact 23: 1231–1247 [DOI] [PubMed] [Google Scholar]
- Verchot-Lubicz J, Ye CM, Bamunusinghe D. (2007) Molecular biology of potexviruses: recent advances. J Gen Virol 88: 1643–1655 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Lederer C, Baulcombe DC. (2000) A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103: 157–167 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Yang Y, Ding B, Baulcombe DC, Verchot J. (2000) Cell-to-cell movement of the 25K protein of Potato virus X is regulated by three other viral proteins. Mol Plant Microbe Interact 13: 599–605 [DOI] [PubMed] [Google Scholar]
- Zayakina O, Arkhipenko M, Kozlovsky S, Nikitin N, Smirnov A, Susi P, Rodionova N, Karpova O, Atabekov J. (2008) Mutagenic analysis of potato virus X movement protein (TGBp1) and the coat protein (CP): in vitro TGBp1-CP binding and viral RNA translation activation. Mol Plant Pathol 9: 37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zettler FW, Edwardson JR, Purcifull DE. (1968) Ultramicroscopic differences in inclusions of papaya mosaic virus and papaya ringspot virus correlated with differential aphid transmission. Phytopathology 58: 332–335 [Google Scholar]