Abstract
Recent advances in -omics technologies such as transcriptomics, metabolomics, and proteomics along with genotypic profiling have permitted dissection of the genetics of complex traits represented by molecular phenotypes in nonmodel species. To identify the genetic factors underlying variation in primary metabolism in potato (Solanum tuberosum), we have profiled primary metabolite content in a diploid potato mapping population, derived from crosses between S. tuberosum and wild relatives, using gas chromatography-time of flight-mass spectrometry. In total, 139 polar metabolites were detected, of which we identified metabolite quantitative trait loci for approximately 72% of the detected compounds. In order to obtain an insight into the relationships between metabolic traits and classical phenotypic traits, we also analyzed statistical associations between them. The combined analysis of genetic information through quantitative trait locus coincidence and the application of statistical learning methods provide information on putative indicators associated with the alterations in metabolic networks that affect complex phenotypic traits.
The variation observed in phenotypic trait values in plants is often of quantitative nature, and it remains challenging to unravel the genetic basis of these traits. Quantitative trait locus (QTL) mapping is currently the most commonly used approach to dissect the genetic factors underlying complex traits. The goal of QTL mapping is to identify genomic regions associated with a specific complex phenotype by statistical analysis of the associations between genetic markers and phenotypic variation (Doerge, 2002). Recently, advances in high-throughput analysis and analytical detection methods have facilitated more integrated approaches to measure global phenotypic variation at the molecular level. Metabolite profiling is a rapidly evolving technology that has significantly increased the possibilities of performing high-throughput analysis of hundreds to thousands of compounds in a range of plants, including complex crop species. Metabolite composition is of great importance in crop plants, as a number of important traits such as biotic and abiotic stress resistance, postharvest processing, and nutritional value are largely dependent on the metabolic content (Fernie and Schauer, 2009).
In potato (Solanum tuberosum) breeding, metabolomic studies have progressively increased in importance, as many potato tuber traits such as content and quality of starch, chipping quality, flesh color, taste, and glycoalkaloid content have been shown to be linked to a wide range of metabolites (Coffin et al., 1987; Dobson et al., 2008). As a result, tuber quality can be assessed by assaying a range of metabolites. Gas chromatography-time of flight-mass spectrometry (GC-TOF-MS) has been shown to be useful for the rapid and highly sensitive detection of a large fraction of plant metabolites covering the central pathways of primary metabolism (Roessner et al., 2000; Lisec et al., 2006). In potato, untargeted metabolomic approaches by GC-MS have been successfully applied to assess changes in metabolites under different conditions (Roessner et al., 2000; Urbanczyk-Wochniak et al., 2005), to evaluate the metabolic response to various genetic modifications (Roessner et al., 2001; Szopa et al., 2001; Davies et al., 2005), and to explore the phytochemical diversity among potato cultivars and landraces (Dobson et al., 2008, 2009). Additionally, metabolite profiling has been applied to monitor changes during key stages in the tuber life cycle (Davies, 2007). Untargeted approaches have thus generated a substantial amount of data providing important information concerning compositional metabolite changes upon perturbation and phytochemical diversity in potato. However, so far, these studies have focused on applications of metabolite profiling as an evaluation and comparative tool. Technological developments and improved data-processing techniques now also allow the use of metabolite profiling to obtain further insight into the genetic factors controlling metabolic traits (Keurentjes, 2009). Exploration of the genetic factors underlying metabolite variation in mapping populations is particularly advantageous. The genetic variation between related individuals in a segregating mapping population can be exploited to locate the genomic regions involved in the regulation of the observed metabolite variation (Keurentjes et al., 2006).
Here, we report on the metabolic profiling of a segregating diploid potato population (C × E). This population is highly heterozygous and has been analyzed in a number of studies to investigate the genetic architecture of quantitative traits (van Eck et al., 1994; Werij et al., 2007; Kloosterman et al., 2010; Wolters et al., 2010). For a number of traits, candidate genes and their allelic variants have been identified from these studies, including tuber flesh color and cooking type (Kloosterman et al., 2010), tuber shape (van Eck et al., 1994), carotenoid biosynthesis (Wolters et al., 2010), and Met content (Kloosterman et al., 2010).
In this study, we have used the C × E population to explore the genetic basis of primary metabolites. To this end, untargeted GC-TOF-MS metabolic profiling was carried out on a core set of individuals of the C × E population. In order to investigate the variation in the detected metabolite levels in individuals of the population, we applied a genetical genomics approach (Jansen and Nap, 2001). QTL analysis of metabolite levels resulted in the identification of genomic regions associated with the metabolic variation. In addition, we performed a parallel QTL analysis for starch- and cold sweetening-related traits, and genetically inferred links were established between these phenotypic traits and primary metabolites. We further applied multivariate analysis to the combined data sets of starch-related traits and metabolic profiles to determine the predictive power of metabolites on a given phenotypic trait. For this, we used a Random Forest (RF; Breiman, 2001) approach to find significant associations between phenotypic and metabolic traits independent of genetic information. Putative predictors were tested and confirmed in an independent collection of potato cultivars.
Our results show the value of combining biochemical profiling with genetic information to identify associations between metabolites and phenotypes. This approach reveals previously unknown links between phenotypic traits and metabolism and thus facilitates the discovery of biomarkers for agronomically important traits.
RESULTS
Metabolite Profiling
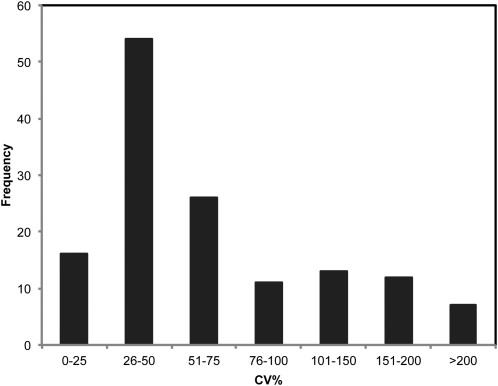
In order to assess the content and variation of polar primary metabolites present in the C × E diploid potato population, untargeted GC-TOF-MS-based metabolite profiling was performed. The GC-TOF-MS method was applied to the polar aqueous methanol extracts of dormant tubers of 97 genotypes and the parental clones of the C × E population. After processing of the raw data, 139 representative masses were obtained, consisting of reconstituted mass spectra (see “Materials and Methods”). The distribution of trait values for the detected compounds across all the genotypes was wide, with coefficients of variation higher than 50% for the majority of metabolites (approximately 52%; Fig. 1). This large variation can in part be explained by the segregation of genetic factors and therefore is amenable to genetic mapping approaches.
Figure 1.
Histogram of the distribution of metabolic variation. Data shown are based on the percentage of Coefficient of Variation (CV%) across the C × E population for the 139 representative masses. The distribution shows that the majority of metabolites have a CV higher than 50%, indicating a high level of variability.
The 139 representative masses were putatively identified by comparing the mass spectra with those of authentic reference standards and mass spectral databases (National Institute of Standards and Technology [NIST08]; Golm metabolome database [http://gmd.mpimp-golm.mpg.de/]). Supplemental Table S1 lists the derived identification (i.e. putative metabolites). Further inspection of the spectra and retention indices of these fragments allowed a more accurate annotation of 58 of them (Table I). Using all samples, we performed a hierarchical cluster analysis using Pearson correlations on the processed data for the abundance of the 139 representative masses. Figure 2 shows the degree of correlation among the detected compounds. The majority of the compounds were identified as amino acids, organic acids, or carbohydrates (Table I; Supplemental Table S1). The annotation of a number of compounds could not be verified by further inspection of the spectra and retention indices. However, we included these unknown compounds in the cluster analysis to investigate the degree of association with identified metabolites. Compounds from the same class, such as amino acids or carbohydrates, generally clustered together. The correlation coefficients within the identified amino acids ranged between 0.6 and 0.9 (Fig. 2), and two metabolites were considered to be highly correlated if the absolute correlation coefficient had a value of 0.6 or greater. Such high correlations have been reported before in potato between amino acids (Roessner et al., 2001; Dobson et al., 2008). It has been suggested that this correlation may reflect the mechanism of general amino acid control in plants (Halford et al., 2004). Interestingly, within the amino acid cluster, the branched amino acids Ile, Leu, and Val clustered separately from the aromatic amino acids Tyr, Phe, and Trp, as was also reported in earlier metabolomics studies in different potato cultivars (Roessner et al., 2001; Noctor et al., 2002; Dobson et al., 2008). Amino acids that are biosynthetically linked, such as Ser, Gly, and Thr, also show a strong correlation (Fig. 2). This suggests that much of the variation in amino acid content is genetically controlled by just a few master regulators. However, this was not the case for all related amino acids. The Pearson correlation coefficients among γ-aminobutyric acid (GABA), Glu, and Pro were less than 0.2, although they are closely linked biosynthetically as members of the Glu family. Other amino acids, such as Glu and Asn, show weak correlations (less than 0.4) with the major cluster of amino acids. This could suggest that the genetic regulation of these biosynthetic routes occurs independently from that of the main cluster of amino acids. In addition, most of the detected carbohydrates, such as Man and Fru, also form a cluster (Fig. 2). In contrast, Suc clusters with a group of organic acids rather than with the other carbohydrates. The clustering of organic acids, however, is less distinct, and this is likely due to the diverse biochemical origins of these compounds.
Table I. Metabolites putatively identified from the polar phase of methanol extract from potato tubers.
List of metabolites that were putatively identified based upon the similarity of mass spectra and the retention indices published in the literature. Compounds that were detected in more than one derivatized form are listed only once.
| Compound Class | Metabolites |
| Amino acid | Ala, Asn, Gln, Gly, Asp, Glu, Ile, Leu, Met, Phe, Pro, Ser, Thr, Trp, Tyr, Val, Lys, pyro-Glu, β-Ala, GABA |
| Organic acid | 2-Piperidinecarboxylic acid (pipecolic acid), ascorbic acid, butanoic acid, citric acid, quinic acid, fumaric acid, glyceric acid, malic acid, phosphoric acid, succinic acid, dehydro-l-(+)-ascorbic acid dimer, lactic acid, threonic acid |
| Sugar | Allose, Fru, galactiric acid, galactinol, Man, Suc, glucopyranose |
| Sugar alcohol | Myoinositol |
| Amino alcohol | Ethanolamine |
| Other | Calystegine B2, 5-aminocarboxy-4,6-dihydroxypyrimidine, allantoin |
Figure 2.
Correlation matrix and cluster analysis of detected polar metabolites. Pearson correlation between metabolites is indicated by color intensity. A total of 58 out of the 139 metabolites could be identified based upon spectral libraries and retention indices. Metabolites belonging to the same biochemical class (color coded) tend to cluster together.
Identification of Metabolic QTLs
To determine if the variation observed in metabolite levels could indeed be explained by allelic differences in genetic factors, we performed metabolic quantitative trait locus (mQTL) analysis on the metabolic profiles. The software package MetaNetwork was used to map the metabolite variation. MetaNetwork applications (Fu et al., 2007) were designed from data collected from recombinant inbreed lines; hence, in order to adjust the software applications to a cross-pollinated species like potato, we used two separate linkage maps: one for the female parent C and one for the male parent E. Overall, the variation in abundance of approximately 72% of the metabolites could be mapped in at least one of the two linkage maps. In total, we found 187 mQTLs for 121 metabolites, of which 58 could be putatively annotated. A complete list and a description of the detected mQTLs are given in Supplemental Tables S2 and S3.
C-Parent Map
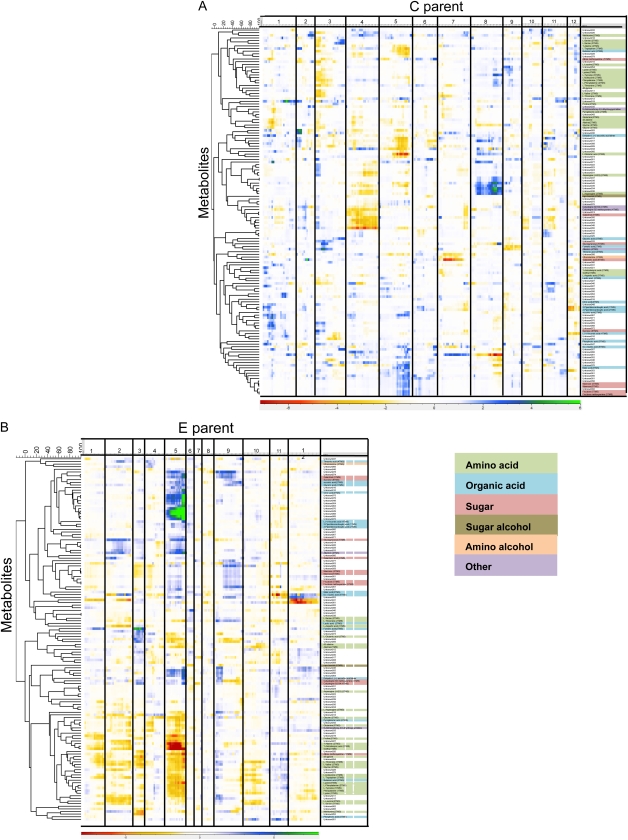
Forty-five significant mQTLs were detected in this map for 39 masses representing unique metabolites. For 33 metabolites, only one QTL was identified, and a maximum of two QTLs were found for six different metabolites (identified as galactiric acid and five unknown metabolites [nos. 012, 034, 044, 078, and 081]). The largest number of mQTLs on a single chromosome was 11 for chromosome 8, where mQTLs were detected for Asn, Tyr, and other, unidentified metabolites. The mQTL for Tyr on chromosome 8 was also detected in a previous study on the same population (Werij et al., 2007). On chromosome 5, we found eight mQTLs for Glu, Man, Trp, and a number of unknown compounds. On chromosomes 3, 6, and 10, no significant mQTLs were detected. Figure 3A shows the QTL profiles of all the metabolites mapped to the C-parent map in a heat map.
Figure 3.
Heat map of mQTL profiles of the detected compounds in the C × E population. A, Using the genetic map of the C parent. B, Using the genetic map of the E parent. Metabolites are clustered according to −10log(p) values across significantly associated markers. Vertical lines indicate chromosomal borders. Colors indicate the sign of the additive effect and the significance of the mQTL.
E-Parent Map
In this map, 160 significant mQTLs were detected for 85 representative masses (Fig. 3B). For 33 masses, only one QTL was detected, and a maximum of six mQTLs was detected for one metabolite (identified as quinic acid). The highest number of mQTLs on a single chromosome was 71 on chromosome 5. This chromosome also contributed the most to the total explained variation of all detected mQTLs. A single genomic region, spanning three adjacent markers, accounted for the highest density of detected mQTLs (34). This region is known to be involved in plant maturity (van Eck and Jacobsen, 1996; Collins et al., 1999; Oberhagemann et al., 1999) and as such exerts many pleiotropic effects on development-related traits. The majority of compounds mapping to this region were classified as amino acids, organic acids, and carbohydrates. This is not unexpected, as rapid changes in primary metabolism are known to occur during the later stages of maturation. Interestingly, similar to observations in the C-parent map, some amino acids that are biochemically related shared an mQTL at the plant maturity locus (e.g. Gly and Thr). Other amino acids, like Phe, Lys, Val, and Met, also mapped to the plant maturity region. Some of the identified compounds mapping to this region also showed significant mQTLs on other chromosomes, both in the C and the E maps. For example, four more mQTLs were detected for l-Thr, on chromosomes 1, 2, and 10 in the E-parent map and chromosome 7 in the C-parent map. For Met, another mQTL was detected on chromosome 2 of the C parent, and for Val, one additional mQTL was detected on chromosome 3 of the E parent, indicating complex regulation of these traits.
Association between Phenotypic and Metabolic Traits
Traits of agronomic importance in potato, such as starch and cold sweetening, are expected to be associated with primary metabolites. To investigate this relationship in more detail, we carried out a parallel QTL analysis for phenotypic starch- and cold sweetening-related traits determined for this population for two years of harvest: 2002 and 2003 (Supplemental Table S5; J.S. Werij, H.F. van Eck, C.W.B. Bachem, and R.G.F. Visser, unpublished data). Having mapped both metabolic and phenotypic QTLs to the two parental maps, we investigated the level of coregulation of these two sets of traits by determining colocalizing QTLs. As expected, a substantial number of the phenotypic traits mapped to the plant maturity locus at chromosome 5. However, a number of significant QTLs for essential metabolites were also detected outside this region, indicating a possible regulation independent of the developmental stage; therefore, these mQTLs are of particular interest.
From a total of 26 phenotypic traits, nine showed QTLs colocalizing with mQTLs outside the plant maturity region on chromosome 5 (Supplemental Table S4). Five phenotypic traits mapped to the C map, of which three mapped to the same position on chromosome 8: specific gravity of starch (also mapping to chromosome 10 in the E-parent map) and discoloration of tuber flesh after cooking at 5 and 30 min. This position coincided with mQTLs for two unknown compounds (nos. 044 and 081). A fourth trait, starch grain particle size, mapped to a different region of chromosome 8 on the C map that colocalized with mQTLs for amino acids, organic acids, and carbohydrates. Chip color difference between reconditioning and harvest mapped to chromosome 6 on the C-parent map and chromosomes 3 and 9 on the E-parent map, but only the latter two regions colocalized with a limited number of mQTLs.
Two traits, chip color after harvest and chip color difference between storage and harvest, mapped to chromosomes 9 and 3 of the E-parent map. These positions coincide with genomic regions where mQTLs for succinic acid, fumaric acid, butanoic acid, and unknown compound 044 were detected.
The strongest association, however, was detected between starch phosphorylation and a number of metabolites. Starch phosphorylation maps to positions on chromosomes 2 and 9 of the E-parent map in both harvest years 2002 and 2003. The detection of identical QTLs in independent experiments suggests a strong and robust genetic control, although a third QTL on chromosome 5 was detected in the 2003 harvest that was only suggestive in 2002. This QTL colocalizes with mQTLs for Ala, butanoic acid, β-Ala, GABA, succinic acid, pyro-Glu, Phe, Gln, Tyr, Trp, and seven unknown compounds (nos. 011, 027, 033, 048, 061, 068, and 069). The QTL for starch phosphorylation on chromosome 2 colocalizes with mQTLs for Ser, Thr, Asp, GABA, Gln, and four unknown metabolites (nos. 015, 027, 041, and 043). The QTL on chromosome 9 colocalizes with a mQTL for galactinol.
RF Analysis Reveals a Link between Primary Metabolites and Starch Phosphorylation
To further investigate the strength of the colocalizations, we focused on the phenotypic trait starch phosphorylation (i.e. the degree of phosphorylation per milligram of starch). Potato starch is characterized by a relatively high content of phosphate groups compared with, for example, cereal starches (Rooke et al., 1949; Hizukuri et al., 1970). This level of phosphate groups influences the viscosity and the chemical properties of starch and therefore is important for the different uses of potato starch for food and industrial applications. Among the evaluated starch-related traits, the measurements for starch phosphorylation in 2002 and 2003 showed the highest correlation coefficient (r2 = 0.8; Supplemental Table S6), indicating a general high reproducibility of the expression of this trait. The observation that two to three QTLs could be mapped in each year together with a high broad sense heritability (0.5) further indicates that a substantial part of the trait variation can be attributed to genetic factors. In addition, a number of colocalizing mQTLs were identified (see above) that suggest links between starch phosphorylation and metabolic processes. To rank the associations between starch phosphorylation and the 139 representative masses, we used a multivariate RF regression approach (Breiman, 2001). The starch phosphorylation measurements of the two years were used separately, as a response variable regressed against the 139 representative masses over all population individuals and significantly associated metabolites were recorded.
Using starch phosphorylation measurements of the 2002 harvest and all of the detected compounds, the RF model explained 16% of the variance at a permutation threshold of α = 0.001. Twelve metabolites were significantly associated with phosphate content at this threshold (Table II). Univariate correlation analyses between these significant metabolites and starch phosphorylation yielded absolute r2 values ranging between 0.07 and 0.26 (of which two have a negative Pearson correlation value). For the 2003 harvest, the RF model explained 33% of the variance at a permutation threshold of α = 0.001. In this case, eight metabolites were found to be significantly associated with starch phosphorylation (Table II). The correlations ranged from 0.12 to 0.39 (of which one has a negative Pearson correlation value). Interestingly, all the significantly contributing metabolites from the 2003 model were also identified using the 2002 data, again illustrating the high reproducibility between the two years. From these eight compounds, seven showed colocalizing QTLs with at least one of the starch phosphorylation QTLs: β-Ala, GABA, l-Asp, Ala, butanoic acid, unknown 027, and unknown 033 (Supplemental Table S4).
Table II. List of associated metabolites ranked after RF and significance permutation tests.
The first column shows the putative annotation of the representative mass. The second column shows whether a colocalizing mQTL with starch phosphorylation was detected. The values in the remaining columns indicate ranking order after RF analysis. Compounds with two numbers represent the same compound with different trimethylsilyl (TMS) groups (derivatization groups).
| Rank of Metabolites |
||||
| Metabolite Putative Identification | Colocalizing Starch Phosphorylation QTL C × E Population | 2002 Harvest | 2003 Harvest | Potato Cultivars |
| β-Ala (2TMS) | Yes | 1 | 1 | 7 |
| γ-Aminobutyric acid (2TMS), (3TMS) | Yes | 2, 10 | 2 | |
| Ala (2TMS) | Yes | 3 | 4 | |
| Gly (2TMS) | 4 | |||
| Unknown 027 | Yes | 5 | 7 | |
| Glyceric acid (3TMS) | 6 | |||
| Unknown 033 | Yes | 7 | 6 | |
| d3-Ala | 8 | 8 | ||
| Unknown 044 | 9 | |||
| Lys (3TMS), (4TMS) | 3, 5 | |||
| l-Asp (3TMS) | Yes | 11 | 3 | |
| Butanoic acid (2TMS) | Yes | 12 | 5 | |
| Unknown 082 | 1 | |||
| Putrescine (4TMS) | 2 | |||
| Unknown 083 | 4 | |||
| Myoinositol (6TMS) | 6 | |||
| Glucopyranose (5TMS) | 8 | |||
| Unknown 084 | 9 | |||
As a third independent line of investigation, we performed RF regression on a subset of cultivars from a potato collection available in our laboratory (year of harvest 2007). All 214 cultivars of this collection were analyzed for starch phosphorylation, and from these, 30 cultivars were selected covering the whole distribution range of this trait (Supplemental Fig. S1) These 30 cultivars were analyzed for metabolite content with the same analytical procedure used for the C × E population. RF regression was performed using the starch phosphorylation measurements and the metabolites detected in this set of cultivars (data not shown). The resulting RF model explained 30% of the variation in starch phosphorylation, and nine compounds were found to contribute significantly at a permutation threshold of α = 0.001 (Table II). The univariate correlations between these significant compounds and starch phosphorylation ranged between 0.09 and 0.41 (of which four have a negative Pearson correlation value).
A comparison of the significant predictive compounds after RF analysis in the two sets of potato material (i.e. C × E [2002 and 2003] and cultivar collection) revealed one compound in common, β-Ala. In the C × E population, β-Ala showed a positive correlation with starch phosphorylation in both years. This positive trend was also observed in the selected cultivar set (Supplemental Fig. S2). Because a robust metabolic predictor of a phenotypic feature is preferably valid across different potato sources, we consider β-Ala as a reliable metabolite significantly linked to the level of phosphorylation of starch in potato tubers.
DISCUSSION
The use of an untargeted metabolomics approach permits a quantitative assessment of a wide range of metabolites and allows the detection of known and unknown metabolites. Untargeted metabolomics approaches have been successfully applied to experimental plant populations to uncover loci controlling the variation of metabolites (Overy et al., 2005; Keurentjes et al., 2006; Morreel et al., 2006; Schauer et al., 2006; Tieman et al., 2006; Lisec et al., 2008; Rowe et al., 2008).
In this study, we used untargeted GC-TOF-MS metabolite profiling to assess the quantitative variation in polar metabolites present in dormant tubers of a diploid potato population. The observed variation in this population enabled us to locate genomic regions involved in the regulation of a range of polar primary metabolites. Primary metabolites, consisting mainly of carbohydrates, amino acids, and organic acids, have an essential role in plant growth and development. In potato, carbohydrates are important for a number of agronomic traits related to tuber quality, such as starch content and cold sweetening. In this study, the same genetic material was used to detect QTLs for starch- and cold sweetening-related traits and metabolic traits.
We investigated the associations between phenotypic and metabolic traits through QTL colocalization, correlation analysis, and RF analyses. The detection of a QTL identifies the existence of at least one polymorphic locus that is contributing to the variation observed for a given trait (Causse et al., 2004). When QTLs for two different traits colocalize, this might indicate the existence of a common regulator that controls the variation of both traits. This is of special value in the search of candidate genes for traits with complex modes of inheritance or for which most of the genetic basis is unknown. However, it cannot be excluded that colocalizing genomic regions contain genes that are closely linked but are involved in different biological processes. Due to the limited resolution of QTL mapping and a finite number of markers, colocalization of (unrelated) QTLs is inevitable when large data sets are involved (Lisec et al., 2008). Therefore, we performed independent statistical tests to confirm true positive associations between metabolites and phenotypes. In addition, we have validated putative metabolic biomarkers in an independent set of potato varieties. Our stringent selection criteria resulted in the determination of a strong relationship between potato starch phosphorylation and primary metabolism. Furthermore, our analyses resulted in the identification of β-Ala as an important predictor for the degree of phosphorylation of starch in potato tubers.
Mapping of Metabolic Variation in Potato Tubers
Mapping populations are very suitable to identify loci controlling the variation of a given trait. In this study, we aimed to explore the variation of metabolic and phenotypic traits present in dormant potato tubers. Our results show that we could assign genomic regions involved in metabolite variation for approximately 72% of the detected metabolites.
The abundances of metabolites that share an mQTL, especially for major loci of qualitative traits, are expected to correlate because they cosegregate in a mapping population. For instance, l-Leu and Lys share an mQTL on chromosome 9 and are positively correlated. Metabolites sharing an mQTL often belong to the same biochemical pathway. For example, Phe and Tyr share a common biosynthetic pathway and hence are found to be coregulated. Alternatively, colocating QTLs can be the result of closely linked independent regulators (Lisec et al., 2008). In the case of a shared regulator, a direct, or even causal, relationship can be expected between traits, whereas in the latter case, the two traits are independently controlled. This distinction should be reflected in correlation analysis with higher values expected for coregulated traits. In contrast, high correlation values between traits can also be expected when environmental factors that affect multiple traits simultaneously are in play. The resulting decrease in signal-to-noise ratio would be reflected in low heritabilities and QTL detection power. Therefore, we have applied independent lines of investigation, including genetic, correlation, and RF analyses, to reliably identify biologically meaningful relationships between metabolites and complex phenotypes.
In targeted studies, QTLs were found for some of the metabolites that were also detected in our analyses. In a previous study, an mQTL for Tyr was detected on chromosome 8 in the C-parent map (Werij et al., 2007). This amino acid has been reported to be associated with the level of enzymatic discoloration (Corsini et al., 1992), although other studies have reported otherwise (Mondy and Munshi, 1993). In agreement with the results of Werij et al. (2007), we did not find overlapping QTLs for Tyr and enzymatic discoloration. In addition, we confirmed the QTL detected by Werij et al. (2007) and also detected two more QTLs for Tyr at chromosomes 5 and 11 of the E-parent map. This difference is likely explained by differences in the analytical techniques used in the two studies. Additionally, revisions in the linkage map that was used in our study may have influenced the power of detection of QTLs.
Another interesting metabolite that was also mapped in previous studies is Met. The level of this amino acid is important for the nutritional value of potato. Moreover, it is the precursor of metabolites important to potato flavor (e.g. methional), and attempts have been made to enhance the Met content to improve flavor (Di et al., 2003; Dancs et al., 2008). In earlier work on the C × E population, two QTLs, on chromosomes 3 and 5, were detected underlying the variation of this amino acid content in tubers (Kloosterman et al., 2010). In agreement with that study, we detected QTLs for Met that mapped on chromosomes 3 and 5 of the E-parent map and an additional QTL on chromosome 2 of the C-parent map.
The significant mQTLs detected in both parental maps were unequally distributed over the genome, indicating hot and cold spots for metabolite regulation. A well-known locus involved in plant maturity is located at chromosome 5, where a major QTL for this trait has been detected in the C × E population (van Eck and Jacobsen, 1996). Plant maturity has been shown to be closely linked to a number of traits, including resistance and developmental traits (Collins et al., 1999; Oberhagemann et al., 1999; Bormann et al., 2004), although the underlying genetic factor has not been identified thus far. Products of primary metabolism, such as carbohydrates and amino acids, are expected to influence the degree of plant maturity and vice versa. Therefore, we anticipated that a large number of metabolic traits would show an association with the plant maturity region on chromosome 5. Nonetheless, a substantial number of mQTLs for amino acids, organic acids, and carbohydrates have not been reported before and were identified outside this region. This finding highlights the importance of other genomic regions in the regulation of primary metabolite accumulation despite the pleiotropic effects displayed by the plant maturity region.
In addition, a number of mQTLs mapped to multiple positions, which indicates complex regulation. Among the multiple loci detected for these metabolites, at least one mapped to the plant maturity region. This raises the question of whether metabolites are under developmental control or whether development is under metabolic control. In this study many metabolites map to the maturity locus in addition to multiple other loci. Plant maturity, however, maps only to a single locus and this may indicate that the metabolism is at least partly under developmental control or another factor upstream.
Putative Predictors of Starch Phosphorylation
QTL colocalizations can be useful to identify metabolites involved in the regulation of phenotypic traits. This is of special importance for traits for which the genetic basis is unknown, providing a valuable tool to search for candidate genes. However, one should be cautious when making such assumptions, because two different traits that share the same regulatory region are not necessarily involved in the same molecular or biological process. In a specific genomic region, genes might be present that are linked but that have different enzymatic functions. The phenotypic traits evaluated in this study are known to be related to carbohydrate metabolism; consequently, metabolites involved in this pathway are likely to be linked to these traits. Nevertheless, QTL colocalizations can disclose unknown associations and moreover identify candidate predictors of trait variation (Lisec et al., 2008).
One of the aims of this study was to test to what extent phenotypic and metabolic QTLs colocalize in order to identify metabolites associated with phenotypic features. We focused on starch phosphorylation as a phenotypic case study. Potato starch has a particularly high content of phosphate groups in comparison with other plant species. The degradation of starch is dependent on reversible phosphorylation of the glucans at the surface of the starch granule (Zeeman et al., 2010), and although a direct link between the content of phosphate groups and starch degradation has not been found, it has been shown that alterations in the starch-phosphorylating enzymes lead to an excess of starch accumulation in the plant (Caspar et al., 1991; Zeeman and Rees, 1999; Yu et al., 2001). In potato, the high phosphate content of starch affects the viscosity and formation of stable starch pastes (Wiesenborn et al., 1994; Viksø-Nielsen et al., 2001), which is important for the diversified uses of starch in industry.
Here, we show that a number of amino acid mQTLs colocalize with trait QTLs for starch phosphorylation. To measure the strength of the genetically inferred links between the detected metabolic and phenotypic QTL colocalizations, we examined the associations and predictive power of the metabolite data for starch phosphorylation using RF regression analysis.
The application of multivariate statistical methods to assess associations between metabolites and phenotypic traits has been successfully applied in a number of studies. An approach using canonical correlation analysis to test the predictive power of metabolic composition for biomass traits in Arabidopsis revealed a number of metabolites related to biomass and growth (Meyer et al., 2007). In potato, a partial least-squares analysis was used to discover metabolites that function as predictors for susceptibility to black spot bruising and chip quality (Steinfath et al., 2010). The validity of these results was tested in a collection of potato cultivars and in a set of individuals of a segregating population where metabolic and phenotypic information obtained from a first environment was used to predict phenotypic properties from the metabolic data obtained from a second environment. Those results demonstrate the application of multivariate data analysis and the value of independent validation to discover a small set of metabolites that can be used as biomarkers for a phenotypic trait of interest.
We used RF analyses to predict starch phosphorylation from a GC-MS data set. A similar approach was used to predict flesh color and enzymatic discoloration from transcriptomics and liquid chromatography-MS data sets (Acharjee et al., 2011). This study resulted in the successful identification of associated genes and metabolites, of which some were known to be involved in the regulation of the traits under study. Correspondingly, in our study, the application of RF regression resulted in a list of highly ranked metabolites representing the most important compounds associated with starch phosphorylation. Inspection of the annotation of these included a number of unknown metabolites and, more interestingly, a few amino acids for which we also detected mQTLs coinciding with starch phosphorylation QTLs. Among these relevant metabolites, β-Ala was of particular interest because it consistently ranked in the top metabolites in the different potato materials used for the analysis.
Starch phosphorylation is mainly driven by the action of two glucan-water dikinases (i.e. glucan water dikinase [GWD] and phosphoglucan water dikinase [PWD]). These enzymes are critical in the transfer of phosphate groups within amylopectin (Smith et al., 2005; Zeeman et al., 2010). Analysis of Arabidopsis (Arabidopsis thaliana) mutants also showed that GWD is required for phosphorylation (Yu et al., 2001). The sex1 (loss of GWD activity) and pwd (loss of PWD activity) mutants lead to excess- and reduced-starch content phenotypes, respectively. Interestingly, transgenic potato plants with reduced GWD expression also displayed a starch-excess phenotype (Lorberth et al., 1998). In our results, the amino acid β-Ala was highly ranked after RF analysis for both sets of potato material (i.e. the C × E segregating population [2002 and 2003] and the set of potato cultivars). What is more, an mQTL for β-Ala was detected in the C × E population colocalizing with a phenotypic QTL for starch phosphorylation measurements of 2003 and a suggestive QTL in 2002.
It is known that GWD follows a dikinase-type reaction catalyzing the transfer of the β-phosphate of ATP to either the C6 or C3 position of the glucosyl residue (Ritte et al., 2002). In this type of reaction, the formation of an autophosphorylated intermediate precedes the transfer of the phosphate to the glucosyl residues. The autophosphorylation of this GWD intermediate depends on a conserved His residue that, when replaced by Ala, results in a mutant phenotype without phosphorylating activity (Mikkelsen et al., 2004). Ala isomers were further suggested as phosphate carriers when reacting with cyclotriphosphate to form orthophosphate derivatives in high-pH conditions (Tsuhako et al., 1985). These studies suggest a role for Ala in phosphorylation reactions, although further research is needed to confirm these relationships. β-Ala, as a substrate for pantothenate (vitamin B5) biosynthesis, is the only naturally occurring β-amino acid in plants (Chakauya et al., 2006). Little is known about the formation of β-Ala in plants, while in bacteria, β-Ala is synthesized from the decarboxylation of l-Asp in a reaction catalyzed by Asp decarboxylase (Chakauya et al., 2006). Interestingly, we observed a shared mQTL for β-Ala and l-Asp, suggesting common genetic regulation through shared biosynthetic pathways.
After a dormant phase, potatoes develop from a sink to a source organ that will subsequently support the growth and development of the new sprout. Owing to a higher content of phosphate groups, starch may be more easily mobilized and converted into resources for the growing sprout. Vitamin B5 is used in the synthesis of CoA, an acyl carrier protein. CoA is required in many central metabolic processes, and it is essential in the conversion of pyruvate to acetyl-CoA to enter the tricarboxylic acid cycle (Chakauya et al., 2006). In addition, CoA is fundamental in the biosynthesis of fatty acids, polyketides, depsipeptides, and peptides (Kleinkauf, 2000). β-Ala constitutes an important part in the biosynthesis pathway of vitamin B5, and the presence of this amino acid may be indicative for the formation of many essential metabolites for plant development; furthermore, it may act as an indicator of the mobilization of storage resources. In this study, we identified β-Ala associated with starch phosphorylation as well as a number of other metabolites for which it also might be predictive. Our approach has been shown to be instrumental in generating hypotheses about functional relationships between metabolites and phenotypes. In addition, it may help for a gradual understanding of metabolic processes contributing to the observed phenotypic features of interest.
Our data here demonstrate the benefits of the applied methods for a broad untargeted metabolomics approach in potato. In this study, we combined genetic information through mQTL and phenotypic QTL analysis and nongenetic information through the regression of trait values to predict phenotypic traits from metabolomics analysis. We identified candidate metabolites that can be informative for phenotypic traits of interest.
Advances in metabolomics have opened up the way to high-throughput approaches, allowing the analysis of variation of a large number of samples in a reasonable amount of time. In addition, advanced statistical methods enable us to explore and monitor different profiling techniques in nonmodel species. A multilevel integrative approach to study organisms as a system of genetic, proteomic, and metabolic events may enable us to achieve a higher level of understanding of the interactions occurring in a biological system of interest. In potato, although this field is still in its infancy, some examples have already shown the advantages of such approaches to identify, and hypothesize about, the components in biologically relevant pathways (Acharjee et al., 2011). Furthermore, the genome sequence of potato (Potato Genome Sequencing Consortium, 2011) has now revealed genes specific to this highly heterozygous crop, bringing a platform that will ultimately facilitate the elucidation of the genetic basis of complex traits of high importance in breeding for tuber quality.
MATERIALS AND METHODS
Plant Material
The C × E Population
The diploid population (C × E) consisting of a total of 251 individuals was obtained from a cross between two heterozygous diploid potato clones, USW5337.3 (coded C: Solanum phureja × Solanum tuberosum) and 77.2102.37 (coded E: Solanum vernei × S. tuberosum). This population has been of special use to study the inheritance and genetic mapping of traits related to tuber quality (i.e. tuber shape, tuber size, eye depth, flesh color, among others). The development and characteristics of the population and the parental lines are described in detail (Celis-Gamboa, 2002; Werij et al., 2011). For starch- and cold sweetening-related traits, the values for the parental lines lie very close to each other centered on these normally distributed traits (data not shown), demonstrating the large amount of transgression present in this population. In addition, the female and male parents show very similar plant maturity phenotypes, with the female C showing a slightly earlier maturity phenotype than the male E.
A subset of 97 genotypes of this population was grown in two subsequent years (2002 and 2003) during the normal potato growing season (April–September) in Wageningen, The Netherlands. For each genotype, tubers were collected from three plants. Harvested tubers were either used for phenotypic analysis or mechanically peeled and immediately frozen in liquid nitrogen before being ground into fine powder and stored at −80°C.
Phenotypic analyses for 26 starch- and cold sweetening-related traits were performed for both years of harvest (Supplemental Table S5). Metabolite profiling was carried out on the ground material of tubers of the 2003 harvest.
Potato Cultivars
Potato cultivars used for independent confirmation and further statistical analysis were part of the potato collection available at Wageningen University & Research centre Plant Breeding. This collection consists of 221 tetraploid potato cultivars that were provided by Dutch breeding companies and gene banks. Phosphate measurements were carried out for 214 potato cultivars. In accordance with the distribution of the trait values (Supplemental Fig. S2), we selected 30 cultivars representing high, medium, and low phosphate contents.
Determination of Starch Phosphorylation
The degree of phosphorylation of starch was determined in a colorimetric assay. A total of 20 mg of starch was mixed with 250 μL of 70% HClO4 and incubated at 250°C for 25 min. Then, 50 μL of 30% hydrogen peroxide (w/v) was added and incubated for another 5 min. After cooling, 2 mL of water was added to reach a final concentration of HClO4 of 8.75% (w/v).
The color reagent consisted of 0.75% (w/v) (NH4)6Mo7O240.4H2O, 3% (w/v) FeSO40.7H2O, and 0.75% SDS (w/v) dissolved in 0.375 m H2SO4. A total of 100 μL of the sample extract, or a standard solution, was mixed with 200 μL of the color reagent solution on a microtiter plate and incubated for 10 min at room temperature. The absorbance was measured at 750 nm in a microplate reader using 8.75% HClO4 as a blank. A calibration curve of PO4 dissolved in HClO4 (0–500 μm) was used to determine the phosphate content.
Extraction and Derivatization of Potato Tuber Metabolites for GC-MS Analysis
Relative metabolite content was determined as described (Weckwerth et al., 2004) with modifications specific to the potato material. Briefly, polar metabolite fractions were extracted from approximately 100 mg fresh weight of tuber powder. A total of 1.4 mL of a single-phase solvent mixture of methanol:chloroform:water (2.5:1:1) was added to the ground tuber powder in a 2-mL Eppendorf tube. d3-Ala was used as a deuterated internal standard and ribitol was used as a representative internal standard, and they were all mixed in one solution. In the water phase, 25 μL of a solution containing the aforementioned internal standard solution was added. After vortexing, the closed tubes were sonicated for 15 min. After 5 min of centrifugation, the supernatant was transferred into a new Eppendorf tube and 400 μL of water was added. The mixture was thoroughly mixed by vortexing and centrifuged for 10 min at 21,000 rcf (relative centrifugal force). The methanol/water supernatant (polar phase) was carefully transferred into a new Eppendorf tube. Aliquots of the polar phase (100 μL) were dried by vacuum centrifugation for 12 to 16 h.
The dried samples were derivatized online as described by Lisec et al. (2006) using a Combi PAL autosampler (CTC Analytics). First, 12.5 μL of O-methylhydroxylamine hydrochloride (20 mg mL−1 pyridine) was added to the samples and incubated for 30 min at 40°C with agitation. Then, the samples were derivatized with 17.5 μL of N-methyl-N-trimethylsilyltrifluoroacetamide for 60 min. An alkane mixture (C9–C17 and C20–C34) was added to determine the retention indices of metabolites. The derivatized samples were analyzed by a GC-TOF-MS system consisting of an Optic 3 high-performance injector (ATAS) and an Agilent 6890 gas chromatograph (Agilent Technologies) coupled to a Pegasus III time-of-flight mass spectrometer (Leco Instruments).
Two microliters of each sample was introduced to the injector at 70°C using a split flow of 19 mL min−1.The injector was rapidly heated with 6°C s−1 to 240°C. The chromatographic separation was performed using a VF-5ms capillary column (Varian; 30 m × 0.25 mm × 0.25 μm) including a 10-m guardian column with helium as carrier gas at a column flow rate of 1 mL min−1. The temperature was isothermal for 2 min at 70°C, followed by a 10°C min−1 ramp to 310°C, and was held at this temperature for 5 min. The transfer line temperature was set at 270°C. The column effluent was ionized by electron impact at 70 eV. Mass spectra were acquired at 20 scans s−1 within a mass-to-charge ratio range of 50 to 600 at a source temperature of 200°C. A solvent delay of 295 s was set. The detector voltage was set to 1,400 V.
GC-MS Data-Processing Methods
Data Preprocessing
Raw data were processed by ChromaTOF software 2.0 (Leco Instruments) and MassLynx software (Waters), and further analysis was performed using MetAlign software (Lommen, 2009) to extract and align the mass signals (signal-to-noise ratio ≥ 2). Mass signals that were present in fewer than two samples were discarded. Signal redundancy per metabolite was removed by means of clustering, and mass spectra were reconstructed (Tikunov et al., 2005, 2011). This resulted in 139 reconstructed polar metabolites (representative masses).
Compound Identification
The mass spectra of the representative masses were subjected to tentative identification by matching to the NIST08 and Wiley spectral libraries and by comparison with retention indices calculated using a series of alkanes and fitted with a second-order polynomial function (Strehmel et al., 2008). Library hits were manually curated, and a series of commercial standards were used to check annotation. Compound identification is limited to the availability of spectra in the libraries used. The identities of the detected compounds are listed in Supplemental Table S1.
Data Normalization and Multivariate Analysis
Mass intensity values of the representative masses were normalized using isotope-labeled d3-Ala as an internal standard. Relative amounts of the compounds were obtained by normalizing the intensity of individual masses to the response of the internal standard. The ratio between the mass intensity value of the putative compound and the d3-Ala internal standard was then scaled by multiplying the resulting value by the average of the d3-Ala mass intensity across all samples.
Normalized values were log transformed in GeneMaths XT version 2.12 software (www.applied-maths.com). These data were used for cluster analysis using Pearson’s correlation coefficient and Unweighted Pair Group Method with Arithmetic Mean (UPGMA) for hierarchical clustering.
Metabolic and Phenotypic QTL Analyses
Metabolite QTL analyses were performed using the software package MetaNetwork (Keurentjes et al., 2006; Fu et al., 2007). MetaNetwork applies a two-part model, and a P value is determined for each part of the model. In this study, P values and QTL thresholds were determined as described (Keurentjes et al., 2006). Since MetaNetwork is not designed for cross-pollinated species, two separate linkage maps were used in our analysis: one for the female parent C and one for the male parent E. The number of markers specific to the C-parent map is 218 and that for the E-parent map is 178, with an average spacing between markers of 6.1 and 3.9 centimorgan, respectively. The significance QTL threshold value was estimated by MetaNetwork. Empirical thresholds for significant mQTLs were calculated separately for both parental maps: C-parent map, −10log(p) = 3.43 (P = 0.00037); E-parent map, −10log(p) = 3.19 (P = 0.00065).
Phenotypic measurements containing missing data cannot be analyzed by MetaNetwork; hence, QTL analyses for phenotypic data were performed using the software package MapQTL version 6.0. QTL log of the odds thresholds were calculated per trait using a permutation test (n = 10,000) provided in MapQTL.
Broad sense heritability was estimated for starch phosphorylation measurements over the two years (2002 and 2003) according to the formula H2 = VG/(VG + VE + VG×E), where VG is the variance among genotypes and VE is the year variation. One phosphate content measurement per year was used in a mixed model to calculate variance components for genotypes, years, and residual (= genotype × year).
RF
RF (Breiman, 2001) was used for regression of the phenotypic trait starch phosphorylation on the GC-TOF-MS signals. RF constructs a predictive model for the response using all predictors but quantifies the importance of each, here the metabolites, in explaining the variation present in the starch phosphorylation. RF by itself does not provide significance levels of individual metabolites and does not perform a variable selection to choose a possible subset of associated metabolites. Therefore, we included a permutation test to indicate the significance of the association of a metabolite with a trait. In each of 1,000 permutations of the trait values, we estimated the variance explained by the RF model (R2) and the variable importance of each metabolite in terms of the decrease in node impurities (Breiman, 2001). We ordered node purity values from the permuted data sets and took the 95th percentile from the distribution of impurity values as the significance threshold of the individual metabolites. The same procedure was done for R2 values of the model: the 95th percentile was taken as a significance threshold for the RF model. RF regression of starch phosphorylation on metabolite values was conducted using the “randomForest” package of the R statistical software. R2 in RF is not just a measure of goodness of fit of the data at hand but is determined on left-out samples (the “out-of-bag” samples), so it should be interpreted as a measure for predictive quality (here considered as prediction R2) of the RF on independent samples that have the same properties as the in-bag samples (Breiman, 2001).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Frequency distribution of phosphate content in potato collection.
Supplemental Figure S2. Correlation between beta-alanine and phosphate content.
Supplemental Table S1. Polar metabolites detected in dormant tubers by GC-TOF-MS analysis.
Supplemental Table S2. Metabolic QTL results of untargeted GC-TOF-MS metabolite profiling for C-parent linkage map.
Supplemental Table S3. Metabolic QTL results of untargeted GC-TOF-MS metabolite profiling for E-parent linkage map.
Supplemental Table S4. List of metabolic QTLs and phenotypic QTLs colocalizations outside plant maturity region.
Supplemental Table S5. List of starch and cold sweetening related traits of the C x E population.
Supplemental Table S6. Correlation values between starch related traits in two years of harvest.
Acknowledgments
We gratefully acknowledge the technical and intellectual input of our laboratory colleagues, Francel Verstappen, Anna Undas, Desalegn Etalo, and Benyamin Houshyani Hassanzadeh. We thank Aaron Velez-Ramirez for help in the preparation of the figures. We also thank Thierry Delatte for valuable comments and suggestions on the “Discussion.”
References
- Acharjee A, Kloosterman B, de Vos RCH, Werij JS, Bachem CWB, Visser RGF, Maliepaard C. (2011) Data integration and network reconstruction with -omics data using Random Forest regression in potato. Anal Chim Acta 705: 56–63 [DOI] [PubMed] [Google Scholar]
- Bormann CA, Rickert AM, Ruiz RA, Paal J, Lübeck J, Strahwald J, Buhr K, Gebhardt C. (2004) Tagging quantitative trait loci for maturity-corrected late blight resistance in tetraploid potato with PCR-based candidate gene markers. Mol Plant Microbe Interact 17: 1126–1138 [DOI] [PubMed] [Google Scholar]
- Breiman L. (2001) Random Forests. Mach Learn 45: 5–32 [Google Scholar]
- Caspar T, Lin T-P, Kakefuda G, Benbow L, Preiss J, Somerville C. (1991) Mutants of Arabidopsis with altered regulation of starch degradation. Plant Physiol 95: 1181–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causse M, Duffe P, Gomez MC, Buret M, Damidaux R, Zamir D, Gur A, Chevalier C, Lemaire-Chamley M, Rothan C. (2004) A genetic map of candidate genes and QTLs involved in tomato fruit size and composition. J Exp Bot 55: 1671–1685 [DOI] [PubMed] [Google Scholar]
- Celis-Gamboa BC. (2002) The life cycle of the potato (Solanum tuberosum L.): from crop physiology to genetics. PhD thesis. Wageningen University, Wageningen, The Netherlands [Google Scholar]
- Chakauya E, Coxon KM, Whitney HM, Ashurst JL, Abell C, Smith AG. (2006) Pantothenate biosynthesis in higher plants: advances and challenges. Physiol Plant 126: 319–329 [Google Scholar]
- Coffin RH, Yada RY, Parkin KL, Grodzinski B, Stanley DW. (1987) Effect of low temperature storage on sugar concentrations and chip color of certain processing potato cultivars and selections. J Food Sci 52: 639–645 [Google Scholar]
- Collins A, Milbourne D, Ramsay L, Meyer R, Chatot-Balandras C, Oberhagemann P, De Jong W, Gebhardt C, Bonnel E, Waugh R. (1999) QTL for field resistance to late blight in potato are strongly correlated with maturity and vigour. Mol Breed 5: 387–398 [Google Scholar]
- Corsini D, Pavek J, Dean B. (1992) Differences in free and protein-bound tyrosine among potato genotypes and the relationship to internal blackspot resistance. Am J Potato Res 69: 423–435 [Google Scholar]
- Dancs G, Kondrák M, Bánfalvi Z. (2008) The effects of enhanced methionine synthesis on amino acid and anthocyanin content of potato tubers. BMC Plant Biol 8: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies HV. (2007) Metabolomics: applications in functional biodiversity analysis in potato. Acta Hortic 745: 471–484 [Google Scholar]
- Davies HV, Shepherd LVT, Burrell MM, Carrari F, Urbanczyk-Wochniak E, Leisse A, Hancock RD, Taylor M, Viola R, Ross H, et al. (2005) Modulation of fructokinase activity of potato (Solanum tuberosum) results in substantial shifts in tuber metabolism. Plant Cell Physiol 46: 1103–1115 [DOI] [PubMed] [Google Scholar]
- Di R, Kim J, Martin MN, Leustek T, Jhoo J, Ho C-T, Tumer NE. (2003) Enhancement of the primary flavor compound methional in potato by increasing the level of soluble methionine. J Agric Food Chem 51: 5695–5702 [DOI] [PubMed] [Google Scholar]
- Dobson G, Shepherd T, Verrall SR, Conner S, McNicol JW, Ramsay G, Shepherd LVT, Davies HV, Stewart D. (2008) Phytochemical diversity in tubers of potato cultivars and landraces using a GC-MS metabolomics approach. J Agric Food Chem 56: 10280–10291 [DOI] [PubMed] [Google Scholar]
- Dobson G, Shepherd T, Verrall SR, Griffiths WD, Ramsay G, McNicol JW, Davies HV, Stewart D. (2009) A metabolomics study of cultivated potato (Solanum tuberosum) groups Andigena, Phureja, Stenotomum, and Tuberosum using gas chromatography-mass spectrometry. J Agric Food Chem 58: 1214–1223 [DOI] [PubMed] [Google Scholar]
- Doerge RW. (2002) Mapping and analysis of quantitative trait loci in experimental populations. Nat Rev Genet 3: 43–52 [Google Scholar]
- Fernie AR, Schauer N. (2009) Metabolomics-assisted breeding: a viable option for crop improvement? Trends Genet 25: 39–48 [DOI] [PubMed] [Google Scholar]
- Fu J, Swertz MA, Keurentjes JJB, Jansen RC. (2007) MetaNetwork: a computational protocol for the genetic study of metabolic networks. Nat Protoc 2: 685–694 [DOI] [PubMed] [Google Scholar]
- Halford NG, Hey S, Jhurreea D, Laurie S, McKibbin RS, Zhang Y, Paul MJ. (2004) Highly conserved protein kinases involved in the regulation of carbon and amino acid metabolism. J Exp Bot 55: 35–42 [DOI] [PubMed] [Google Scholar]
- Hizukuri S, Tabata S, Kagoshima , Nikuni Z. (1970) Studies on starch phosphate. Part 1. Estimation of glucose-6-phosphate residues in starch and the presence of other bound phosphate(s). Starch-Stärke 22: 338–343 [Google Scholar]
- Jansen RC, Nap JP. (2001) Genetical genomics: the added value from segregation. Trends Genet 17: 388–391 [DOI] [PubMed] [Google Scholar]
- Keurentjes JJB. (2009) Genetical metabolomics: closing in on phenotypes. Curr Opin Plant Biol 12: 223–230 [DOI] [PubMed] [Google Scholar]
- Keurentjes JJB, Fu JY, de Vos CHR, Lommen A, Hall RD, Bino RJ, van der Plas LHW, Jansen RC, Vreugdenhil D, Koornneef M. (2006) The genetics of plant metabolism. Nat Genet 38: 842–849 [DOI] [PubMed] [Google Scholar]
- Kleinkauf H. (2000) The role of 4′-phosphopantetheine in the biosynthesis of fatty acids, polyketides and peptides. Biofactors 11: 91–92 [DOI] [PubMed] [Google Scholar]
- Kloosterman B, Oortwijn M, uitdeWilligen J, America T, de Vos R, Visser RG, Bachem CW. (2010) From QTL to candidate gene: genetical genomics of simple and complex traits in potato using a pooling strategy. BMC Genomics 11: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Meyer RC, Steinfath M, Redestig H, Becher M, Witucka-Wall H, Fiehn O, Törjék O, Selbig J, Altmann T, et al. (2008) Identification of metabolic and biomass QTL in Arabidopsis thaliana in a parallel analysis of RIL and IL populations. Plant J 53: 960–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Lommen A. (2009) MetAlign: interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Anal Chem 81: 3079–3086 [DOI] [PubMed] [Google Scholar]
- Lorberth R, Ritte G, Willmitzer L, Kossmann J. (1998) Inhibition of a starch-granule-bound protein leads to modified starch and repression of cold sweetening. Nat Biotechnol 16: 473–477 [DOI] [PubMed] [Google Scholar]
- Meyer RC, Steinfath M, Lisec J, Becher M, Witucka-Wall H, Törjék O, Fiehn O, Eckardt A, Willmitzer L, Selbig J, et al. (2007) The metabolic signature related to high plant growth rate in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 4759–4764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen R, Baunsgaard L, Blennow A. (2004) Functional characterization of alpha-glucan,water dikinase, the starch phosphorylating enzyme. Biochem J 377: 525–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondy NI, Munshi CB. (1993) Effect of maturity and storage on ascorbic acid and tyrosine concentrations and enzymic discoloration of potatoes. J Agric Food Chem 41: 1868–1871 [Google Scholar]
- Morreel K, Goeminne G, Storme V, Sterck L, Ralph J, Coppieters W, Breyne P, Steenackers M, Georges M, Messens E, et al. (2006) Genetical metabolomics of flavonoid biosynthesis in Populus: a case study. Plant J 47: 224–237 [DOI] [PubMed] [Google Scholar]
- Noctor G, Novitskaya L, Lea PJ, Foyer CH. (2002) Co-ordination of leaf minor amino acid contents in crop species: significance and interpretation. J Exp Bot 53: 939–945 [DOI] [PubMed] [Google Scholar]
- Oberhagemann P, Chatot-Balandras C, Schäfer-Pregl R, Wegener D, Palomino C, Salamini F, Bonnel E, Gebhardt C. (1999) A genetic analysis of quantitative resistance to late blight in potato: towards marker-assisted selection. Mol Breed 5: 399–415 [Google Scholar]
- Overy SA, Walker HJ, Malone S, Howard TP, Baxter CJ, Sweetlove LJ, Hill SA, Quick WP. (2005) Application of metabolite profiling to the identification of traits in a population of tomato introgression lines. J Exp Bot 56: 287–296 [DOI] [PubMed] [Google Scholar]
- Potato Genome Sequencing Consortium (2011) Genome sequence and analysis of the tuber crop potato. Nature 475: 189–195 [DOI] [PubMed] [Google Scholar]
- Ritte G, Lloyd JR, Eckermann N, Rottmann A, Kossmann J, Steup M. (2002) The starch-related R1 protein is an alpha-glucan, water dikinase. Proc Natl Acad Sci USA 99: 7166–7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie AR. (2001) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L. (2000) Technical advance: simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J 23: 131–142 [DOI] [PubMed] [Google Scholar]
- Rooke HS, Lampitt LH, Jackson EM. (1949) The phosphorus compounds of wheat starch. Biochem J 45: 231–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HC, Hansen BG, Halkier BA, Kliebenstein DJ. (2008) Biochemical networks and epistasis shape the Arabidopsis thaliana metabolome. Plant Cell 20: 1199–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Roessner U, Gur A, Balbo I, Carrari F, Pleban T, Perez-Melis A, Bruedigam C, Kopka J, et al. (2006) Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat Biotechnol 24: 447–454 [DOI] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC, Smith SM. (2005) Starch degradation. Annu Rev Plant Biol 56: 73–98 [DOI] [PubMed] [Google Scholar]
- Steinfath M, Strehmel N, Peters R, Schauer N, Groth D, Hummel J, Steup M, Selbig J, Kopka J, Geigenberger P, et al. (2010) Discovering plant metabolic biomarkers for phenotype prediction using an untargeted approach. Plant Biotechnol J 8: 900–911 [DOI] [PubMed] [Google Scholar]
- Strehmel N, Hummel J, Erban A, Strassburg K, Kopka J. (2008) Retention index thresholds for compound matching in GC-MS metabolite profiling. J Chromatogr B Analyt Technol Biomed Life Sci 871: 182–190 [DOI] [PubMed] [Google Scholar]
- Szopa J, Wróbel M, Matysiak-Kata I, Swiedrych A. (2001) The metabolic profile of the 14-3-3 repressed transgenic potato tubers. Plant Sci 161: 1075–1082 [Google Scholar]
- Tieman DM, Zeigler M, Schmelz EA, Taylor MG, Bliss P, Kirst M, Klee HJ. (2006) Identification of loci affecting flavour volatile emissions in tomato fruits. J Exp Bot 57: 887–896 [DOI] [PubMed] [Google Scholar]
- Tikunov YM, Laptenok S, Hall RD, Bovy A, de Vos RCH. (October 15, 2011) MSClust: a tool for unsupervised mass spectra extraction of chromatography-mass spectrometry ion-wise aligned data. Metabolomics http://dx.doi.org/10.1007/s11306-011-0368-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikunov Y, Lommen A, de Vos CHR, Verhoeven HA, Bino RJ, Hall RD, Bovy AG. (2005) A novel approach for nontargeted data analysis for metabolomics: large-scale profiling of tomato fruit volatiles. Plant Physiol 139: 1125–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuhako M, Nakajima A, Miyajima T, Ohashi S, Nariai H, Motooka I. (1985) The reaction of cyclo-triphosphate with l-alpha-alanine or beta-alanine. Bull Chem Soc Jpn 58: 3092–3098 [Google Scholar]
- Urbanczyk-Wochniak E, Baxter C, Kolbe A, Kopka J, Sweetlove LJ, Fernie AR. (2005) Profiling of diurnal patterns of metabolite and transcript abundance in potato (Solanum tuberosum) leaves. Planta 221: 891–903 [DOI] [PubMed] [Google Scholar]
- van Eck HJ, Jacobs JM, Stam P, Ton J, Stiekema WJ, Jacobsen E. (1994) Multiple alleles for tuber shape in diploid potato detected by qualitative and quantitative genetic analysis using RFLPs. Genetics 137: 303–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eck HJ, Jacobsen E. (1996) Application of molecular markers in the genetic analysis of quantitative traits. In PC Struik, J Hoogendoorn, JK Kowenhoven, LJ Mastenbroek, LJ Turkensteen, A Veerman, J Vos eds, Abstracts of Conference Papers, Posters and Demonstrations of the 13th Triennial Conference of the EAPR. European Association for Potato Research, Wageningen, The Netherlands, pp 130–131 [Google Scholar]
- Viksø-Nielsen A, Blennow A, Jørgensen K, Kristensen KH, Jensen A, Møller BL. (2001) Structural, physicochemical, and pasting properties of starches from potato plants with repressed r1-gene. Biomacromolecules 2: 836–843 [DOI] [PubMed] [Google Scholar]
- Weckwerth W, Wenzel K, Fiehn O. (2004) Process for the integrated extraction, identification and quantification of metabolites, proteins and RNA to reveal their co-regulation in biochemical networks. Proteomics 4: 78–83 [DOI] [PubMed] [Google Scholar]
- Werij JS, Kloosterman B, Celis-Gamboa C, de Vos CH, America T, Visser RG, Bachem CW. (2007) Unravelling enzymatic discoloration in potato through a combined approach of candidate genes, QTL, and expression analysis. Theor Appl Genet 115: 245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesenborn DP, Orr PH, Casper HH, Tacke BK. (1994) Potato starch paste behavior as related to some physical/chemical properties. J Food Sci 59: 644–648 [Google Scholar]
- Wolters A-M, Uitdewilligen JG, Kloosterman BA, Hutten RC, Visser RG, van Eck HJ. (2010) Identification of alleles of carotenoid pathway genes important for zeaxanthin accumulation in potato tubers. Plant Mol Biol 73: 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TS, Kofler H, Häusler RE, Hille D, Flügge UI, Zeeman SC, Smith AM, Kossmann J, Lloyd J, Ritte G, et al. (2001) The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. Plant Cell 13: 1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman SC, Kossmann J, Smith AM. (2010) Starch: its metabolism, evolution, and biotechnological modification in plants. Annu Rev Plant Biol 61: 209–234 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Rees TA. (1999) Changes in carbohydrate metabolism and assimilate export in starch-excess mutants of Arabidopsis. Plant Cell Environ 22: 1445–1453 [Google Scholar]