Abstract
TETRATRICOPEPTIDE THIOREDOXIN-LIKE (TTL) proteins are characterized by the presence of six tetratricopeptide repeats in conserved positions and a carboxyl-terminal region known as the thioredoxin-like domain with homology to thioredoxins. In Arabidopsis (Arabidopsis thaliana), the TTL gene family is composed by four members, and the founder member, TTL1, is required for osmotic stress tolerance. Analysis of sequenced genomes indicates that TTL genes are specific to land plants. In this study, we report the expression profiles of Arabidopsis TTL genes using data mining and promoter-reporter β-glucuronidase fusions. Our results show that TTL1, TTL3, and TTL4 display ubiquitous expression in normal growing conditions but differential expression patterns in response to osmotic and NaCl stresses. TTL2 shows a very different expression pattern, being specific to pollen grains. Consistent with the expression data, ttl1, ttl3, and ttl4 mutants show reduced root growth under osmotic stress, and the analysis of double and triple mutants indicates that TTL1, TTL3, and TTL4 have partially overlapping yet specific functions in abiotic stress tolerance while TTL2 is involved in male gametophytic transmission.
Root growth is dependent on proper meristem activity and cell cycle regulation. This is particularly important under drought conditions, in which plant roots need to get deep into soils to access water supply. Extensive efforts have been devoted to the elucidation of sensory and signal transduction mechanisms that perceive osmotic stress and control cellular homeostasis in plants (Xiong and Zhu, 2001; Borsani et al., 2002, 2003; Yamaguchi-Shinozaki and Shinozaki, 2006; Munns and Tester, 2008). However, little is known about the mechanism(s) coordinating osmosensing and plant cell cycle regulation as well as the adaptive processes required to maintain meristem function under osmotic stress conditions.
In Arabidopsis (Arabidopsis thaliana), several genes required for cell cycle progression under osmotic and drought stresses have been identified using genetic approaches. For instance, the salt overly sensitive5 (sos5) characterized by root tip swelling and root growth arrest under salt stress was isolated in a screen for Arabidopsis salt-hypersensitive mutants (Shi et al., 2003). The predicted SOS5 protein contains an N-terminal signal sequence for plasma membrane localization, two arabinogalactan protein-like domains, two fasciclin-like domains, and a C-terminal glycosylphosphatidylinositol lipid anchor signal sequence. The presence of fasciclin-like domains, which typically are found in animal cell adhesion proteins, suggests a role for SOS5 in cell-to-cell adhesion in plants (Shi et al., 2003). The Arabidopsis STT3 encodes an essential subunit of the oligosaccharyltransferase complex that is involved in protein N-glycosylation. Osmotic stress disturbs the cell cycle progression in stt3a root meristematic cells, indicating the need of proper protein glycosylation and folding for osmotic stress adaptation (Koiwa et al., 2003). Mutations in the Arabidopsis TETRATRICOPEPTIDE THIOREDOXIN-LIKE1 gene (TTL1) cause reduced tolerance to NaCl and osmotic stress that is characterized by reduced root elongation, disorganization of the root meristem, and impaired osmotic responses during germination and seedling development (Rosado et al., 2006b).
TTL1 is the first characterized member of a novel protein family defined by the presence of six tetratricopeptide repeat (TPR) motifs and a region in the C terminus with homology to class h thioredoxins but lacking essential Cys residues required for thioredoxin activity (Rosado et al., 2006b; Ceserani et al., 2009). The 34-amino acid TPR motif is conserved in all organisms studied and is present in proteins involved in numerous cellular processes (D’Andrea and Regan, 2003; Cliff et al., 2005; Schapire et al., 2006; Schreiber et al., 2011). TPR domains have been described as protein-protein interaction modules defined by a pattern of small and large amino acids forming an all-helical secondary structure (Blatch and Lässle, 1999; D’Andrea and Regan, 2003; Yang et al., 2005).
In Arabidopsis, more than 235 proteins containing TPR motifs, usually arranged in tandem repeats of three to 16, have been identified (D’Andrea and Regan, 2003; Prasad et al., 2010). Many of these proteins show a modular structure containing additional domains that mediate interactions with downstream proteins acting as cochaperones or have enzymatic activities that amplify and transduce signals (D’Andrea and Regan, 2003; Davies and Sánchez, 2005). Proteins containing TPR domains are involved in protein folding and mRNA stability, processing, and translation and have also been found to be essential for responses to hormones such ethylene, cytokinin, gibberellins, auxin, and abscisic acid (Prodromou et al., 1999; Gounalaki et al., 2000; Fedoroff, 2002; Wang et al., 2004; Greenboim-Wainberg et al., 2005; Yang et al., 2005; Silverstone et al., 2007). Thus, TPR proteins are emerging as essential components of signal transduction pathways mediated by most plant hormones.
Of all proteins containing TPR domains, Hsp90 is arguably the best studied (Sangster and Queitsch, 2005; Whitesell and Lindquist, 2005; Pratt et al., 2008; Taipale et al., 2010). The highly conserved and abundant molecular chaperone Hsp90 is distinct from other chaperones in that it plays a key role in signal transduction networks, cell cycle control, protein degradation, and genomic silencing (Taipale et al., 2010). Hsp90 lies at the center of a multiprotein chaperone complex that facilitates the folding of client proteins into their stable or active conformations (Krishna and Gloor, 2001). In Arabidopsis, TTL proteins have been identified as putative chaperone receptors based on the identification of conserved amino acids in their TPR motifs known as carboxylate clamps, opening the possibility that TTL proteins act in concert with Hsp90 chaperone complexes (Prasad et al., 2010). In addition, TTL3 was identified as an interacting partner of the activated (phosphorylated) cytoplasmic domain of VASCULAR HIGHWAY1/BRI1-LIKE RECEPTOR KINASE2, a receptor-like kinase of the BRI1 family with a role in vascular development (Ceserani et al., 2009).
In this study, we have performed a functional analysis of the four Arabidopsis TTL genes. Genetic analysis indicates that TTLs are essential for plant viability. TTL expression patterns and physiological characterization of ttl mutants indicate that TTLs have overlapping and yet distinct functions in the maintenance of root meristem integrity under osmotic stress as well as in male gametophyte and vasculature development.
RESULTS
TTL Proteins Are Characteristic of Land Plants
The TTLs constitute a family of plant-specific proteins with a common modular architecture containing six TPR domains distributed in specific positions throughout the sequence and a C-terminal sequence displaying homology to thioredoxins termed the thioredoxin-like (TRXL) motif (Rosado et al., 2006b). A distinctive feature of the TRXL motif is the lack of essential Cys residues conserved in all thioredoxins and required for reducing activity (Rosado et al., 2006b; Cesarani et al., 2009).
Extensive searches in a variety of sequenced genomes using the typical distribution of TPRs and TRXL domains of the Arabidopsis TTLs failed to identify any members of this protein family in microorganisms and animals but showed that they are present in all plant genomes analyzed. Next, we investigated the phylogeny of TTL proteins using the genomic data available from the Phytozome database version 7.0 (Goodstein et al., 2011) and genomic data from barley (Hordeum vulgare) and tomato (Solanum lycopersicum; Supplemental Table S1). Using the strict criteria detailed in “Materials and Methods,” three and one genes encoding TTLs were identified in the primitive land plant species Physcomitrella patens and Selaginella moellendorffii, respectively. The absence of TTL genes in algae indicated that this gene class is restricted to land plants. We also identified TTL-like proteins containing the TRXL motif and seven to eight TPR motifs that could possibly function as TTL proteins but were not considered in our analysis.
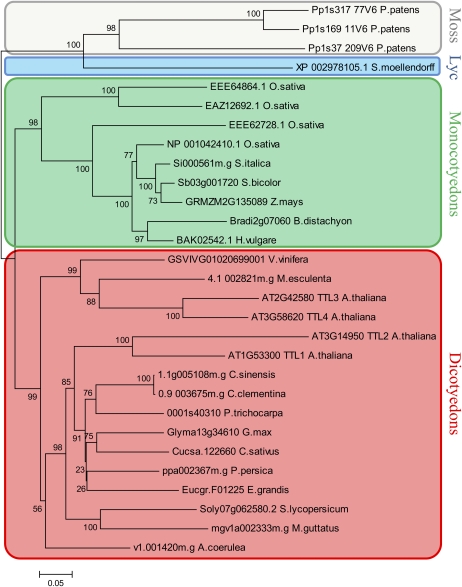
Figure 1 shows the phylogenetic analysis of the four Arabidopsis TTLs, the four TTLs from rice (Oryza sativa; model species for dicots and monocots, respectively), and TTL1 homologs from selected species listed in Supplemental Table S1. The analysis separately grouped monocot and dicot TTLs, and, as expected, the ancestral TTLs from P. patens and S. moellendorffii were outgrouped.
Figure 1.
Phylogenetic tree of 29 TTL proteins from 21 plant species. TTL proteins from moss, a lycophyte (Lyc), monocots, and dicots were used in the analysis. All TTL proteins identified in P. patens, S. moellendorffii, rice, and Arabidopsis were included in the analysis. From the rest of the species, only the TTL protein most homologous to Arabidopsis TTL1 was included. The phylogenetic tree was generated using ClustalW software, and bootstrap values are shown. [See online article for color version of this figure.]
In Arabidopsis, at least four different large-scale duplication events occurred 100 to 200 million years ago favoring the diversification of this species (Vision et al., 2000). To test the contribution of the Arabidopsis segmental duplications in the number of TTL genes, we searched the Arabidopsis genome for duplicated chromosomal segments containing TTLs using the Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/). As shown in Supplemental Figure S1, two independent duplicate blocks containing the genomic pairs TTL1/TTL2 and TTL3/TTL4 were identified. Considering the function of Arabidopsis TTL1, a protein required for osmotic stress tolerance, the large-scale duplication and retention of TTL genes in plants suggests that TTL genes may be a source of diversity important for proper responses to stressful environments.
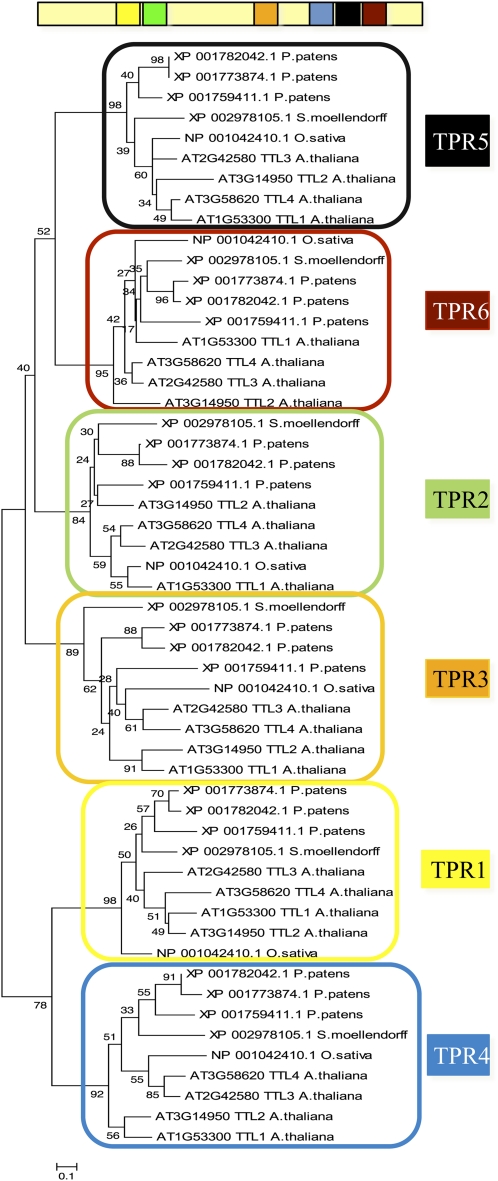
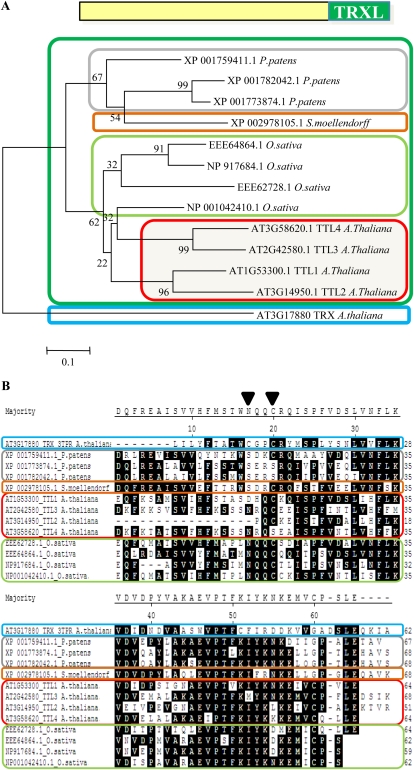
We next investigated whether TTL genes originated from an ancestral gene in land plants and subsequent duplications or resulted from convergent evolution in species with several TTL genes. Because TPRs are located in similar positions in all TTL proteins identified, we aligned the six TPR motifs of TTLs from phylogenetically distant species and obtained the corresponding cladogram. As shown in Figure 2, TPRs in equivalent positions are more homologous among them than TPRs within a TTL protein. Thus, TPR1 from Arabidopsis TTL1 is more closely related to TPR1 from P. patens than to any other TPR within TTL1. This indicates that an original TTL protein expanded among different species by duplication and that TPRs have been relatively well conserved throughout evolution, probably due to functional constraints. A phylogenetic analysis using the TRXL sequences provides similar results to those using the complete TTL sequence (Fig. 3A). We did not find any TTL protein containing the consensus sequence required for thioredoxin activity (Fig. 3B).
Figure 2.
Phylogenetic relationship among individual TPR motifs. An unrooted tree was built using the six TPR sequences of the four Arabidopsis proteins and TPR sequences from TTL orthologs from phylogenetically distant species such moss, lycophytes, and monocots. [See online article for color version of this figure.]
Figure 3.
Phylogenetic and sequence analyses of TRXL domains. A, Phylogenetic analysis of the TRXL domain of the TTLs from Arabidopsis, rice, P. patens, and S. moellendorffii. In the analysis, the protein sequence of At3g17880, a genuine thioredoxin, was included. B, Detail of the alignment of the TRXL domains, including the two-Cys motif (WCGPC) required for thioreductase activity. As shown, homologous regions from the TRXL domains of TTLs lack at least one of the two Cys residues that are essential for reductase activity. [See online article for color version of this figure.]
Arabidopsis TTL Genes Display Overlapping and Divergent Functions in Abiotic Stress Tolerance
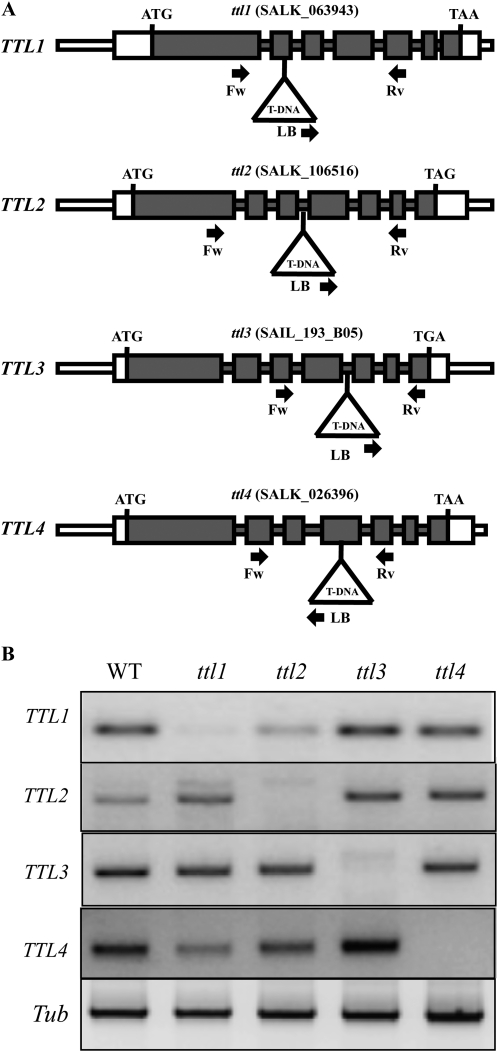
Plant genes that respond to environmental stimuli tend to have a higher rate of retention after duplication than nonresponsive genes; however, it is not clear which mechanisms contribute to their retention (Hanada et al., 2008). After duplication, the predominant fate of duplicated genes is pseudogenization (Taylor and Raes, 2004); still, a significant fraction of gene duplicates in plants are preserved and follow different evolutionary scenarios, including retention, loss, gain, and switch of functions. To investigate the specific evolutionary path(s) followed by the different members of the TTL family in Arabidopsis, we studied T-DNA mutants for the four Arabidopsis TTL genes and evaluated their specific roles in abiotic stress responses. The previously characterized ttl1-2 mutant allele (SALK_063943; hereafter referred to as ttl1) in the Columbia (Col-0) background was used in this study (Rosado et al., 2006b). In addition, homozygous mutant lines for TTL2 (SALK_106516), TTL3 (SALK_193805), and TTL4 (SALK_026396) in the Col-0 background were isolated (Fig. 4A). In every case, transcripts for the TTL gene in its corresponding mutant background were not detected by reverse transcription (RT)-PCR analysis, indicating that none of the ttl mutant alleles were able to produce intact full-length mRNAs (Fig. 4B).
Figure 4.
Molecular characterization of ttl mutants. A, ttl1, ttl2, ttl3, and ttl4 mutant alleles. The corresponding T-DNA line from SALK is indicated at the top. Gray boxes represent exons, and lines represent introns. ATG and stop codons are indicated. White boxes indicate the 5′ and 3′ untranslated regions. T-DNA insertion sites (not drawn to scale) are represented by triangles. The primers used for PCR genotyping are indicated by arrows (see “Materials and Methods” for primer sequences). LB, Left border. B, Expression analysis of the four TTL genes in the wild type (WT) and ttl mutants by RT-PCR. RNA was extracted from flowers, which is the only tissue where TTL2 expression was detected. The gene-specific primers designed to amplify cDNA fragments are detailed in “Materials and Methods.” The tubulin gene (Tub) was used as a positive control for the RT-PCR.
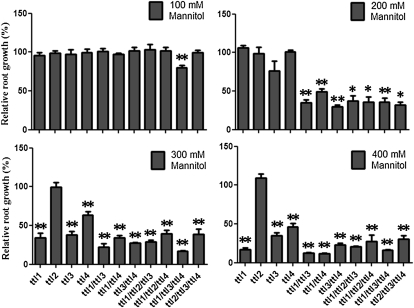
Mutations in TTL1 caused reduced tolerance to NaCl and osmotic stress, characterized by altered seedling development, reduced root elongation, disorganization of the root meristem, and impaired responses during germination. However, no differences between ttl1 and wild-type plants were observed under nonstressing conditions (Rosado et al., 2006b). Similarly, we did not detect abnormalities in any of the ttl single mutants based on detailed analyses of root elongation under nonstressing conditions (Supplemental Fig. S2). Next, we investigated seedling development of the ttl mutant genotypes at increasing concentrations of mannitol. As shown in Figure 5, while roots of ttl1, ttl3, and ttl4 single mutants showed hypersensitivity to mannitol at high concentrations (300 and 400 mm), ttl2 did not show any difference compared with the wild type. These results suggest that the ancestral TTL had a role in osmotic stress tolerance and that after the TTL1 segmental duplication TTL2 changed its original function.
Figure 5.
Root growth responses of ttl mutants to mannitol stress. Root elongation of single, double, and triple ttl mutants was measured, and root growth is expressed as the percentage relative to wild-type seedlings grown in the same conditions. Results are means of three independent experiments ± sd. Asterisks indicate significant differences between samples as determined by t test (* P < 0.1, ** P < 0.05).
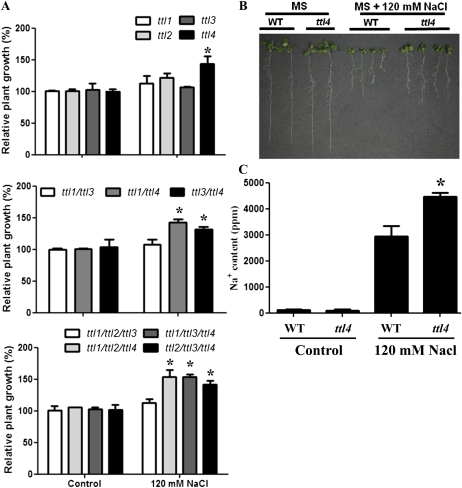
In addition, ttl1, ttl3, and ttl4 mutants also exhibited hypersensitivity to NaCl at concentrations above 150 mm, known to also generate osmotic stresses (Borsani et al., 2002). Surprisingly, at moderate NaCl concentrations (120 mm), ttl4 seedlings displayed improved root growth and increased cotyledon fresh weight compared with the wild type, while no differences were observed in nonstress conditions (Fig. 6, A and B). Next, we investigated whether the higher cotyledon fresh weight in the ttl4 mutant correlated with differences in Na+ accumulation. For that purpose, Na+ content was analyzed in wild-type and ttl4 seedlings grown for 1 week in Murashige and Skoog (MS) medium with or without 120 mm NaCl. As shown in Figure 6C, no differences in Na+ content were detected in wild-type and ttl4 seedlings in control medium, while ttl4 accumulated significantly more Na+ than the wild type in medium supplemented with NaCl. This result suggests that ttl4 improved seedling tolerance to NaCl by either improving long-distance Na+ transport or increasing the seedling capacity to compartmentalize Na+. Finally, an extended analysis of the ttl mutants and wild-type responses against oxidative, sugar, pH, and nutrient deprivation stresses using root elongation bioassays showed no significant differences (Supplemental Fig. S3). These results indicate that TTL genes are specifically involved in the response to osmotic and NaCl stresses.
Figure 6.
TTL4 loss of function increases tolerance to mild NaCl stress. A, Relative fresh weight of single ttl mutants and selected double and triple ttl mutants in MS medium or MS medium supplemented with 120 mm NaCl. All genotypes containing the ttl4 mutation displayed improved growth in medium supplemented with NaCl. Asterisks indicate significant differences between samples as determined by t test (* P < 0.05). B, Images of seedlings grown on MS agar medium for 1 week and transferred to MS agar medium for an additional 7 d without (left) or with (right) 120 mm NaCl. C, Sodium content in the shoot of the seedlings depicted in B. The asterisk indicates a significant difference between samples as determined by t test (* P < 0.05). WT, Wild type. [See online article for color version of this figure.]
TTL Promoters Differentially Respond to Abiotic Stress Treatments
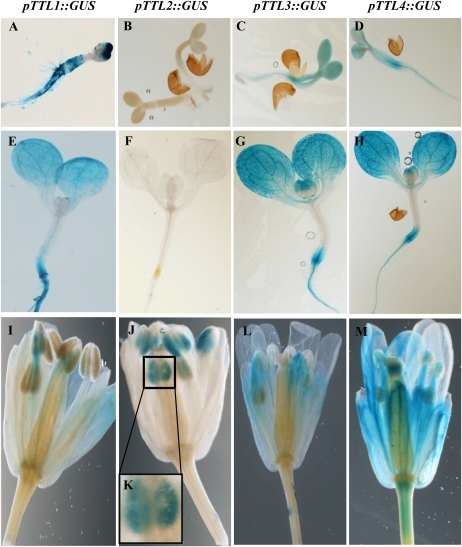
Based on the phenotypic analysis, TTL genes have different roles in NaCl and osmotic stress tolerance. While ttl1, ttl3, and ttl4 display hypersensitivity to osmotic stress in roots, ttl4 shows increased tolerance to NaCl and ttl2 does not show altered responses to abiotic stresses. Mutations in cis-regulatory elements in the promoter regions are a common mechanism for gene subfunctionalization (Van de Peer et al., 2001; Haberer et al., 2004), and differences in gene expression are mainly the result of divergence in cis-regulatory motifs (Evangelisti and Wagner, 2004). Therefore, we determined whether the different roles of the TTL genes in stress responses correlate with differential expression patterns. We first compiled expression data for the four TTL Arabidopsis genes using the Genevestigator database (Hruz et al., 2008). As shown in Supplemental Figure S4, the analysis revealed two distinct gene expression patterns for TTL genes. Whereas TTL1, TTL3, and TTL4 were ubiquitously expressed throughout most tissues, TTL2 expression was confined to pollen/stamen. GUS reporter gene fusions (pTTL::GUS) were constructed using approximately 2 kb of the upstream regulatory region for each TTL gene. Seedlings of at least 10 independent transgenic lines per construct were stained for GUS activity, and lines with single insertions harboring homozygous pTTL::GUS were used to analyze expression patterns.
Figure 7 shows representative expression profiles for all pTTL::GUS transgenic lines in 2-d-old seedlings, 10-d-old seedlings, and flowers. The first phylogenetic pair, TTL1/TTL2, showed very different expression patterns, with pTTL1 showing ubiquitous expression in seedlings while pTTL2 expression was restricted to pollen. The second phylogenetic pair, TTL3/TTL4, displayed similar expression patterns, with both genes ubiquitously expressed in vegetative tissues and strong signal in the vasculature. Thus, the expression patterns obtained from the promoter-GUS constructs corroborate the available microarray data (Supplemental Fig. S5).
Figure 7.
Tissue-specific expression pattern of TTL genes by promoter-reporter (GUS) fusions. GUS histological staining in transgenic Arabidopsis lines containing pTTL::GUS constructs is shown. The samples depicted are 2-d-old seedlings (A–D), 10-d-old seedlings (E–H), and flowers (I–M).
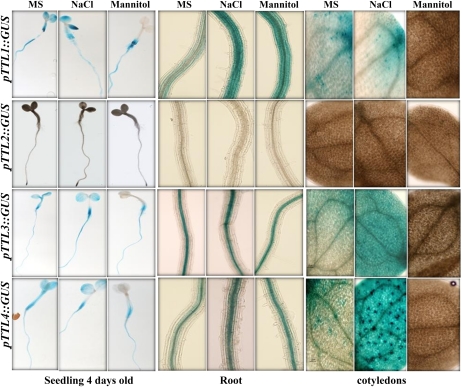
Next, we analyzed the TTL expression patterns in response to abiotic stresses (Fig. 8). For the first phylogenetic pair, TTL1/TTL2, NaCl treatments induced pTTL1::GUS expression in roots and cotyledons, while mannitol stress induced pTTL1::GUS expression in roots but suppressed it in cotyledons. TTL2 expression was never detected in vegetative tissues. The second phylogenetic pair, TTL3/TTL4, showed an expression pattern very similar to TTL1, with strong induction by NaCl and drastic repression by mannitol in cotyledons.
Figure 8.
TTL genes show differential expression responses to NaCl and mannitol stresses. Transgenic lines harboring TTL promoter::GUS fusions were grown for 4 d and then transferred to control medium or medium supplemented with 120 mm NaCl or 300 mm mannitol for 24 h before staining for GUS activity. The images show whole seedlings, roots, and cotyledons.
These results indicate that individual members of the TTL family have differential abiotic stress responses and that changes in cis-regulatory elements might have been instrumental in the subfunctionalization within the TTL genes.
TTL2 Functions in Male Gametophyte Development
We next examined whether TTL genes act redundantly or synergically, and for that purpose, we aimed to produce all possible mutant combinations of the TTL genes (Fig. 4A). In total, six double mutants (ttl1/2, ttl1/3, ttl1/4, ttl2/3, ttl2/4, ttl3/4) and four triple mutants (ttl1/2/3, ttl1/2/4, ttl1/3/4, ttl2/3/4) were obtained. We were unable to obtain plants containing homozygous mutations in all TTL genes (quadruple ttl1/2/3/4 mutant) despite numerous attempts, suggesting that TTL function is essential for plant viability.
During the generation of the different mutant combinations, we noticed that ttl2 homozygous mutants in F2 segregant populations appeared at lower frequencies than theoretically expected for a Mendelian recessive trait. Considering that TTL2:GUS was strongly and specifically expressed in pollen grains, we hypothesized that TTL2 might be required for male gametophytic transmission. Reciprocal crosses between the wild type and ttl2 were performed, and our results indicate that no significant differences in the number of seeds per silique existed when ttl2 was used as a female parent, but the number of seeds per silique was significantly reduced when ttl2 was used as the male parent (Table I). This result indicates that ttl2 has defects in the male gametophyte and confirms that ttl2 mutant pollen is responsible for the reduced fertility.
Table I. Effect of mutation in TTL2 on the number of seeds per silique.
| Genotype: Pollen Donor/Recipient | Obtained | t Test | No. of Siliques |
| TTL2/ttl2 | 13.3 ± 6.34 | P < 0.001 | 10 |
| ttl2/TTL2 | 22.8 ± 9.41 | P > 0.001 | 10 |
| TTL2/TLL2 | 29.5 ± 10.61 | – | 10 |
We next analyzed the progeny derived from wild-type plants, ttl2 plants, and reciprocal crosses between wild-type and heterozygous (TTL2/ttl2) plants. According to Mendelian inheritance for recessive traits, the result of the cross between a wild-type plant and a heterozygous TTL2/ttl2 plant should render 50% of the population with a wild-type (TTL2) genotype and 50% with a heterozygous TTL2/ttl2 genotype. Since TTL2/ttl2 plants are resistant to kanamycin due to the presence of this selectable marker in the T-DNA of the ttl2 allele, we carried out the genetic analysis of F2 seeds on MS plates supplemented with kanamycin. As shown in Table II, approximately 50% of the seeds were resistant to kanamycin when the wild type was used as the pollen donor, suggesting that there are no defects in the transmission of the ttl2 female gametophyte. However, when pollen from heterozygous (TTL2/ttl2) plants was used, only approximately 10% of the progeny was resistant to kanamycin, further confirming that mutations in TTL2 decrease the efficiency of the male gametophyte transmission.
Table II. Genotypic analysis using kanamycin resistance.
Expected and experimental percentages of germination in kanamycin selection medium of the indicated genotypes are shown.
| Genotype: Pollen Donor/Recipient | Expected | Obtained | No. of Seeds |
| % | |||
| ttl2 | 100 | 100 ± 0 | 116 |
| TTL2 | 0 | 0 ± 0 | 113 |
| TTL2/ttl2 × TTL2 | 50 | 45 ± 7.07 | 128 |
| TTL2 × TLL2/ttl2 | 50 | 9.7 ± 8.4 | 110 |
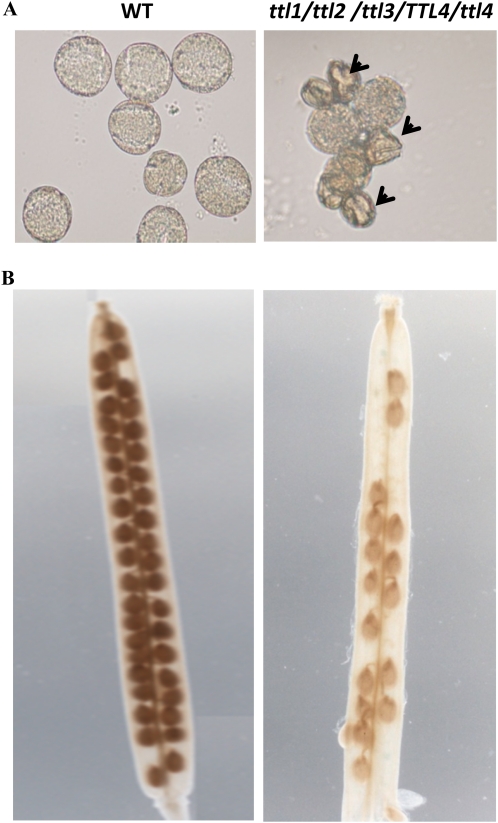
As previously indicated, we were unable to obtain plants that contained homozygous mutations in all four TTL genes; however, we were able to obtain viable plants with a ttl1/ttl2/ttl3/TTL4/ttl4 genotype, albeit at very low rate. To further investigate the role of TTL genes in embryogenesis, we analyzed the progeny of ttl1/ttl2/ttl3/TTL4/ttl4 plants in more detail. Based on Mendelian inheritance, a frequency of 25% quadruple homozygous mutants was expected; however, a large proportion of the pollen obtained from ttl1/ttl2/ttl3/TTL4/ttl4 plants showed striking morphological distortions (Fig. 9A), and the proportion of aborted seeds on the ttl1/ttl2/ttl3/TTL4/ttl4 progeny was approximately 42%, which is significantly higher than the expected 25% (Fig. 9B). This result indicates that a single copy of TTL4 can, in some cases, provide enough TTL4 protein for proper seed development. The requirement of TTL4 for seed development is consistent with the large progeny required to obtain an individual ttl1/ttl2/ttl3/TTL4/ttl4 plant from a ttl1/2/3 × ttl1/2/4 cross. Interestingly, once the ttl1/ttl2/ttl3/TTL4/ttl4 plant was obtained, it did not show obvious differences in vegetative growth, indicating that TTL genes are particularly important for gametophytic transmission.
Figure 9.
TTL genes are required for male gametophytic development. Pollen and siliques from wild-type (WT) and ttl1/ttl2/ttl3/TTL4/ttl4 plants were analyzed by light microscopy. ttl1/ttl2/ttl3/TTL4/ttl4 plants showed abnormal, shrunken pollen grains (indicated by arrowheads). ttl1/ttl2/ttl3/TTL4/ttl4 plants do not support normal seed development, showing approximately 50% aborted seeds. [See online article for color version of this figure.]
The Function of TTL1, TTL3, and TTL4 in Abiotic Stress Responses Is Not Fully Redundant
To determine whether the contribution of TTL genes to abiotic stress responses is fully redundant, partially redundant, or additive, we examined the responses of the different ttl mutant combinations against mannitol and NaCl stresses. We did not find differences in germination or seedling development between the wild type and any of the mutant combinations under control conditions (Supplemental Fig. S2). As indicated previously, ttl1, ttl3, and ttl4 but not ttl2 single mutants were hypersensitive to high mannitol concentrations (Fig. 5). The double mutant combinations ttl1/ttl3, ttl1/ttl4, and ttl3/ttl4 displayed higher sensitivity to 200 mm mannitol than their respective single mutant counterparts, suggesting that those genes have an additive effect in their tolerance to osmotic stress. This was confirmed by the triple ttl1/ttl3/ttl4 mutant being the most sensitive genotype analyzed, showing reduced root growth at mannitol concentrations as low as 100 mm. The enhanced sensitivity observed in the triple mutant relative to the double mutants, and in double mutants relative to single mutants, indicates that TTLs have similar roles in osmotic stress tolerance but that they are not fully redundant.
We further investigated the redundancy and/or additive effects of TTL genes by generating triple-heterozygous mutants for the TTL1, TTL3, and TTL4 genes. We reasoned that if the responses of the different TTLs to osmotic stress were dependent on a dosage effect, we should observe a certain degree of hypersensitivity in triple heterozygous plants. As shown in Supplemental Figure S6, the response of the triple heterozygous plants to mannitol was identical to the wild type, suggesting that the osmotic stress response is not controlled by total TTL gene dosage but by the individual contributions of the different TTL genes.
Finally, ttl4 but not ttl1, ttl2, and ttl3 seedlings displayed improved cotyledon development and higher fresh weight than the wild type at low and moderate NaCl concentrations. To determine whether the mutation in TTL4 was the only contributor to this increased NaCl tolerance, we evaluated the different ttl mutant combinations in the presence of 120 mm NaCl. As shown in Figure 6, only those genotypes carrying the ttl4 mutation showed improved growth in NaCl comparable to the improvement observed in the ttl4 single mutant. This result confirms that TTL4 is the only one affecting NaCl tolerance; therefore, we can assign a nonredundant function to TTL4 in response to high NaCl.
Root Meristem and Vascular Defects Are Enhanced in Plants with Multiple Mutations in TTL Genes
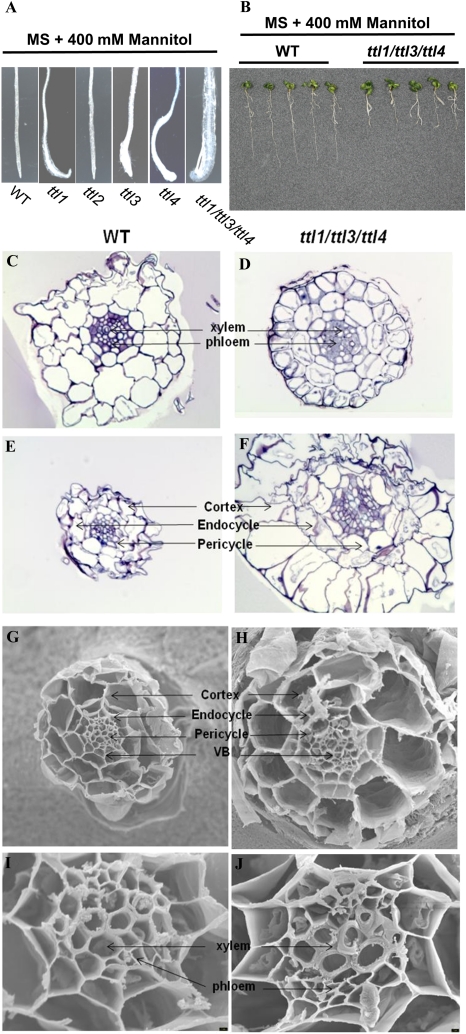
The main feature caused by osmotic stress in ttl1 mutants was swollen root tips and meristem disorganization (Rosado et al., 2006b); therefore, we analyzed in detail the root developmental defects in plants harboring different ttl mutant combinations. Thus, under mannitol stress, all mutants (with the expected exception of ttl2) showed root meristem disorganization similar to that previously reported for ttl1 (Fig. 10A). Correlating with the osmotic sensitivity previously reported, the degree of root meristem disorganization increased with the number of ttl mutations, being maximal in the triple ttl1/ttl3/ttl4 mutant (Fig. 10B).
Figure 10.
Osmotic stress effects on root morphology of the triple mutant ttl1/ttl3/ttl4. A, ttl mutant root swelling induced by a 7-d treatment in growth medium supplemented with 400 mm mannitol. B, Root growth hypersensitivity of the triple ttl1/ttl3/ttl4 mutant after 400 mm mannitol treatment. C to F, Transverse sections of the roots of wild-type (WT) and triple ttl1/ttl3/ttl4 mutant plants in control conditions (C and D) or after treatment with mannitol (E and F). Magnification = 40×. G to J, Scanning electron microscopy of wild-type (G and I) and ttl1/ttl3/ttl4 mutant (H and J) root meristematic tissue after osmotic stress treatments. The mutant shows vascular bundle (VB) disorganization, large endodermis and cortex cells, altered pericycle, and damage in vascular tissue with highly lignified xylem. For G and H, magnification = 700×; for I and J, magnification = 3,000×. [See online article for color version of this figure.]
To gain insight into the changes that caused the root tip swelling at the cellular level, we analyzed transverse sections of the primary roots in the wild type and the triple ttl1/ttl3/ttl4 mutant grown on control medium or medium supplemented with 400 mm mannitol. No differences in radial organization between the wild type and ttl1/ttl3/ttl4 were observed in control conditions; however, a reduced differentiation of vascular tissues was observed in ttl1/ttl3/ttl4 mutants, suggesting a role for TTL genes in vasculature development (Fig. 10, C and D). After mannitol treatment, wild-type roots decreased in diameter, and the typical shrink effect induced by low water potential was observed (Fig. 10E). In contrast, roots of ttl1/ttl3/ttl4 showed a significant increase in the size of all cell layers, indicating that all root cell types were affected by the ttl mutations (Fig. 10F). We further analyzed the radial organization of roots grown in control medium or medium supplemented with 400 mm mannitol using scanning electron microscopy (Fig. 10, G–J). The analysis confirmed the disorganization in vascular bundles and the aberrant thickness and diameter of the meristematic cells in the ttl1/ttl3/tt4 background compared with the wild type. Thus, cell diameters for cortex, endodermis, pericycle, xylem, and phloem in ttl1/ttl3/ttl4 under osmotic stress conditions were approximately twice bigger than wild-type cells after mannitol treatment (Supplemental Fig. S7). These results indicate a role for TTLs in the control of meristematic cell size during osmotic stress.
DISCUSSION
The Role of TTL Duplication in Arabidopsis
In Arabidopsis, the TTL family is composed of four members derived from two syntenous genomic regions with collinear features suggesting the existence of a common ancestor. This aspect is further supported by the evolutionary conservation of the internal modular domains among Arabidopsis TTLs and phylogenetically separated TTL proteins and by the identification of a single TTL gene in S. moellendorffii (SmTTL1), an important model organism in comparative genomics representing the oldest extant division of the vascular plants. The presence of a single TTL gene in S. moellendorffii strongly suggests that an ancestral TTL protein can perform all TTL functions required for plant viability. Therefore, we propose that successive duplications of an ancestral TTL gene led to the neofunctionalization and subfunctionalization of the TTL gene family in Arabidopsis, contributing to the fine-tuning of the Arabidopsis responses against abiotic stresses. Thus, the combination of gene duplication and selection pressure imposed by stress conditions likely led to substantial changes in TTL gene expression and functional changes of the TTL gene duplicates over time that likely contributed to the physiological complexity of the Arabidopsis TTL response against abiotic stress. Our results are a clear example supporting the hypothesis that gene duplication is an important evolutionary mechanism in the generation of novel functions and phenotypes, contributing to the adaptation of land plants to stressful environments (Hanada et al., 2008; Dassanayake et al., 2011).
Neofunctionalization and Subfunctionalization in the Arabidopsis TTL Family
Phylogenetic studies within the Arabidopsis TTL family indicate that TTL1 and TTL2 proteins cluster together and form a separate cluster from TTL3 and TTL4 (Fig. 1). Although closely related to TTL1 based on amino acid sequence, the observed differences in TTL2 expression patterns compared with the other TTL genes suggests that TTL2 subfunctionalization is driven by mutations in its regulatory sequence. Moreover, TTL2 expression is confined to pollen grains, and our genetic studies confirmed that TTL2 performs a distinct function among the TTLs in male gametophyte development.
The TTL1 expression pattern closely resembles that of TTL3 and TTL4, based not only on microarray analysis but also on promoter-GUS studies. Accordingly, the phenotypic consequences of mutations in TTL1, TTL3, and TTL4 affect similar Arabidopsis responses to abiotic stress. A plausible explanation for the TTL1, TTL3, and TTL4 gene duplications is that TTL duplications are required to increase the bulk of TTL protein amounts without changing protein function. This seems to be the case for mannitol tolerance, in which an increment in the number of TTL mutations results in additive hypersensitivity to mannitol. In this scenario, the increased hypersensitivity to mannitol could be explained as a result of a decrease in gene dosage, independently of the TTL genes mutated. However, the triple heterozygous mutant for TTL1, TTL3, and TTL4 is not hypersensitive to mannitol in conditions where the roots of single mutants are clearly affected. Thus, our results suggest that although some redundancy within the TTL family may occur, the roles of TTL genes cannot be simply explained as TTL gene dosage defects and suggest specific functions. Furthermore, the fact that TTL4 increased the shoot growth at moderate NaCl indicates that TTL4 followed a process of neofunctionalization that differentiates it from the rest of the TTLs.
In this study, we have shown that independent TTL genes are essential for root growth and integrity under osmotic stress, having a role in root meristem organization and vascular genesis, and that TTL4 controls the osmotic/ionic stress responses by regulating Na+ homeostasis. Since the colonization of different habitats by plants is determined by morphological and physiological adaptation designed to deal with multiple abiotic stresses such as drought and salinity, it is very attractive to hypothesize that the specialization of TTL function contributed to Arabidopsis adaptation to very different environmental conditions. Still, an important question that needs to be addressed is how TTL proteins are mechanistically involved in these adaptive processes.
Protein-Protein Interactions Mediating TTL Function
TTL proteins play important roles in different aspects of plant development, being essential for both gametophytic viability and root growth and integrity under osmotic stress. The TPR units are common modules in molecular chaperones and are required for the establishment of protein-protein interaction during the formation of multiprotein complexes (Blatch and Lässle, 1999).
Recently, TTL proteins have been proposed as potential interactors of the Hsp90 and Hsp70 chaperone complexes (Prasad et al., 2010). This is based on the finding that TTLs, like confirmed Hsp90 cochaperones, such as Hsp70-Hsp90 organizing protein, high-Mr immunophilins (cyclophilin 40 and FK506-binding proteins 51 and 52), protein phosphatase 5, and the C terminus of Hsc70-interacting protein, contain a subset of basic residues in the TPR-binding pocket known as the carboxylate clamp. The carboxylate clamp domain interacts with the acidic side chains of the highly conserved EEVD motif at the C-terminal ends of Hsp90 and Hsp70 (Carrigan et al., 2006; Prasad et al., 2010).
Hsp90 and Hsp70 complexes perform key roles in signal transduction by regulating maturation, localization, stability, and protein interactions of a large number of signaling proteins in eukaryotes (Pearl et al., 2008). Alterations in these complexes lead to morphological abnormalities, such as altered leaf shape, organ number, and pigment accumulation, indicating an important role for these chaperone complexes in plant development (Sangster et al., 2007). Since the overexpression of the cytosolic Hsp90 complex alters the sensitivity to drought and NaCl in Arabidopsis (Song et al., 2009), and the HSP70/HSP90 machinery is important in plants to integrate signals from their biotic and abiotic environments through stomatal regulation (Clément et al., 2011), we hypothesize that TTL proteins could regulate abiotic stress responses through their specific expression under stress conditions and their interaction with the Hsp90 or Hsp70 complexes.
Chaperone Function of TTL Proteins
In Arabidopsis, the proteins with highest homology to TTL are AtTPR12, AtTPR13, and AtTPR14. These proteins show similar TPR domain localization to TTL proteins but lack the C-terminal region with homology to thioredoxin. Another protein with high homology is AtTDX (Vignols et al., 2003; Lee et al., 2009), a protein that contains three TPR domains and a C-terminal bona fide thioredoxin domain. AtTDX exhibits multiple functions, acting as a disulfide reductase, foldase chaperone, and holdase chaperone (Lee et al., 2009). The disulfide reductase and foldase chaperone functions predominate when AtTDX occurs in the low-Mr form, whereas the holdase chaperone function predominates in the high-Mr complexes. Chaperones are largely classified into two groups: foldase chaperones, which support the folding of denatured proteins to their native state; and holdase chaperones, which bind to folding intermediates, thereby preventing their nonspecific aggregation (Beissinger and Buchner, 1998).
This study reveals that TTL genes are specific for land plants and play a regulatory role in different aspects of plant development and stress responses. Although the molecular mechanisms regulated by TTLs need to be elucidated, this study provides important insights for the understanding of a novel and essential protein family with very little available functional information.
MATERIALS AND METHODS
Sequence Analysis
Genes encoding TTL proteins in different plant species were identified using the Phytozome Web page (www.phytozome.net), with the exception of tomato (Solanum lycopersicum) and barley (Hordeum vulgare), where their genome databases were employed. We used the BLASTP algorithm against the putative proteome derived from the predicted genes. Later, the TPR motif (Pfam:00505) and the TRLX motif (Pfam:00085) were analyzed individually in each sequence using both the tool available in Phytozome and the Conserved Domain Database at the National Center for Biotechnology Information (NCBI; www.ncbi.nlm.nih.gov/structure/ccd/ccd.shtml) and discarded those sequences without six TPR motifs in conserved positions or lacking the TRLX domain. Protein data matrices were aligned using the ClustalX multiple sequence alignment program with default gap penalties.
Phylogenetic Analysis and Genomic Structure
Multiple sequence alignments were performed with the ClustalW program. An unrooted phylogenetic tree was generated with the ClustalX2 program and visualized using Treeview. Alignment analysis of Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), Physcomitrella patens, and Selaginella moellendorffii TTLs was obtained using MegAlign 4 software. TTL in silico analysis was performed using DNASTAR Lasergene 8 software. All parameter values correspond to default definitions.
Plant Material
The plants used were Arabidopsis ecotype Col-0 wild type and the T-DNA insertion lines Salk_063943 (for TTL1), Salk_106516 (for TTL2), Sail_193_B05 (for TTL3), and Salk_026396 (for TTL4). The Col-0 T-DNA insertion mutants were identified using the SIGnal Web site at http://signal.salk.edu. Seeds of the T-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center.
Analysis of the T-DNA Insertion Lines, and Generation of Mutant Combinations
PCR-based genotypic analysis was performed as described (Koiwa et al., 2006) using the following primers: LBSAIL (5′-TAGCATCTGAATTTCATAACCAATCTCGATACAC-3′), LBSALK (5′-TGGTTCACGTAGTGGGCCATCG-3′), TTL1DPCRF (5′-TGGACTCACCACCACCACTA-3′), TTL1DPCRR (5′-ACCGAGTCTGCGAACAAGAT-3′), TTL2DPCRF (5′-GAGCTCTCCCACATTCCAAC-3′), TTL2DPCRR (5′-CCGTTTAGCCTCATTGCTTC-3′), TTL3DPCRF (5′-AGAGAGCTGCGATGCTTGAT-3′), TTL3DPCRR (5′-ATGCTC-TCCTCCACATCCAC-3′), TTL4DPCRF (5′-AGATCGGTGATTGGAGAACG-3′), and TTL4DPCRR (5′-TCGATAGAGCGTTCCTTGCT-3′). Mutant combinations generated in this work were obtained by classical genetic crosses and further PCR genotypic analysis.
Arabidopsis Seed Germination and Root Elongation Measurements
Arabidopsis seeds were surface sterilized with 70% (v/v) ethanol for 5 min, followed with 20% sodium hypochlorite for 15 min, and then washed three times with sterile water. For in vitro germination, seeds were stratified for 2 d at 4°C in the dark and plated onto petri dishes containing basal MS medium (Murashige and Skoog, 1962). Seedlings were grown 4 d in MS medium and then transferred to petri dishes containing different concentrations of the stress agent or hormone (Rosado et al., 2006a). Plates were located under a long-day regime (16 h of light/8 h of dark) of 25 μE at 23°C (day/night). For root elongation measurements, eight seeds were used per replicate, and three replicates were made for each treatment. Seedlings with 1- to 1.5-cm-long roots were transferred from vertical agar plates containing MS medium onto a second agar medium supplemented with different concentrations of salts and osmotic stress agents as described above. Root length and plant weight were scored 7 d after transfer to the plate with the different treatments.
Ion Content Determination
Seedlings were grown as indicated previously. For ion content determination, 15-d-old ttl4 and wild-type seedlings grown in 120 mm NaCl after the transfer from MS medium were used. The shoot was removed and washed three times with bidistilled water. To determine tissue Na+ concentrations, the dried plant material was boiled for 20 min in 1 mL of 100 mm nitric acid and analyzed using a flame photometer at the Laboratorio de Ionómica (Centro de Edafología y Biología Aplicada del Segura-Consejo Superior de Investigaciones Científicas).
Promoter TTL::GUS Analysis
Approximately 2.0 kb of the genomic sequence upstream of the TTL translation start site was amplified by PCR. The following primers were used: PROTTL1F (5′-TGGTACCTTGAGTGGAAGAAGGAA-3′) and PROTTL1R (5′-ACCATGGTGAGTGTTGTGGTGAGTGAA-3′) for TTL1::GUS; PROTTL2F (5′-AGGTACCTTGTTATTTAAATCCGA-3′) and PROTTL2R (5′-ACCATGGTTAGCAAGATTACAAAAAAG-3′) for TTL2::GUS; PROTTL3F (5′-AGGTACCGCAACACCCTTCTATTT-3′) and PROTTL3R (5′-ACCATGGTCGTCACTT-CCTCGTGAGCT-3′) for TTL3::GUS; PROTTL4F (5′-TGGTACCCAAATCTTGTTATTTTG-3′) and PROTTL4R (5′-ACCATGGTCTCGAAACCAAAGCCTCAG-3′) for TTL4::GUS. The amplified fragments were cloned into the binary vector pCAMBIA1303. The recombinant plasmids were introduced into Agrobacterium tumefaciens strain GV3101 and used to transform wild-type plants. Control plants consisted of the wild type transformed with the pCAMBIA 1303 empty vector. GUS activity was detected in situ as described previously (Jefferson et al., 1987). Histochemical analysis for TTL promoter-GUS constructs under stress treatments was performed using 4-d-old seedlings treated with 120 mm NaCl and 300 mm mannitol for 24 h. Special care was taken to ensure that controls and transformed plants were mannitol and NaCl treated and stained for GUS activity simultaneously.
TTL Expression Analysis Using RT-PCR
Two-week-old plants were transferred from MS agar plates to soil and grown at 23°C under a long-day regime in a growth chamber during 1 month until flowering. Whole flowers were collected from each mutant for RNA isolation, immediately frozen in liquid nitrogen, and stored at −80°C until use. RNA extraction and RT-PCR were performed as described previously (Rosado et al., 2006b).
Microscopy Analysis
Arabidopsis roots were fixed overnight at 4°C in 4% (v/v) paraformaldehyde in 0.1 m phosphate buffer saline buffer, pH 7.4. Samples were dehydrated gradually in an ethanol series subsequently followed by dry acetone, 1:1 Araldite 502:dry acetone during 1 h, and Araldite 502 overnight (ethyl methanesulfonate). The Araldite 502 resin is a mixture of 9.2 g of Araldite 502, 10.8 g of dodecenyl succinic anhydride, and 0.4 g of 2,4,6-tri-(dimethylaminemethyl) phenol. After embedding in Araldite, blocks were left for polymerization at 60°C during 72 h. Transverse sections of the roots (1 μm thick) were obtained using an ultramicrotome (Reichert-Jung Ultracut E), stained with 0.1% (w/v) toluidine blue, and visualized with a Leica DM 1000 microscope. For each line, an average of 10 plants were used for analysis.
Vascular bundles were visualized using a Ranvier-type microtome. Sections were dehydrated in absolute ethanol (99.9%, v/v), which was removed in a critical point dryer system (BAL-TEC CPD030). Then, samples were mounted on specimen holders and covered with a fine gold coat in a metallization unit (JEOL JCC 1100). A scanning electron microscope equipped with a digital acquisition system (JEOL JSM-840) was used for visualization of the samples.
Quantification of the diameter of vascular cells, root apex cells, and number of cells was performed using ImageJ software (http://rsb.info.nih.gov/ij/). Measurements were performed on eight different cells for each tissue (cortex, endocycle, pericycle, xylem, and phloem) from three independent plants.
Silique examination was performed by clearing them in 95% ethanol for 1 h, hydrating them in 50% glycerol, and examining them with a magnifying glass.
Bioinformatics Analysis
Bioinformatics analysis was performed using the following databases: Genevestigator (www.genevestigator.com), The Bio-Array Resource for Arabidopsis Functional Genomics (www.bar.utoronto.ca), The Arabidopsis Information Resource (www.arabidopsis.org), NCBI (www.ncbi.nlm.nih.gov), and European Bioinformatics Institute (www.ebi.ac.uk/Tools/clustalw2/index.html).
TTL Analysis
The mining of TTL genes was performed by searching sequences homologous to the TTL1 protein using the Phytozome database (www.phytozome.net) Redundant sequences as well as those sequences that did not present six TPRs and the TRLX domain were discarded. Sequences were also analyzed using the NCBI Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). Generation of the unrooted phylogenetic tree was performed by alignment of the full-length amino acid sequences using ClustalW and the BIOEDIT software, followed by manual adjustments.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Schematic representation of TTL containing duplicated segments identified in the Arabidopsis genome.
Supplemental Figure S2. ttl mutants show identical root growth in control medium.
Supplemental Figure S3. Root growth of triple ttl mutants is not affected by pH, low nutrients, or reactive oxygen species generators.
Supplemental Figure S4. Expression pattern of Arabidopsis TTL genes based on data available from the Genevestigator database.
Supplemental Figure S5. Expression pattern of Arabidopsis TTL genes in response to NaCl stress based on data available from the Genevestigator database.
Supplemental Figure S6. Root growth inhibition is not caused by reduced gene dosage of TTL genes.
Supplemental Figure S7. Root cell diameters of the wild type and the ttl1/ttl3/ttl4 triple mutant after 400 mm mannitol stress.
Acknowledgments
We thank the Nottingham Arabidopsis Stock Centre. We thank P. Martín for excellent technical assistance and José R. Botella for comments on the manuscript.
References
- Beissinger M, Buchner J. (1998) How chaperones fold proteins. Biol Chem 379: 245–259 [PubMed] [Google Scholar]
- Blatch GL, Lässle M. (1999) The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21: 932–939 [DOI] [PubMed] [Google Scholar]
- Borsani O, Cuartero J, Valpuesta V, Botella MA. (2002) Tomato tos1 mutation identifies a gene essential for osmotic tolerance and abscisic acid sensitivity. Plant J 32: 905–914 [DOI] [PubMed] [Google Scholar]
- Borsani O, Valpuesta V, Botella MA. (2003) Developing salt tolerant plants in a new century: a molecular biology approach. Plant Cell Tissue Organ Cult 73: 101–115 [Google Scholar]
- Carrigan PE, Sikkink LA, Smith DF, Ramirez-Alvarado M. (2006) Domain:domain interactions within Hop, the Hsp70/Hsp90 organizing protein, are required for protein stability and structure. Protein Sci 15: 522–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceserani T, Trofka A, Gandotra N, Nelson T. (2009) VH1/BRL2 receptor-like kinase interacts with vascular-specific adaptor proteins VIT and VIK to influence leaf venation. Plant J 57: 1000–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clément M, Leonhardt N, Droillard M-J, Reiter I, Montillet J-L, Genty B, Laurière C, Nussaume L, Noël LD. (2011) The cytosolic/nuclear HSC70 and HSP90 molecular chaperones are important for stomatal closure and modulate abscisic acid-dependent physiological responses in Arabidopsis. Plant Physiol 156: 1481–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliff MJ, Williams MA, Brooke-Smith J, Barford D, Ladbury JE. (2005) Molecular recognition via coupled folding and binding in a TPR domain. J Mol Biol 346: 717–732 [DOI] [PubMed] [Google Scholar]
- D’Andrea LD, Regan L. (2003) TPR proteins: the versatile helix. Trends Biochem Sci 28: 655–662 [DOI] [PubMed] [Google Scholar]
- Dassanayake M, Oh D-H, Haas JS, Hernandez A, Hong H, Ali S, Yun D-J, Bressan RA, Zhu J-K, Bohnert HJ, et al. (2011) The genome of the extremophile crucifer Thellungiella parvula. Nat Genet 43: 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies TH, Sánchez ER. (2005) FKBP52. Int J Biochem Cell Biol 37: 42–47 [DOI] [PubMed] [Google Scholar]
- Evangelisti AM, Wagner A. (2004) Molecular evolution in the yeast transcriptional regulation network. J Exp Zoolog B Mol Dev Evol 302: 392–411 [DOI] [PubMed] [Google Scholar]
- Fedoroff NV. (2002) RNA-binding proteins in plants: the tip of an iceberg? Curr Opin Plant Biol 5: 452–459 [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Richard D, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, et al. (2011) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res Database Issue 40: 1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gounalaki N, Tzamarias D, Vlassi M. (2000) Identification of residues in the TPR domain of Ssn6 responsible for interaction with the Tup1 protein. FEBS Lett 473: 37–41 [DOI] [PubMed] [Google Scholar]
- Greenboim-Wainberg Y, Maymon I, Borochov R, Alvarez J, Olszewski N, Ori N, Eshed Y, Weiss D. (2005) Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17: 92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberer G, Hindemitt T, Meyers BC, Mayer KFX. (2004) Transcriptional similarities, dissimilarities, and conservation of cis-elements in duplicated genes of Arabidopsis. Plant Physiol 136: 3009–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu S-H. (2008) Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol 148: 993–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformat 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiwa H, Bressan RA, Hasegawa PM. (2006) Identification of plant stress-responsive determinants in Arabidopsis by large-scale forward genetic screens. J Exp Bot 57: 1119–1128 [DOI] [PubMed] [Google Scholar]
- Koiwa H, Li F, McCully MG, Mendoza I, Koizumi N, Manabe Y, Nakagawa Y, Zhu J, Rus A, Pardo JM, et al. (2003) The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell 15: 2273–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna P, Gloor G. (2001) The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperones 6: 238–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JR, Lee SS, Jang HH, Lee YM, Park JH, Park S-C, Moon JC, Park SK, Kim SY, Lee SY, et al. (2009) Heat-shock dependent oligomeric status alters the function of a plant-specific thioredoxin-like protein, AtTDX. Proc Natl Acad Sci USA 106: 5978–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R, Tester M. (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiol 15: 473–497 [Google Scholar]
- Pearl LH, Prodromou C, Workman P. (2008) The Hsp90 molecular chaperone: an open and shut case for treatment. Biochem J 410: 439–453 [DOI] [PubMed] [Google Scholar]
- Prasad BD, Goel S, Krishna P. (2010) In silico identification of carboxylate clamp type tetratricopeptide repeat proteins in Arabidopsis and rice as putative co-chaperones of Hsp90/Hsp70. PLoS ONE 5: e12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt WB, Morishima Y, Osawa Y. (2008) The Hsp90 chaperone machinery regulates signaling by modulating ligand binding clefts. J Biol Chem 283: 22885–22889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodromou C, Siligardi G, O’Brien R, Woolfson DN, Regan L, Panaretou B, Ladbury JE, Piper PW, Pearl LH. (1999) Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J 18: 754–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado A, Amaya I, Valpuesta V, Cuartero J, Botella MA, Borsani O. (2006a) ABA- and ethylene-mediated responses in osmotically stressed tomato are regulated by the TSS2 and TOS1 loci. J Exp Bot 57: 3327–3335 [DOI] [PubMed] [Google Scholar]
- Rosado A, Schapire AL, Bressan RA, Harfouche AL, Hasegawa PM, Valpuesta V, Botella MA. (2006b) The Arabidopsis tetratricopeptide repeat-containing protein TTL1 is required for osmotic stress responses and abscisic acid sensitivity. Plant Physiol 142: 1113–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangster TA, Bahrami A, Wilczek A, Watanabe E, Schellenberg K, McLellan C, Kelley A, Kong SW, Queitsch C, Lindquist S. (2007) Phenotypic diversity and altered environmental plasticity in Arabidopsis thaliana with reduced Hsp90 levels. PLoS ONE 2: e648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangster TA, Queitsch C. (2005) The HSP90 chaperone complex, an emerging force in plant development and phenotypic plasticity. Curr Opin Plant Biol 8: 86–92 [DOI] [PubMed] [Google Scholar]
- Schapire AL, Valpuesta V, Botella MA. (2006) TPR proteins in plant hormone signaling. Plant Signal Behav 1: 229–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber A, Stengel F, Zhang Z, Enchev RI, Kong EH, Morris EP, Robinson CV, da Fonseca PCA, Barford D. (2011) Structural basis for the subunit assembly of the anaphase-promoting complex. Nature 470: 227–232 [DOI] [PubMed] [Google Scholar]
- Shi H, Kim Y, Guo Y, Stevenson B, Zhu J-K. (2003) The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15: 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Tseng T-S, Swain SM, Dill A, Jeong SY, Olszewski NE, Sun T-P. (2007) Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol 143: 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Zhao R, Fan P, Wang X, Chen X, Li Y. (2009) Overexpression of AtHsp90.2, AtHsp90.5 and AtHsp90.7 in Arabidopsis thaliana enhances plant sensitivity to salt and drought stresses. Planta 229: 955–964 [DOI] [PubMed] [Google Scholar]
- Taipale M, Jarosz DF, Lindquist S. (2010) HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol 11: 515–528 [DOI] [PubMed] [Google Scholar]
- Taylor JS, Raes J. (2004) Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet 38: 615–643 [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Taylor JS, Braasch I, Meyer A. (2001) The ghost of selection past: rates of evolution and functional divergence of anciently duplicated genes. J Mol Evol 53: 436–446 [DOI] [PubMed] [Google Scholar]
- Vignols F, Mouaheb N, Thomas D, Meyer Y. (2003) Redox control of Hsp70-co-chaperone interaction revealed by expression of a thioredoxin-like Arabidopsis protein. J Biol Chem 278: 4516–4523 [DOI] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD. (2000) The origins of genomic duplications in Arabidopsis. Science 290: 2114–2117 [DOI] [PubMed] [Google Scholar]
- Wang KL-C, Yoshida H, Lurin C, Ecker JR. (2004) Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428: 945–950 [DOI] [PubMed] [Google Scholar]
- Whitesell L, Lindquist SL. (2005) HSP90 and the chaperoning of cancer. Nat Rev Cancer 5: 761–772 [DOI] [PubMed] [Google Scholar]
- Xiong L, Zhu JK. (2001) Abiotic stress signal transduction in plants: molecular and genetic perspectives. Physiol Plant 112: 152–166 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57: 781–803 [DOI] [PubMed] [Google Scholar]
- Yang J, Roe SM, Cliff MJ, Williams MA, Ladbury JE, Cohen PTW, Barford D. (2005) Molecular basis for TPR domain-mediated regulation of protein phosphatase 5. EMBO J 24: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]