Abstract
Despite the widespread use of Arabidopsis (Arabidopsis thaliana) as a model plant, a curated dataset of Arabidopsis genes with mutant phenotypes remains to be established. A preliminary list published nine years ago in Plant Physiology is outdated, and genome-wide phenotype information remains difficult to obtain. We describe here a comprehensive dataset of 2,400 genes with a loss-of-function mutant phenotype in Arabidopsis. Phenotype descriptions were gathered primarily from manual curation of the scientific literature. Genes were placed into prioritized groups (essential, morphological, cellular-biochemical, and conditional) based on the documented phenotypes of putative knockout alleles. Phenotype classes (e.g. vegetative, reproductive, and timing, for the morphological group) and subsets (e.g. flowering time, senescence, circadian rhythms, and miscellaneous, for the timing class) were also established. Gene identities were classified as confirmed (through molecular complementation or multiple alleles) or not confirmed. Relationships between mutant phenotype and protein function, genetic redundancy, protein connectivity, and subcellular protein localization were explored. A complementary dataset of 401 genes that exhibit a mutant phenotype only when disrupted in combination with a putative paralog was also compiled. The importance of these genes in confirming functional redundancy and enhancing the value of single gene datasets is discussed. With further input and curation from the Arabidopsis community, these datasets should help to address a variety of important biological questions, provide a foundation for exploring the relationship between genotype and phenotype in angiosperms, enhance the utility of Arabidopsis as a reference plant, and facilitate comparative studies with model genetic organisms.
Identifying genes responsible for mutant phenotypes has long been the focus of human genetics and of basic research with model genetic organisms. Large-scale functional genomics projects have expanded the amount of phenotype data available and increased the need to catalog and compare gene disruptions obtained with different species. As a result, biologists face the daunting task of locating and evaluating phenotype of interest. For heritable human traits, the Online Mendelian Inheritance in Man database (McKusick, 2007) illustrates the value, to biologists and physicians alike, of a central repository of curated genotype and phenotype information. Databases for model genetic organisms, including mouse, Caenorhabditis elegans, Drosophila, zebrafish, and yeast, contain detailed phenotype information as well, although different database structures and the lack of controlled vocabulary often limit the utility of this information outside of selected research communities. In response to these concerns, several groups have developed cross-species methodologies (Hoehndorf et al., 2011) and databases (Groth et al., 2011) to support the emergent field of comparative phenomics and to facilitate gene function annotation and human disease gene discovery. Efforts have also begun to standardize phenotype descriptions in animal systems using controlled vocabularies (Mabee et al., 2007; Washington et al., 2009).
By comparison, large-scale genotype-to-phenotype associations in model plants have lagged behind. Phenotype descriptions are an integral part of public databases for model plants such as Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), maize (Zea mays), and tomato (Solanum lycopersicum), but none of these databases can be readily queried for complete datasets of sequenced genes known to be associated with a mutant phenotype of interest or with a loss-of-function phenotype in general. This deficiency will only become more severe as global approaches to plant phenotyping are expanded in the future (Sozzani and Benfey, 2011). Attempts to document plant phenotypes on a large scale have been described before (Kuromori et al., 2009). But finding a simple, comprehensive dataset of cloned genes with mutant phenotypes in a reference plant remains elusive.
Nine years ago, we began to address this issue by assembling a physical map of 620 sequenced genes with a known mutant phenotype in Arabidopsis (Meinke et al., 2003). In that project, we emphasized the need to move beyond the classical genetic map of phenotypic markers, which contains regions inconsistent with the sequenced genome, to a sequence-based map of genes with mutant phenotypes, which can be readily updated with additional genotype-to-phenotype associations. We also suggested, based on comparisons with other model eukaryotes, that at least 10% of protein-coding genes in Arabidopsis would eventually be shown to be associated with a loss-of-function mutant phenotype detected through suitable genetic screens.
In this report, we describe the results of an expanded, genome-wide literature curation effort to identify known genes associated with a loss-of-function mutant phenotype in Arabidopsis. Throughout this project, we have been motivated by a conviction that quick access to information on genes associated with mutant phenotypes should be a defining feature of model genetic systems and that curated phenotype information in plants represents a valuable tool for functional genomics, comparative phenomics, and gene discovery relevant to agriculture, bioenergy, and the environment. We began by reviewing our initial dataset published nine years ago, obtained lists of candidate phenotype genes from The Arabidopsis Information Resource (TAIR; www.arabidopsis.org), performed extensive PubMed searches of the literature through the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/pubmed), and incorporated a wide range of essential genes (Meinke et al., 2008) from our SeedGenes database (www.seedgenes.org). The result is a robust collection of 2,400 Arabidopsis genes with a documented loss-of-function mutant phenotype, along with a complementary dataset of 401 Arabidopsis genes that exhibit a mutant phenotype only when disrupted in combination with a redundant paralog.
These datasets provide a benchmark for future research and a valuable resource for addressing basic questions in plant biology. To underscore this point, we begin to evaluate here whether protein function, protein localization, genetic redundancy, and the number of protein interactors correlate with mutant phenotype, whether all genes might be expected to have a knockout phenotype of some kind, and whether knockouts of orthologous genes in different plant species tend to produce similar phenotypes. We also note the practical benefits of having a comprehensive dataset readily available to serve as a reference point for phenotype information genome wide, to highlight the diversity of phenotypic markers associated with specific chromosomal regions, to help uncover genes responsible for natural variation in wild-type accessions, and to improve the criteria used to identify functionally redundant paralogs.
Because updating and curating a definitive phenotype dataset is beyond the resources of a single laboratory, we focused here not simply on finding suitable genes to include but also on establishing a framework for acquiring and organizing information in the future. We highlight the need for members of the Arabidopsis community to assist with future curation efforts to edit, improve, and expand the initial dataset and to provide additional phenotype details. In addition, we encourage those familiar with other plant species to establish similar datasets to enhance their research communities, enable comparative studies, and realize the full potential of Arabidopsis as a reference organism for plant biology.
RESULTS AND DISCUSSION
Definition and Classification of Mutant Phenotypes
In order to establish a comprehensive dataset of genes with mutant phenotypes in Arabidopsis, we first needed to determine what constitutes a mutant phenotype and what types of genetic changes to include. Unlike our past work (Meinke et al., 2003), where genes with dominant gain-of-function phenotypes were evaluated because they were part of the classical genetic map, we limited the current dataset to genes with a documented loss-of-function phenotype. That focused attention on determining the biological significance of each gene following the reduction or elimination of its function. Loss of gene function was confirmed in some publications by measuring the amount of residual gene product detected in homozygotes. In other cases, it was simply inferred from the nature and location of the mutation combined with a recessive pattern of inheritance. We included some loci for which loss-of-function phenotypes were demonstrated only through antisense or RNA interference experiments. Because these methods can also result in the down-regulation of redundant genes that affect the phenotype, future curation efforts will need to develop more consistent guidelines for when to consider loci for which phenotype information is limited to gene silencing.
We defined a mutant phenotype as a heritable change that could be detected through visual inspection, cellular characterization, or biochemical analysis under standard greenhouse or specialized laboratory conditions. This covered alterations in morphology, physiology, and biochemistry, including responses to pathogens and changes in plant metabolites or storage products. Several genes with mutant phenotypes detected only in specific accessions were also included. Enhancers with no phenotype of their own were added to the multiple mutant dataset when redundant genes were involved but were otherwise excluded from the single gene dataset. Heritable changes in gene expression profiles; protein activity, accumulation, or complex formation; and RNA modification without an associated morphological or biochemical defect were not considered mutant phenotypes. These criteria made it possible to focus on changes that most geneticists would agree constitute a phenotype while disregarding the types of subtle changes noted above that are likely to be characteristic of all gene knockouts.
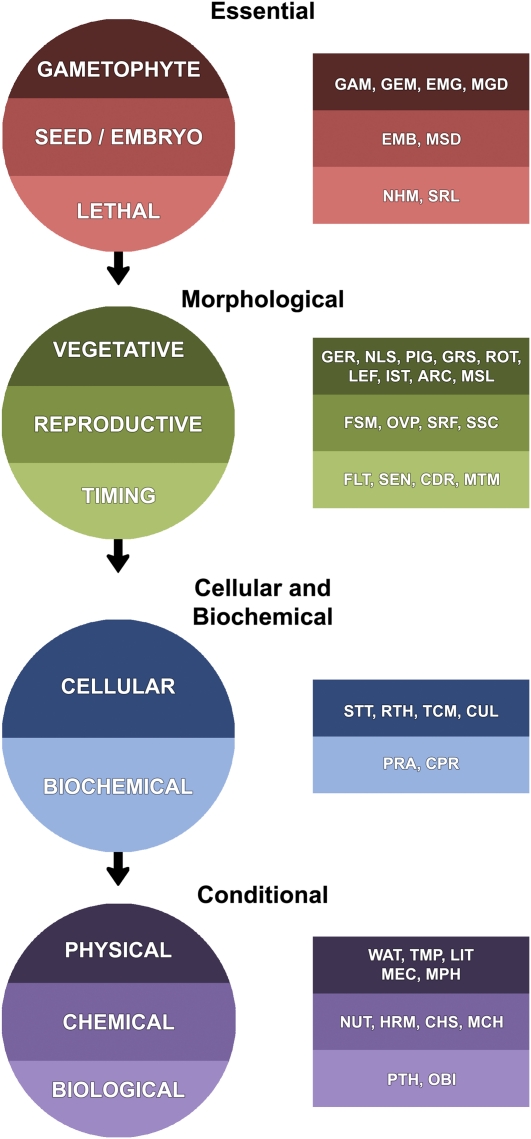
Because phenotypes are features of alleles that can differ in strength, and because mutant alleles can exhibit multiple phenotypes at different stages of development, in different parts of the plant, in distinct genetic backgrounds, and under different growth conditions, we devised a multitiered, prioritized classification system (Fig. 1) to place genes into categories based on their known loss-of-function phenotypes. Each gene was assigned to one of four groups (essential, morphological, cellular-biochemical, or conditional) and one of 11 classes within those groups based on the phenotype of the strongest mutant allele. Each class was further divided into subsets, 42 in total, to reflect additional phenotype details. The full classification system is described in Supplemental Table S1. The ranking of groups and classes illustrated in Figure 1 was designed to categorize genes based on the stage of development when the phenotype is first observed and, to some extent, what methods or conditions are required to detect the phenotype. Genes can be associated with multiple subsets, with the exception of most within the essential group, and assignments of weak and strong alleles to different subsets are addressed with different symbols. Genes with defects in stomata, trichomes, or root hairs were placed in the cellular class, along with genes that exhibit other cellular or ultrastructural defects requiring magnification for detection, because that provided the most seamless method of classifying the range of phenotypes observed. Sporophytic defects in ovule or pollen morphology were included in the reproductive class regardless of the methods required for detection, because that emphasized the stage of development when those defects first became apparent.
Figure 1.
Classification system for Arabidopsis genes with mutant phenotypes based on a series of unique, prioritized phenotype groups (black headings; complete circles) and classes (circle segments), along with nonexclusive phenotype subsets (abbreviated in rectangles). Phenotype subsets are described in more detail in Supplemental Table S1.
When a striking phenotype was accompanied by a subtle phenotype of higher priority, the distinctive phenotype of lower rank was usually chosen. In other words, a knockout mutant with a prominent defect in floral morphology and a subtle change in leaf morphology was often assigned to the reproductive class instead of the vegetative class. However, relevant phenotypes were still captured through association with the appropriate subset categories. Individuals wishing to identify known mutants with altered leaf or floral morphology, therefore, may query the dataset for phenotype subsets of interest. We suspect that genes with borderline group and class assignments will increase as more mutants are grown under standardized conditions and phenotyped throughout the life cycle. Some genes were classified as essential to reflect the known null phenotype, even though most publications focused on phenotypes of weak alleles. A good example is the flowering time gene, FY, which encodes an essential protein required for RNA processing. A complete loss of function of this gene results in embryo lethality (Henderson et al., 2005). Precise boundaries between related phenotype subsets were in some cases difficult to establish and justify, although they often made sense in the context of the entire dataset. For example, we assigned dwarf mutants to one subset (GRS) and plants with small leaves to another (LEF); some mutants with cotyledon defects at the seedling stage were assigned to a subset (NLS) different from that used for cotyledon defects observed during seed development (EMB); mutants defective in the accumulation of a cellular product (PRA) could also be viewed as defective in a cellular process (CPR); responses to micronutrients such as iron and zinc were distinguished from responses to other metals such as cadmium and cobalt, which are not considered essential for growth; and pigment mutants with subtle changes in pigmentation were placed in the vegetative class, whereas albino mutants were typically assigned to the lethal class. All phenotype subsets are defined in Supplemental Table S1. We acknowledge that our dataset is incomplete and imperfect, consistent with the challenges we faced. But it represents an important step forward and a prerequisite for ultimately documenting the consequences of gene disruption across the genome.
Importance of Defining Gene Identity Confidence Levels
Genes in the phenotype dataset are differentiated by the level of confidence that the correct gene responsible for the mutant phenotype has been identified. This feature helps to balance preliminary studies and contributions from large-scale projects, where many genes are involved and supporting data for individual mutants are limited, with efforts focused on a small number of genes examined in considerable detail. We recognized two categories of genes in the dataset: those labeled as confirmed, usually through molecular complementation, the analysis of additional mutant alleles, supporting cellular or biochemical data, or phenotype reversion following the excision of a transposable element; and those labeled as not confirmed, where a single mutant allele was typically involved, formal confirmation is lacking, and robust genetic data alone often indicated close linkage between the gene and mutant phenotypes. Including genes with solid but unconfirmed phenotype associations allows a broad sample of genes to be analyzed and can be justified given that most of the associations are likely to be correct. Documenting identity confidence levels, on the other hand, provides valuable information for future experiments, along with appropriate cautions for data analysis.
A Comprehensive Dataset of Genes with Mutant Phenotypes
Information presented in the phenotype dataset is summarized in Table I. The primary dataset (Supplemental Table S2) contains 2,400 genes arranged by locus number and 19 columns with gene and mutant information. The dataset includes about 9% of the protein-coding sequences in the genome. An expanded version of the dataset (second tabbed spreadsheet in Supplemental Table S2) allows genes to be sorted by phenotype subset, thereby simplifying the process of identifying features of interest. Phenotype descriptions are based on SeedGenes curation and personal observations for genes required for embryo development and on information presented in the literature for genes with other mutant phenotypes. Although we attempted to use consistent language in describing similar phenotypes, we did not establish a definitive, controlled vocabulary for all possible phenotypes in Arabidopsis. This remains a challenge for future studies.
Table I. Information presented in the Arabidopsis phenotype dataset.
| Dataset Columns | Nature of Information Presented |
| 4 | Locus number; gene name, symbol, aliases |
| 1 | Confirmation status of gene-to-phenotype association |
| 3 | Phenotype group, class, and subset assignments |
| 1 | Brief, curated description of mutant phenotype |
| 1 | Method of gene identification |
| 2 | Reference laboratory and year of publication |
| 3 | Closest BLASTP match within Arabidopsis |
| 2 | Limited protein function information, classification |
| 2 | Mitochondrial and plastid localization information |
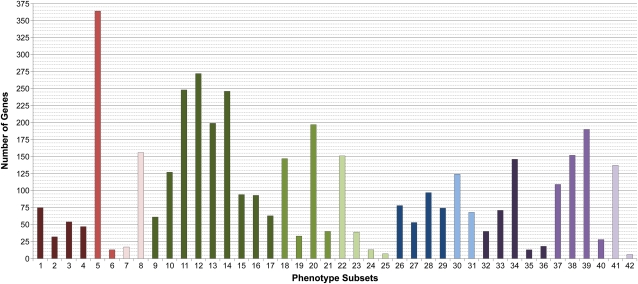
Gene distributions among different phenotype groups and classes are presented in Table II, along with information on confidence levels for gene identities. About 30% of the genes are essential for early development or survival, 36% are assigned to the morphological group, 12% cellular and biochemical, and 22% conditional. Overall, 84% of gene identities have been confirmed. Essential genes are least often confirmed, in part because the results of several large-scale insertional mutagenesis projects are included. Some “conditional” genes may have visible phenotypes under standard growth conditions that were overlooked when the conditional response was first described. Three common phenotype classes (embryo-seed, vegetative, and conditional-chemical) account for 53% of gene assignments in the dataset. The remaining eight classes (gametophyte, lethal, reproductive, timing, cellular, biochemical, conditional-physical, and conditional-biological) range from 3% to 8% each. Gene distributions among the 42 phenotype subsets are shown in Figure 2. The abundance of each subset reflects both the number of target genes involved and the effort devoted to identifying those phenotypes. For example, the large number of EMB genes without a known gametophyte defect (subset 5) is consistent with estimates of 1,000 genes required for embryo development (Meinke et al., 2009) but also reflects a long-term project (McElver et al., 2001) designed to saturate for this class of mutants (www.seedgenes.org). The current level of saturation for gene knockouts with any phenotype remains to be determined.
Table II. Phenotype groups and classes in the Arabidopsis phenotype dataset.
| Phenotype Category |
Genes in Dataset |
Gene Identity Confirmed |
|||
| Groupa | Class | No. | Percentage | No. | Percentage |
| ESN | Subtotal | 719 | 29.9 | 540 | 75.1 |
| Gametophyte | 197 | 8.2 | 136 | 69.0 | |
| Embryo/seed | 370 | 15.4 | 281 | 75.9 | |
| Lethal | 152 | 6.3 | 123 | 80.9 | |
| MRP | Subtotal | 862 | 35.9 | 775 | 89.9 |
| Vegetative | 640 | 26.7 | 572 | 89.2 | |
| Reproductive | 152 | 6.3 | 141 | 92.8 | |
| Timing | 70 | 2.9 | 62 | 88.6 | |
| CLB | Subtotal | 297 | 12.4 | 261 | 87.9 |
| Cellular | 124 | 5.2 | 111 | 89.5 | |
| Biochemical | 173 | 7.2 | 150 | 86.7 | |
| CND | Subtotal | 522 | 21.8 | 445 | 85.2 |
| Physical | 157 | 6.6 | 126 | 80.3 | |
| Chemical | 257 | 10.7 | 229 | 89.1 | |
| Biological | 108 | 4.5 | 90 | 83.3 | |
| Total | All combined | 2,400 | 100.0 | 2,021 | 84.2 |
ESN, Essential; MRP, morphological; CLB, cellular and biochemical; CND, conditional.
Figure 2.
Distribution of phenotype subset assignments for Arabidopsis genes with a loss-of-function mutant phenotype. Subsets are colored according to phenotype class (Fig. 1) and numbered as described in Supplemental Table S1. Most essential genes are assigned to a single phenotype subset. Many other genes have more than one subset assignment. Phenotypes of weak alleles and semidominant features observed in heterozygotes are included.
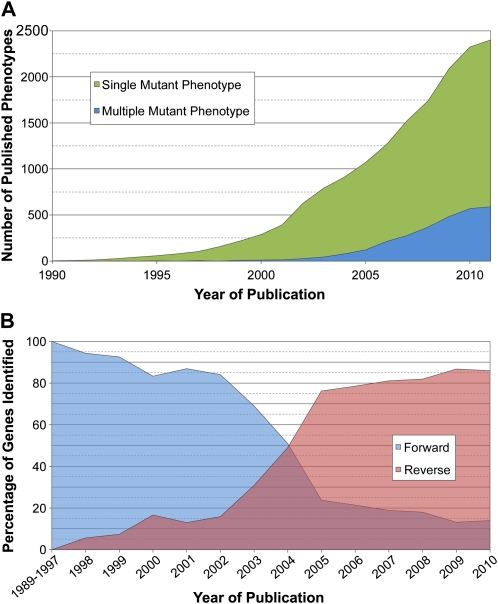
Ninety percent of the Arabidopsis phenotype genes have been identified over the past 12 years (Fig. 3A). Most recent additions were the result of reverse genetics, a trend that will likely continue. The opposite was true 12 years ago, when more than 80% of the entries were identified through forward genetics (Fig. 3B). The change in preferred approach happened shortly after the genome was sequenced (Arabidopsis Genome Initiative, 2000) and large populations of sequenced insertion lines were established (Sessions et al., 2002; Alonso et al., 2003; Rosso et al., 2003). Overall, reverse genetics was used to analyze 60% of total genes in the dataset. When forward genetics was involved, map-based cloning accounted for 50% of the gene identities revealed, compared with 44% for T-DNA insertions and 6% for transposon tagging. The number of redundant genes associated with multiple mutant phenotypes also increased in recent years, reflecting enhanced utilization of public resources for reverse genetics (O’Malley and Ecker, 2010).
Figure 3.
Historical perspective on the identification of Arabidopsis genes with a loss-of-function mutant phenotype through forward and reverse genetics. The year of publication in some cases refers to the date of inclusion in a public database. Additional details are presented in Supplemental Table S2.
Chromosome distributions of genes in the dataset are presented in Figure 4. With the exception of centromeric regions, phenotype genes are widely dispersed throughout the genome. The ends of chromosomes are also well represented, with an average of 3.4 phenotype genes identified within the first and last 25 locus numbers for each chromosome. This frequency is marginally above the average (2.2) genome wide (χ2 test; P = 0.02). There are 191 documented cases of two adjacent genes with mutant phenotypes and 34 cases of three adjacent genes. The largest cluster (TRP3, FBL17, EMB2360, TTN8) contains four essential genes on chromosome 3. Knowledge of the chromosome locations of genes with mutant phenotypes should facilitate ongoing map-based cloning efforts, contribute to the design of additional genetic markers, assist with the analysis of mapped traits in accessions and related species, and reveal whether duplicated chromosomal regions are deficient in phenotype loci, as might be expected for genes with redundant functions.
Figure 4.
Chromosomal locations of 2,400 phenotype genes of Arabidopsis (black lines) placed on a sequence-based physical map of the genome. This figure was generated using the map visualization tool available through TAIR (www.arabidopsis.org/jsp/ChromosomeMap/tool.jsp).
Protein Function and Mutant Phenotypes
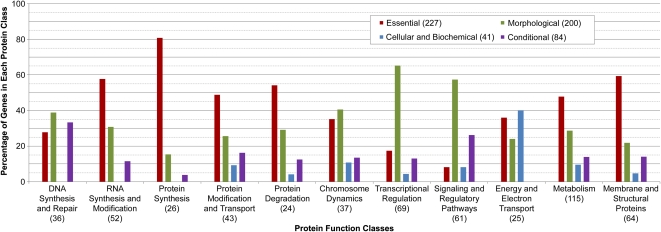
For protein function classification, we utilized a system (Supplemental Table S3) developed for past work on mutant phenotypes in Arabidopsis (Meinke et al., 2003; Bryant et al., 2011; Muralla et al., 2011). To streamline this effort and facilitate comparisons of greatest interest, we limited function assignments to unique genes, essential seed and gametophyte genes, and proteins localized to chloroplasts and mitochondria. Protein function assignments for unique genes are presented in Figure 5. Overall, interfering with a specific function often results in a variety of phenotypes, and phenotype groups often include proteins with a variety of cellular functions. However, different patterns of protein function assignments can be identified. The essential group is enriched for genes involved with RNA and protein synthesis and modification, as might be expected, but deficient in transcription factors, whereas the morphological group is enriched for genes involved in transcriptional regulation and signaling networks and deficient in genes required for protein synthesis. The conditional group is often associated with disruptions in signaling and regulatory pathways. The abundance of chlorophyll fluorescence and nonphotochemical quenching phenotypes described in the literature contributes to the large number of energy and electron transport proteins included in the cellular-biochemical group.
Figure 5.
Distribution of phenotype groups among single-copy Arabidopsis phenotype genes with different protein functions. The total numbers of genes analyzed are noted in parentheses.
Differences in protein function also extend to phenotype classes and subsets. A recent comparison of functions associated with defects in embryo and gametophyte development revealed that although interfering with DNA replication and RNA modification typically disrupts embryo rather than gametophyte development, and blocking cytosolic translation often leads to gametophyte defects, extensive overlap exists between protein functions required for embryo and gametophyte development (Muralla et al., 2011). Therefore, one cannot explain the difference between embryo and gametophyte mutants on the basis of protein function alone. Even when genes with discrete subset assignments such as flowering time and root hair development are examined, a wide range of protein functions is found.
A fundamental question related to protein function and mutant phenotype concerns the origin of phenotypic diversity through evolution. Although changes in coding regions have long been thought to be the principal factor involved, the importance of variations in linked regulatory regions has recently been emphasized (Carroll, 2008; Frankel et al., 2011). Protein functions known to be associated with a loss-of-function phenotype of interest can nevertheless help to illustrate the diversity of genes and cellular processes that might contribute to morphological evolution. Our phenotype dataset includes 938 genes associated with viable morphological defects (including changes in stomata, trichomes, and root hairs) that are observed under standard growth conditions. Whether changes in these genes frequently underlie different phenotypes and patterns of growth and development in related species and in natural accessions of Arabidopsis remains to be determined. With recent advances in the molecular analysis of natural variation in Arabidopsis (Weigel, 2012), such questions will likely soon be addressed.
Protein Localization and Mutant Phenotypes
More than 350 nuclear genes encoding chloroplast-localized proteins are known to be associated with a mutant phenotype in Arabidopsis (Bryant et al., 2011). Embryo development frequently becomes arrested when amino acid, vitamin, or nucleotide biosynthesis is disrupted and when chloroplast translation is blocked, but it proceeds when photosynthesis is compromised and when levels of chlorophyll, carotenoids, or terpenoids are reduced. Interfering with other plastid-localized metabolic pathways typically leads to a mutant phenotype detected at the seedling stage. With the phenotype dataset presented here, the consequences of disrupting mitochondrial and chloroplast functions can be compared. More than 60% of the 123 mitochondria-localized proteins included in the phenotype dataset (Supplemental Table S4) are classified as essential, compared with 30% for the dataset overall. The most striking difference involves defects in gametophyte development, which account for 25% of mitochondrial proteins with a mutant phenotype, less than 2% of chloroplast proteins, and 8% of all dataset entries combined. Whereas interfering with chloroplast translation frequently results in embryo lethality, disruptions in mitochondrial translation tend to be more severe, often resulting in gametophyte lethality. The difference is that plant mitochondrial genomes encode critical subunits of essential respiratory chain complexes required for gametophyte survival. Disruption of mitochondria-localized PPR proteins and metabolic enzymes is often associated with defects in embryo development, whereas morphological defects are more broadly distributed among different protein functions. Datasets of mitochondria- and chloroplast-localized proteins with mutant phenotypes, therefore, have distinctive but overlapping function profiles, consistent with differences in the underlying biological processes involved.
Protein Interactions and Mutant Phenotypes
Recent advances in the global analysis of protein interactions in humans and model organisms have made it possible to address the relationship between mutant phenotype and the degree of protein connectivity within the cell. Early studies with yeast appeared to indicate that essential genes are overrepresented among highly interacting (hub) proteins, consistent with the idea that disrupting genes associated with multiple protein networks often has severe consequences (Jeong et al., 2001). More recent work with yeast, however, suggests that protein connectivity is correlated instead with the level of pleiotropy, defined as the number of different phenotypes exhibited (Yu et al., 2008). Related studies with humans have also been contradictory. Initial reports seemed to indicate that hub proteins were overrepresented among human disease genes (Xu and Li, 2006). A subsequent analysis found that this correlation held for early-lethal disease genes but not for the more common, nonessential disease genes (Goh et al., 2007).
We explored the phenotype-to-interactome relationship in Arabidopsis by comparing our phenotype dataset with a comprehensive, published dataset of known protein interactors in Arabidopsis, which includes a fraction of the total interactions thought to occur within the cell (Arabidopsis Interactome Mapping Consortium, 2011). We established an edited dataset of 10,856 binary interacting pairs involving 4,785 different proteins by combining their experimental and literature-curated interactions and eliminating self-interactors and pairs that included chloroplast- and mitochondria-encoded proteins. Approximately 3.0% of the 4,785 proteins in this interactome dataset and 4.4% of the 923 known interacting proteins included in our phenotype dataset, percentages that differ only marginally (χ2 test; P = 0.03), represent hubs that interact with at least 20 other proteins. We then focused on unique genes in both datasets, because they are most likely to exhibit an informative phenotype not influenced by genetic redundancy. Approximately 1.7% of unique genes in the interactome dataset and an equivalent 1.2% (χ2 test; P = 0.54) of unique genes in the phenotype dataset encode hub20 proteins. Based on current datasets, therefore, it does not appear that highly connected proteins in Arabidopsis are more likely than the proteome as a whole to exhibit a loss-of-function phenotype. Factors other than the degree of protein connectivity, therefore, must often determine whether gene knockouts exhibit an obvious phenotype.
Among the known interacting proteins in our phenotype dataset, hub20 proteins are not more likely to be essential than those with a single known interactor. In fact, the distribution of phenotype groups among the 923 interacting proteins in our dataset is not significantly different (χ2 test) when genes encoding proteins with one and 10 or more known interactors (P = 0.15) and those with one and 20 or more known interactors (P = 0.17) are compared. Similar conclusions are reached when comparisons are limited to unique genes. These results, which provide an interesting contrast to work on yeast and humans, indicate that the degree of protein connectivity in Arabidopsis is not a reliable predictor of the likelihood that a given knockout will exhibit a loss-of-function phenotype or that the observed phenotype will be lethal. In addition, these results contrast with the analysis of a plant defense interactome map, where a small number of highly connected (hub50) proteins were found to be preferred targets for effector molecules associated with two divergent pathogens (Mukhtar et al., 2011). Whether eliminating the functions of nonredundant genes encoding hub proteins in Arabidopsis frequently results in 100% male and female gametophyte lethality, thus preventing the recovery of null alleles, remains to be determined.
A second question that we addressed relative to protein interactions is whether the phenotype of one gene knockout is a good predictor of the phenotype that results from disrupting its protein interactor. Such a strategy underlies ongoing searches for human disease genes (Chen et al., 2011; Yang et al., 2011) and might have important applications to plant biology as well. Our phenotype dataset, along with the Arabidopsis interactome dataset, provide a suitable platform for addressing this question. Once again, we focused on unique genes to limit the impact of genetic redundancy. Assignments to phenotype groups for 70 pairs of protein interactors found in our phenotype dataset (Table III) demonstrated that the phenotype group of one member of the pair is a better predictor of the phenotype group of the paired knockout than expected by chance (χ2 test; P < 0.001). The same conclusion was reached when phenotype classes were compared (χ2 test; P < 0.001). With this in mind, we assembled a list of 155 unique Arabidopsis genes that are not found in our phenotype dataset but encode a protein that interacts with another unique gene product in the dataset (Supplemental Table S5). These genes represent promising candidates for future, reverse genetic screens designed to enhance the existing collection of genes with mutant phenotypes.
Table III. Phenotypes of pairs of mutants disrupted in genes encoding protein interactors.
| Phenotype Group | Percentage of Total Interactorsa | Matched Pairsb | Expected Matched Pairsc | Percentage of Pairs Matchedd | Expected Percentage of Pairs Matchedcd |
| Essential | 45.7 | 22 | 14.6 | 31.4 | 20.9 |
| Morphological | 42.1 | 18 | 12.4 | 25.7 | 17.7 |
| Cellular and biochemical | 3.6 | 1 | 0.1 | 1.4 | 0.1 |
| Conditional | 8.6 | 3 | 0.5 | 4.3 | 0.7 |
| Total | 100.0 | 44 | 27.6 | 62.9 | 39.4 |
Among 140 total interactors from 70 interacting protein pairs encoded by unique genes in the phenotype dataset.
Paired interactors with the same (matched) group assignment among the 70 pairs.
For each phenotype group, Expected Matched Pairs = Expected Percentage of Pairs Matched [or (Percentage of Total Interactors)2/100] × 70 total pairs/100.
Paired interactors have matched group assignments more often than expected based on the frequency of each phenotype group.
Genetic Redundancy and Mutant Phenotypes
The role of gene duplications in modulating the phenotypes of loss-of-function mutations has been examined in a variety of eukaryotes, including yeast (Gu et al., 2003; Ihmels et al., 2007), C. elegans (Conant and Wagner, 2004), mouse (Liao and Zhang, 2007; Makino et al., 2009), and humans (Hsiao and Vitkup, 2008). Two fundamental questions have frequently been raised. (1) How often and completely do gene duplicates compensate for the inactivation of a paralog? (2) How do the knockout phenotypes of genes with paralogs compare with those of genes without paralogs? With yeast, many duplicated genes do not contribute noticeably to genetic robustness against knockouts of paralogs, and duplicates that compensate under optimal growth conditions often fail to extend that effect to other conditions (Ihmels et al., 2007). This inability to provide complete compensation may help to explain the evolutionary stability of duplicated genes in yeast and elsewhere. Yeast knockouts with a strong or lethal phenotype often represent genes without a paralog, whereas knockouts with a weaker effect on growth often identify duplicated genes (Gu et al., 2003). Similar patterns have been noted for RNA interference phenotypes in C. elegans (Conant and Wagner, 2004). In humans, genes without a potential homolog in the genome are several times more likely to be associated with a disease mutation than genes with a homolog (Hsiao and Vitkup, 2008). In mouse, duplicated and unique genes at first appeared to be equally represented among knockout collections of essential genes (Liao and Zhang, 2007). But a subsequent study suggested that the relationship was more complex, with developmental genes and those impacted by whole genome duplication more likely to be essential than other duplicated genes (Makino et al., 2009).
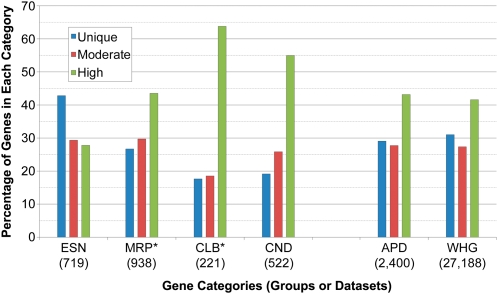
We established three categories of genetic redundancy for evaluating our phenotype dataset: unique genes without a similar sequence in the genome (BLASTP e-30 cutoff); genes with moderate similarity to another Arabidopsis sequence (BLASTP e-30 to e-80, or BLASTP > e-80 if less than 80% of protein length aligned); and genes with high similarity to another Arabidopsis sequence (BLASTP > e-80 and more than 80% aligned). Based on these criteria, 31.0% of all Arabidopsis genes are unique, 27.4% identify a gene with moderate similarity, and 41.6% identify a gene with high similarity. Distributions of these categories for the phenotype groups in our dataset are presented in Figure 6. Compared with the genome as a whole, essential genes are more likely to be unique and to lack a close paralog (χ2 test; P < 0.001). This result is consistent with past work on EMB genes of Arabidopsis (Tzafrir et al., 2004) and on knockouts of essential genes in yeast (Gu et al., 2003) and C. elegans (Conant and Wagner, 2004). By contrast, genes in the morphological group (P < 0.02), and especially those in the cellular-biochemical (P < 0.001) and conditional (P < 0.001) groups, are more likely to have a close paralog than the genome as a whole. This pattern likely reflects a variety of factors, including the recruitment of duplicated genes to function in specialized developmental programs, biochemical pathways, and growth conditions, as well as differences in the size and complexity of protein families associated with specific cellular processes. Surprisingly, the frequencies of phenotype genes assigned to different categories of redundancy are not significantly different (P = 0.12) from those of the genome as a whole. The level of genetic redundancy in Arabidopsis, therefore, is not a good predictor of the likelihood that a loss-of-function mutant will exhibit a discernible phenotype.
Figure 6.
Levels of protein sequence redundancy (defined in the text) for Arabidopsis genes assigned to different phenotype groups (left side), all genes in the Arabidopsis phenotype dataset (APD), and the whole Arabidopsis genome (WHG). * For this analysis, genes associated with visible defects in epidermal features (trichomes, stomata, root hairs) were moved from the cellular-biochemical (CLB) group to the morphological (MRP) group. The total numbers of genes evaluated are noted in parentheses.
Redundant Genes with Multiple Mutant Phenotypes
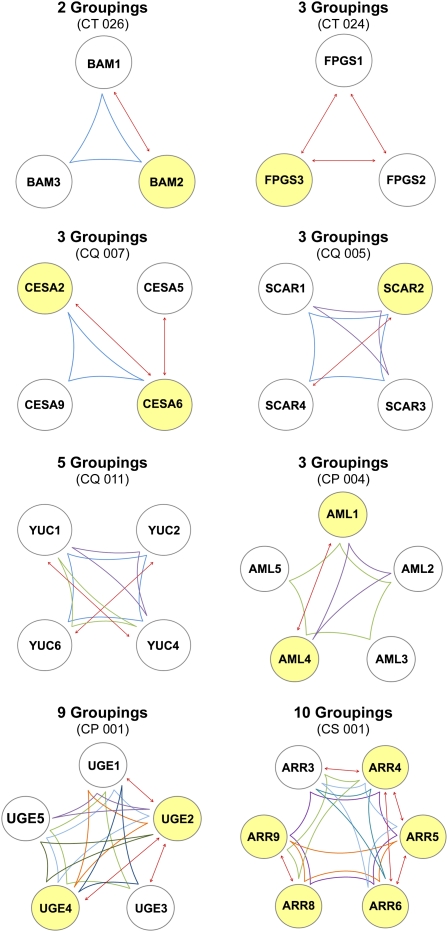
Because genetic redundancy can often mask the phenotypes of single gene disruptions in Arabidopsis, we established a complementary dataset of genes associated with multiple mutant phenotypes, which identified additional proteins required for plant function. The resulting collection of 591 genes (Supplemental Table S6) includes 401 genes not associated with a single mutant phenotype (Supplemental Fig. S1) and 190 genes from the single mutant dataset that exhibit a distinctive phenotype, typically more severe, when combined with mutations in potential paralogs (BLASTP e-30 cutoff). Sets of paralogous genes associated with a multiple mutant phenotype were defined as clusters (Table IV). Three types of simple gene clusters were evaluated: (1) exclusive clusters, where the single mutants all lack an established phenotype; (2) asymmetric clusters, where one (or more) member of the cluster is included in the single gene dataset but others are not; and (3) symmetric doubles, where all members exhibit single mutant phenotypes that differ from that of the multiple mutant. We identified 203 simple gene clusters: 96 exclusive (87 doubles, seven triples, two quadruples), 76 asymmetric (70 doubles, five triples, one quintuple), and 31 symmetric (all doubles). We also analyzed 45 complex gene clusters that involved three or more genes and exhibited phenotypes with two or more groupings of mutants within a single cluster. Overall, 144 different groupings involving 166 genes were recorded for these 45 complex clusters. Several examples are illustrated in Figure 7. Details for the entire dataset of 248 clusters, 347 groupings, and 591 genes are found in Supplemental Table S6.
Table IV. Features of gene clusters in the multiple mutant dataset.
| Genes in Cluster | Cluster Featuresa |
Cluster Phenotype Groupsb |
|||||
| Type | Examples | Percentage Completec | ESN | MRP | CLB | CND | |
| 2 | EXC | 87 | 33 | 35 | 34 | 8 | 10 |
| ASY | 70 | 39 | 30 | 27 | 6 | 7 | |
| SYM | 31 | 10 | 17 | 9 | 2 | 3 | |
| 3 | EXC | 7 | 43 | 0 | 2 | 0 | 5 |
| ASY | 5 | 20 | 0 | 3 | 0 | 2 | |
| CPX | 26 | 8 | 6 | 13 | 4 | 3 | |
| 4+ | EXC | 2 | 100 | 0 | 2 | 0 | 0 |
| ASY | 1 | 0 | 0 | 1 | 0 | 0 | |
| CPX | 19 | 0 | 8 | 8 | 2 | 1 | |
EXC, Exclusive, both single mutants have no phenotype; ASY, asymmetric, one single mutant has a phenotype but the multiple mutant is more severe; SYM, symmetric, both single mutants have a phenotype but the multiple mutant is more severe; CPX, complex, phenotype information available for two or more combinations of genes within a cluster.
ESN, Essential; MRP, morphological; CLB, cellular and biochemical; CND, conditional.
Complete clusters disrupt all potential paralogs in Arabidopsis.
Figure 7.
Examples of complex clusters of three or more paralogous genes with two or more groupings of genes associated with a multiple mutant phenotype. Genes with a single mutant phenotype are highlighted in yellow. Lines indicate groupings that produce a documented phenotype more severe than that of the corresponding single mutants or multiple mutants with fewer members. Cluster identification numbers are noted in parentheses. Supplemental Table S6 presents additional information on the genes and phenotypes involved.
The distribution of phenotype groups differs (χ2 test; P < 0.001) between the single and multiple mutant datasets. Essential genes and those associated with morphological defects constitute a higher percentage of genes in the multiple mutant dataset than in the single mutant dataset. The opposite is true for the cellular-biochemical and conditional groups, which are more characteristic of single gene disruptions. However, no significant difference is found (χ2 test; P = 0.81) when phenotypes for unique genes in the single mutant dataset are compared with those for complete clusters, where all potential paralogs (BLASTP e-30 cutoff) are disrupted. Because neither dataset is saturated, and some genes and mutant phenotypes have been studied in more detail than others, the patterns observed here may also be influenced by differences in the levels of saturation for certain phenotypes (single mutant dataset) and genes (multiple mutant dataset) of special interest. However, the enrichment of essential genes in the multiple mutant dataset raises the intriguing possibility that some gene duplications have been maintained in natural populations to protect against the severe, deleterious effects of single gene disruptions.
Exclusive gene clusters and double mutants within complex clusters where single mutants have no documented phenotype provide an informative set of redundant genes that merit further evaluation, because each gene fully compensates for the disruption of a functional paralog. Gene pairs where both members compensate in part (symmetric clusters and some groupings in complex clusters) or one member compensates more fully than the other (asymmetric clusters and some groupings in complex clusters) might be expected to have more divergent protein sequences or patterns of expression because they are not fully redundant. When the extent of protein similarity was compared between different types of clusters, 65% of 214 fully redundant genes, including those associated with exclusive doubles and complex clusters, exhibited an especially high level of protein identity (BLASTP > e-100; more than 95% aligned length), compared with 49% for 328 partially redundant genes, including those associated with symmetric, asymmetric, and complex pairs, and 40% for all protein-coding genes in Arabidopsis, excluding those without a significant match (BLASTP e-30 cutoff). Average BLASTP scores also differed (Student’s t test; P = 0.01) between the fully and partially redundant categories and, most notably, when all protein-coding genes with a significant match were compared (P < 0.001). Redundant gene pairs without single mutant phenotypes, therefore, are enriched for genes with highly similar protein sequences.
With respect to gene expression, we expected that exclusive gene pairs would exhibit similar patterns and levels of expression, because each gene compensates for the disruption of the other. By contrast, genes with single mutant phenotypes in asymmetric pairs were expected to be expressed more highly or broadly than those without a single mutant phenotype. To compare expression patterns for gene pairs throughout the life cycle, we averaged the ratios of transcript levels for nine different stages of development from public microarray datasets (www.genevestigator.com). This provided a measure of the abundance and to some extent the localization of transcripts throughout the plant. Expression data for 23 exclusive doubles (10 ESN and 13 MRP groups) and 21 asymmetric doubles (15 ESN and six MRP groups) were evaluated by comparing the number of pairs that differed in transcript abundance by less than 2-fold, 2- to 3-fold, and more than 3-fold. Asymmetric pairs showed greater divergence in transcript levels than symmetric pairs (χ2 = 7.50; P = 0.024). In 18 of 21 asymmetric pairs examined, the gene with the higher averaged transcript level also exhibited the single mutant phenotype, consistent with our expectation. This included all 12 pairs with levels that differed by more than 2-fold. In nine asymmetric pairs, however, the difference in transcript abundance was less than 2-fold, and in some cases, it was difficult to reconcile global transcript data with the phenotypes observed. Members of exclusive pairs often exhibited different patterns of expression as well. For example, 61% of these pairs had transcript levels that differed by more than 2-fold, and in some cases, the lack of single mutant phenotypes was difficult to reconcile with the expression pattern. These results indicate that functionally redundant paralogs often have expression patterns that are less similar than expected based on genetic analysis alone, and paralogs that exhibit functional redundancy in one direction but not in the other frequently have expression patterns that are more similar than might otherwise be expected.
Phenotypes of Orthologous Gene Knockouts in Other Angiosperms
Identifying candidate genes responsible for phenotypes in other plant species represents a promising application of the Arabidopsis phenotype dataset. In the absence of functional redundancy, one might expect that disrupting essential genes, and those with conserved metabolic or physiological roles, should often result in similar phenotypes in different species, because interfering with basic cellular functions should have equivalent consequences. In some cases, this assumption seems to be valid. For example, knockouts of the orthologous maize DEK1, Arabidopsis AtDEK1/EMB1275, and rice ADL1 loci all exhibit striking defects in seed development (Becraft et al., 2002; Johnson et al., 2005; Hibara et al., 2009; Meinke et al., 2009), although the mutant phenotypes are not identical. Whether eliminating an essential function results in embryo, gametophyte, or seedling lethality, however, can be difficult to predict. For instance, interfering with amino acid biosynthesis in Arabidopsis often leads to embryo lethality, but for species with long pollen tubes or haploid spores with reduced contributions from parental sporocytes, the consequence might be gametophyte lethality instead (Muralla et al., 2011). In other cases, genetic redundancy may prolong the growth of homozygotes until the seedling stage. Interfering with chloroplast translation can also produce different phenotypes: embryo lethality in Arabidopsis and albino seedlings in maize and Brassica. In this case, the difference seems to result from redundant genes that support chloroplast fatty acid biosynthesis when heteromeric acetyl-CoA carboxylase activity is disrupted (Bryant et al., 2011).
To compare mutant phenotypes of plant orthologs on a broad scale, we evaluated a recent dataset of 112 classical genes of maize (Schnable and Freeling, 2011) and searched journal publications and genome databases to identify phenotype genes of rice and tomato. This uncovered more than 100 candidate genes with mutant phenotypes in rice, maize, or tomato, where the top Arabidopsis BLASTP match is included in our single mutant dataset. We limited our analysis to gene pairs with reciprocal top BLASTP matches, using an e-40 cutoff, and nonreciprocal pairs (e-60 cutoff) where the Arabidopsis phenotype gene identifies a different potential ortholog in the other plant species. We then excluded duplicated maize genes (white pollen1, Zea floricaula, orange pericarp, alternative discordia1) with a loss-of-function phenotype limited to double mutants (www.maizegdb.org; Wright et al., 1992, 2009), dominant mutants of maize (Gnarley, Rough sheath1, White cap) and tomato (e.g. Green-ripe, Delta, Curl) with a gain-of-function mutant phenotype (Schneeberger et al., 1995; Parnis et al., 1997; Foster et al., 1999; Ronen et al., 1999; Barry and Giovannoni, 2006), and semidominant mutants of maize (Rolled1, Tasselseed6) with altered mRNA-binding sites for inhibitory microRNAs (Nelson et al., 2002; Juarez et al., 2004; Chuck et al., 2007).
The resulting dataset (Supplemental Table S7) contains 82 pairs of Arabidopsis phenotype genes and matched orthologs with a loss-of-function phenotype in rice (37 cases), maize (26), or tomato (19). Seven Arabidopsis genes (GAI, UFO, LAS, SVP, AtCWINV4, BRI1, ABA1) are the top match to phenotype genes in both rice and tomato. Mutants with morphological (MRP) defects are more common in this ortholog dataset (82%) than in the Arabidopsis phenotype dataset as a whole (36%), reflecting a longstanding emphasis on studying mutants with visible phenotypes in crop plants. Transcriptional regulators and components of signaling pathways are also well represented, accounting for more than half of the entries in the ortholog dataset. Paired genes in the dataset are frequently assigned to the same phenotype group (66%) and class (51%) but often do not exhibit the same phenotype. These differences are not explained by genetic redundancy alone. Other potential factors include variations in plant structure, physiological processes, patterns of intracellular protein localization, downstream targets for transcription factors, and roles of signaling molecules in growth and development. To assess the reliability of Arabidopsis phenotype information for predicting the identities of phenotype genes in other plants, we classified phenotype similarities as high (26%), moderately high (23%), moderate (22%), or low (29%), with only the final category providing little useful information. Based on these frequencies and the size of the Arabidopsis phenotype dataset, we conclude that maintaining a comprehensive dataset of genes with mutant phenotypes in Arabidopsis has considerable potential to facilitate ongoing efforts to identify candidate genes responsible for phenotypes of interest in a wide range of flowering plants.
Should Every Gene Exhibit a Null Phenotype?
In some respects, documenting the loss-of-function phenotype of a single locus is more straightforward than demonstrating that a null allele has no phenotype. In fact, the point can be made that all genes should exhibit a null phenotype of some kind, because otherwise they would escape natural selection and degenerate. From this perspective, knockouts that appear to lack a mutant phenotype have simply not been grown under the appropriate conditions or been examined with the appropriate methods. Two factors are frequently invoked to explain the absence of a notable phenotype in some knockouts: genetic redundancy and compensating metabolic pathways or cellular processes. Over time, duplicated, redundant genes are thought to (1) diverge through the evolution of new functions (neofunctionalization), (2) diverge by dividing ancestral functions among duplicates (subfunctionalization), or (3) retain all functions in both duplicates through a process known as gene conservation (Ohno, 1970; Walsh, 1995; Hahn, 2009). Alternatively, one or more of the duplicated genes may accumulate deleterious mutations over time and become nonfunctional (i.e. a pseudogene). Surprisingly, some duplicated genes with overlapping functions appear to derive from ancient duplication events, where sufficient time was available for functions to diverge (Dean et al., 2008; Vavouri et al., 2008). One proposed mechanism that could enable the stable maintenance of functional redundancy in duplicated genes involves a reduction in the expression level of each duplicate, such that both genes are needed to provide the original, required level of gene product (Qian et al., 2010). Although the long-term maintenance of functional redundancy is still not fully understood, duplicated genes that retain overlapping, redundant functions for extended periods of time are likely to both be required for optimal fitness under different conditions.
There are several possible explanations for the apparent absence of a phenotype in a known mutant. Residual protein function in a weak allele can mask the phenotype, redundant genes or related pathways can compensate for gene disruption, mutant alleles can exhibit a subtle phenotype that is not readily distinguished from the wild type, and mutant phenotypes can be overlooked or missed because special growth conditions or genetic backgrounds were not employed. This final scenario is frequently invoked and may in fact be most common. In addition, genes can be excluded from phenotype datasets because suitable mutants and gene silencing results are not available, publications or databases describing the phenotype are overlooked, or the mutation cannot be transmitted through either male or female gametes. We considered this final possibility when analyzing aminoacyl-tRNA synthetase mutants defective in cytosolic translation (Berg et al., 2005). The phenotype datasets presented here do not conflict with the conclusion, based on many independent studies, that a sizable number of gene disruptions in Arabidopsis have a minimal impact on plants grown in the laboratory. We believe that this underscores the value of identifying and evaluating those genes that by comparison have an obvious mutant phenotype. A curated list of Arabidopsis genes that appear to lack a null phenotype would provide an interesting contrast to the dataset presented here. However, such information would be difficult to evaluate without additional molecular data and phenotype information from plants grown under standardized conditions.
Practical Applications and Future Curation of the Phenotype Dataset
In addition to providing a valuable resource for addressing fundamental questions in plant biology, a curated, comprehensive dataset of Arabidopsis phenotype genes should facilitate a wide range of future experimental studies. Most importantly, such a dataset serves as a quick reference for phenotype information for a model plant genome. By searching the dataset for subsets of interest, a broad spectrum of genes known to be associated with a desired phenotype can be readily identified. Such a dataset can also be used to obtain phenotype information on genes in chromosome regions of interest to assist with the identification of candidate loci responsible for phenotypes in mapped mutants and natural accessions. This information is currently scattered throughout multiple databases and hundreds of publications. A phenotype dataset that includes protein function and localization information can help to illustrate the range of cellular disruptions that lead to a phenotype of interest and, conversely, the range of phenotypes that result from disrupting a cellular process, organelle, or protein family of interest.
A major challenge for the future concerns the development of a curation infrastructure that distributes responsibility for maintaining and updating a phenotype dataset throughout the community. Because the primary database for Arabidopsis research (TAIR) is undergoing a period of transition and limited funding is available to support manual curation, we believe a new approach is needed to encourage community involvement in the curation of basic phenotype information associated with single and multiple gene disruptions. One approach would require journal involvement and author input at the time of publication. Recently, several journals adopted such a strategy in conjunction with TAIR. A second approach would require establishing a central database portal for the input of basic information on gene-to-phenotype associations by each investigator. We already helped to establish one such portal at TAIR to facilitate the assignment of gene class symbols. A similar approach could enable the rapid collection of gene-to-phenotype information using a format based upon the data presented in this report. However, both strategies are likely to have only limited success without extensive oversight by curators charged with reviewing phenotype information. Input from seed stock centers, large-scale phenotyping projects, and participants at international conferences would also be critical. We believe that all of these approaches should be pursued in order to make information on mutant phenotypes readily available to a broad spectrum of research biologists and ultimately to realize the full potential of Arabidopsis as a model genetic organism.
MATERIALS AND METHODS
Establishment and Analysis of Phenotype Datasets
To establish the primary dataset of Arabidopsis (Arabidopsis thaliana) genes with a single mutant phenotype, we started with a published list of 620 Arabidopsis genes included in our sequence-based map of genes with mutant phenotypes (Meinke et al., 2003), removed problematic loci with questionable genotype-to-phenotype associations, eliminated suppressors and genes with a dominant gain-of-function mutant phenotype but no apparent loss-of-function phenotype, and further curated the phenotype and gene function information. Several classical genetic loci with well-characterized dominant phenotypes (e.g. GAI, ETR1, ABI1) were retained in the dataset because they are also associated with a distinctive loss-of-function phenotype (Peng et al., 1997; Cancel and Larsen, 2002; Ludwików et al., 2009). We then requested from Eva Huala at TAIR a list of genes that appeared to be associated with phenotype information in the TAIR database. Each locus on the list was evaluated. Most entries yielded useful information, but many candidate genes were eliminated because no suitable phenotype information was found or because the locus did not code for a protein. To complement these efforts, we initiated extensive PubMed searches of the scientific literature, using a combination of the following keywords: Arabidopsis, mutant(s), mutation(s), knockout, and null. Several thousand articles were retrieved and analyzed to obtain the information presented in Supplemental Table S2. Information on genes with multiple mutant phenotypes was also retrieved with this approach. In order to proceed with further analysis of these datasets, no literature searches were performed for publications added to the PubMed database after December 31, 2010.
We then updated the information on essential genes based on the eighth release of the SeedGenes database (www.seedgenes.org). Additional updates were obtained from a recent publication on embryo and gametophyte essentials of Arabidopsis (Muralla et al., 2011). We classified essential genes as being required for early development or survival. A locus was considered to be essential when knockout heterozygotes segregated for defective embryos or gametophytes, regardless of whether the resulting homozygotes remained viable to the seedling stage or beyond. To be consistent with the prioritized classification system established here, EMB loci with defects in gametophyte function were assigned to the gametophyte class instead of the seed/embryo class, regardless of whether the locus was classified elsewhere as being required for seed development, because that is when the mutant phenotype was first detected. The criteria used to differentiate between the GAM, GEM, EMG, and EMB subsets of essential genes are detailed elsewhere (Muralla et al., 2011). Mutants with defective gametes that produced viable homozygotes were typically assigned to the MGD subset. The criteria used to make other phenotype subset assignments, listed in Supplemental Table S1, can be gauged by accessing the second tabbed spreadsheet in Supplemental Table S2, sorting for the subset of interest, and evaluating the diversity of phenotypes represented.
Gene Symbols and Reference Laboratories
The reference laboratories and publication dates listed in Supplemental Table S2 reflect resources that we used to obtain information for the dataset. These columns were retained in the final dataset to facilitate data tracking. In general, we listed the final author of the publication that identified the gene responsible for the mutant phenotype. This portion of the dataset could be replaced in the future with PubMed identification numbers for the most relevant publications, once issues of priority have been resolved. Determining which individuals should be credited with identifying the gene responsible for a mutant phenotype can sometimes be problematic, and when a single locus has been associated with more than one gene symbol in publications from different laboratories, determining which symbol should be given priority can lead to vigorous disagreements. We do not claim to have resolved these differences. That remains an important topic of discussion for future updates. Even the list of alias gene symbols for some loci is likely to be incomplete. Symbols for genes analyzed through reverse genetics are most problematic, because the locus is often named for the function of the protein product, which frequently does not conform to community standards for Arabidopsis genetics and nomenclature (Meinke and Koornneef, 1997). With respect to the capitalization of gene symbols, we retained some atypical examples of lowercase letters that were presented in publications, but otherwise we designated gene symbols in uppercase letters, consistent with community standards.
Identification of Chloroplast and Mitochondrial Proteins
The chloroplast localization data presented in Supplemental Tables S2 and S4 are based on the predicted chloroplast proteome of Richly and Leister (2004), the curated Plant Proteome Database of Sun et al. (2009), and experimental evidence presented in the SUBA (http://suba.plantenergy.uwa.edu.au) database (Heazlewood et al., 2007), as described previously (Bryant et al., 2011). Information on mitochondrial localization of gene products was also obtained from SUBA. The mitochondrial rank was calculated by adding a point each time a protein was predicted to be localized to mitochondria based on the following programs: (1) TargetP and Predotar; (2) Ipsort and Predotar; (3) TargetP and Ipsort; and (4) TargetP, Predotar, and Ipsort. Single points were also added for experimental evidence based on mass spectrometry or GFP analysis. The maximal rank score was 5 for chloroplasts and 6 for mitochondria. After manual curation, some candidate proteins with low scores were retained, because published work confirmed localization to mitochondria, whereas others were removed when a publication confirmed localization elsewhere in the cell.
Protein Sequence Comparisons
BLASTP (Blastall 2.2.23) was downloaded from the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/blast/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=Download) and run locally to analyze genetic redundancy and protein sequence similarity. For Arabidopsis sequences, we chose the longest gene model from TAIR 10 (www.arabidopsis.org/download/index.jsp). Rice (Oryza sativa) sequences were obtained from the Rice Genome Annotation Project, version 7 (http://rice.plantbiology.msu.edu/downloads.shtml); tomato (Solanum lycopersicum) sequences (ITAG2.3) were obtained from the Sol Genomics Network (www.solgenomics.net/organism/Solanum_lycopersicum/genome); and maize (Zea mays) sequences (Schnable et al., 2009) were obtained from the Phytozome database (www.phytozome.net/maize.php). Because unique locus numbers for maize genes could not be found, maize protein sequences identified through a reciprocal match with an Arabidopsis gene in the phenotype dataset were aligned with the original maize sequence using EMBOSS Needle (www.ebi.ac.uk/Tools/psa/) to determine whether the two sequences likely derived from a single locus.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Chromosome locations of redundant genes with multiple mutant phenotypes.
Supplemental Table S1. Detailed classification system of phenotype groups, classes, and subsets.
Supplemental Table S2. Comprehensive dataset of Arabidopsis genes with a loss-of-function mutant phenotype.
Supplemental Table S3. Protein functional classification system for genes with mutant phenotypes.
Supplemental Table S4. Phenotypes of genes encoding mitochondria-localized proteins.
Supplemental Table S5. Nonredundant protein interactors of unique genes with mutant phenotypes.
Supplemental Table S6. Comprehensive dataset of redundant genes with multiple mutant phenotypes.
Supplemental Table S7. Orthologous genes with mutant phenotypes in tomato, rice, and maize.
Supplementary zip file supplied by authors.
Acknowledgments
We thank Eva Huala at TAIR for contributing lists of candidate Arabidopsis genes thought to contain phenotype information, Lukas Mueller at the Boyce Thompson Institute for information on tomato genes with mutant phenotypes, and Rosanna Muralla, Nicole Bryant, Krista McGuire, and Kyle Mackin at Oklahoma State University for assistance with phenotype curation.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry CS, Giovannoni JJ. (2006) Ripening in the tomato Green-ripe mutant is inhibited by ectopic expression of a protein that disrupts ethylene signaling. Proc Natl Acad Sci USA 103: 7923–7928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becraft PW, Li K, Dey N, Asuncion-Crabb Y. (2002) The maize dek1 gene functions in embryonic pattern formation and cell fate specification. Development 129: 5217–5225 [DOI] [PubMed] [Google Scholar]
- Berg M, Rogers R, Muralla R, Meinke D. (2005) Requirement of aminoacyl-tRNA synthetases for gametogenesis and embryo development in Arabidopsis. Plant J 44: 866–878 [DOI] [PubMed] [Google Scholar]
- Bryant N, Lloyd J, Sweeney C, Myouga F, Meinke D. (2011) Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis. Plant Physiol 155: 1678–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancel JD, Larsen PB. (2002) Loss-of-function mutations in the ethylene receptor ETR1 cause enhanced sensitivity and exaggerated response to ethylene in Arabidopsis. Plant Physiol 129: 1557–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. (2008) Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36 [DOI] [PubMed] [Google Scholar]
- Chen Y, Jiang T, Jiang R. (2011) Uncover disease genes by maximizing information flow in the phenome-interactome network. Bioinformatics 27: i167–i176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Meeley R, Irish E, Sakai H, Hake S. (2007) The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat Genet 39: 1517–1521 [DOI] [PubMed] [Google Scholar]
- Conant GC, Wagner A. (2004) Duplicate genes and robustness to transient gene knock-downs in Caenorhabditis elegans. Proc Biol Sci 271: 89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean EJ, Davis JC, Davis RW, Petrov DA. (2008) Pervasive and persistent redundancy among duplicated genes in yeast. PLoS Genet 4: e1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T, Yamaguchi J, Wong BC, Veit B, Hake S. (1999) gnarley1 is a dominant mutation in the knox4 homeobox gene affecting cell shape and identity. Plant Cell 11: 1239–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N, Erezyilmaz DF, McGregor AP, Wang S, Payre F, Stern DL. (2011) Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature 474: 598–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabási AL. (2007) The human disease network. Proc Natl Acad Sci USA 104: 8685–8690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth P, Leser U, Weiss B. (2011) Phenotype mining for functional genomics and gene discovery. Methods Mol Biol 760: 159–173 [DOI] [PubMed] [Google Scholar]
- Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH. (2003) Role of duplicate genes in genetic robustness against null mutations. Nature 421: 63–66 [DOI] [PubMed] [Google Scholar]
- Hahn MW. (2009) Distinguishing among evolutionary models for the maintenance of gene duplicates. J Hered 100: 605–617 [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. (2007) SUBA: the Arabidopsis subcellular database. Nucleic Acids Res 35: D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Liu F, Drea S, Simpson GG, Dean C. (2005) An allelic series reveals essential roles for FY in plant development in addition to flowering-time control. Development 132: 3597–3607 [DOI] [PubMed] [Google Scholar]
- Hibara K, Obara M, Hayashida E, Abe M, Ishimaru T, Satoh H, Itoh J, Nagato Y. (2009) The ADAXIALIZED LEAF1 gene functions in leaf and embryonic pattern formation in rice. Dev Biol 334: 345–354 [DOI] [PubMed] [Google Scholar]
- Hoehndorf R, Schofield PN, Gkoutos GV. (2011) PhenomeNET: a whole-phenome approach to disease gene discovery. Nucleic Acids Res 39: e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao TL, Vitkup D. (2008) Role of duplicate genes in robustness against deleterious human mutations. PLoS Genet 4: e1000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J, Collins SR, Schuldiner M, Krogan NJ, Weissman JS. (2007) Backup without redundancy: genetic interactions reveal the cost of duplicate gene loss. Mol Syst Biol 3: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabási AL, Oltvai ZN. (2001) Lethality and centrality in protein networks. Nature 411: 41–42 [DOI] [PubMed] [Google Scholar]
- Johnson KL, Degnan KA, Ross Walker J, Ingram GC. (2005) AtDEK1 is essential for specification of embryonic epidermal cell fate. Plant J 44: 114–127 [DOI] [PubMed] [Google Scholar]
- Juarez MT, Twigg RW, Timmermans MC. (2004) Specification of adaxial cell fate during maize leaf development. Development 131: 4533–4544 [DOI] [PubMed] [Google Scholar]
- Kuromori T, Takahashi S, Kondou Y, Shinozaki K, Matsui M. (2009) Phenome analysis in plant species using loss-of-function and gain-of-function mutants. Plant Cell Physiol 50: 1215–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao BY, Zhang J. (2007) Mouse duplicate genes are as essential as singletons. Trends Genet 23: 378–381 [DOI] [PubMed] [Google Scholar]
- Ludwików A, Kierzek D, Gallois P, Zeef L, Sadowski J. (2009) Gene expression profiling of ozone-treated Arabidopsis abi1td insertional mutant: protein phosphatase 2C ABI1 modulates biosynthesis ratio of ABA and ethylene. Planta 230: 1003–1017 [DOI] [PubMed] [Google Scholar]
- Mabee PM, Ashburner M, Cronk Q, Gkoutos GV, Haendel M, Segerdell E, Mungall C, Westerfield M. (2007) Phenotype ontologies: the bridge between genomics and evolution. Trends Ecol Evol 22: 345–350 [DOI] [PubMed] [Google Scholar]
- Makino T, Hokamp K, McLysaght A. (2009) The complex relationship of gene duplication and essentiality. Trends Genet 25: 152–155 [DOI] [PubMed] [Google Scholar]
- McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman MA, et al. (2001) Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159: 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKusick VA. (2007) Mendelian Inheritance in Man and its online version, OMIM. Am J Hum Genet 80: 588–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke D, Koornneef M. (1997) Community standards for Arabidopsis genetics. Plant J 12: 247–253 [Google Scholar]
- Meinke D, Muralla R, Sweeney C, Dickerman A. (2008) Identifying essential genes in Arabidopsis thaliana. Trends Plant Sci 13: 483–491 [DOI] [PubMed] [Google Scholar]
- Meinke D, Sweeney C, Muralla R. (2009) Integrating the genetic and physical maps of Arabidopsis thaliana: identification of mapped alleles of cloned essential (EMB) genes. PLoS ONE 4: e7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke DW, Meinke LK, Showalter TC, Schissel AM, Mueller LA, Tzafrir I. (2003) A sequence-based map of Arabidopsis genes with mutant phenotypes. Plant Physiol 131: 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar MS, Carvunis AR, Dreze M, Epple P, Steinbrenner J, Moore J, Tasan M, Galli M, Hao T, Nishimura MT, et al. (2011) Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333: 596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralla R, Lloyd J, Meinke D. (2011) Molecular foundations of reproductive lethality in Arabidopsis thaliana. PLoS ONE 6: e28398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JM, Lane B, Freeling M. (2002) Expression of a mutant maize gene in the ventral leaf epidermis is sufficient to signal a switch of the leaf’s dorsoventral axis. Development 129: 4581–4589 [DOI] [PubMed] [Google Scholar]
- Ohno S. (1970) Evolution by Gene Duplication. Springer-Verlag, New York [Google Scholar]
- O’Malley RC, Ecker JR. (2010) Linking genotype to phenotype using the Arabidopsis unimutant collection. Plant J 61: 928–940 [DOI] [PubMed] [Google Scholar]
- Parnis A, Cohen O, Gutfinger T, Hareven D, Zamir D, Lifschitz E. (1997) The dominant developmental mutants of tomato, Mouse-ear and Curl, are associated with distinct modes of abnormal transcriptional regulation of a Knotted gene. Plant Cell 9: 2143–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP. (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Liao BY, Chang AYF, Zhang J. (2010) Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet 26: 425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly E, Leister D. (2004) An improved prediction of chloroplast proteins reveals diversities and commonalities in the chloroplast proteomes of Arabidopsis and rice. Gene 329: 11–16 [DOI] [PubMed] [Google Scholar]
- Ronen G, Cohen M, Zamir D, Hirschberg J. (1999) Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant Delta. Plant J 17: 341–351 [DOI] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Schnable JC, Freeling M. (2011) Genes identified by visible mutant phenotypes show increased bias toward one of two subgenomes of maize. PLoS ONE 6: e17855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Schneeberger RG, Becraft PW, Hake S, Freeling M. (1995) Ectopic expression of the knox homeo box gene rough sheath1 alters cell fate in the maize leaf. Genes Dev 9: 2292–2304 [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R, Benfey PN. (2011) High-throughput phenotyping of multicellular organisms: finding the link between genotype and phenotype. Genome Biol 12: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zybailov B, Majeran W, Friso G, Olinares PD, van Wijk KJ. (2009) PPDB, the plant proteomics database at Cornell. Nucleic Acids Res 37: D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, et al. (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135: 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavouri T, Semple JI, Lehner B. (2008) Widespread conservation of genetic redundancy during a billion years of eukaryotic evolution. Trends Genet 24: 485–488 [DOI] [PubMed] [Google Scholar]
- Walsh JB. (1995) How often do duplicated genes evolve new functions? Genetics 139: 421–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington NL, Haendel MA, Mungall CJ, Ashburner M, Westerfield M, Lewis SE. (2009) Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biol 7: e1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D. (2012) Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiol 158: 2–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AD, Moehlenkamp CA, Perrot GH, Neuffer MG, Cone KC. (1992) The maize auxotrophic mutant orange pericarp is defective in duplicate genes for tryptophan synthase β. Plant Cell 4: 711–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AJ, Gallagher K, Smith LG. (2009) discordia1 and alternative discordia1 function redundantly at the cortical division site to promote preprophase band formation and orient division planes in maize. Plant Cell 21: 234–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Li Y. (2006) Discovering disease-genes by topological features in human protein-protein interaction network. Bioinformatics 22: 2800–2805 [DOI] [PubMed] [Google Scholar]
- Yang P, Li X, Wu M, Kwoh CK, Ng SK. (2011) Inferring gene-phenotype associations via global protein complex network propagation. PLoS ONE 6: e21502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, et al. (2008) High-quality binary protein interaction map of the yeast interactome network. Science 322: 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary zip file supplied by authors.