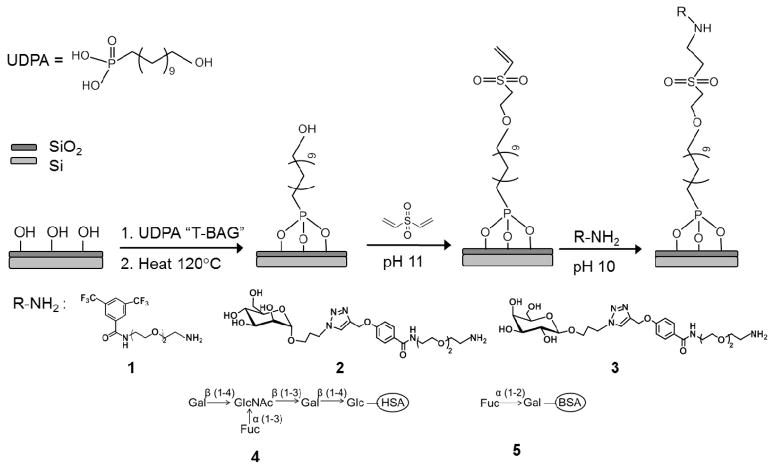
Abstract
Silicon photonic microring resonators have established their potential for label-free and low-cost biosensing applications. However, the long-term performance of this optical sensing platform requires robust surface modification and biofunctionalization. Herein, we demonstrate a conjugation strategy based on an organophosphonate surface coating and vinyl sulfone linker to biofunctionalize silicon resonators for biomolecular sensing. To validate this method, a series of glycans, including carbohydrates and glycoconjugates, were immobilized on divinyl sulfone (DVS)/organophosphonate-modified microrings and used to characterize carbohydrate-protein and norovirus particle interactions. This biofunctional platform was able to orthogonally detect multiple specific carbohydrate-protein interactions simultaneously. Additionally, the platform was capable of reproducible binding after multiple regenerations by high-salt, high-pH or low-pH solutions and after 1-month storage in ambient conditions. This remarkable stability and durability of the organophosphonate immobilization strategy will facilitate the application of silicon microring resonators in various sensing conditions, prolong their lifetime, and minimize the cost for storage and delivery; these characteristics are requisite for developing biosensors for point-of-care and distributed diagnostics and other biomedical applications. In addition, the platform demonstrated its ability to characterize carbohydrate-mediated host-virus interactions, providing a facile method for discovering new anti-viral agents to prevent infectious disease.
INTRODUCTION
Biosensors allow sensitive and rapid detection of a variety of biomolecular interactions, facilitating basic biomedical research, drug discovery, food and environmental monitoring, and diagnostics.1,2 Among the emerging biosensing technologies, silicon photonics – specifically the silicon microring resonator – has gained increasing attention due to demonstrated capabilities in sensitive multiplexed detection, chip-scale integration, and the potential of low-cost mass production using existing silicon fabrication processes.3–6 The optical microring resonator platform consists of an array of planar ring-shaped silicon waveguides optically coupled to linear bus waveguides on a silicon oxide insulator. Binding of biomolecules to the ligand-functionalized microring sensor causes small changes in the effective refractive index, resulting in a detectable shift in resonance wavelength.7 The feasibility of microring resonators for label-free detection of various biomolecules and cells, including proteins, oligonucleotides, and bacteria has been previously demonstrated in the literature.3,7
The dominant strategy for functionalizing silicon devices, including microring resonators, is based on common siloxane chemistries.5,8 However, the moisture-sensitivity of silanization and the instability of bound silanes limit the real world use of silicon-based biosensors.9 Silanized surface coating quality strongly depends on the atmospheric moisture content, making standardization and reproducibility difficult.10 Low surface coverage and hydrolytic instability of silane layers also limit ligand conjugation to, and reproducible detection by, silicon-based biosensors.9,11 Furthermore, formation of multi-layer silane networks attenuates the sensitivity and reduces the stability of functional surfaces for biosensing.12 Therefore, more robust surface functionalization strategies could result in stable and reliable silicon-based biosensors. Recently, organophosphonate self-assembled monolayers (SAMs) have been employed successfully to modify various inorganic oxide surfaces, such as Al2O313, TiO214 and SiO215. The “T-BAG” method developed by Hanson et al., involves adsorbing organophosphonic acid to a solid surface, which converts to surface-bound phosphonate at 120–140 °C.16,17 These organophosphonates have superior physicochemical properties. Relative to silanes, phosphonate SAMs can form densely-packed monolayers with higher surface coverage,16,17 and are much more stable in both acidic and alkaline solutions.12,14,18 Previous studies have demonstrated the efficacy of phosphonate chemistry in the fabrication of complementary circuits and transistors,19,20 modification of DNA biosensors9,17 and preparation of cell adhesion substrates15,21,22. Towards the development of stable and reproducible silicon microring biosensors, we applied organophosphonate SAMs in the modification of this biosensing platform.
The suitability of organophosphonate-modified microring resonators for biosensing applications was demonstrated by examining carbohydrate-mediated host-virus interactions. Carbohydrates play an essential role in various pathogenic processes.23 Pathogenesis is frequently mediated via the adhesion of pathogens to glycans on the host cell surface. For example, norovirus (NV), a major cause of acute gastroenteritis, recognizes human histo-blood group antigens, which contain well-defined carbohydrate epitopes.24 Inhibition of these glycan-dependent host-pathogen interactions has been established as a valuable target for drug development. For instance, human milk glycans containing fucosylated carbohydrate moieties can protect infants against diarrhea caused by a variety of bacterial and viral pathogens, including NV.25,26 These glycans represent a promising new class of anti-microbial agents to prevent infectious disease.27 The structure and the inhibitory mechanism of many of the human milk glycans are under active investigation.28 To meet the urgent need for new anti-infective agents based on human milk glycans, it is necessary to unravel the specific binding patterns between these glycans and pathogens. We propose that the silicon photonic microring resonator provides a versatile label-free and sensitive platform that can advance the study of carbohydrate-mediated host-pathogen interactions.
Herein, an 11-hydroxyundecylphosphonic acid (UDPA) modified silicon microring resonator biosensor was examined for its ability to reproducibly detect glycan-protein/virus interactions. Glycans were immobilized on the organophosphonate-modified sensors via a facile and versatile divinyl sulfone (DVS) conjugation method; DVS can conjugate biomolecules containing nucleophile groups (e.g. hydroxyl, amine and thiol) on hydroxyl-terminated surfaces.29 In the present study, amine-bearing saccharides and glycoproteins were conjugated to DVS-activated organophosphonates on silicon microrings through an amine-vinyl sulfone. Each surface functionalization step was characterized by X-ray photoelectron spectroscopy (XPS). Time-of-flight secondary ion mass spectrometry (ToF-SIMS) imaging further verified the attachment of a phosphonate film on the silicon microrings. These glycan-functionalized microring resonators were evaluated by their response to well-characterized lectins (glycan-binding proteins); the reproducibility and stability of the bioactive surfaces was subsequently examined under different regeneration and storage conditions. Finally, this biosensing platform was used successfully to screen interactions between glycans and NV-like particles, demonstrating the utility of organophosphonate-modified microring resonators for studying carbohydrate-mediated host-virus interactions.
EXPERIMENTAL SECTION
Materials
All chemical reagents and bovine serum albumin (BSA) were from Sigma-Aldrich (St. Louis, MO) and were used without further purification. 11-hydroxyundecylphosphonic acid was synthesized as described.30 Hexafluorobenzamide 1, mannosyl amino OEG (oligo(ethylene glycol)) 2 and galactosyl amino OEG 3 were provided by Justin Kaplan and Prof. Rodrigo B. Andrade at Temple University (see Supporting Information for synthetic details). Lacto-N-fucopentaose III-human serum albumin (LNFPIII-HSA, 4) and H disaccharide-bovine serum albumin (H2-BSA, 5) were provided by Dr. David Newburg. Virus-like particles of norovirus strain VA387 were derived as described previously.31 Millipore-filtered water was used for all aqueous solutions and rinsing. Concanavalin A (Con A), Ricinus Communis Agglutinin II (RCA60), Ulex Europaeus Agglutinin I (UEA I) and Lotus Tetragonolobus lectin (LTL) were from Vector Laboratories (Burlingame, CA). Con A was dissolved in HEPES pH 7.4 buffer (10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 150 mM NaCl, 1 mM CaCl2, 1 mM MnCl2). RCA60 and UEA I were dissolved in PBS pH 7.4 buffer (phosphate buffered saline, 10 mM phosphate, 2.7 mM KCl, 137 mM NaCl). LTL was dissolved in HEPES pH 7.5 buffer (10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 150 mM NaCl, 0.1 mM CaCl2). Culture-well silicone sheets were from Grace Biolaboratories (Bend, OR). Silicon wafers were from Silicon Valley Microelectronics (San Jose, CA).
Silicon microring resonator chips and instrumentation
Silicon microring resonator biosensors and the corresponding analysis instrumentation were manufactured by Genalyte, Inc. (San Diego, CA), as described.8 Briefly, each biosensor chip contains an array of 32 individually addressable microring resonators (30 μm in diameter). Of the 32 microring sensors, 24 are exposed for biosensing and 8 are coated with a fluoropolymer-cladding to serve as temperature and vibration reference controls. Figure 1 shows a scanning electron micrograph of a microring and an illustration of the microring cross-section. An external cavity diode laser with a center frequency of 1560 nm was rastered across the biosensor surface to rapidly interrogate individual microring resonators, measuring the resonance wavelength shift for each sensor with ~250 ms time resolution. The transmission spectrum of the ring is captured at the photodetector and digitized via data acquisition system. The Genalyte sensing system has a bulk refractive index response of 163 nm/RIU (refractive index unit) with a bulk refractive index sensitivity of 7.6 × 10−7, corresponding to a wavelength noise of 0.22 pm.32
Figure 1.
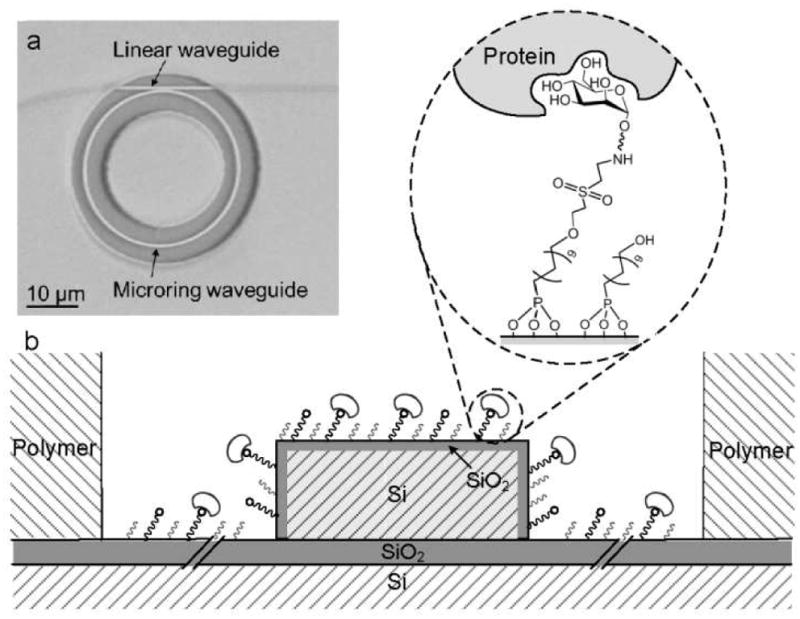
(a) SEM image of a microring resonator; (b) Cross-section of a glycan-modified organophosphonate/DVS microring resonator.
Functionalization of the silicon microring resonator chips
Modification with organophosphonic acid
Chips were cleaned by immersion in freshly prepared piranha solution (1:1 30% H2O2: 98% H2SO4) for 15min, rinsed with water, and dried in a stream of nitrogen. (Caution! Piranha solution is extremely dangerous; it can explode or catch fire when contacted by organic compounds.) Chips were modified with 11-hydroxyundecylphosphonic acid (UDPA) using the “T-BAG” method.16 Briefly, silicon chips were held vertically by clips and immersed in a 1 mM solution of UDPA in anhydrous tetrahydrofuran. The solvent was allowed to evaporate at room temperature over 3 h, until the level of the solution fell below the silicon chips. The coated silicon chips were transferred to an oven and heated at 120 °C overnight. Following heating, the chips were washed with sonication in 0.5 M K2CO3 in 2:1 ethanol/water for 20 min to remove any multilayer UDPA, rinsed with water and dried under a stream of nitrogen.
Immobilization of glycans
Glycan immobilization was accomplished using DVS chemistry.29 The organophosphonate modified silicon chips were immersed in 10% DVS (v/v, 50 mM carbonate buffer, pH 11) solution for 1 h at ambient temperature. After activation, the silicon chips were rinsed with water and dried by a stream of nitrogen. A silicone mask with four wells was placed on top of the silicon chips, exposing four biosensing microrings in each well for functionalization. 16 of the 24 microrings were functionalized with glycans, the remaining 8 rings were passivated with BSA and used as controls for non-specific binding to the surface. One microliter of each glycan solution (5 mM for 2 and 3, 0.5 mg/ml for 4 and 5, Scheme 1) in pH 10 carbonate buffer were spotted into each well and incubated in a 75% relative humidity chamber. After 16 h incubation, solutions in each well were removed, and the chips were immersed in a BSA solution (0.1% w/v, pH 8.5) for 2 h to passivate the surface. Finally, the chips were rinsed with water and dried under a stream of nitrogen. The silicone mask-based procedure for functionalizing a DVS-organophosphonate-modified chip is illustrated in Scheme S1 (Supporting Information).
Scheme 1.
Functionalization of silicon substrate using 11-hydroxyundecylphosphonic acid (UDPA) and divinyl sulfone (DVS) linking chemistry. The molecules immobilized on silicon microrings include: Hexafluorobenzamide (1), mannosyl amino OEG (2), galactosyl amino OEG (3), LNFPIII-HSA (4), H2-BSA (5). Abbreviation of carbohydrate residues in 4 and 5 represents: Gal, galactose; Fuc, fucose; GlcNAc, N-acetylglucosamine; Glc, glucose.
Surface characterization
XPS analysis
To facilitate surface analysis, silicon wafers were cleaned, modified with UDPA and activated by DVS using the same procedure described above. Hexafluorobenzamide 1, containing six fluorine atoms and a primary amine, was immobilized on the DVS-activated silicon wafer to mimic the glycan immobilization procedure. XPS composition data were acquired on a Kratos AXIS Ultra DLD instrument equipped with a monochromatic Al-Kα X-ray source (hv = 1486.6 eV). XPS data were collected at 0° take-off angle in the hybrid mode with approximately 10 nm sampling depth, using a pass energy of 80 eV. Three spots on two or more duplicate samples were analyzed. Reported compositional data were averaged over multiple spots. Data analysis was performed on the CasaXPS software (Casa Software Ltd.).
ToF-SIMS analysis
ToF-SIMS data was acquired on an ION-TOF 5-100 instrument (ION-TOF GmbH, Münster, Germany) equipped with a 25 KV liquid metal ion gun source. Ion images were acquired using Bi3+ primary ions in the high current bunched mode (i.e., high mass resolution mode). These images were collected by rastering the beam over a 100 × 100 μm2 area on the sample surface and keeping the fluence below the SIMS static limit of 1 × 1012 ions/cm2. All images contained 128 × 128 pixels. A low-energy electron beam was used for charge compensation. The mass resolution (m/Δm) of negative secondary ion spectra was typically between 6000 and 7500 for the m/z = 25 peak. Mass calibration was applied by using CH−, OH− and C2H− peaks.
Detection of glycan-mediated interactions by organophosphonate-modified silicon microring resonators
Glycan-containing chips, functionalized by the DVS/organophosphonate method, were loaded into a custom two-channel microfluidic flow cell defined by a laser cut gasket (Genalyte, Inc.; San Diego, CA). Negative-pressure syringe pumps were used to draw analyte solutions over the sensor surface. The flow rate was set to 10 μl/min. A typical binding experiment was preceded by flowing buffer solution over the surface for 10 minutes to stabilize the sensor response. Lectins or virus-particle solutions were passed over the sensor for 20 min, followed by another 20 min of buffer to examine dissociation. After acquisition of association/dissociation curves, regeneration solutions were passed over the analyte-bound surface for 5 minutes, and then buffer solution was flowed over the surface to reestablish the baseline. To validate the bioactivity of the immobilized glycans, a panel of the following lectins were used: Con A (binds to α-linked mannose)33, RCA60 (binds to β-linked galactose)34, UEA I (binds to α-1,2-linked fucose)35 and LTL (binds to both α-1,2- and α-1,3-linked fucose)36. HEPES pH 7.4 buffer was used for Con A binding, PBS (pH 7.4) was used for RCA60 and UEA I binding, and HEPES pH 7.5 buffer was used for LTL binding. Unless otherwise stated, lectins were used at 500 nM. Norovirus particles were used in pH 7.4 PBS at 0.25 μg/ml to 20 μg/ml. Three regeneration solutions were used to strip proteins from lectin-bound chips: 8 M urea; pH 2.0 glycine buffer (10 mM glycine, 160 mM NaCl, HCl was used to adjust pH); and 10 mM NaOH. Virus particle-bound chips were regenerated by 8 M urea. The binding curves are normalized to temperature reference control microrings and referenced to the BSA-passivated microrings to control for thermal effects and non-specific binding. The data are presented as the average ± standard deviation for a set of 4 identical microrings.
RESULTS AND DISCUSSION
Surface modification of silicon microring resonators
The stepwise organophosphonate-based procedure to modify a silicon oxide surface is illustrated in Scheme 1. Piranha-cleaned silicon substrates were modified with UDPA using the “T-BAG” method,16 phosphonate head groups form a tri-dentate bond with silicon oxide on the substrate.37 By keeping the concentration of organophosphonic acid below its critical micelle concentration (CMC), the “T-Bag” method allows the phosphonic acid amphiphile to aggregate at the air-solvent interface as the solvent meniscus slowly traverses the substrate, resulting in a uniform monolayer on the surface.12,16 As shown in the XPS elemental composition results (Table 1), a phosphorous signal was detected on the organophosphonate SAM-modified sample, but was absent in the bare silicon substrate. As compared to previous organophosphonate SAM studies, the relative percentage of phosphorous observed is lower than the reported value (0.8 atom %).12 However, this result was expected, as we adopted an expedient one-cycle SAM deposition approach, instead of three cycles as previously reported. Using the “T-BAG” method, the organophosphonic acid-adsorbed silicon substrates must be heated in order to covalently deposit onto silicon oxide;37 however, the repeated thermal cycling may cause functional loss of silicon waveguide-based biosensors. Thus, the single-cycle deposition was selected to minimize this possible damage. It has been noted that one-cycle deposition may not be sufficient for 100% surface coverage.9 But for the purpose of carbohydrate-protein binding studies, a low density organophosphonate film is sufficient for surface modification.38
Table 1.
Relative compositions of organophosphonate-functionalized silicon substrates determined by XPS.
| Silicon | PO3-C11-OH | PO3-C11-OH + DVS | PO3-C11-OH + DVS + 1 | |
|---|---|---|---|---|
| Si 2p | 48.9 ± 0.8 | 46.6 ± 0.9 | 47.4 ± 1.4 | 33.0 ± 0.3 |
| C 1s | 12.4 ± 1.6 | 20.8 ± 0.4 | 18.2 ± 1.6 | 28.4 ± 0.4 |
| O 1s | 38.7 ± 0.8 | 32.5 ± 0.5 | 34.1 ± 0.4 | 29.6 ± 0.4 |
| P 2s | n/d | 0.3 ± 0.1 | 0.3 ± 0.1 | 0.2 ± 0.1 |
| F 1s | n/d | n/d | n/d | 6.6 ± 0.5 |
| N 1s | n/d | n/d | n/d | 2.2 ± 0.1 |
ToF-SIMS imaging analysis was conducted to directly characterize organophosphonate-modified microrings. In the negative ion spectra, we observed Si−, SiO− and SiO2– peaks as well as the peaks representing organophosphonate fragments, including PO2– and PO3–. As shown in Figure 2, the ring structure of the device is clearly distinguished on the SiO2– and PO3–ion images. In combination with the XPS composition analysis, the ToF-SIMS data further verified the successful deposition of organophosphonate onto the silicon microrings.
Figure 2.
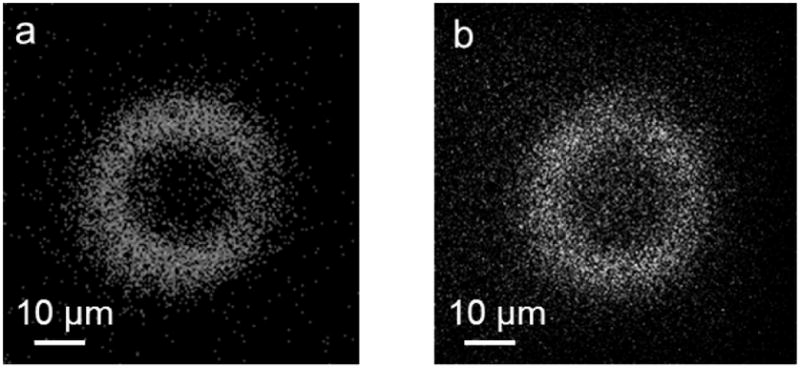
ToF-SIMS images of organophosphonate-modified silicon microrings. (a) SiO2– ions; (b) PO3– ions.
After deposition of the organophosphonate layer, we utilized DVS, a homobifunctional linker, to link one end to the UDPA hydroxyl moiety, and the other end to the amine-bearing monosaccharides and glycoconjugates, thereby fixing them covalently to the silicon microrings. In the XPS spectra, sulfur from the DVS molecule cannot be used for demonstrating the immobilization of DVS on the surface, due to the proximity of S2p peak to the Plasmon peak of silicon. Therefore, we used a fluorine tagged molecule 1 to facilitate characterization. Hexafluorobenzamide 1 with a pendant amino functional group was used to mimic the reaction between amine-bearing glycans and the DVS-activated surface. Following immobilization of 1 on the DVS-activated silicon substrate, fluorine and nitrogen peaks (Table 1) appeared with a compositional ratio of 3:1, which is in good agreement with their stoichiometric ratio in one fluorine-tagged molecule. To examine the possibility of nonspecific adsorption of aminated ligands on the DVS-organophosphonate-modified surface, we analyzed two control samples treated with either DVS or organophosphonic acid and subsequently exposed to 1. The XPS results demonstrated a trivial amount of fluorine on these two samples (< 0.3 atom %), which is significantly lower than the fluorine composition on the DVS-organophosphonate-modified surface (~6.6 atom %). These results suggest that the nonspecific adsorption of 1 on the surface is negligible; both DVS and organophosphonate are required for covalent immobilization of biomolecules. Due to the signal attenuation of phosphorous, which is buried under a long alkane chain, it is difficult to quantify the coupling efficiency of this method. However, based on our previous study on DVS chemistry,29 we estimated that ~6% of the phosphonate moieties are functionalized with aminated molecules. Thus, silicon oxide can be functionalized with amine-bearing biomolecules through phosphonate modification and DVS conjugation.
Carbohydrate-protein interactions detected by silicon microring resonators
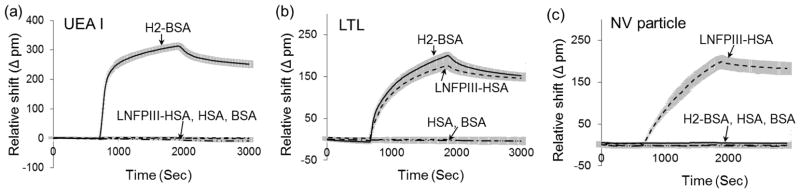
Two functionalized monosaccharides (mannose 2 and galactose 3) were immobilized onto DVS/organophosphonate-modified silicon microrings to assess the bioactivity of the functional biosensor. On one sensor chip, four micorings were selectively functionalized with mannose and another four with galactose to realize simultaneous dual detection. The availability of immobilized sugars to ligands was tested by protein-binding experiments. Con A lectin specifically binds to α-mannose33 while RCA60 recognizes β-galactose34. When Con A is passed over the sensor surface, only the microrings functionalized with mannose exhibited binding to this lectin while the microrings with galactose did not show a specific response (Figure 3(a)). Conversely, in the presence of RCA60, only the microrings functionalized with galactose exhibited a positive response (Figure 3(b)). Thus, silicon microring resonators functionalized with bioactive carbohydrates through organophosphonate modification and DVS activation are able to detect specific and multiple binding events on a single chip.
Figure 3.
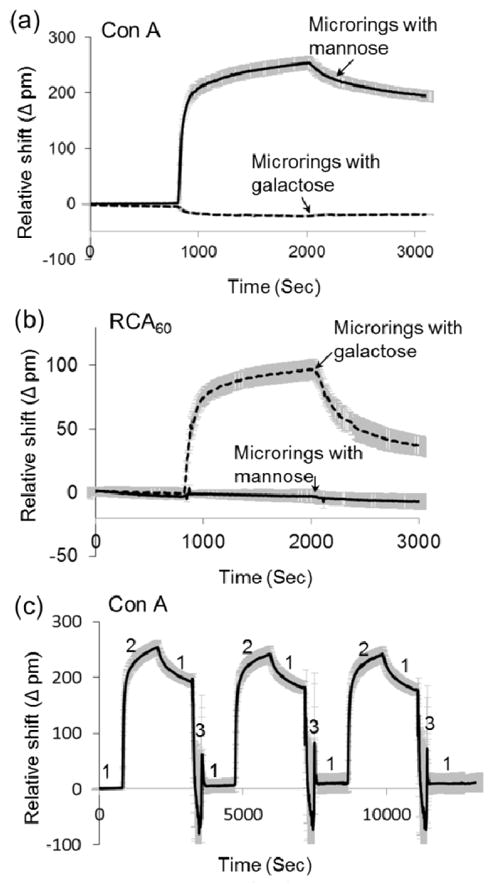
Binding response of microrings functionalized with 2 and 3 to (a) Con A and (b) RCA60 . (c) Reproducible binding response of the microrings functionalized with mannose to Con A after urea regeneration. 1. HEPES buffer; 2. 500 nM Con A in HEPES; 3. 8 M urea. All data are presented as the average ± standard deviation (n = 4) and are referenced to BSA-passivated microrings.
Stability and reproducibility of carbohydrate-modified silicon microring resonators
A robust and reusable biosensing system requires the ability to remove surface-bound analytes (e.g., proteins and cells) after each binding experiment without losing the underlying ligand functionality. Many common regeneration solutions rely on stringent conditions or reagents to disrupt analyte-ligand interactions, including high- or low-pH buffers and high-concentration chaotropic salts, which can damage the ligand-functionalized layer on silicon chips and subsequently deteriorate the performance of the biosensor.39 To assess the potential degradation of performance after many cycles of use, Con A binding to mannose-functionalized microring resonators was measured after a series of binding-regeneration cycles. Three commonly-used solutions (8M urea, pH 2.0 glycine buffer and 10 mM NaOH), which represent typical high-salt, low-pH and high-pH regeneration conditions, were used to strip surface-bound proteins following analyte capture. The sensing response (maximum relative wavelength shift after 20 min exposure to Con A) of a mannose-functionalized microring sensor was examined after each binding-regeneration cycle. Binding between Con A and immobilized mannose is highly reproducible after the 8 M urea wash (Figure 3(c)). After one binding-regeneration cycle, the microring resonator retained ~98% bioactivity (Table S1), and after 20 binding-regeneration cycles over 80% of the initial response. Similar binding curves were also obtained after pH 2.0 glycine and 10 mM NaOH regeneration (See Figure S1). The sensing response remained 88% after regeneration by pH 2.0 glycine and 97% after 10 mM NaOH (Table S1). High-pH solutions are frequently required to regenerate whole-cell bound surfaces, and often damage silane-modified surfaces.14,40 Remarkably, 10 mM NaOH is not harmful to the organophosphonate-modified sensors.
Long-term stability of the bioactive sensor surface under ambient conditions is critical for the utility of a distributed reusable biosensor. To explore the ability of the biofunctional organophosphonate-modified sensor to retain bioactivity, a mannose-functionalized microring chip was exposed to the atmosphere for 1-month at room temperature and tested for its Con A-binding. The relative response of these stored microring resonators to Con A was ~94% of the original value (Table S1).
The organophosphonate layer, already established as a robust modification method for silicon oxide,15 confers remarkable stability of carbohydrate/organophosphonate-modified sensors towards both concentrated-salt/extreme-pH, and prolonged storage under ambient conditions. The data validate the DVS/organophosphonate approach towards producing biofunctional sensors capable of reproducible detection, prolonged lifetime and adaptability to a variety of experimental conditions. These capabilities support the future application of silicon-based biosensors for point-of-care diagnostics, environmental monitoring and other applications.
Glycoconjugate-virus interactions measured by the microring resonator biosensor
The ability of the DVS/organophosphonate microring resonator to measure glycan-virus interactions was evaluated. Carbohydrates on host cell surface are used by many pathogens as initial recognition and attachment receptors.41 Human histo-blood group antigens (HBGAs) are complex glycans present on the surface of red blood cells and epithelial cells, or as free antigens in biological fluids.42 These HBGAs can be recognized by noroviruses (NVs), initiating binding and pathogenesis.31 Previous studies have demonstrated that human milk glycans possessing HBGAs can block the binding of NVs to their host-cell receptors.26 However, the specific molecules in human milk responsible for this anti-infectious activity have not been identified.
To elucidate these norovirus-glycan interactions, the organophosphonate-modified silicon microring resonators were employed to investigate the specificity of norovirus adhesion to select human milk glycans. Virus-like particles constructed from the recombinant outer capsid proteins of NV strain VA387 were screened for binding to two neoglycoproteins, LNFPIII-HSA (4) and H2-BSA (5). The glycoproteins used for this study were prepared from a core albumin protein (human serum or bovine serum, respectively), to which covalently-bound carbohydrate moieties from human milk (lacto-N-fucopentaose III (LNFPIII) and H disaccharide (H2)) are attached. LNFPIII contains α-1,3-linked fucose while H2 possesses α-1,2-linked fucose.43 We selected glycoprotein conjugates for this study over unmodified oligosaccharides due to their multivalent presentation of multiple sugar moieties per protein, mimicing the natural multivalent interactions between virus and its host receptors.44 Previous studies have demonstrated that a macromolecular carrier displaying a high density of oligosaccharide is required for norovirus-receptor binding.45 Conjugation of these glycoproteins onto the DVS/organophosphonate-modified sensor was accomplished through the reaction between the surface-bound vinyl sulfone and an amine on the protein carriers. Two fucose-binding lectins, UEA I and LTL, were used to evaluate the bioactivity of the immobilized glycoconjugates. UEA I recognizes α-1,2-fucose residues35 while LTL binds to both α-1,3- and α-1,2-linked fucose residues36. As expected, the microrings functionalized with H2-BSA but not LNFPIII-HSA responded to UEA I, while both types of functionalized microrings bound to LTL (Figure 4(a) and (b)). Control microrings functionalized with HSA or BSA did not respond to either lectin.
Figure 4.
Binding response of microring resonators functionalized with H2-BSA, LNFPIII-HSA, HSA and BSA to (a) UEA I (500 nM), (b) LTL (500 nM) and (c) NV VA387 particles (10 μg/ml). The data are presented as the mean ± standard deviation (n = 4) and are referenced to BSA-passivated microrings.
By presenting glycans on microring resonators, the carbohydrate-functionalized biosensing platform is able to model the host cell surface for characterizing host-virus interactions. The norovirus-like particles used in this study are a complex of 24 protruding domains of VA387 capsid protein, with a total molecular weight of ~850 KDa.31 The NV capsids are organized in octahedral symmetry and exhibit the native virus-receptor binding specificity.31 Microrings functionalized with LNFPIII-HSA bound to NV particles (Figure 4(c)), while microrings with H2-BSA, HSA and BSA showed no binding. Thus, glycan-functionalized microring resonators are able to detect specific binding of NV particles to the glycans with LNFPIII moieties. Previous studies have detected the binding of VA 387 particles to oligosaccharides possessing an α-1,2-fucose residue, but haven’t reported binding to α-1,3-fucosylated glycans.46 This is the first report of LNFPIII-HSA with α-1,3-linked fucose residues binding to NV particles from VA387 strain, leading to the hypothesis that human milk glycans containing LNFPIII motifs could act as soluble inhibitors and block the adhesion of NV VA387 to their host cell receptors. NVs include more than 20 strains, each of which recognizes different glycan epitopes.47 Our preliminary study of NV-glycan interactions suggests that expanded glycan libraries coupled to orthogonal multiplex silicon resonators could potentially unravel complex NV-host interactions. Future efforts will focus on using the glycan-functionalized microring resonators to screen the binding specificities of additional strains of NVs. Human milk glycans could then be screened to identify potential anti-viral components in human milk that block specific NV strains.
Glycan-modified biosensors are capable of interrogating carbohydrate-mediated host-pathogen interactions, facilitating the study of fundamental biochemical and molecular mechanisms of pathogenesis. Antibody-functionalized sensors contrast this approach, where detection is the primary goal of the functional device. The limit of detection (LOD) of antibody-functionalized microring resonators, similar to those used in this study, have been reported in sub-ng/mL range, which is comparable to other label-free biosensors, such as surface plasmon resonance.48 However, NV-glycan binding affinities are several orders of magnitude weaker than antibody-virus binding. As a result, the carbohydrate-functionalized sensors are expected to have only modest LOD. Within the detection range in this study, we observed a linear relationship between the initial slope of binding and the concentration of NV particles (Figure S2). The experimental LOD for NV particles is estimated to be 250 ng/mL, which is sufficient for the analysis of carbohydrate-mediated NV binding. However, if detection were the objective, antibody-mediated capture should be employed.
CONCLUSIONS
This study demonstrated the versatility of an organophosphonate-based strategy for covalent modification of silicon photonic microring resonators, and validated the capability of this robust biosensing platform for label-free detection of glycan-protein and glycan-virus interactions. Glycans, including monosaccharides and glycoprotein conjugates, were immobilized on organophosphonate-modified microring sensors using a divinyl sulfone (DVS) linking chemistry, and the conjugation strategy was validated by XPS and ToF-SIMS analysis. Using this method, an array of microrings were functionalized with multiple glycans and used to detect simultaneous orthogonal binding events. This organophosphonate-modified biosensing platform exhibited remarkable stability and reproducibility after concentrated salt, high- and low-pH regeneration and long-term storage under ambient conditions. These results strongly support the conclusion of multiple prior studies that demonstrated that organophosphonate chemistry is a more stable and reliable strategy compared to siloxanes for the modification of biosensors and other biomedical surfaces.14,18,49 The robustness of this surface functionalization method will advance silicon microring resonators towards practical applications as portable and reusable devices and also facilitate multiplexed detection of biomolecules and cells for drug discovery, environmental monitoring as well as basic biomedical research. This study presents the first example of using organophosphonate-modified microring resonators to characterize glycan-virus particle interactions, demonstrating the potential of this platform for screening carbohydrate-mediated host-pathogen interactions and discovering new carbohydrate-based prophylactics and therapeutics.
Supplementary Material
Acknowledgments
This work was supported by NSF CBET (award no. 0930411), the Washington Research Foundation, the University of Washington Royalty Research Fund, and NIH grants HD061930, HD013021, and AI075563. Surface analysis was performed by NESAC/BIO (NIH Grant P41 EB002027). The authors would like to thank Mr. Jeff Chamberlain and Mr. James Kirk for their instruction on Genalyte instrument, Dr. Jim Hull and Dr. Daniel Graham for SIMS analysis.
Supporting Information Available. Scheme illustrating the functionalization of silicon microring chips, table of retained bioactivity of mannose-functionalized microrings after one binding-regeneration cycle and 1-month storage, figure of reproducible binding response after glycine and NaOH regeneration, linear curve-fitting of the initial slope of LNFPIII-HSA—norovirus particle binding curves, synthetic schemes and characterization of hexafluorobenzamide, mannosyl amino OEG and galactosyl amino OEG. This information is available free of charge via the Internet at http://pubs.acs.org/.
Contributor Information
Jing Shang, Email: shangj@u.washington.edu.
Fang Cheng, Email: ffcheng@gmail.com.
Manish Dubey, Email: manish.dubey@intel.com.
Justin M. Kaplan, Email: jkaplan1@temple.edu.
Meghana Rawal, Email: rawal@uw.edu.
Xi Jiang, Email: jason.jiang@cchmc.org.
David S. Newburg, Email: david.newburg@bc.edu.
Philip A. Sullivan, Email: psull76@u.washington.edu.
Rodrigo B. Andrade, Email: randrade@temple.edu.
Daniel M. Ratner, Email: dratner@uw.edu.
References
- 1.Lazcka O, Del Campo FJ, Munoz FX. Biosens Bioelectron. 2007;22:1205–1217. doi: 10.1016/j.bios.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 2.Lee H, Sun E, Ham D, Weissleder R. Nat Med. 2008;14:869–874. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Washburn AL, Luchansky MS, Bowman AL, Bailey RC. Anal Chem. 2010;82:69–72. doi: 10.1021/ac902451b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirk JT, Fridley GE, Chamberlain JW, Christensen ED, Hochberg M, Ratner DM. Lab Chip. 2011;11:1372–1377. doi: 10.1039/c0lc00313a. [DOI] [PubMed] [Google Scholar]
- 5.Byeon JY, Bailey RC. Analyst. 2011;136:3430–3433. doi: 10.1039/c0an00853b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luchansky MS, Bailey RC. Anal Chem. 2010;82:1975–1981. doi: 10.1021/ac902725q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramachandran A, Wang S, Clarke J, Ja SJ, Goad D, Wald L, Flood EM, Knobbe E, Hryniewicz JV, Chu ST, Gill D, Chen W, King O, Little BE. Biosens Bioelectron. 2008;23:939–944. doi: 10.1016/j.bios.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Washburn AL, Gunn LC, Bailey RC. Anal Chem. 2009;81:9499–9506. doi: 10.1021/ac902006p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cattani-Scholz A, Pedone D, Dubey M, Neppl S, Nickel B, Feulner P, Schwartz J, Abstreiter G, Tornow M. ACS Nano. 2008;2:1653–1660. doi: 10.1021/nn800136e. [DOI] [PubMed] [Google Scholar]
- 10.Lapin NA, Chabal YJ. J Phys Chem B. 2009;113:8776–8783. doi: 10.1021/jp809096m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asenath-Smith E, Chen W. Langmuir. 2008;24:12405–12409. doi: 10.1021/la802234x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubey M, Weidner T, Gamble LJ, Castner DG. Langmuir. 2010;26:14747–14754. doi: 10.1021/la1021438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hauffman T, Blajiev O, Snauwaert J, van Haesendonck C, Hubin A, Terryn H. Langmuir. 2008;24:13450–13456. doi: 10.1021/la801978a. [DOI] [PubMed] [Google Scholar]
- 14.Silverman BM, Wieghaus KA, Schwartz J. Langmuir. 2005;21:225–228. doi: 10.1021/la048227l. [DOI] [PubMed] [Google Scholar]
- 15.Midwood KS, Carolus MD, Danahy MP, Schwarzbauer JE, Schwartz J. Langmuir. 2004;20:5501–5505. doi: 10.1021/la049506b. [DOI] [PubMed] [Google Scholar]
- 16.Hanson EL, Schwartz J, Nickel B, Koch N, Danisman MF. J Am Chem Soc. 2003;125:16074–16080. doi: 10.1021/ja035956z. [DOI] [PubMed] [Google Scholar]
- 17.Cattani-Scholz A, Pedone D, Blobner F, Abstreiter G, Schwartz J, Tornow M, Andruzzi L. Biomacromolecules. 2009;10:489–496. doi: 10.1021/bm801406w. [DOI] [PubMed] [Google Scholar]
- 18.Hoque E, DeRose JA, Kulik G, Hoffmann P, Mathieu HJ, Bhushan B. J Phys Chem B. 2006;110:10855–10861. doi: 10.1021/jp061327a. [DOI] [PubMed] [Google Scholar]
- 19.Klauk H, Zschieschang U, Pflaum J, Halik M. Nature. 2007;445:745–748. doi: 10.1038/nature05533. [DOI] [PubMed] [Google Scholar]
- 20.Zschieschang U, Halik M, Klauk H. Langmuir. 2008;24:1665–1669. doi: 10.1021/la703818d. [DOI] [PubMed] [Google Scholar]
- 21.Luo W, Westcott NP, Pulsipher A, Yousaf MN. Langmuir. 2008;24:13096–13101. doi: 10.1021/la802775v. [DOI] [PubMed] [Google Scholar]
- 22.Adden N, Gamble LJ, Castner DG, Hoffmann A, Gross G, Menzel H. Langmuir. 2006;22:8197–8204. doi: 10.1021/la060754c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oyelaran O, Gildersleeve JC. Curr Opin Chem Biol. 2009;13:406–413. doi: 10.1016/j.cbpa.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shirato H, Ogawa S, Ito H, Sato T, Kameyama A, Narimatsu H, Xiaofan Z, Miyamura T, Wakita T, Ishii K, Takeda N. J Virol. 2008;82:10756–10767. doi: 10.1128/JVI.00802-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrow AL, Ruiz-Palacios GM, Jiang X, Newburg DS. J Nutr. 2005;135:1304–1307. doi: 10.1093/jn/135.5.1304. [DOI] [PubMed] [Google Scholar]
- 26.Jiang X, Huang P, Zhong W, Tan M, Farkas T, Morrow AL, Newburg DS, Ruiz-Palacios GM, Pickering LK. J Infect Dis. 2004;190:1850–1859. doi: 10.1086/425159. [DOI] [PubMed] [Google Scholar]
- 27.Newburg DS. J Nutr. 2005;135:1308–1312. doi: 10.1093/jn/135.5.1308. [DOI] [PubMed] [Google Scholar]
- 28.Newburg DS. J Anim Sci. 2009;87:26–34. doi: 10.2527/jas.2008-1347. [DOI] [PubMed] [Google Scholar]
- 29.Cheng F, Shang J, Ratner DM. Bioconjugate Chem. 2011;22:50–57. doi: 10.1021/bc1003372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Putvinski TM, Schilling ML, Katz HE, Chidsey CED, Mujsce AM, Emerson AB. Langmuir. 1990;6:1567–1571. [Google Scholar]
- 31.Tan M, Fang P, Chachiyo T, Xia M, Huang P, Fang Z, Jiang W, Jiang X. Virology. 2008;382:115–123. doi: 10.1016/j.virol.2008.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iqbal M, Gleeson MA, Spaugh B, Tybor F, Gunn WG, Hochberg M, Baehr-Jones T, Bailey RC, Gunn LC. Ieee J Sel Top Quant. 2010;16:654–661. [Google Scholar]
- 33.Tagawa K, Sendai N, Ohno K, Kawaguchi T, Kitano H, Matsunaga T. Bioconjugate Chem. 1999;10:354–360. doi: 10.1021/bc980083x. [DOI] [PubMed] [Google Scholar]
- 34.Newton DL, Wales R, Richardson PT, Walbridge S, Saxena SK, Ackerman EJ, Roberts LM, Lord JM, Youle RJ. J Biol Chem. 1992;267:11917–11922. [PubMed] [Google Scholar]
- 35.Farr AG, Anderson SK. J Immunol. 1985;134:2971–2977. [PubMed] [Google Scholar]
- 36.Pereira ME, Kabat EA. Biochemistry (Mosc) 1974;13:3184–3192. doi: 10.1021/bi00712a029. [DOI] [PubMed] [Google Scholar]
- 37.Gouzman I, Dubey M, Carolus MD, Schwartz J, Bernasek SL. Surf Sci. 2006;600:773–781. [Google Scholar]
- 38.Dhayal M, Ratner DA. Langmuir. 2009;25:2181–2187. doi: 10.1021/la8031122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seitz O, Fernandes PG, Tian RH, Karnik N, Wen HC, Stiegler H, Chapman RA, Vogel EM, Chabal YJ. J Mater Chem. 2011;21:4384–4392. [Google Scholar]
- 40.Marcinko S, Fadeev AY. Langmuir. 2004;20:2270–2273. doi: 10.1021/la034914l. [DOI] [PubMed] [Google Scholar]
- 41.Liang PH, Wu CY, Greenberg WA, Wong CH. Curr Opin Chem Biol. 2008;12:86–92. doi: 10.1016/j.cbpa.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao S, Lou ZY, Tan M, Chen YT, Liu YJ, Zhang ZS, Zhang XJC, Jiang X, Li XM, Rao ZH. J Virol. 2007;81:5949–5957. doi: 10.1128/JVI.00219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newburg DS, Ruiz-Palacios GM, Altaye M, Chaturvedi P, Meinzen-Derr J, Guerrero Mde L, Morrow AL. Glycobiology. 2004;14:253–263. doi: 10.1093/glycob/cwh020. [DOI] [PubMed] [Google Scholar]
- 44.Mammen M, Choi SK, Whitesides GM. Angew Chem Int Edit. 1998;37:2755–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 45.Huang P, Morrow AL, Jiang X. Glycoconjugate J. 2009;26:1085–1096. doi: 10.1007/s10719-009-9229-x. [DOI] [PubMed] [Google Scholar]
- 46.Huang P, Farkas T, Zhong W, Tan M, Thornton S, Morrow AL, Jiang X. J Virol. 2005;79:6714–6722. doi: 10.1128/JVI.79.11.6714-6722.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan M, Jiang X. Expert Rev Mol Med. 2007;9:1–22. doi: 10.1017/S1462399407000348. [DOI] [PubMed] [Google Scholar]
- 48.Luchansky MS, Washburn AL, Martin TA, Iqbal M, Gunn LC, Bailey RC. Biosens Bioelectron. 2010;26:1283–1291. doi: 10.1016/j.bios.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeRose JA, Hoque E, Bhushan B, Mathieu HJ. Surf Sci. 2008;602:1360–1367. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.