Abstract
We investigated whether mutations in the KLF1 gene are associated with increased Hb F levels in ethnically diverse patients referred to our laboratory for hemoglobinopathy investigation. Functionally effective KLF1 mutations were identified in 11 out of 131 adult samples with an elevated Hb F level (1.5–25.0%). Eleven different mutations were identified, 9 of which were previously unreported. KLF1 mutations were not identified in a matched cohort of 121 samples with normal Hb F levels (<1.0%). A further novel KLF1 mutation was also found in a sickle cell disease patient with a Hb F level of 20.3% who had a particularly mild phenotype. Our results indicate KLF1 mutations could make a significant contribution to Hb F variance in malarial regions where hemogobinopathies are common. All the mutations identified were heterozygous providing further in vivo evidence that a single altered KLF1 allele is sufficient to increase Hb F levels.
Keywords: KLF1, mutations, Hb F, malarial regions, thalassemia
Introduction
Hereditary persistence of fetal hemoglobin (HPFH) is defined as an increased proportion of fetal hemoglobin which persists beyond infancy. The condition has virtually no adverse clinical effects but is of considerable interest because increased Hb F levels are known to ameliorate the severity of some hemoglobin disorders. HPFH has been identified in a diverse range of ethnic groups.1 However, higher frequencies are observed in populations with a high prevalence of hemoglobinopathies and thalassemias, and consequently it is often observed as a coexisting feature of these conditions. It is well established that HPFH can be caused by deletions within the HBB (β-globin) gene cluster on chromosome 112 and point mutations in the promoters of the HBG1 and HBG2 (γ-globin) genes.3,4 There are also single nucleotide polymorphisms (SNPs) or oligonucleotide motifs within the β-globin gene cluster which are associated with higher Hb F levels under conditions of erythroid stress such as sickle cell disease or beta thalassemia. The best known of these is the C-T polymorphism at position −158 of the G-γ promoter which creates an Xmn1 restriction site.5 More recently, loci unlinked to the β-globin gene cluster that affect Hb F levels have been identified. Two major sites are the HBS1L-MYB intergenic region on chromosome 6q236 and BCL11A on chromosome 2p16.1.7 Known loci have been shown to account for 50% of the variance seen in Hb F levels indicating that additional loci must be involved.8
Recently, an additional potential locus was identified when point mutations in the KLF1 gene were found to be associated with HPFH in a Maltese9 family and in a family from Sardinia.10 KLF1 is an essential erythroid transcription factor first identified in 1993 that binds to an important DNA binding site, the CACCC motif in the β-globin gene.11 The gene comprises a proline rich N-terminal region containing a transactivation domain and a C-terminal region containing three zinc finger domains essential for DNA binding. Recent studies have shown that KLF1 could play a critical role in regulating the switch between fetal and adult hemoglobin expression both by direct activation of β-globin and indirect repression of γ-globin gene expression in adult erythroid progenitors via regulation of BCL11A.12 The finding of HPFH associated with mutations in KLF1 is of significance as it may be in vivo evidence that controlled reduction of KLF1 expression could activate fetal hemoglobin production and, therefore, provide a potential therapeutic target.13
This study investigates whether KLF1 mutations are involved in the increased Hb F levels observed in blood samples referred to our laboratory for hemoglobinopathy investigation.
Design and Methods
Study subjects
Elevated Hb F level
A total of 131 blood samples referred for hemoglobinopathy investigation (subjects aged 1–81 years) with an elevated Hb F level (range 1.5–25%) were tested for KLF1 mutations. These samples were then compared against a matched control group of 121 samples that had also been referred for hemoglobinopathy investigation but had normal Hb F levels (<1%). Hemoglobinopathy phenotypes in the study group with an elevated Hb F level consisted of 55 samples with co-existing α-thalassemia trait, 6 carriers for sickle cell trait, 28 β-thalassemia carriers, one case of Hb E disease, and 41 samples with no evidence of any other hemoglobinopathy apart from the elevated Hb F level. Hemoglobinopathy phenotypes in the control group with normal Hb F levels consisted of 41 α-thalassemia carriers, 9 sickle cell carriers, 29 carriers for a hemoglobin variant, 20 β-thalassemia carriers, and 22 samples with no evidence of any hemoglobinopathy.
Sickle cell disease
A cohort of 55 patient samples with sickle cell disease (all homozygous for the sickle cell mutation) were studied to investigate whether KLF1 mutations could be involved in Hb F level variation in this group. Twenty patients had Hb F levels below 10% and 35 had Hb F levels above 10%.
Laboratory procedures
Peripheral blood erythrocyte indices were determined using an automated cell counter. (Sysmex XE 2100™). Hemoglobin identification and quantifications were carried out using a high performance liquid chromatography system (VARIANT™, Bio-Rad Laboratories, USA) and isoelectric focusing gel electrophoresis (RESOLVE®, PerkinElmer, USA). DNA was extracted from peripheral blood leukocytes by conventional phenol chloroform extraction or on an automated DNA extractor (Chemagen, Baesweiler, Germany). Genomic DNA samples underwent PCR to amplify the human KLF1 gene (NT_086897.1: 4090501–4093981) using previously published primers.14 PCR products were sequenced using the ABI-PRISM 3100 automated DNA sequencer (Applied Biosystems). Samples containing a mutation in the KLF1 gene had the promoter regions of both the Aγ and Gγ genes amplified using previously published primers15 and the PCR products sequenced as above. The common 3.7 and 4.2 Kb single α+ thalassemia globin gene deletion mutations were identified by Gap-PCR16 and β-globin gene cluster deletions excluded by MLPA17 in all samples. PolyPhen-2 and SIFT were used to predict the effects of any mutations on protein structure and function.18,19 This study was approved by the Oxford Research Ethics Committee.
Results and Discussion
Elevated Hb F subjects
KLF1 mutations that are predicted to effect gene function (PolyPhen-2 and SIFT) were identified in 11 out of 131 (8.4%) subjects with increased Hb F levels (Table 1). Ten had a single heterozygous mutation and one individual was compound heterozygous for two mutations. In total, eleven different KLF1 mutations were identified (Table 1). Beta cluster deletion mutations and mutations in the γ-globin gene promoter sequences were excluded as a cause of the increased Hb F level in all 11 subjects. Functionally effective KLF1 mutations were not identified in the matched cohort of 121 samples with normal Hb F levels. Nine of the 11 mutations identified were previously unreported.
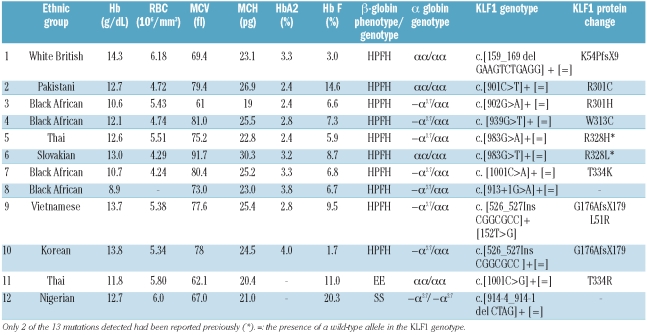
Table 1.
Hematologic parameters and genotypes in subjects with mutations detected in the KLF1 gene. Cases 1–11 are subjects from the elevated Hb F cohort and case 12 was from the sickle cell anemia cohort.
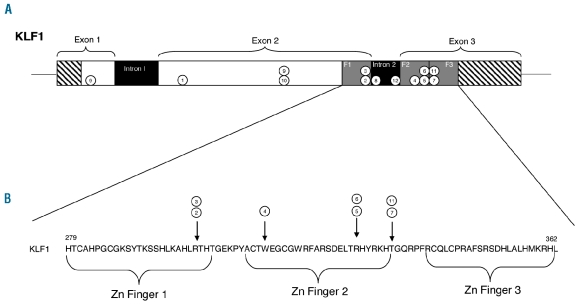
Eight were missense mutations, one in exon 1 (L51R) and seven in the zinc finger domains (R301C, R301H, W313C, R328H, R328L, T334K and T334R) which would be expected to disrupt DNA binding (Figure 1). Two were frame shift mutations in exon 2, an 11bp deletion (K54PfsX9) producing a new stop codon 8 nucleotides downstream and a 7bp insertion (G176AfsX179) producing a stop codon 178 nucleotides downstream. The latter mutation was identified in 2 patients, singly and compound heterozygous with the L51R missense mutation. The final mutation identified in this group was a 1bp nucleotide substitution (c.913+1G>A) at the 3’end of exon 2 which would be predicted to disrupt splicing.
Figure 1.
(A) Diagram showing the position of the mutations identified in the KLF1 gene. Numbered circles correspond to the case number in Table 1. Hatched: untranslated regions; white: coding regions; grey: zinc finger domains; black: introns. (B) Amino acid sequence of the three zinc fingers in KLF1 (NCBI’s Homologene 24) in Homo sapiens. Arrows show the positions of missense mutations within the zinc finger domains.
Sickle cell anemia subjects
Out of the 55 sickle cell disease patients studied, one further unreported functionally effective KLF1 mutation was identified (c.914-4_914-1 del CTAG ) in a sickle cell disease patient with an elevated Hb F level of 20.3%. Mutations in the γ-globin gene promoter sequences and the Xmn1 polymorphism were excluded as the cause of the increased Hb F in this patient. This KLF1 mutation is a 4bp deletion and is located close to the start of the second zinc finger domain in exon 3 and is likely to result in aberrant splicing.
Tolerated SNPs
Two patients in the HPFH cohort and one patient in the sickle cell disease cohort were found to have SNPs in the coding regions of the KLF1 gene (G5K, G160K, and G250A). PolyPhen-2 and SIFT analyses suggest that they are neutral substitutions which are tolerated and, therefore, not pathogenic. In support of this, G5K and G160K were also found in 2 samples in the normal F level control group.
Recent reports have identified mutations in the KLF1 gene that are associated with a variety of phenotypes in humans. These include the Lutheran blood group,14 congenital dyserythropoietic anemia,20 hereditary spherocytosis,21 high levels of zinc protoporphyrin,10 HPFH in two families,9,10 and most recently, borderline increases in Hb A2 levels.22 The UK population is ethnically diverse and our laboratory receives requests for hemoglobinopathy investigations for individuals who originate from all the malarial regions of the world. Our study identified KLF1 mutations in a significant proportion of these referrals with increased Hb F levels. KLF1 mutations predicted to have an effect were found in 11 out of 131 referrals with increased Hb F levels, but KLF1 mutations were not identified in 121 hemoglobinopathy referrals with normal Hb F levels. This strongly suggests that the KLF1 mutations are associated with the observed increased Hb F levels in these patients. All the mutations identified were heterozygous, indicating that a single altered KLF1 allele can elevate Hb F.
The increased Hb F levels observed in our KLF1 mutation positive subjects ranged from 1.7 to 14.4%. Well established factors known to cause HPFH (deletions in the β-globin cluster or mutations in the γ-globin gene promoters) were excluded in all cases. An interesting finding was that 4 out of our 11 cases also had borderline increased Hb A2 levels (3.3–4.0%). All of these samples had normal β-globin gene sequences, most likely excluding β-thalassemia as the cause of the elevated Hb A2 level. This finding concurs with a recent report that shows that mutations in the KLF1 gene are associated with elevated HBD (δ-globin) gene expression which gives rise to borderline increased Hb A2 levels.22 Significantly, a proportion of the cohort in that study had increased Hb F levels as well as increased Hb A2 levels. The proposed mechanism for KLF1 mutations increasing Hb F levels is reduced activation of BCL11A by KLF1 that in turn results in inefficient repression of γ-globin synthesis. The δ-globin gene has no KLF1 binding sites, therefore the increase in δ-globin gene expression is most likely to be due to indirect effects; probably impaired looping of the LCR with the β-globin gene that results in increased expression of the competing δ-globin gene.22 Whether a KLF1 mutation produces a HPFH phenotype or an increased Hb A2 level (or a combination of both phenotypes) will possibly depend on the balance between these two effects, which in turn will most likely depend on factors specific to a particular KLF1 mutation and other interacting factors.
The majority of our subjects with a KLF1 mutation had hypochromic red cells (MCH <27pg). However, this could mostly be explained by the co-existing presence of the extremely common 3.7kb single α-globin gene deletion (Table 1). Exceptions to this were cases 3, 5 and 1. Cases 3 and 5 had an MCH lower than normally observed with a single α-globin gene deletion. Case 1 was of White British descent and had markedly thalassemic indices but had tested negative for all types of α and β-thalassemia mutations. The KLF1 mutation (K54PfsX9) identified in this individual would be predicted to be more severe than the other mutations in that it results in the loss of all three zinc finger domains and most of exon 2. It is possible that this could result in severe impairment of the β-globin gene’s association with the LCR, resulting in a marked reduction in β-globin expression producing a β-thalassemia type phenotype. Only one mutation was found more than once (G176AfsX179): in case 10 it was associated with a Hb F level of 1.7% while in case 9, where it was found in combination with the L51R missense mutation, the Hb F level was 9.5%, suggesting that the effects of these mutations may be additive.
A previously unreported KLF1 mutation (c.914-4_914-1 del CTAG ) was identified in one of the 55 patients investigated who were homozygous for the sickle cell mutation, which suggests KLF1 mutations are not particularly common in sickle disease. However, interestingly, the Nigerian sickle cell disease patient identified with this mutation was completely asymptomatic and maintained a hemoglobin level of 12.7g/dL with a Hb F level of 20.3%. It is possible that the KLF1 mutation is ameliorating the phenotype by increasing the Hb F level via reduced γ-globin gene suppression. However, the patient also has homozygous α+-thalassemia that is known to have a complex interaction with sickle cell disease but does increase the overall hemoglobin level slightly.23 It is, therefore, likely that complex mechanisms including multiple gene interactions are involved in the maintenance of this patient’s robust hemoglobin level and asymptomatic phenotype.
In summary, KLF1 mutations were found in 8.4% of our elevated Hb F cohort, predominantly in individuals of African, Indian and Southeast Asian descent. This indicates KLF1 mutations could be a widespread cause of HPFH in malarial regions where hemogobinopathies are common, possibly making a significant contribution to Hb F variance in these populations. Also, the identification of KLF1 mutations in individuals with a thalassemia carrier phenotype and a particularly mild form of sickle cell disease indicates the effects of these mutations are likely to be heterogeneous and complex.
Footnotes
Funding: this work was supported by the NIHR Biomedical Research Centre, Oxford with funding from the Department of Health’s NIHR Biomedical Research Centre’s funding scheme. The views expressed in this publication are those of the authors and not necessarily those of the Department of Health.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Giardine B, van Baal S, Kaimakis P, Riemer C, Miller W, Samara M, et al. HbVar database of human hemoglobin variants and thalassemia mutations: 2007 update. Hum Mutat. 2007;28(2):206. doi: 10.1002/humu.9479. [DOI] [PubMed] [Google Scholar]
- 2.Henthorn PS, Smithies O, Mager DL. Molecular analysis of deletions in the human beta-globin gene cluster: deletion junctions and locations of breakpoints. Genomics. 1990;6(2):226–37. doi: 10.1016/0888-7543(90)90561-8. [DOI] [PubMed] [Google Scholar]
- 3.Ottolenghi S, Mantovani R, Nicolis S, Ronchi A, Giglioni B. DNA sequences regulating human globin gene transcription in nondeletional hereditary persistence of fetal hemoglobin. Hemoglobin. 1989;13(6):523–41. doi: 10.3109/03630268908993104. [DOI] [PubMed] [Google Scholar]
- 4.Wood WG. Increased HbF in adult life. Baillieres Clin Haematol. 1993;6(1):177–213. doi: 10.1016/s0950-3536(05)80070-8. [DOI] [PubMed] [Google Scholar]
- 5.Gilman JG, Huisman TH. DNA sequence variation associated with elevated fetal G gamma globin production. Blood. 1985;66(4):783–7. [PubMed] [Google Scholar]
- 6.Thein SL, Menzel S, Peng X, Best S, Jiang J, Close J, et al. Intergenic variants of HBS1L-MYB are responsible for a major quantitative trait locus on chromosome 6q23 influencing fetal hemoglobin levels in adults. Proc Natl Acad Sci USA. 2007;104(27):11346–51. doi: 10.1073/pnas.0611393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uda M, Galanello R, Sanna S, Lettre G, Sankaran VG, Chen W, et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc Natl Acad Sci USA. 2008;105(5):1620–5. doi: 10.1073/pnas.0711566105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thein SL, Menzel S, Lathrop M, Garner C. Control of fetal hemoglobin: new insights emerging from genomics and clinical implications. Hum Mol Genet. 2009;18(R2):R216–23. doi: 10.1093/hmg/ddp401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borg J, Papadopoulos P, Georgitsi M, Gutierrez L, Grech G, Fanis P, et al. Haploinsufficiency for the erythroid transcription factor KLF1 causes hereditary persistence of fetal hemoglobin. Nat Genet. 2010;42(9):801–5. doi: 10.1038/ng.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Satta S, Perseu L, Moi P, Asunis I, Cabriolu A, Maccioni L, et al. Compound heterozygosity for KLF1 mutations associated with remarkable increase of fetal hemoglobin and red cell protoporphyrin. Haematologica. 2011;96(5):767–70. doi: 10.3324/haematol.2010.037333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol Cell Biol. 1993;13(5):2776–86. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siatecka M, Bieker JJ. The multifunctional role of EKLF/KLF1 during erythropoiesis. Blood. 2011;118(8):2044–54. doi: 10.1182/blood-2011-03-331371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou D, Liu K, Sun CW, Pawlik KM, Townes TM. KLF1 regulates BCL11A expression and gamma- to beta-globin gene switching. Nat Genet. 2010;42(9):742–4. doi: 10.1038/ng.637. [DOI] [PubMed] [Google Scholar]
- 14.Singleton BK, Burton NM, Green C, Brady RL, Anstee DJ. Mutations in EKLF/KLF1 form the molecular basis of the rare blood group In(Lu) phenotype. Blood. 2008;112(5):2081–8. doi: 10.1182/blood-2008-03-145672. [DOI] [PubMed] [Google Scholar]
- 15.Huang XD, Yang XO, Huang RB, Zhang HY, Zhao HL, Zhao YJ, et al. A novel four base-pair deletion within the Agamma-GLOBin gene promoter associated with slight increase of Agamma expression in adult. Am J Hematol. 2000;63(1):16–9. doi: 10.1002/(sici)1096-8652(200001)63:1<16::aid-ajh4>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 16.Liu YT, Old JM, Miles K, Fisher CA, Weatherall DJ, Clegg JB. Rapid detection of alpha-thalassaemia deletions and alpha-globin gene triplication by multiplex polymerase chain reactions. Br J Haematol. 2000;108(2):295–9. doi: 10.1046/j.1365-2141.2000.01870.x. [DOI] [PubMed] [Google Scholar]
- 17.Gallienne AE, Dreau HM, McCarthy J, Timbs AT, Hampson JM, Schuh A, et al. Multiplex ligation-dependent probe amplification identification of 17 different beta-globin gene deletions (including four novel mutations) in the UK population. Hemoglobin. 2009;33(6):406–16. doi: 10.3109/03630260903344564. [DOI] [PubMed] [Google Scholar]
- 18.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30(17):3894–900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arnaud L, Saison C, Helias V, Lucien N, Steschenko D, Giarratana MC, et al. A dominant mutation in the gene encoding the erythroid transcription factor KLF1 causes a congenital dyserythropoietic anemia. Am J Hum Genet. 2010;87(5):721–7. doi: 10.1016/j.ajhg.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heruth DP, Hawkins T, Logsdon DP, Gibson MI, Sokolovsky IV, Nsumu NN, et al. Mutation in erythroid specific transcription factor KLF1 causes Hereditary Spherocytosis in the Nan hemolytic anemia mouse model. Genomics. 2010;96(5):303–7. doi: 10.1016/j.ygeno.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perseu L, Satta S, Moi P, Demartis FR, Manunza L, Sollaino MC, et al. KLF1 gene mutations cause borderline HbA2. Blood. 2011;118(16):4454–8. doi: 10.1182/blood-2011-04-345736. [DOI] [PubMed] [Google Scholar]
- 23.Ballas SK. Effect of alpha-globin genotype on the pathophysiology of sickle cell disease. Pediatr Pathol Mol Med. 2001;20(2):107–21. [PubMed] [Google Scholar]
- 24.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2012 Jan;40(Database issue):D13–25. doi: 10.1093/nar/gkr1184. Epub 2011 Dec 2. [DOI] [PMC free article] [PubMed] [Google Scholar]