Abstract
Iron-sulfur (Fe-S) clusters are ubiquitous cofactors composed of iron and inorganic sulfur. They are required for the function of proteins involved in a wide range of activities, including electron transport in respiratory chain complexes, regulatory sensing, photosynthesis and DNA repair. The proteins involved in the biogenesis of Fe-S clusters are evolutionarily conserved from bacteria to humans, and many insights into the process of Fe-S cluster biogenesis have come from studies of model organisms, including bacteria, fungi and plants. It is now clear that several rare and seemingly dissimilar human diseases are attributable to defects in the basic process of Fe-S cluster biogenesis. Although these diseases –which include Friedreich’s ataxia (FRDA), ISCU myopathy, a rare form of sideroblastic anemia, an encephalomyopathy caused by dysfunction of respiratory chain complex I and multiple mitochondrial dysfunctions syndrome – affect different tissues, a feature common to many of them is that mitochondrial iron overload develops as a secondary consequence of a defect in Fe-S cluster biogenesis. This Commentary outlines the basic steps of Fe-S cluster biogenesis as they have been defined in model organisms. In addition, it draws attention to refinements of the process that might be specific to the subcellular compartmentalization of Fe-S cluster biogenesis proteins in some eukaryotes, including mammals. Finally, it outlines several important unresolved questions in the field that, once addressed, should offer important clues into how mitochondrial iron homeostasis is regulated, and how dysfunction in Fe-S cluster biogenesis can contribute to disease.
Introduction
Iron-sulfur (Fe-S) clusters are important prosthetic groups with unusual chemical properties that enable the proteins that contain them (Fe-S proteins) to function in pathways ranging from metabolism to DNA repair. They are evolutionarily ancient and are present in essentially all organisms, including Archaea, bacteria, plants and animals. The high level of evolutionary conservation is consistent with the possibility that Fe-S clusters contributed to the success of early life forms. In the anaerobic atmosphere of ancient earth, Fe-S inorganic metal compounds were probably already present in hydrothermal vents (Martin et al., 2008). Evolving primitive organisms might have used these versatile Fe-S compounds and incorporated them into early metabolic pathways; in fact, Fe-S cluster activities might have contributed to the origin of life itself. Once primitive organisms evolved, they could use the reducing power of their metabolic pathways to generate organic molecules and to spread to less protected environments (Russell and Martin, 2004). However, as plants oxygenated the earth’s atmosphere, Fe-S clusters were no longer as readily assembled or as stable owing to the oxidation of iron and of Fe-S clusters themselves (Imlay, 2006).
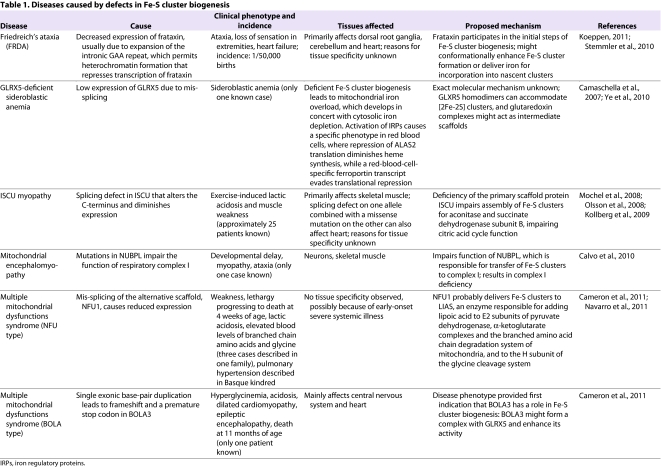
Despite their inherent vulnerability to oxidation and degradation, Fe-S clusters are crucial for facilitating enzyme activities in all kingdoms of life because they can bind electron-rich enzymatic substrates, accept or donate single electrons and stabilize specific protein conformations that are important to the activities of numerous proteins. Their importance for normal mammalian physiology is illustrated by the fact that mutations in proteins involved in Fe-S cluster biogenesis cause at least five distinctive human diseases (Table 1), including Friedreich’s ataxia (FRDA), ISCU myopathy and a specific sideroblastic anemia, all of which seem to be caused by mutations in proteins in the main Fe-S cluster biogenesis pathway (reviewed by Ye and Rouault, 2010). In addition, mutations in NUBPL (Calvo et al., 2010), also known as IND1, a protein involved in transferring Fe-S clusters to complex I (Bych et al., 2008; Sheftel et al., 2009), were recently discovered to cause deficiency of respiratory chain complex I, resulting in mitochondrial encephalomyopathy. Finally, two forms of a disease called multiple mitochondrial dysfunctions syndrome, caused by mutations in either NFU1 or BOLA3, indicate that some Fe-S proteins might acquire their clusters from a specific and essential subset of donor proteins (Cameron et al., 2011; Navarro-Sastre et al., 2011). Careful phenotyping of humans with problems caused by defects in Fe-S cluster biogenesis should help to clarify aspects of the pathway, particularly regarding how target Fe-S proteins are identified, that have remained elusive in studies of bacteria and yeast model systems. Determining why these diseases affect some tissues, but not others, is an important challenge that will probably be resolved by detailed studies of molecular pathophysiology.
Table 1.
Diseases caused by defects in Fe-S cluster biogenesis
Insights into the pathophysiology of these diseases depends on gaining a better understanding of Fe-S cluster biogenesis and its regulation in eukaryotic cells, particularly in the mitochondria, which must synthesize Fe-S clusters for incorporation into the citric acid cycle enzymes aconitase and succinate dehydrogenase, and into respiratory chain complexes I–III. Many studies have been performed to investigate Fe-S cluster biogenesis in the model organism Saccharomyces cerevisiae; however, there are apparently differences between aspects of mammalian and yeast Fe-S cluster biogenesis, including multicellularity, the role of frataxin (an important component of the initial Fe-S biogenesis complex; see below) (Bridwell-Rabb et al., 2011; Schmucker et al., 2011) and differences in the cytosolic Fe-S cluster biogenesis machinery. Notably, in mammalian cells, accumulating evidence suggests that several key Fe-S cluster biogenesis proteins are present not only in mitochondria but also in cytosolic and/or nuclear compartments, whereas their yeast counterparts are alleged to reside only in mitochondria [with the exception of cysteine desulfurase (Biederbick et al., 2006)]. Thus, there is a need to clarify the situation in the mammalian system to obtain a better understanding of the role of the Fe-S synthetic machinery in normal and diseased physiology. This Commentary focuses on how advances in understanding the steps of basic Fe-S cluster biogenesis can enhance our understanding of the pathogenesis of a relatively new class of human diseases. In addition, it highlights important features of the compartmentalization of the Fe-S biogenesis machinery in mammalian cells, which seem to differ from those of the lower eukaryote S. cerevisiae.
Basic biochemistry and function of Fe-S clusters
Fe-S clusters are cofactors that are generally ligated to cysteine residues of proteins, where they facilitate numerous types of reactions. Composed of iron and inorganic sulfur, they are most frequently found in a cubane form that contains four iron and four inorganic sulfur atoms (Meyer, 2008). Fe-S clusters are highly chemically versatile because both iron and sulfur can readily donate or accept multiple electrons (Beinert, 2000), and these chemical features can synergize so that the affinity of an Fe-S cluster for electrons can be fine-tuned across an extremely broad electrochemical range by its surrounding protein residues. For example, in mitochondrial complex I, seven Fe-S clusters with gradually increasing reduction potentials align to form a wire-like pathway along which electrons ascend. Thus, the ability of Fe-S clusters to maintain low reduction potentials (i.e. low affinity for electrons) facilitates efficient capture of chemical energy from NADH as electrons move progressively through respiratory chain complexes (Hirst, 2010). Fe-S clusters are versatile in other ways, because they can directly facilitate chemical reactions by binding to an Fe-S protein’s substrate, as in the enzyme aconitase, which binds and interconverts citrate and isocitrate in the citric acid cycle. In addition, Fe-S proteins have been found to function as sensors, as in the bacterial FNR and IscR proteins (Kiley and Beinert, 2003), and in mammalian IRP1, which regulates cytosolic iron metabolism in mammalian cells (Rouault, 2006; Muckenthaler et al., 2008).
Fe-S cluster biogenesis: the basic pathway
The highly conserved general Fe-S cluster biogenesis pathway has been the subject of intense study in numerous species of bacteria, plants, yeast and mammals since it was first described in bacteria (reviewed by Frazzon and Dean, 2003). Many of the general steps of the pathway are common to all kingdoms of life, but it seems that the situation is more complicated in eukaryotes, in which Fe-S proteins are functional and necessary in multiple subcellular compartments, including mitochondria, plastids, cytosol and nucleus.
The initial stage of Fe-S cluster biogenesis is accomplished by a multimeric protein complex in which a dimer of a cysteine desulfurase [called IscS (Escherichia coli), Nfs1 (S. cerevisiae) or NFS1 (mammals)] forms a core to which two monomers of a dedicated scaffold protein (IscU in bacteria, Isu1 or Isu2 in yeast and ISCU in humans) bind at either end (Shi et al., 2010) (Fig. 1). Aided by its cofactor, pyridoxal phosphate, NFS1 supplies sulfur by removing it from cysteine residues, and ISCU provides the backbone structure and the cysteine ligands upon which a new cluster consisting of covalently bound iron and inorganic sulfur is synthesized (Raulfs et al., 2008; Py and Barras, 2010; Bandyopadhyay et al., 2008; Lill, 2009). The source of the iron in the nascent Fe-S cluster has not been clearly identified, but suggested sources include iron that is bound to acidic patches on frataxin (see below) (Stemmler et al., 2010), or iron that is donated from a complex of glutathione and glutaredoxin that tethers an Fe-S cluster (Qi and Cowan, 2011). In eukaryotes, the stability of the cysteine desulfurase depends on its binding to a small partner protein called ISD11 (Wiedemann et al., 2006) (Adam et al., 2006), which is found in the mitochondrial matrix of S. cerevisiae, but has been detected in both the mitochondrial matrix and the cytosolic and/or nuclear compartments of mammalian cells (Shi et al., 2009). ISD11 apparently became an indispensable binding partner for the eukaryotic cysteine desulfurase early in the evolution of eukaryotes (Richards and van der Giezen, 2006). The discovery that the ISD11 protein is present in the mammalian nucleus supported previous reports that human NFS1 (Land and Rouault, 1998), ISCU (Tong and Rouault, 2000) and an alternative scaffold protein called NFU1 (Tong et al., 2003) are present and active in the cytosolic and/or nuclear compartments of mammalian cells (Tong and Rouault, 2006).
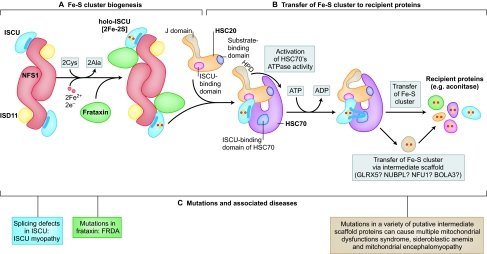
Fig. 1.
A general scheme for biogenesis of Fe-S clusters in mammalian cells. (A) NFS1 is a cysteine desulfurase that forms a dimer to which monomers of the primary scaffold protein ISCU bind near the top and bottom of the complex. In eukaryotes, ISD11 is an obligate binding partner for NFS1. NFS1 also binds the cofactor pyridoxal phosphate (not shown). Structural and biochemical studies suggest that frataxin forms part of the initial Fe-S cluster biogenesis complex, potentially occupying a pocket between NFS1 and ISCU. NFS1 donates inorganic sulfur, and cysteines from ISCU provide the sulfur ligands that directly bind iron in the nascent Fe-S cluster. A highly reduced protein such as ferredoxin probably provides needed electrons. (B) Once the Fe-S cluster is assembled, it must be transferred to recipient proteins. Work in bacteria and yeast model systems suggests that a dedicated chaperone–co-chaperone pair of proteins participates in cluster transfer from the primary scaffold, ISCU, to recipient Fe-S proteins. The co-chaperone is known to be HSC20 (a DNAJ protein), whereas the chaperone is an HSP70 homolog that has not yet been clearly identified in mammalian cells. The role of a putative HSC70 protein is proposed here. HSC20 binds ISCU, and the HSC20-ISCU complex probably then binds to its HSC70 partner through two different binding sites: HSC20 contacts the N-terminus of HSC70 and its binding partner, ISCU, binds to the C-terminal substrate-binding domain region of HSC70. The J domain region of HSC20 contains three residues [His (H) Pro (P) and Asp (D); HPD] that activate the ATPase activity of HSC70. Upon activation, a conformational change is proposed to occur in the substrate-binding domain of HSC70 that affects bound ISCU, resulting in extrusion of a peptide containing the residues LPPVK from the ISCU globular protein. The LPPVK peptide then binds to a groove in the substrate-binding domain of HSC70, which consolidates or perhaps further enhances the conformational change in ISCU, which might convert it to a conformation that facilitates donation of its cluster to recipient proteins. In this model, HSC20 helps protect the vulnerable Fe-S cluster bound to ISCU as it dissociates from the multimeric assembly complex, and HSC20 then escorts ISCU to form a trimeric complex with HSC70. The consumption of ATP probably provides a powerful impetus to drive conformational changes of ISCU and the substrate-binding domain of HSC70; these changes might facilitate release of the Fe-S cluster from ISCU. By capturing the energy released by ATP hydrolysis and coupling it to conformational changes, the chaperone–co-chaperone pair help the Fe-S cluster to reach its target proteins. Target proteins could include some direct targets, or proteins such as NFU1, BOLA3, NUBPL or GLRX5 that might function as intermediary scaffolds that then donate Fe-S clusters to specific subsets of recipient proteins. (C) Mutations in proteins acting at different points in the biogenesis pathway cause diseases with markedly different phenotypes (described in Table 1).
Structural modeling suggests that the protein frataxin (Yfh1 in yeast) then binds in a pocket-like region between NFS1 and ISCU (Fig. 1), where it might either repress, stabilize or enhance activity of the Fe-S cluster biogenesis core complex (Prischi et al., 2010; Schmucker et al., 2011; Tsai and Barondeau, 2010). Structural data demonstrating the inclusion of frataxin in the initial multimeric complex detracts somewhat from previous hypotheses that the role of frataxin was to deliver bound iron to the complex (Cook et al., 2010). Rather, it now seems that frataxin binding to the NFS1-ISCU complex drives a useful conformational change that enhances initial cluster formation (Bridwell-Rabb et al., 2011).
Assembly of the nascent clusters from the iron and sulfur building blocks available in cells relies on a source of electrons to achieve the electronic configurations that are observed in Fe-S clusters. Candidates for the provision of electrons to nascent Fe-S clusters include glutaredoxin 5 (GLRX5) (Lillig et al., 2008) and ferredoxin (Ewen et al., 2011; Shi et al., 2011); ferredoxin has been shown to facilitate reductive coupling of two distinct [2Fe-2S] clusters into a single [4Fe-4S] cluster on bacterial IscU in vitro (Chandramouli et al., 2007). Glutathione, which reduces glutaredoxins, might also serve as a previously unrecognized iron source (Hider and Kong, 2011). Notably, a mutation in human mitochondrial GLRX5 causes sideroblastic anemia and impairs Fe-S cluster biogenesis, but the function of GLRX5 is still not clear. GLRX5 can dimerize and ligate a single bridging Fe-S cluster in vitro, using a cysteine from each GLRX5 monomer and two glutathiones as the ligands of a [2Fe-2S] cluster (Ye et al., 2010).
After an Fe-S cluster is formed by the core complex (composed of NFS1, ISD11, frataxin and ISCU in mammalian cells) (Tsai and Barondeau, 2010; Schmucker et al., 2011; Bridwell-Rabb et al., 2011), the Fe-S cluster must be transferred to recipient proteins. Although many details of this transfer process remain undefined, much evidence indicates that a highly conserved chaperone–co-chaperone system participates in the next steps. A co-chaperone known as HSC20 in mammalian cells (Uhrigshardt et al., 2010), HscB in bacteria (Vickery and Cupp-Vickery, 2007) and Jac1 in S. cerevisiae (Craig and Marszalek, 2002) binds to ISCU. In bacteria, HscB has been shown to form a complex with IscU and a co-chaperone partner, HscA (Ssq1 in S. cerevisiae), a member of the HSP70 heat shock protein family. The mammalian HSP70 homolog with this function has not yet been identified, so references to mammalian HSC70 below and in Fig. 1 are speculative and based on experimental data from bacteria and yeast. HSP70 homologs use the energy released by hydrolysis of ATP to drive conformational changes and the refolding of target proteins. Recent studies indicate that bacterial HscB binds to IscU using highly conserved residues in a hydrophobic patch in the C-terminal portion of HscB (Fuzery et al., 2011). Together, HscA (HSC70) and HscB (HSC20) might facilitate Fe-S cluster transfer from ISCU to target apoproteins or to other secondary scaffolds by facilitating dissociation from the initial scaffold protein complex (Vickery and Cupp-Vickery, 2007; Bonomi et al., 2011) (Fig. 1). It remains unclear how Fe-S cluster target proteins are identified, because the process seems to be selective and might depend on specific interactions between apoprotein targets and binding surfaces of the chaperone–co-chaperone complex, particularly along the extensive potential interaction area provided by the unusually large and distinctive C-terminus of HSC20 (Kampinga and Craig, 2010).
Because Fe-S clusters are notoriously vulnerable to oxidative stress, the biogenesis system might have evolved to protect and enshroud Fe-S clusters during most of the biogenesis and transfer process. Interestingly, mammalian HSC20 contains a cysteine-rich N-terminus that binds zinc in the crystal structure (Bitto et al., 2008), which could potentially bind an Fe-S cluster (Uhrigshardt et al., 2010). On the basis of studies in bacterial systems, it is likely that binding of human HSC20 protects the nascent cluster on ISCU, and leads ISCU into a new multisubunit complex that contains the HSP70 homolog along with HSC20 and ISCU. Using the energy derived from ATPase activity of the HSP70 homolog, a conformational change in this complex might facilitate protected transfer of the Fe-S cluster from ISCU directly to recipient apoproteins or to secondary scaffold proteins that then facilitate delivery of the Fe-S cluster to a defined group of recipient proteins. Target apoproteins are likely to be tethered to the chaperone–co-chaperone complex when the Fe-S cluster of ISCU is released so that cluster release can be coupled to Fe-S acquisition by apoproteins.
Defects in Fe-S cluster biogenesis as causes of human disease
As discussed, the Fe-S cluster biogenesis pathway is fundamental to a variety of cellular processes: Fe-S proteins are required for the function of two proteins of the citric acid cycle, succinate dehydrogenase and aconitase; for respiratory chain complexes I–III; and for numerous other proteins in the mitochondria, cytosol and nucleus of mammalian cells. Thus, it is not surprising that disruption of Fe-S cluster biogenesis causes human disease. However, defects in different parts of the pathway cause very different disease phenotypes (Fig. 1, Table 1). For example, markedly diminished expression of frataxin causes FRDA, which is characterized by cardiac failure and the death of specific neuronal cell types, including the dorsal root ganglia, which are responsible for sensory perception and maintenance of balance [see Table 1 and the review by Martelli et al. in this issue of Disease Models & Mechanisms (Martelli et al., 2012)]. By contrast, ISCU myopathy, a disease resulting from a splicing mutation of ISCU, causes pathology almost exclusively in skeletal muscles, and cardiac involvement is rare, developing only when there is a specific inactivating mutation on one allele (Kollberg et al., 2009). Thus far, the reasons why these two diseases affect specific tissues are unclear. Tissue-specific factors might exacerbate the loss of protein function in affected tissues; indeed, it has been suggested that tissue-specific splicing factors drive abnormal splicing of ISCU in the skeletal muscles of individuals with ISCU myopathy (Nordin et al., 2011). It seems likely that the cells in affected tissues in FRDA express less frataxin than unaffected tissues, and that the Fe-S cluster biogenesis pathway is therefore more compromised at those sites. However, this observation has not been closely examined, possibly because of challenges associated with obtaining tissues from patients or autopsies. Because there has only been a single patient with sideroblastic anemia caused by GLRX5 mutation, it is not possible to verify proposed causes of tissue specificity for this disease (Table 1).
Studies of three newly described diseases caused by mutations in NUBPL, NFU1 or BOLA3 suggest that transfer of Fe-S clusters to specific target proteins downstream of the ISCU chaperone–co-chaperone complex depends on specific pathways that shepherd Fe-S clusters to subsets of recipient proteins (also discussed below). For instance, mutations in the complex-I-dedicated chaperone protein NUBPL have been reported to cause mitochondrial encephalomyopathy, a severe multisystem infantile disease attributable to loss of complex I function. By contrast, mutations in NFU1 or BOLA3 interfere quite specifically with activity of an Fe-S enzyme, lipoate synthase, which provides lipoic acid modifications to a small group of specific enzymes, including the E2 subunits of pyruvate dehydrogenase and α-ketoglutarate dehydrogenase, and to enzymes involved in branched chain amino acid metabolism and glycine degradation (Cameron et al., 2011; Navarro-Sastre et al., 2011). The small number of individuals with NFU1 or BOLA3 mutations died within the first year of life, and their main symptoms of lactic acidosis and failure to thrive have thus far not been reported to be highly tissue specific (see Table 1). More patients must be characterized to clearly define the features of these diseases.
Subcellular distribution of Fe-S cluster biogenesis proteins in eukaryotic cells
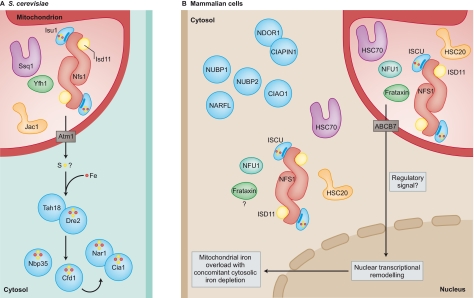
To understand the diseases that result from defects in the Fe-S cluster biogenesis machinery, it is important to fully understand how this process occurs in mammalian cells. Insights into mammalian Fe-S cluster biogenesis have been confounded by the fact that a dominating paradigm has been developed in the model system S. cerevisiae, in which it was proposed that Fe-S cluster biogenesis occurred or was initiated solely in the mitochondria, but not cytosolic or nuclear compartments (Lill, 2009). Specifically, it was proposed that the mitochondrial exporter Atm1 exported either fully formed Fe-S clusters or a special form of sulfur that was required for cytosolic Fe-S cluster biogenesis (Lill, 2009), and that loss of Atm1 transporter activity led to the accumulation of iron within the mitochondrial matrix. In S. cerevisiae, export of a ‘sulfur compound’ from mitochondria has been proposed to contribute to cytosolic Fe-S cluster biogenesis by proteins known as Tah18 and Dre2 (Netz et al., 2010). It was proposed that a cluster is transferred from Dre2 to other members of the cytosolic Fe-S cluster assembly (CIA) proteins (Sharma et al., 2010). There are mammalian homologs of yeast CIA proteins, including NDOR1 and CIAPIN1 (homologs of Tah18 and Dre2, respectively), NUBP1 and NUBP2 [homologs of yeast Nbp35 and Cfd1, respectively (Stehling et al., 2008)], NARFL (homolog of yeast Nar1) and CIAO1 (homolog of yeast Cia1) (Srinivasan et al., 2007). However, the model purporting that cytosolic Fe-S cluster biogenesis depends on export of a mitochondrial product was recently called into question because genetic manipulations that were initially used to inactivate Atm1 also introduced wild-type Leu2 into a Leu2-null strain, which unknowingly resulted in repressed transcription of Leu1 relative to the control strain. Leu1 encodes a cytosolic Fe-S protein that was being used as a readout to quantify cytosolic Fe-S cluster biogenesis. Because Leu1 activity was assessed as being low in Atm1-null strains, this was misinterpreted as evidence that the mitochondrial exporter, Atm1, contributed an essential component to cytosolic Fe-S cluster biogenesis. Recently, when the experiment was performed with appropriate controls, there was no evidence that loss of Atm1 adversely affected Leu1 activity (Bedekovics et al., 2011). Thus, the model that Fe-S clusters are assembled solely in mitochondria of yeast cells lacks crucial experimental support, and some models based on work in S. cerevisiae must be reconsidered.
In any case, there is now substantial evidence that the paradigm that Fe-S clusters are assembled or initiated solely in mitochondria is not true in mammalian cells, an issue that must be clarified to allow insights into disease pathophysiology. It is well accepted that the basic Fe-S cluster biosynthetic proteins are highly expressed in mammalian mitochondria, where they supply Fe-S clusters to proteins of the citric acid cycle, the respiratory chain complexes and other pathways (described above). Less appreciated and somewhat controversial, however, is the idea that some of these proteins are also present in the cytosolic and/or nuclear compartments of mammalian cells (Fig. 2). It has been reported that small amounts of the cysteine desulfurase NFS1 and its binding partner ISD11 are expressed in the cytosolic and/or nuclear compartments of mammalian cells (Land and Rouault, 1998; Shi et al., 2009), where small amounts of the scaffold proteins ISCU and NFU1 are also present (Tong et al., 2003; Tong and Rouault, 2006). The observed extra-mitochondrial localization of these proteins is consistent with the fact that there are nuclear localization signals in mammalian NFS1 (Land and Rouault, 1998) and ISD11 (Shi et al., 2009), as well as in Nfs1 of S. cerevisiae (Nakai et al., 2001); however, S. cerevisiae Nfs1 is not believed to participate in extra-mitochondrial Fe-S cluster biogenesis (Biederbick et al., 2006). The extra-mitochondrial isoforms of mammalian ISCU and NFU1 are encoded by alternatively spliced transcripts that lack a recognizable mitochondrial targeting signal (Tong et al., 2003; Tong and Rouault, 2006), and there is functional evidence that the cytosolic ISCU functions as a scaffold and source for extra-mitochondrial Fe-S clusters in mammalian cells (Tong and Rouault, 2006). Moreover, the co-chaperone HSC20 has been detected in cytosolic fractions (Uhrigshardt et al., 2010). However, because proteins are synthesized in the cytosol, it is possible that some of the Fe-S cluster biogenesis proteins found in this compartment represent proteins that were inefficiently imported into and processed by mitochondria. Notably, there is almost always much more of each protein in the mitochondrial matrix than in the extra-mitochondrial fractions.
Fig. 2.
Proposed differences in the localization of Fe-S cluster biogenesis proteins in S. cerevisiae versus mammalian cells. (A) Yeast mitochondria contain the basic Fe-S cluster biogenesis proteins, whereas cytosolic Fe-S cluster biogenesis has been proposed to depend on the export of a sulfur compound from mitochondria through the transporter Atm1 (ABCB7 in human cells). Further biogenesis in the cytoplasm depends on a distinct set of proteins, including Tah18, Dre2, Nbp35, Cfd1, Nar1 and Cia1, which are collectively referred to as CIA (cytosolic iron-sulfur assembly) proteins. (B) By contrast, in mammalian cells, it seems that most of the basic Fe-S cluster biogenesis proteins are expressed in mitochondria as well as in the cytosolic and/or nuclear compartment (nuclear localization of these proteins not represented in the figure). In addition, there are human counterparts to the proteins implicated in cytosolic Fe-S cluster biogenesis in yeast, including NDOR1 (homolog of Tah18), CIAPIN1, (homolog of Dre2), NUBP1 and NUBP2 (homologs of Nbp35 and Cfd1, respectively), and NARFL and CIAO1 (homologs of Nar1 and Cia1, respectively). To explain mitochondrial iron overload (which occurs as a consequence of disrupted Fe-S cluster biogenesis), it is possible that mitochondria depend on Fe-S cluster biogenesis to synthesize a molecule that functions as a regulatory signal. In the absence of this signal (i.e. when Fe-S cluster biogenesis is impaired), remodeling of nuclear transcription leads to mitochondrial iron overload, as the cell attempts to compensate for possible mitochondrial iron deficiency. Because mitochondrial iron overload probably contributes to disease pathogenesis in FRDA, ISCU myopathy and possibly several other diseases, elucidation of the details of this pathway might pave the way to new therapeutic interventions.
Understanding the pathway by which Fe-S clusters could be assembled outside the mitochondrial compartment is becoming more important as the list of recognized cytosolic and nuclear Fe-S proteins grows. Indeed, there might be many as-yet-undiscovered Fe-S proteins in the mitochondrial, cytosolic and nuclear compartments of mammalian cells. However, Fe-S proteins lack a distinctive motif that permits easy recognition, and they tend to lose their sensitive prosthetic groups during purification. Importantly, a significant number of DNA repair enzymes are Fe-S proteins, including the protein responsible for excision-repair of UV damage that causes the disease xeroderma pigmentosum (XPD) when defective, and a related helicase that causes Fanconi anemia when defective (Rudolf et al., 2006). In addition, telomere maintenance depends on the nuclear Fe-S protein RTEL-1 (Uringa et al., 2011), and eukaryotic DNA polymerases depend on Fe-S clusters for function (Netz et al., 2011). Some of the cytosolic Fe-S ‘assembly’ proteins previously identified in yeast might function in mammalian cells as secondary scaffold proteins that transfer clusters to specific classes of protein targets such as DNA repair enzymes. It is well known that fully formed Fe-S clusters can readily move from one protein to another; this mobility of transfer between proteins is one of the most distinctive features of Fe-S clusters (Beinert et al., 1997). Thus, it is perhaps not surprising that mutations of NUBP1 and NUBP2 impair cell division (Christodoulou et al., 2006), because these proteins are implicated in cytosolic Fe-S cluster biogenesis and some of their likely targets are involved in chromosomal duplication and in maintenance of nuclear integrity, processes that are necessary for proper cell division (Netz et al., 2010).
In summary, although the scenario in yeast remains in dispute, the accumulating evidence that Fe-S cluster biogenesis can occur de novo in the mitochondrial matrix as well as in extra-mitochondrial compartments of mammalian cells is strong and consistent. It is crucial to correctly analyze the basics of the mammalian Fe-S cluster biogenesis pathway, because it sets the stage for understanding one of the most common complications of impaired Fe-S cluster biogenesis: the propensity to develop marked mitochondrial iron overload and subsequent iron-related mitochondrial damage.
A unifying hypothesis to integrate defective Fe-S cluster biogenesis with mitochondrial iron overload
Evidence suggests that interruption of mitochondrial Fe-S cluster biogenesis leads to mitochondrial iron overload and cytosolic iron depletion in mammalian cells, which is associated with disease pathology. For example, mitochondrial iron overload is a distinctive feature of the heart and some neurons of FRDA patients (Koeppen, 2011), of the skeletal muscle cells of individuals with ISCU myopathy (Mochel et al., 2008), and of the immature red blood cells of a patient with sideroblastic anemia attributable to deficiency of GLRX5 (Ye et al., 2010). In yeast, mutations of frataxin (Babcock et al., 1997), of the chaperone Ssq1 (HSP70 homolog) (Knight et al., 1998) or the J-domain co-chaperone Jac1 (HSC20 homolog) (Kim et al., 2001) compromise the biogenesis of Fe-S clusters, and also cause mitochondrial iron overload. In mammalian cells, mitochondrial iron overload has been observed to occur in concert with cytosolic iron depletion, as judged by activities of cytosolic iron regulatory proteins (Li et al., 2008; Shi et al., 2009). In both yeast and human cells, this mitochondrial iron overload can be readily reversed by restoration of the missing Fe-S cluster biogenesis protein (Babcock et al., 1997; Ye et al., 2010).
Why does defective Fe-S cluster biogenesis cause mitochondrial iron overload? One early hypothesis referred to above was that iron was able to leave mitochondria only when it had been incorporated into an Fe-S cluster (Kispal et al., 1999). Subsequently, this hypothesis was modified to suggest that ABCB7 (the mammalian homolog of yeast Atm1) exports a special type of sulfur, but not iron, from mitochondria (Lill, 2009; Netz et al., 2010). However, the revised theory does not offer an explanation for why iron accumulates in mitochondria: the idea that iron accumulates in mitochondria passively because it cannot be incorporated into Fe-S clusters does not take into account the possibility that mitochondrial iron metabolism is a highly regulated process that might be imbalanced in cells with defective Fe-S cluster biogenesis. In support of this idea, transcriptional expression of the mitochondrial iron importer mitoferrin is increased in frataxin-deficient mouse hearts (Huang et al., 2009). Remodeling transcription of this nuclear gene might represent a response to signals of iron deficiency from mitochondria. How might mitochondria generate a signal that could instruct the nucleus to upregulate the expression of mitoferrin? One possibility is that mitochondria export a product that is made only when the Fe-S cluster biogenesis pathway is intact, and absence of this signal triggers a coordinated nuclear transcriptional response.
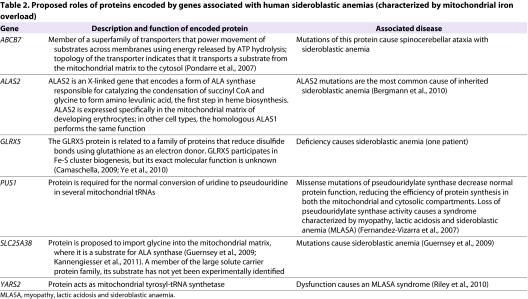
Mitochondrial iron overload also occurs in individuals with X-linked sideroblastic anemia and ataxia (Allikmets et al., 1999) caused by mutations in the ABCB7 mitochondrial iron transporter, as well as in individuals with sideroblastic anemia caused by mutations in the red-blood-cell-specific amino-levulinate synthase ALAS2, a mitochondrial protein that is involved in heme biosynthesis (Table 2). It is possible that both ALAS2 and ABCB7 participate in a signaling pathway that regulates mitochondrial iron homeostasis, and, furthermore, that ABCB7 transports a crucial regulatory molecule related to the product of ALAS2. Because the ABCB7 substrate (or that of its yeast homolog Atm1) (Kuhnke et al., 2006) has not yet been identified, it might be time to think more broadly about the regulation of mitochondrial iron homeostasis and to integrate the phenotypes observed upon mutation of ALAS2 and ABCB7 into a single overarching model for mitochondrial iron regulation.
Table 2.
Proposed roles of proteins encoded by genes associated with human sideroblastic anemias (characterized by mitochondrial iron overload)
For instance, it will be important to determine whether mitochondrial iron overload occurs in cells from individuals with multiple mitochondrial dysfunctions syndrome caused by mutations in NFU1 or BOLA3 (Cameron et al., 2011) (Table 1). NFU1 probably acts as a scaffold downstream of ISCU during Fe-S cluster biogenesis (Navarro-Sastre et al., 2011), whereas BOLA3 is related to proteins that bind glutaredoxins, which play an unknown role in Fe-S cluster biogenesis. Mutations in either NFU1 or BOLA3 disrupt the function of lipoic acid synthetase (LIAS), which depends on a [4Fe-4S] cluster for activity. LIAS adds a lipoate moiety to key subunits of pyruvate dehydrogenase and several other enzymes (Table 1). The fact that a specific subset of Fe-S proteins is affected in these newly described diseases suggests that target specificity is achieved through use of specific scaffold proteins that function as intermediates between ISCU and target proteins. Because loss of GLRX5 causes mitochondrial iron overload, and GLRX5 might function as a BOLA3 partner (Huynen et al., 2005), it seems likely that the mitochondrial iron overload phenotype will also be observed in this newly described disease.
Mitochondrial iron overload develops in several other diseases that do not involve defects in Fe-S cluster biogenesis. These diseases include those caused by mutations in an enzyme known as PUS1, which is expressed in both mitochondria and the nucleus and is required for pseudouridylate processing on transfer RNAs (tRNAs; see Table 2), and by mutations in YARS2, a gene that encodes a mitochondrial tyrosine tRNA synthetase. These results imply that a molecule produced by the mitochondrial protein synthesis apparatus also contributes to regulation of mitochondrial iron homeostasis.
Perspective
The list of diseases attributable to defects in Fe-S cluster biogenesis is growing. In the human genome, many of the genes encoding Fe-S cluster biogenesis proteins, including HSC20 and ISCU, are represented only once. The lack of duplication of these essential genes might enhance the likelihood that deleterious phenotypes will arise from mutations that affect Fe-S cluster biogenesis proteins. Notably, mutations in NFU1, BOLA3 and NUBPL cause severe disease and mortality in early infancy, and there is no evidence of significant tissue specificity. Identification of new disease genes in humans should accelerate progress in understanding Fe-S cluster biogenesis, and should shed light on how specific target proteins are identified. Better characterization of several newly recognized diseases offers the prospect of making progress on details of the second part of the Fe-S biogenesis pathway, particularly downstream of the chaperone–co-chaperone proteins, when Fe-S clusters are ligated to specific recipient apoproteins. Further work will help to clarify how mitochondrial iron homeostasis is regulated, and learning what drives mitochondrial iron overload should yield better therapeutic approaches for diseases in which this feature contributes to pathology.
Footnotes
COMPETING INTERESTS
The authors declare that they do not have any financial or competing interests.
FUNDING
This research was supported by the intramural program of the National Institute of Child Health and Human Development.
REFERENCES
- Adam A. C., Bornhovd C., Prokisch H., Neupert W., Hell K. (2006). The Nfs1 interacting protein Isd11 has an essential role in Fe/S cluster biogenesis in mitochondria. EMBO J. 25, 174–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allikmets R., Raskind W. H., Hutchinson A., Schueck N. D., Dean M., Koeller D. M. (1999). Mutation of a putative mitochondrial iron transporter gene (ABC7) in X-linked sideroblastic anemia and ataxia (XLSA/A). Hum. Mol. Genet. 8, 743–749 [DOI] [PubMed] [Google Scholar]
- Babcock M., de Silva D., Oaks R., Davis-Kaplan S., Jiralerspong S., Montermini L., Pandolfo M., Kaplan J. (1997). Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science 276, 1709–1712 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S., Chandramouli K., Johnson M. K. (2008). Iron-sulfur cluster biosynthesis. Biochem. Soc. Trans. 36, 1112–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedekovics T., Li H., Gajdos G. B., Isaya G. (2011). Leucine biosynthesis regulates cytoplasmic iron-sulfur enzyme biogenesis in an Atm1-independent manner. J. Biol. Chem. 286, 40878–40888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinert H. (2000). Iron-sulfur proteins: ancient structures, still full of surprises. J. Biol. Inorg. Chem. 5, 2–15 [DOI] [PubMed] [Google Scholar]
- Beinert H., Holm R. H., Munck E. (1997). Iron-sulfur clusters: nature’s modular, multipurpose structures. Science 277, 653–659 [DOI] [PubMed] [Google Scholar]
- Bergmann A. K., Campagna D. R., McLoughlin E. M., Agarwal S., Fleming M. D., Bottomley S. S., Neufeld E. J. (2010). Systematic molecular genetic analysis of congenital sideroblastic anemia: evidence for genetic heterogeneity and identification of novel mutations. Pediatr. Blood Cancer 54, 273–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederbick A., Stehling O., Rosser R., Niggemeyer B., Nakai Y., Elsasser H. P., Lill R. (2006). Role of human mitochondrial Nfs1 in cytosolic iron-sulfur protein biogenesis and iron regulation. Mol. Cell. Biol. 26, 5675–5687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitto E., Bingman C. A., Bittova L., Kondrashov D. A., Bannen R. M., Fox B. G., Markley J. L., Phillips G. N. J. (2008). Structure of human J-type co-chaperone HscB reveals a tetracysteine metal-binding domain. J. Biol. Chem. 283, 30184–30192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomi F., Iametti S., Morleo A., Ta D., Vickery L. E. (2011). Facilitated transfer of IscU-[2Fe2S] clusters by chaperone-mediated ligand exchange. Biochemistry 50, 9641–9650 [DOI] [PubMed] [Google Scholar]
- Bridwell-Rabb J., Winn A. M., Barondeau D. P. (2011). Structure-function analysis of Friedreich’s ataxia mutants reveals determinants for frataxin binding and activation of the Fe-S assembly complex. Biochemistry 50, 7265–7274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bych K., Kerscher S., Netz D. J., Pierik A. J., Zwicker K., Huynen M. A., Lill R., Brandt U., Balk J. (2008). The iron-sulphur protein Ind1 is required for effective complex I assembly. EMBO J. 27, 1736–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo S. E., Tucker E. J., Compton A. G., Kirby D. M., Crawford G., Burtt N. P., Rivas M., Guiducci C., Bruno D. L., Goldberger O. A., et al. (2010). High-throughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency. Nat. Genet. 42, 851–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camaschella C., Campanella A., De Falco L., Boschetto L., Merlini R., Silvestri L., Levi S., Iolascon A. (2007). The human counterpart of zebrafish shiraz shows sideroblastic-like microcytic anemia and iron overload. Blood 110, 1353–1358 [DOI] [PubMed] [Google Scholar]
- Cameron J. M., Janer A., Levandovskiy V., Mackay N., Rouault T. A., Tong W. H., Ogilvie I., Shoubridge E. A., Robinson B. H. (2011). Mutations in iron-sulfur cluster scaffold genes NFU1 and BOLA3 cause a fatal deficiency of multiple respiratory chain and 2-oxoacid dehydrogenase enzymes. Am. J. Hum. Genet. 89, 486–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramouli K., Unciuleac M. C., Naik S., Dean D. R., Huynh B. H., Johnson M. K. (2007). Formation and properties of [4Fe-4S] clusters on the IscU scaffold protein. Biochemistry 46, 6804–6811 [DOI] [PubMed] [Google Scholar]
- Christodoulou A., Lederer C. W., Surrey T., Vernos I., Santama N. (2006). Motor protein KIFC5A interacts with Nubp1 and Nubp2, and is implicated in the regulation of centrosome duplication. J. Cell Sci. 119, 2035–2047 [DOI] [PubMed] [Google Scholar]
- Cook J. D., Kondapalli K. C., Rawat S., Childs W. C., Murugesan Y., Dancis A., Stemmler T. L. (2010). Molecular details of the yeast frataxin-Isu1 interaction during mitochondrial Fe-S cluster assembly. Biochemistry 49, 8756–8765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig E. A., Marszalek J. (2002). A specialized mitochondrial molecular chaperone system: a role in formation of Fe/S centers. Cell. Mol. Life Sci. 59, 1658–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen K. M., Kleser M., Bernhardt R. (2011). Adrenodoxin: the archetype of vertebrate-type [2Fe-2S] cluster ferredoxins. Biochim. Biophys. Acta 1814, 111–125 [DOI] [PubMed] [Google Scholar]
- Fernandez-Vizarra E., Berardinelli A., Valente L., Tiranti V., Zeviani M. (2007). Nonsense mutation in pseudouridylate synthase 1 (PUS1) in two brothers affected by myopathy, lactic acidosis and sideroblastic anaemia (MLASA). J. Med. Genet. 44, 173–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazzon J., Dean D. R. (2003). Formation of iron-sulfur clusters in bacteria: an emerging field in bioinorganic chemistry. Curr. Opin. Chem. Biol. 7, 166–173 [DOI] [PubMed] [Google Scholar]
- Fuzery A. K., Oh J. J., Ta D. T., Vickery L. E., Markley J. L. (2011). Three hydrophobic amino acids in Escherichia coli HscB make the greatest contribution to the stability of the HscB-IscU complex. BMC Biochem. 12, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guernsey D. L., Jiang H., Campagna D. R., Evans S. C., Ferguson M., Kellogg M. D., Lachance M., Matsuoka M., Nightingale M., Rideout A., et al. (2009). Mutations in mitochondrial carrier family gene SLC25A38 cause nonsyndromic autosomal recessive congenital sideroblastic anemia. Nat. Genet. 41, 651–653 [DOI] [PubMed] [Google Scholar]
- Hider R. C., Kong X. L. (2011). Glutathione: a key component of the cytoplasmic labile iron pool. Biometals 24, 1179–1187 [DOI] [PubMed] [Google Scholar]
- Hirst J. (2010). Towards the molecular mechanism of respiratory complex I. Biochem. J. 425, 327–339 [DOI] [PubMed] [Google Scholar]
- Huang M. L., Becker E. M., Whitnall M., Rahmanto Y. S., Ponka P., Richardson D. R. (2009). Elucidation of the mechanism of mitochondrial iron loading in Friedreich’s ataxia by analysis of a mouse mutant. Proc. Natl. Acad. Sci. USA 106, 16381–16386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen M. A., Spronk C. A., Gabaldon T., Snel B. (2005). Combining data from genomes, Y2H and 3D structure indicates that BolA is a reductase interacting with a glutaredoxin. FEBS Lett. 579, 591–596 [DOI] [PubMed] [Google Scholar]
- Imlay J. A. (2006). Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 59, 1073–1082 [DOI] [PubMed] [Google Scholar]
- Kampinga H. H., Craig E. A. (2010). The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannengiesser C., Sanchez M., Sweeney M., Hetet G., Kerr B., Moran E., Fuster Soler J. L., Maloum K., Matthes T., Oudot C., et al. (2011). Missense SLC25A38 variations play an important role in autosomal recessive inherited sideroblastic anemia. Haematologica 96, 808–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiley P. J., Beinert H. (2003). The role of Fe-S proteins in sensing and regulation in bacteria. Curr. Opin. Microbiol. 6, 181–185 [DOI] [PubMed] [Google Scholar]
- Kim R., Saxena S., Gordon D. M., Pain D., Dancis A. (2001). J-domain protein, Jac1p, of yeast mitochondria required for iron homeostasis and activity of Fe-S cluster proteins. J. Biol. Chem. 276, 17524–17532 [DOI] [PubMed] [Google Scholar]
- Kispal G., Csere P., Prohl C., Lill R. (1999). The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 18, 3981–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S. A., Sepuri N. B., Pain D., Dancis A. (1998). Mt-Hsp70 homolog, Ssc2p, required for maturation of yeast frataxin and mitochondrial iron homeostasis. J. Biol. Chem. 273, 18389–18393 [DOI] [PubMed] [Google Scholar]
- Koeppen A. H. (2011). Friedreich’s ataxia: pathology, pathogenesis, and molecular genetics. J. Neurol. Sci. 303, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollberg G., Tulinius M., Melberg A., Darin N., Andersen O., Holmgren D., Oldfors A., Holme E. (2009). Clinical manifestation and a new ISCU mutation in iron-sulphur cluster deficiency myopathy. Brain 132, 2170–2179 [DOI] [PubMed] [Google Scholar]
- Kuhnke G., Neumann K., Muhlenhoff U., Lill R. (2006). Stimulation of the ATPase activity of the yeast mitochondrial ABC transporter Atm1p by thiol compounds. Mol. Membr. Biol. 23, 173–184 [DOI] [PubMed] [Google Scholar]
- Land T., Rouault T. A. (1998). Targeting of a human iron-sulfur cluster assembly enzyme, nifs, to different subcellular compartments is regulated through alternative AUG utilization. Mol. Cell 2, 807–815 [DOI] [PubMed] [Google Scholar]
- Li K., Besse E. K., Ha D., Kovtunovych G., Rouault T. A. (2008). Iron-dependent regulation of frataxin expression: implications for treatment of Friedreich ataxia. Hum. Mol. Genet. 17, 2265–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R. (2009). Function and biogenesis of iron-sulphur proteins. Nature 460, 831–838 [DOI] [PubMed] [Google Scholar]
- Lillig C. H., Berndt C., Holmgren A. (2008). Glutaredoxin systems. Biochim. Biophys. Acta 1780, 1304–1317 [DOI] [PubMed] [Google Scholar]
- Martelli A., Napierala M., Puccio H. (2012). Understanding the genetic and molecular pathogenesis of Friedreich’s ataxia through animal and cellular models. Dis. Model. Mech. 5, 165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W., Baross J., Kelley D., Russell M. J. (2008). Hydrothermal vents and the origin of life. Nat. Rev. Microbiol. 6, 805–814 [DOI] [PubMed] [Google Scholar]
- Meyer J. (2008). Iron-sulfur protein folds, iron-sulfur chemistry, and evolution. J. Biol. Inorg. Chem. 13, 157–170 [DOI] [PubMed] [Google Scholar]
- Mochel F., Knight M. A., Tong W. H., Hernandez D., Ayyad K., Taivassalo T., Andersen P. M., Singleton A., Rouault T. A., Fischbeck K. H., et al. (2008). Splice mutation in the iron-sulfur cluster scaffold protein ISCU causes myopathy with exercise intolerance. Am. J. Hum. Genet. 82, 652–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenthaler M. U., Galy B., Hentze M. W. (2008). Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu. Rev. Nutr. 28, 197–213 [DOI] [PubMed] [Google Scholar]
- Nakai Y., Nakai M., Hayashi H., Kagamiyama H. (2001). Nuclear localization of yeast Nfs1p is required for cell survival. J. Biol. Chem. 276, 8314–8320 [DOI] [PubMed] [Google Scholar]
- Navarro-Sastre A., Tort F., Stehling O., Uzarska M. A., Arranz J. A., Del Toro M., Labayru M. T., Landa J., Font A., Garcia-Villoria J., et al. (2011). A fatal mitochondrial disease is associated with defective NFU1 function in the maturation of a subset of mitochondrial Fe-S proteins. Am. J. Hum. Genet. 89, 656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netz D. J., Stumpfig M., Dore C., Muhlenhoff U., Pierik A. J., Lill R. (2010). Tah18 transfers electrons to Dre2 in cytosolic iron-sulfur protein biogenesis. Nat. Chem. Biol. 6, 758–765 [DOI] [PubMed] [Google Scholar]
- Netz D. J., Stith C. M., Stumpfig M., Kopf G., Vogel D., Genau H. M., Stodola J. L., Lill R., Burgers P. M., Pierik A. J. (2011). Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat. Chem. Biol. 8, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordin A., Larsson E., Holmberg M. (2011). The defective splicing caused by the ISCU intron mutation in patients with myopathy with lactic acidosis is repressed by PTBP1 but can be de-repressed by IGF2BP1. Hum. Mutat. doi: 10.1002/humu.22002 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Olsson A., Lind L., Thornell L. E., Holmberg M. (2008). Myopathy with lactic acidosis is linked to chromosome 12q23.3-24.11 and caused by an intron mutation in the ISCU gene resulting in a splicing defect. Hum. Mol. Genet. 17, 1666–1672 [DOI] [PubMed] [Google Scholar]
- Pondarre C., Campagna D. R., Antiochos B., Sikorski L., Mulhern H., Fleming M. D. (2007). Abcb7, the gene responsible for X-linked sideroblastic anemia with ataxia, is essential for hematopoiesis. Blood 109, 3567–3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prischi F., Konarev P. V., Iannuzzi C., Pastore C., Adinolfi S., Martin S. R., Svergun D. I., Pastore A. (2010). Structural bases for the interaction of frataxin with the central components of iron-sulphur cluster assembly. Nat. Commun. 1, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Py B., Barras F. (2010). Building Fe-S proteins: bacterial strategies. Nat. Rev. Microbiol. 8, 436–446 [DOI] [PubMed] [Google Scholar]
- Qi W., Cowan J. A. (2011). Mechanism of glutaredoxin-ISU [2Fe-2S] cluster exchange. Chem. Commun. 47, 4989–4991 [DOI] [PubMed] [Google Scholar]
- Raulfs E. C., O’Carroll I. P., Dos Santos P. C., Unciuleac M. C., Dean D. R. (2008). In vivo iron-sulfur cluster formation. Proc. Natl. Acad. Sci. USA 105, 8591–8596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards T. A., van der Giezen M. (2006). Evolution of the Isd11-IscS complex reveals a single alpha-proteobacterial endosymbiosis for all eukaryotes. Mol. Biol. Evol. 23, 1341–1344 [DOI] [PubMed] [Google Scholar]
- Riley L. G., Cooper S., Hickey P., Rudinger-Thirion J., McKenzie M., Compton A., Lim S. C., Thorburn D., Ryan M. T., Giege R., et al. (2010). Mutation of the mitochondrial tyrosyl-tRNA synthetase gene, YARS2, causes myopathy, lactic acidosis, and sideroblastic anemia-MLASA syndrome. Am. J. Hum. Genet. 87, 52–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault T. A. (2006). The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2, 406–414 [DOI] [PubMed] [Google Scholar]
- Rudolf J., Makrantoni V., Ingledew W. J., Stark M. J., White M. F. (2006). The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol. Cell 23, 801–808 [DOI] [PubMed] [Google Scholar]
- Russell M. J., Martin W. (2004). The rocky roots of the acetyl-CoA pathway. Trends Biochem. Sci. 29, 358–363 [DOI] [PubMed] [Google Scholar]
- Schmucker S., Martelli A., Colin F., Page A., Wattenhofer-Donze M., Reutenauer L., Puccio H. (2011). Mammalian frataxin: an essential function for cellular viability through an interaction with a preformed ISCU/NFS1/ISD11 iron-sulfur assembly complex. PLoS ONE 6, e16199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A. K., Pallesen L. J., Spang R. J., Walden W. E. (2010). Cytosolic iron-sulfur cluster assembly (CIA) system: factors, mechanism, and relevance to cellular iron regulation. J. Biol. Chem. 285, 26745–26751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheftel A. D., Stehling O., Pierik A. J., Netz D. J., Kerscher S., Elsasser H. P., Wittig I., Balk J., Brandt U., Lill R. (2009). Human ind1, an iron-sulfur cluster assembly factor for respiratory complex I. Mol. Cell. Biol. 29, 6059–6073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Proteau A., Villarroya M., Moukadiri I., Zhang L., Trempe J. F., Matte A., Armengod M. E., Cygler M. (2010). Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions. PLoS Biol. 8, e1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Ghosh M. C., Tong W. H., Rouault T. A. (2009). Human ISD11 is essential for both iron-sulfur cluster assembly and maintenance of normal cellular iron homeostasis. Hum. Mol. Genet. 18, 3014–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Ghosh M., Kovtunovych G., Crooks D. R., Rouault T. A. (2011). Both human ferredoxins 1 and 2 and ferredoxin reductase are important for iron-sulfur cluster biogenesis. Biochim. Biophys. Acta 1823, 484–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan V., Netz D. J., Webert H., Mascarenhas J., Pierik A. J., Michel H., Lill R. (2007). Structure of the yeast WD40 domain protein Cia1, a component acting late in iron-sulfur protein biogenesis. Structure 15, 1246–1257 [DOI] [PubMed] [Google Scholar]
- Stehling O., Netz D. J., Niggemeyer B., Rosser R., Eisenstein R. S., Puccio H., Pierik A. J., Lill R. (2008). Human Nbp35 is essential for both cytosolic iron-sulfur protein assembly and iron homeostasis. Mol. Cell. Biol. 28, 5517–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmler T. L., Lesuisse E., Pain D., Dancis A. (2010). Frataxin and mitochondrial FeS cluster biogenesis. J. Biol. Chem. 285, 26737–26743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong W. H., Rouault T. A. (2000). Distinct iron-sulfur cluster assembly complexes exist in the cytosol and mitochondria of human cells. EMBO J. 19, 5692–5700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong W. H., Rouault T. A. (2006). Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis. Cell Metab. 3, 199–210 [DOI] [PubMed] [Google Scholar]
- Tong W. H., Jameson G. N., Huynh B. H., Rouault T. A. (2003). Subcellular compartmentalization of human Nfu, an iron-sulfur cluster scaffold protein, and its ability to assemble a [4Fe-4S] cluster. Proc. Natl. Acad. Sci. USA 100, 9762–9767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C. L., Barondeau D. P. (2010). Human frataxin is an allosteric switch that activates the Fe-S cluster biosynthetic complex. Biochemistry 49, 9132–9139 [DOI] [PubMed] [Google Scholar]
- Uhrigshardt H., Singh A., Kovtunovych G., Ghosh M., Rouault T. A. (2010). Characterization of the human HSC20, an unusual DnaJ type III protein, involved in iron-sulfur cluster biogenesis. Hum. Mol. Genet. 19, 3816–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uringa E. J., Youds J. L., Lisaingo K., Lansdorp P. M., Boulton S. J. (2011). RTEL1: an essential helicase for telomere maintenance and the regulation of homologous recombination. Nucleic Acids Res. 39, 1647–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickery L. E., Cupp-Vickery J. R. (2007). Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in iron-sulfur protein maturation. Crit. Rev. Biochem. Mol. Biol. 42, 95–111 [DOI] [PubMed] [Google Scholar]
- Wiedemann N., Urzica E., Guiard B., Muller H., Lohaus C., Meyer H. E., Ryan M. T., Meisinger C., Muhlenhoff U., Lill R., et al. (2006). Essential role of Isd11 in mitochondrial iron-sulfur cluster synthesis on Isu scaffold proteins. EMBO J. 25, 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H., Rouault T. A. (2010). Human iron-sulfur cluster assembly, cellular iron homeostasis, and disease. Biochemistry 49, 4945–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H., Jeong S. Y., Ghosh M. C., Kovtunovych G., Silvestri L., Ortillo D., Uchida N., Tisdale J., Camaschella C., Rouault T. A. (2010). Glutaredoxin 5 deficiency causes sideroblastic anemia by specifically impairing heme biosynthesis and depleting cytosolic iron in human erythroblasts. J. Clin. Invest. 120, 1749–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]