SUMMARY
BTN1, the yeast homolog to human CLN3 (which is defective in Batten disease), has been implicated in the regulation of vacuolar pH, potentially by modulating vacuolar-type H+-ATPase (V-ATPase) activity. However, we report that Btn1p and the V-ATPase complex do not physically interact, suggesting that any influence that Btn1p has on V-ATPase is indirect. Because membrane lipid environment plays a crucial role in the activity and function of membrane proteins, we investigated whether cells lacking BTN1 have altered membrane phospholipid content. Deletion of BTN1 (btn1-Δ) led to a decreased level of phosphatidylethanolamine (PtdEtn) in both mitochondrial and vacuolar membranes. In yeast there are two phosphatidylserine (PtdSer) decarboxylases, Psd1p and Psd2p, and these proteins are responsible for the synthesis of PtdEtn in mitochondria and Golgi-endosome, respectively. Deletion of both BTN1 and PSD1 (btn1-Δ psd1-Δ) led to a further decrease in levels of PtdEtn in ER membranes associated to mitochondria (MAMs), with a parallel increase in PtdSer. Fluorescent-labeled PtdSer (NBD-PtdSer) transport assays demonstrated that transport of NBD-PtdSer from the ER to both mitochondria and endosomes and/or vacuole is affected in btn1-Δ cells. Moreover, btn1-Δ affects the synthesis of PtdEtn by the Kennedy pathway and impairs the ability of psd1-Δ cells to restore PtdEtn to normal levels in mitochondria and vacuoles by ethanolamine addition. In summary, lack of Btn1p alters phospholipid levels and might play a role in regulating their subcellular distribution.
INTRODUCTION
We have previously reported that the Saccharomyces cerevisiae BTN1 gene encodes a nonessential protein that is 39% identical and 59% similar to human CLN3 (Pearce and Sherman, 1997). Mutations in human CLN3, which codes for a lysosomal transmembrane protein, causes Batten disease, the juvenile form of neuronal ceroid lipofuscinosis (NCL) (The International Batten Disease Consortium, 1995; Jarvela et al., 1998; Kyttala et al., 2004; Storch et al., 2004; Phillips et al., 2005). Btn1p is located in the vacuole, the yeast membrane compartment that is equivalent to the mammalian lysosomal organelle (Croopnick et al., 1998; Pearce et al., 1999), although several recent studies in a S. cerevisiae model found Btn1p associated with punctate membrane structures (Vitiello et al., 2010; Wolfe et al., 2011). Furthermore, localization studies of overexpressed Btn1p in Schizosaccharomyces pombe associated this protein to Golgi membranes (Codlin and Mole, 2009). The primary function of this protein remains unknown; however, Btn1p has been implicated in several cellular pathways. In a S. cerevisiae yeast model, lack of Btn1p (btn1-Δ) led to a decreased vacuolar pH during the logarithmic phase of growth (Pearce et al., 1999). Furthermore, btn1-Δ cells downregulate ATP hydrolysis by vacuolar-type H+-ATPase (V-ATPase) most likely as a cell response to control H+ transport across the vacuolar membrane and compensate for pH imbalance caused by the lack of Btn1p (Padilla-Lopez and Pearce, 2006). This alteration at the vacuole has been implicated in deficient regulation of intracellular L-arginine levels, with btn1-Δ cells showing a lower ability to transport this amino acid into isolated vacuoles in vitro (Kim et al., 2003). However, this latter defect could be a secondary consequence from the primary vacuolar pH alteration observed in btn1-Δ cells.
Little is known about the mechanisms regarding lipid transport and distribution within eukaryotic cells. PtdSer is synthesized by PtdSer synthase in a specialized region of the endoplasmic reticulum (ER) membrane that is associated to mitochondria [mitochondria-associated membrane (MAM)] (Gaigg et al., 1995). PtdSer is transported to the locus of PtdSer decarboxylases Psd1p or Psd2p, where is decarboxylated to form phosphatidylethanolamine (PtdEtn). Psd1p is a well-known inner mitochondrial protein (Kuchler et al., 1986). However, whereas previous studies associated Psd2p to Golgi-vacuole membranes (Trotter et al., 1993; Trotter and Voelker, 1995), a recent study refined Psd2p localization to the endosome, arguing that this enzyme controls vacuolar membrane phospholipid content (Gulshan et al., 2010). Following its formation, PtdEtn is transported back to the ER for the synthesis of phosphatidylcholine (PtdCho) (Atkinson et al., 1980; Trotter et al., 1993). Alternatively, PtdEtn can be synthesized through the cytidyldiphosphate (CDP)-ethanolamine branch of the Kennedy pathway (Kennedy and Weiss, 1956). PtdEtn is essential for growth of the yeast S. cerevisiae and is required for the function and integrity of mitochondrial membranes; a lack of the major PtdSer decarboxylase, Psd1p, leads to a substantial decrease of PtdEtn in total cellular and mitochondrial membranes, conferring a petite phenotype (Birner et al., 2001). Moreover, depletion of PtdEtn causes defects in the assembly of mitochondrial protein complexes and loss of mitochondrial DNA (Birner et al., 2001; Storey et al., 2001; Birner et al., 2003). PtdEtn also plays a crucial role in GPI anchor biosynthesis (Menon and Stevens, 1992) and seems to be involved in the autophagy and cytosol-to-vacuole-targeting delivery pathways (Huang and Klionsky, 2002; Wang and Klionsky, 2003; Yoshimori, 2004; Nebauer et al., 2007).
We report a potential newly identified role for Btn1p: being necessary for the distribution of phospholipids in cell membranes. We report that yeast cells lacking BTN1 show decreased transport of PtdSer from ER membranes to either mitochondria or endosome and/or vacuole membranes, resulting in a significant decrease of PtdEtn synthesis in cells lacking Psd1p (psd1-Δ). Furthermore, a lack of Btn1p (btn1-Δ) also partially impairs the synthesis of PtdEtn by the Kennedy pathway. Thus, the multiple cellular phenotypes associated to a lack of Btn1p, or indeed CLN3, could be a consequence of altered membrane phospholipid content.
RESULTS
Btn1p is not a component of the V-ATPase complex
Our previous study (Padilla-Lopez and Pearce, 2006) indicated that Btn1p has the ability to modulate the activity of the V-ATPase complex. However, evidence was lacking in support of a direct role for Btn1p in the regulation of V-ATPase. An attractive explanation for the role of Btn1p in V-ATPase regulation would be to demonstrate either that Btn1p physically interacts with one or more subunits of the V-ATPase complex, or that Btn1p could be part of the macrocomplex formed by the V-ATPase-complex subunits and other proteins that associate with them; for example, those involved in the glycolytic pathway (Lu et al., 2001; Su et al., 2003; Lu et al., 2004; Sautin et al., 2005; Lu et al., 2007).
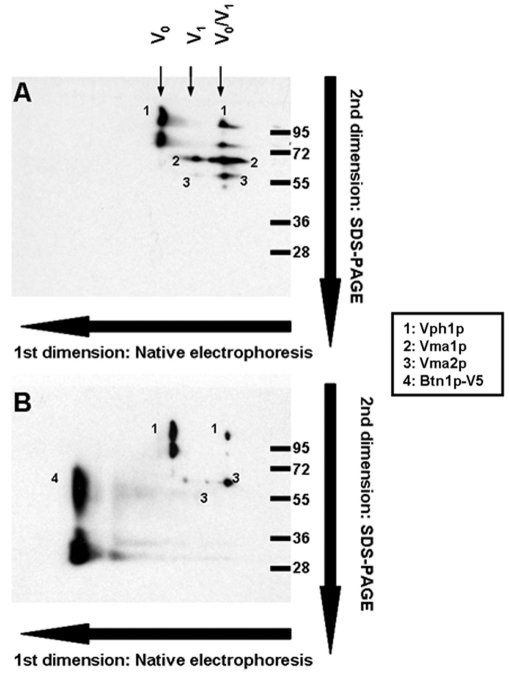
Because specific antibodies against all 13 of the V-ATPase subunits are not available to test Btn1p interaction with them all, we decided to test whether Btn1p is a component of the V-ATPase complex by analyzing the vacuolar proteome using two dimensional (2D) blue-native/SDS-PAGE electrophoresis and western blotting. Vacuolar fractions isolated from wild-type strains harboring the empty plasmid pRS426 or pmBTN1-V5 were used to perform a blue-native electrophoresis. This technique allowed us to resolve the different protein complexes of the vacuolar proteome. The individual components of these complexes were separated in the second dimension by SDS-PAGE, transferred to a nitrocellulose membrane and probed with antibodies against the V5 epitope and three V-ATPase subunits, two components of the V1 sector (Vma1p and Vma2p) and one component of the membrane V0 sector (Vph1p). Fig. 1A shows the normal profile for V-ATPase complexes in a wild-type strain. Three kinds of V-ATPase complexes were detected, which corresponds to the V0, V1 and complete V0–V1 protein complex. Isolating complexes from cells expressing Btn1p-V5 (Fig. 1B) clearly shows that Btn1p-V5 was not part of any of the three V-ATPase complexes detected.
Fig. 1.
Analysis of the protein complexes of the vacuolar proteome by native electrophoresis and second dimension SDS-PAGE. Yeast cells were inoculated at initial A660 0.1 in SC medium and cultured for 24 hours; vacuoles were isolated as indicated in the Methods, and 100 μg samples were analyzed by blue-native electrophoresis to resolve protein complexes followed by a second dimension SDS-PAGE and western blotting using monoclonal antibodies that recognize the V-ATPase subunits Vma1p (69 kDa; V1), Vph1p (100 kDa; V0) and 60-kDa Vma2p (60 kDa; V1), and V5 epitope (46 kDa; Btn1p-V5). (A) Analysis performed in isolated vacuoles from a wild-type strain harboring the empty plasmid pRS426. (B) Vacuoles isolated from a wild-type strain harboring pmBTN1-V5.
In conclusion, there is no evidence in support of a direct physical interaction of Btn1p with subunits that form the V-ATPase complex in S. cerevisiae.
Changes in Btn1p levels alter the membrane phospholipid profile in yeast cells
Different organelles in the cell have a specific composition of membrane phospholipids that is tightly regulated in order to maintain an optimal membrane protein function. Alterations of the phospholipid profile could easily change the way membrane proteins interact with each other and with the phospholipids themselves, leading to the loss of protein activity and causing a number of pleiotropic effects on the cell. We investigated whether the observed effect on V-ATPase activity by Btn1p could be a consequence of alterations in the lipid environment of the vacuolar membrane.
In S. cerevisiae, the different phospholipids are synthesized de novo from CDP-diacylglycerol (CDP-DAG). Addition of serine to CDP-DAG synthesizes PtdSer in the MAMs (Gaigg et al., 1995) that is subsequently transported to other organelles. In the mitochondria, PtdSer is decarboxylated to form PtdEtn by Psd1p. In the endosomes and vacuole compartments, PtdEtn is formed by the action of Psd2p (Gulshan et al., 2010). PtdEtn in the mitochondrial and vacuolar compartments can be subsequently exported from those organelles back to the ER to be transformed to PtdCho by the action of PtdEtn methyltransferases. Phosphatidylinositol (PtdIns) is also synthesized from CDP-DAG by the addition of inositol molecule (Fig. 2). Therefore, lipid extracts of whole cell extracts, vacuolar membrane, mitochondria and MAMs were obtained from wild-type and btn1-Δ cells, and phospholipid content was analyzed by thin layer chromatography (TLC; see Methods). Mitochondrial and vacuolar extracts obtained from btn1-Δ cells showed a slight but significant decrease of PtdEtn levels compared with the wild-type strain (Fig. 3). To further understand the observed changes, potential genetic interaction between BTN1 and either PSD1 or PSD2 was investigated by analyzing the phospholipid profile in psd1-Δ and psd2-Δ cells carrying an additional BTN1 deletion (btn1-Δ psd1-Δ and btn1-Δ psd2-Δ).
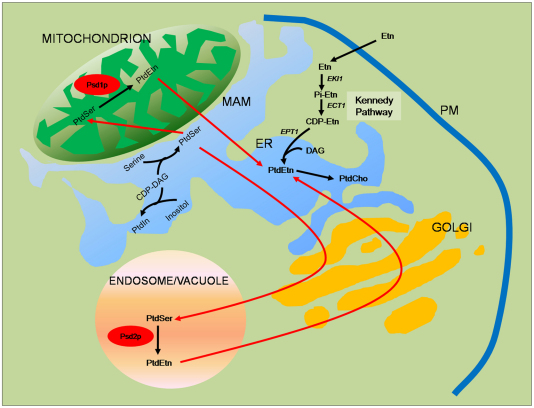
Fig. 2.
Phospholipid synthesis pathway in S. cerevisiae. PtdSer is synthesized in the ER from serine and CDP-DAG, and is subsequently transported to other organelles. In mitochondria, PtdSer is decarboxylated by Psd1p to form PtdEtn. In the endosomes or vacuole compartment, PtdEtn is formed by the action of Psd2p. PtdEtn in the mitochondria and endosomes or vacuole compartments can be subsequently exported from those organelles back to the ER to be transformed to PtdCho by the action of PtdEtn methyltransferases. Alternatively, PtdEtn can be synthesized from ethanolamine (Etn) through the Kennedy pathway. In this pathway, ethanolamine needs to be activated to CDP-ethanolamine (CDP-Etn) and fused to diacylglycerol (DAG) to synthesize PtdEtn.
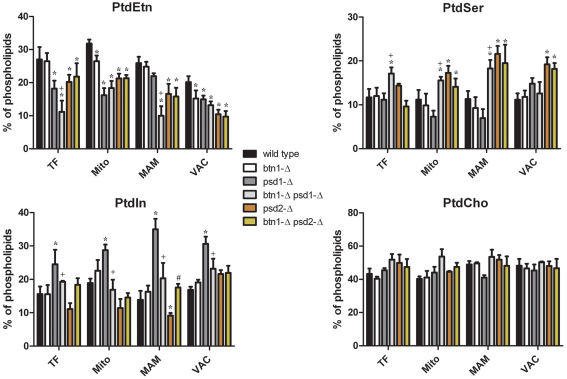
Fig. 3.
Lack of BTN1 leads to alterations in the phospholipid profile of yeast cells. Yeast cells were inoculated at initial A660 0.1 in SC medium and grown for 24 hours, then the indicated cell fractions were isolated and lipid extracts were obtained as described in the Methods. The plots represent the mean and standard errors of the results obtained from five identical experiments. Differences in lipid content are statistically significant with P<0.005 as determined by standard t-test. TF, total fraction; Mito, mitochondrial fraction; MAM, mitochondria-associated membranes fraction; VAC, vacuolar fraction. Symbols denote statistically significant differences to the same membrane fraction of wild-type (*), psd1-Δ (+) or psd2-Δ (#) strains.
The most dramatic alterations were observed in the psd1-Δ genetic background. The PtdSer decarboxylase Psd1p is a component of the inner mitochondrial membrane, which is responsible for the major part of PtdEtn synthesis in yeast cells. It has been described that lack of Psd1p leads to a substantial decrease of PtdEtn in total cellular and mitochondrial membranes, thereby conferring a petite phenotype, which is linked to the loss of respiratory capacity. Thus, this mutant strain requires supplementation with ethanolamine when cultured in nonfermentable carbon sources (Birner et al., 2001). Results obtained in this work confirmed decreased PtdEtn in whole cell, mitochondrial and vacuolar fractions of this strain (Fig. 3). Interestingly, psd1-Δ cells did not show a significant decrease of PtdEtn levels in the MAM fraction. Additionally, deletion of psd1 led to an accumulation of PtdIns that could be observed in all the analyzed fractions, which suggests that blocking the main route for synthesizing PtdEtn is compensated by the overproduction of PtdIns in yeast cells. Surprisingly, combining btn1-Δ with psd1-Δ (btn1-Δ psd1-Δ) led to both PtdSer accumulation in mitochondrial and MAM fractions and a further decrease of PtdEtn levels that was mainly focused in the MAM fraction. Moreover, btn1-Δ psd1-Δ also led to a normalization of PtdIns level.
Analysis of psd2-Δ strains also showed a decrease of PtdEtn in all the studied fractions and, although the mitochondrial fraction was slightly less affected than in psd1-Δ cells, the MAM and vacuole fractions showed lower PtdEtn levels than cells lacking psd1-Δ. In addition, psd2-Δ cells did not show accumulation of PtdIns, but levels of PtdSer were increased in the MAM and vacuole fractions. This suggests that psd2-Δ does not trigger the same compensatory response as the one observed in psd1-Δ cells that leads to the accumulation of PtdIns. In fact, psd2-Δ cells showed a significant decrease of PtdIns in mitochondrial and MAM fractions. btn1-Δ psd2-Δ did not lead to any significant changes on the phospholipid profile as compared with psd2-Δ cells (Fig. 3).
Lack of BTN1 alters phospholipid transport from the ER
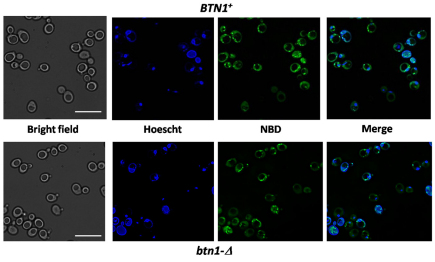
We investigated the potential involvement of Btn1p in phospholipid distribution among cell membranes through a transport assay using fluorescence labeled PtdSer analog, 1-palmitoyl-2-{N-[6(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino]caproyl}-PtdSer (NBD-PtdSer). To determine how NBD-PtdSer is incorporated by yeast cells, wild-type and btn1-Δ yeast strains were grown in synthetic complete (SC) media to logarithmic phase, incubated with 5 μM NBD-PtdSer for 5 minutes and analyzed by fluorescence microscopy. Fig. 4 shows that NBD-PtdSer was incorporated to the same degree in both wild-type and btn1-Δ yeast strains. Moreover, fluorescence was concentrated in similar punctate structures in both strains. It has been described that NBD-PtdEtn is internalized by yeast cells by flip across the plasma membrane and is enriched in ER membranes (Hanson et al., 2002). NBD-PtdSer fluorescence (Fig. 4) accumulates in the same kind of structures as published for NBD-PtdEtn. Therefore, NBD-PtdSer probably accumulates in the ER of yeast cells.
Fig. 4.
NBD-PtdSer is trafficked to ER membranes in yeast. Yeast cells were grown in SC medium to logarithmic phase at 30°C. Cells were then labeled with 5 mM NBD-PtdSer for 5 minutes. After being washed twice with SC medium, yeast cells were incubated with Hoechst dye for 1 minute to label nuclear and mitochondrial DNA, washed again twice with ice-cold SC medium and imaging by fluorescence microscopy. Micrographs were obtained with 1000× magnification. Scale bars: 10 μm.
Transport of NBD-PtdSer from ER to either mitochondria or endosomes and vacuole was determined by the rate of synthesis of NDB-PtdEtn in a psd2-Δ or psd1-Δ genetic background, respectively. As shown in Fig. 5A and 5B, the rate of NBD-PtdEtn synthesis in psd1-Δ cells was significantly lower than in psd2-Δ cells, confirming Psd1p as the main PtdSer decarboxylase in yeast cells. Moreover, combining btn1-Δ in either the psd1-Δ or psd2-Δ genetic background led to a slower rate of NBD-PtdEtn synthesis, suggesting that transport of NBD-PtdSer from the ER to both mitochondria and vacuolar membranes is partially impaired in those strains
Fig. 5.
Cells lacking BTN1 show impaired transport of NBD-PtdSer from ER membranes. Decarboxylation of NBD-labeled PtdSer to NBD-PtdEtn by either Psd1p or Psd2p was used to monitor PtdSer transport from ER to either mitochondria or endosomes and/or vacuole, respectively, as described in the Methods. (A) Transport to endosomes and/or vacuole was determined measuring NBD-PtdEtn synthesis in psd1-Δ yeast protoplasts resuspended in SC media with 1.2 M sorbitol and 5 μM NBD-PtdSer and incubated for the indicated time at 30°C. (B) Transport to mitochondria was determined by performing the same procedure with psd2-Δ yeast protoplasts. Lipid extracts were obtained, analyzed by TLC and the fluorescence associated to NBD-PtdEtn was plotted. The plots represent the mean ± s.e.m. of the results obtained from three identical experiments. Differences between strains are statistically significant with P<0.005 as determined by standard t-test.
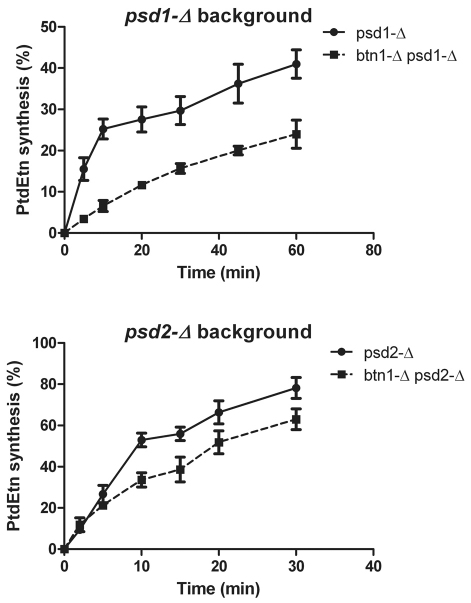
It has been described that lack of Psd1p leads to a substantial decrease of PtdEtn in total cellular and mitochondrial membranes, thereby conferring a petite phenotype and inability to grow on nonfermentable carbon sources. However, exogenous ethanolamine addition allows psd1-Δ cells to synthesize PtdEtn by the Kennedy pathway (Kennedy and Weiss, 1956) in which ethanolamine is activated in the cytosol to CDP-ethanolamine and further bound to diacylglycerol in the ER membrane to synthesize PtdEtn. Thus, PtdEtn synthesized by the Kennedy pathway can be delivered to either the mitochondria or the vacuolar membrane and restore normal levels in those organelles. Because Btn1p seems to be necessary for normal delivery of phospholipids to the mitochondria, we tested whether a lack of Btn1p affected restoration of PtdEtn in the mitochondria and vacuole of psd1-Δ cells when grown in SC-glucose with 5 mM ethanolamine supplementation (Fig. 6). Although ethanolamine supplementation did not significantly raise the level of PtdEtn in total cell fractions of any of the analyzed strains, mitochondrial and vacuolar fractions of psd1-Δ cells grown with ethanolamine supplementation elevated and returned PtdEtn to wild-type levels. No changes in the psd1-Δ MAM fraction of PtdEtn was observed after supplementation with ethanolamine. Interestingly, ethanolamine supplementation of btn1-Δ psd1-Δ did not raise PtdEtn in either mitochondria or vacuoles, whereas a significant increase was observed in MAMs, compared with nonsupplemented media, suggesting that PtdEtn synthesized at the ER by the Kennedy pathway does not reach the other two cellular organelles. However, PtdEtn accumulation in MAMs of btn1-Δ psd1-Δ cells did not reach the levels observed in a single psd1-Δ strain, raising the issue of whether the observed phenotype is caused by an impairment of PtdEtn synthesis through the Kennedy pathway rather than a failure to transport the phospholipid to either mitochondria or vacuoles.
Fig. 6.
Lack of BTN1 leads to alterations in the phospholipids profile of yeast cells. Yeast cells were inoculated at initial A660 0.1 in SC medium with or without 5 mM ethanolamine and grown for 24 hours, then the indicated cell fractions were isolated and lipid extracts were obtained as described in the Methods. The plots represent the mean and standard errors of the results obtained from five identical experiments. Differences in lipid content are statistically significant with P<0.005 as determined by standard t-test. TF, total fraction; Mito, mitochondrial fraction; MAM, mitochondria-associated membrane fraction; VAC, vacuolar fraction. Differences in lipid content are statistically significant with P<0.005 as determined by standard t-test. Symbols denote statistically significant differences to the same membrane fraction of wild-type (*), psd1-Δ (+) or btn1-Δ psd2-Δ (#) strains grown in the absence of ethanolamine.
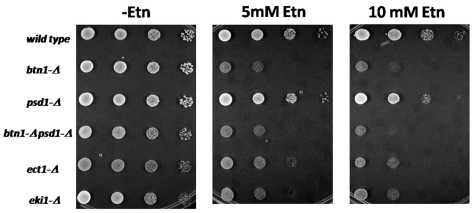
Impairment of the Kennedy pathway could lead to accumulation of free ethanolamine in the cytosol, which potentially could become toxic to cells. Therefore, we wondered whether yeast strains lacking some steps of the salvage pathway for PtdEtn synthesis are sensitive to high concentrations of ethanolamine in the media. Fig. 7 shows that yeast mutant strains lacking either the EKI1 or ECT1 genes, coding for the first and second enzymes of the Kennedy pathway for PtdEtn synthesis, respectively, were unable to grow in the presence of 10 mM ethanolamine. Moreover, ethanolamine was shown to be toxic for btn1-Δ strains, suggesting that lack of BTN1 also impairs the synthesis of PtdEtn through the Kennedy pathway.
Fig. 7.
Yeast cells lacking either BTN1 or genes involved in the Kennedy pathway for PtdEtn synthesis show sensitivity to high concentrations of extracellular ethanolamine. Equal number of cells from freshly grown yeast cultures were deposited as undiluted, 1:10, 1:100 or 1:1000 dilutions (left to right) on SC-glucose plates with or without ethanolamine (Etn) and incubated at 30°C for 48 hours.
DISCUSSION
We report a newly identified genetic interaction for BTN1 and the major PtdSer decarboxylase, Psd1p, suggesting a potential role for Btn1p in phospholipid transport and distribution among yeast membranes.
The idea that the lipid environment modifies the activity of membrane proteins is not new. Crider and Xie demonstrated that phospholipids play a crucial role in the functional coupling of V-ATPases in bovine brain (Crider and Xie, 2003). It also known that Btn1p could have a role in modulating V-ATPase activity (Padilla-Lopez and Pearce, 2006) and importing basic amino acids to the vacuolar lumen (Kim et al., 2003), and that overexpression of the plasma membrane L-arginine transporter Can1p is toxic in btn1-Δ cells (Vitiello et al., 2007). In addition, Opekarová et al. demonstrated that PtdEtn is an essential factor for targeting Can1p to the plasma membrane of yeast (Opekarova et al., 2002). Therefore, taken together, it can be suggested the observed alterations in btn1-Δ cells in V-ATPase activity and L-arginine metabolism might be a secondary consequence of a defect in phospholipid metabolism that we now describe.
The two major alterations in the phospholipid profile in btn1-Δ psd1-Δ yeast include: (1) accumulation of PtdSer in the mitochondrial and MAM fractions, and (2) a further decreased PtdEtn in the MAM fraction. Both alterations suggest an impairment of the transport of PtdSer from ER membranes and/or MAMs to the Psd2p locus at endosomes and/or the vacuole, which would lead to a decreased synthesis of PtdEtn, as observed. However, it is intriguing that only PtdEtn levels of MAMs are diminished in btn1-Δ psd1-Δ cells, whereas mitochondrial and vacuolar fractions did not show any significant differences compared with the psd1-Δ strain. The latter observation might be explained if we assume that PtdEtn transport from vacuolar membranes back to ER is also impaired in the double mutant, whereas its delivery to mitochondria remains unaffected. NBD-PtdSer transport analysis presented in Fig. 5 supports a role for Btn1p in the transport of phospholipids among cells membrane. Because lack of Btn1p does not affect the uptake of the fluorescent analog of PtdSer by yeast cells (Fig. 4), decreased synthesis of NBD-PtdEtn in btn1-Δ cells is probably a result of an impaired delivery of NBD-PtdSer from ER and/or MAMs to the cell organelles in which the two PtdSer decarboxylases are located. Finally, it is also interesting how additional BTN1 deletion normalizes PtdIns accumulation in psd1-Δ cells, raising the issue of whether lack of Btn1p has deeper consequences in the regulation of the phospholipid biosynthetic pathway.
The distribution of PtdEtn synthesized in the ER through the Kennedy pathway further supports a phospholipid transport role for Btn1p (Fig. 6). However, if BTN1 deletion only impaired transport of phospholipids from ER to other cellular membranes, high amounts of PtdEtn accumulated in ER and MAM fraction would be expected. As this is not the case, it is possible that synthesis of PtdEtn through the Kennedy pathway is also impaired in btn1-Δ cells. Interestingly, we show that yeast strains lacking genes of the Kennedy pathway for the synthesis of PtdEtn are sensitive to high concentrations of free ethanolamine in the media. To our knowledge, the present work is the first report describing this phenotype. Importantly, btn1-Δ cells show the same degree of sensitivity to ethanolamine, supporting that the Kennedy pathway is impaired in this strain.
It has been proposed that Btn1p in S. pombe affects the transport of membrane proteins through the Golgi. In this yeast model, deletion of BTN1 leads to secretion of the vacuolar protease Cpyp as a consequence of mistrafficking of its receptor, Vps10p (Codlin and Mole, 2009). Therefore, the findings presented in the current work could partially explain the effect described above as a result of an alteration of the phospholipid content of the cell membranes. However, deletion of BTN1 does not lead to a mistrafficking of Cpyp in S. cerevisiae (supplementary material Fig. S1), suggesting that membrane protein trafficking is not affected by Btn1p in this yeast model.
In summary, this work points to a role for Btn1p in the synthesis and distribution of phospholipids in yeast cells. Although Btn1p has been described mainly as a vacuolar protein in S. cerevisiae, a recent study shows that Btn1p changes its subcellular localization from the vacuole to unidentified punctae in a pH-dependent manner and that the glycosylation state of Btn1p probably regulates this process (Wolfe et al., 2011). It is possible that, although unconfirmed in this study, these unidentified punctae might belong to regions of the ER such as the MAM. Thus, in addition to being highly hydrophobic, Btn1p could be a mobile protein with the potential capacity of transporting different kinds of lipid species among cell membranes.
Phospholipid synthesis is regulated by the transcription repressor Opi1p. Opi1p binds precursor phosphatidic acid in ER membranes. Glucose starvation causes a rapid decrease in intracellular pH that protonates phosphatidic acid and breaks its interaction with Opi1p. Once free in the cytosol, Opi1p moves into the nucleus and represses the expression of several genes involved in phospholipid synthesis (Young et al., 2010). Interestingly, Btn1p changes localization from vacuole to punctate structures at acidic pH (Wolfe et al., 2011). Therefore, it could be that low pH mimics glucose starvation conditions and triggers both repression of phospholipid synthesis through Opi1p action, and Btn1p retention in the MAM fraction, inhibiting any further transport of phospholipids to the vacuole. However, in order to confirm this hypothesis, further study will be required.
Finally, several studies describing CLN3, the human homolog for BTN1, involvement in lipid metabolism has been reported. Thus, it has been proposed that CLN3 is involved in the synthesis of bis(monoacylglycerol) phosphate (BMP), a phospholipid component of lipid rafts (Hobert and Dawson, 2007). Alternatively, an in vitro palmitoyl-protein Δ-9 desaturase activity, which could convert membrane-associated palmitoylated proteins to their respective palmitoylated derivatives, has been associated to this protein (Narayan et al., 2006). Although neither study shows direct involvement of CLN3 in either BMP synthesis or palmitoyl-protein Δ-9 desaturation, respectively, these results might reflect downstream alterations in underlying membrane dynamics owing to the alterations in the phospholipids we report.
In conclusion, we propose a new role for Btn1p in the regulation of phospholipid distribution among the membranes of S. cerevisiae. Because BTN1 and human CLN3 are homologs, the Btn1p-related biochemical alterations presented in the current work might underlie altered lysosomal function in Batten disease and result in the pathological consequences of this disease.
METHODS
Yeast strains and media
The B4742 wild-type (MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), btn1-Δ (MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 btn1::KAN), psd1-Δ (MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 psd1::KAN), psd2-Δ (MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 psd2::KAN), ect1-Δ (MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ect1::KAN) and eki1-Δ (MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 eki1::KAN) yeast strains were purchased from RESGEN. The NATMX4 system was used to delete BTN1 in the psd1-Δ and psd2-Δ backgrounds using homologous recombination (Goldstein and McCusker, 1999). The NATMX4 cassette was obtained by PCR amplification from plasmid p4339 using primers containing 50–55 bp of the sequence upstream and downstream of the BTN1 open reading frame (ORF). Deletions were tested for resistance to Noursethricin (clonNAT) and confirmed by PCR.
Yeast strains were grown as indicated in SC medium (6.7 g/l yeast nitrogen base without amino acids, 2% glucose and complete amino acid supplement). When denoted, 5 and 10 mM ethanolamine was added to the media.
Plasmid pmBTN1 was constructed by ligating the BTN1-containing EcoRI fragment of pAB1793 (Pearce and Sherman, 1998) with an EcoRI-digested 2-μ URA3 multicopy plasmid pRS426. To create the V5-tagged versions of pAB1793 and pmBTN1, the complete BTN1 ORF except the stop codon was amplified by PCR using the oligonucleotide primers 5′-ATGAGTGACAAATCTCATCA-3′ and 5′-TTCCATCCTACAC-CAAAG-3′ and inserted in frame into pYES2.1/V5-His-TOPO to produce a C-terminal V5-His BTN1. This plasmid was used as a template to amplify BTN1-V5 and the downstream region that includes the transcription termination signal for CYC1. For the amplification of this fragment we used the sense primers 5′-TTTTGGCTCTTTGGTTTG-3′ and the two antisense primers 5′-AAAAACTCGAGGTCAGTGAGCGAGGAAGC-3′ and 5′-AAAAAAAGCTTGTCAGTGAGCGAGGAAGC-3′, which generated XhoI and HindIII restriction sites, respectively. To generate pAB1793-V5, the obtained PCR fragment was double-digested with NheI and XhoI and inserted in pAB1793 digested with the same endonucleases. We obtained pmBTN1-V5 by digestion of the PCR fragment with NheI and HindIII and insertion in pmBTN1 digested with the same restriction enzymes.
Preparation of subcellular fractions
Yeast spheroplasts and crude mitochondrial fraction were isolated by published procedures (Gaigg et al., 1995; Glick and Pon, 1995). Mitochondria and MAM were purified using a sucrose gradient. Crude mitochondria were suspended in buffer A {0.6 M sorbitol, 10 mM K-MES [potassium 3-(N-morpholino)ethanesulfonic acid], pH 6.0}, layered on top of a discontinuous gradient consisting of 5 ml 45% (w/v) sucrose and 5 ml 30% (w/v) sucrose in buffer A. After ultracentrifugation for 1 hour at 100,000 g, the purified mitochondria were collected from the 30–45% sucrose interface, diluted in buffer A and collected by centrifugation at 12,000 g for 15 minutes. A fraction containing MAMs was collected from the 0–30% sucrose interface (Gaigg et al., 1995), diluted in buffer A and centrifuged for 1 hour at 100,000 g. For further purification, the membranes were resuspended in buffer A with 2 mM EDTA and 1 mM DTT, adjusted to 50% (w/v) sucrose, loaded on the bottom of a flotation sucrose step gradient, and overlaid with 8 ml 20% (w/v) sucrose in buffer A and 2 ml of buffer A. After ultracentrifugation for 2 hours at 100,000 g, the MAM fraction was collected from the 0–20% sucrose interface, diluted in EM buffer (10 mM MOPS, pH 7.2, 2.5 mM EDTA) and subjected to 1 hour of centrifugation at 100,000 g. The resulting pellet was resuspended in EM buffer.
Yeast vacuoles were obtained by the method of Ohsumi and Anraku with some modifications (Ohsumi and Anraku, 1981) as previously described (Padilla-Lopez and Pearce, 2006).
Analysis of the vacuolar proteome by 2D blue-native/SDS-PAGE
2D blue-native/SDS-PAGE was performed using the NativePAGE Novex Bis-Tris gel system (Invitrogen). Briefly, 100 μg of isolated yeast vacuoles was pelleted by centrifugation at 20,000 g for 30 minutes and resuspended in NativePAGE sample buffer containing 0.5% digitonin, 0.5% n-dodecyl-β-D-maltoside (DDM) and 0.125% Coomassie Blue G-250. For first-dimensional blue-native gel electrophoresis, the solubilized samples were loaded onto a 4–16% Bis-Tris gel and run at 4°C for 1 hour at 150 V followed by 1 hour at 250 V. Individual lanes were then excised from the gel and incubated in SDS buffer (125 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 100 mM DTT) for 30 minutes. The gel strips were then placed horizontally onto 8–16% SDS-PAGE gels, sealed into place with molten agarose and run for 1 hour at 200 V at 10°C until the blue dye front had migrated from the bottom. The separated proteins were then transferred to nitrocellulose membranes and subjected to western blotting. Western blotting was performed using monoclonal antibodies against Vma1p (8B1), Vma2p (13D11) and Vph1p (10D7) from Molecular Probes and V5 epitope antibody from Invitrogen, followed by a horse-radish-peroxidase-conjugated secondary antibody. The obtained blots were analyzed by densitometry using the software Gel-Pro analyzer v. 3.1.
Phospholipid analysis
Total cellular (1 mg of protein) and purified membrane (250 μg protein) fractions were extracted by vigorous mixing with 2 ml chloroform:methanol (2:1, v/v). After 5 minutes centrifugation at 3000 g, the aqueous upper phase was discarded and the lower organic phase was washed three times with 3 ml of 10% methanol solution. The final lipid-containing chloroform phase was evaporated and the dried lipids were resuspended in 20 μl chloroform:methanol and then loaded on silica gel H TLC plates (Analtech). The plates were developed using a solvent system containing chloroform, methanol, 2-propanol, 0.25% aqueous KCl, triethylamine (30:9:25:6:18, v/v/v/v/v). Individual phospholipids were identified under ultraviolet light with authentic standards, by spraying the thin layer plates with 0.1% aqueous 8-anilino-1-naphthalenesulfonic acid. The lipid spots were scraped and quantified using the method of Rouser et al. (Rouser et al., 1966).
NBD-PtdSer transport assays
For the analysis of PtdSer transport to either mitochondria or vacuole, yeast cells lacking Psd1p were cultured in SC medium to logarithmic phase. Yeast protoplasts were generated by Zymoliase 20T incubation in SC media with 1.2 M sorbitol. Protoplasts were harvested by centrifugation, washed and resuspended in SC-1.2M sorbitol medium at 1 unit OD660/ml. Then, 5 μM NBD-PtdSer was added and cells were incubated at 30°C. 0.5 ml aliquots were taken at different time points, mixed with 0.5 ml of 5% SDS and disrupted by vortexing for 30 seconds. Lipids were extracted and resolved by TLC. Transport of PtdSer to the vacuole was determined by measuring the individual fluorescence associated to NBD-PtdEtn (λexc=460 nm; λem=534 nm).
TRANSLATIONAL IMPACT.
Clinical issue
Batten disease, the juvenile form of a group of disorders called neuronal ceroid lipofuscinoses (NCLs), is the most common neurodegenerative disease of childhood, with an incidence of one in 40,000 live births. Children with Batten disease are typically blind by age 7–8 years and suffer a decline in cognitive and motor function, as well as an increasing frequency of seizures, as the disease progresses. Other neurological complications can include schizophrenia, parkinsonism and behavioral problems. There is no cure or treatment, and sufferers commonly die blind, demented and bedridden in their late teens or early twenties.
A hallmark of Batten disease (and of all NCLs) is a lysosomal storage defect involving build up of lipofuscins (autofluorescent pigment granules composed of lipid-containing residues from lysosomal digestion) in the body’s tissues. In line with this phenotype, patients with the disease generally have a defect the CLN3 protein, which localizes to the lysosomal membrane, although other subcellular locations have also been reported. The exact function of CLN3, and how its dysfunction causes Batten disease, is unclear.
Results
In this paper, the authors investigate the potential involvement of BTN1, the yeast homolog of CLN3, in the distribution of membrane phospholipids among different organelles. They show that lack of btn1 (btn1-Δ) leads to changes in the phospholipid profile that are even more pronounced in psd1-Δ cells [which have reduced levels of the essential lipid phosphatidylethanolamine (PtdEtn)]. Specifically, btn1-Δ cells have defects in the transport of phosphatidylserine (a precursor of PtdEtn) from endoplasmic reticulum membranes to mitochondria, as well as to endosomal and vacuolar membranes. Moreover, the Kennedy pathway, which synthesizes PtdEtn, is also impaired when Btn1p is lacking.
Implications and future directions
These findings suggest an important role for BTN1 – and by extension its human homolog CNL3 – in the transport of lipids among cellular membranes. Altered lipid distribution among cell membranes could lead to their accumulation in lysosomes, impairing the normal function of these organelles and thereby triggering pleiotropic cellular defects that might collectively contribute to Batten disease. These findings will help to guide further work on the function of Btn1p/CLN3, and suggest that additional studies of intracellular lipid homeostasis in Batten disease are warranted.
Yeast cell imaging
Cells were visualized under the 100× objective using an epifluorescent microscope (Olympus BX61, Mellville, NY), a CoolSNAP HQ CCD camera (Photometrics, Tucson, AR) and IPLab 4.0 acquisition software (BD Biosciences, Rockville, MD). Image deconvolution was performed using the Autodeblur software package (Media Cybernetics, Bethesda, MD), and overlays of fluorescent images were performed using Image J (NIH) and Photoshop software.
Supplementary Material
Footnotes
COMPETING INTERESTS
The authors declare that they do not have any competing or financial interests.
AUTHOR CONTRIBUTIONS
S.P.-L. performed experiments, analyzed the data, interpreted the data and prepared the manuscript. D.L. and C.-H.C. performed experiments. D.A.P. interpreted the data and prepared the manuscript.
FUNDING
This work was supported by the National Institutes of Health (NIH) [grants R01 NS036610 and R21 060185].
SUPPLEMENTARY MATERIAL
Supplementary material for this article is available at http://dmm.biologists.org/lookup/suppl/doi:10.1242/dmm.008490/-/DC1
REFERENCES
- Atkinson K., Fogel S., Henry S. A. (1980). Yeast mutant defective in phosphatidylserine synthesis. J. Biol. Chem. 255, 6653–6661 [PubMed] [Google Scholar]
- Birner R., Burgermeister M., Schneiter R., Daum G. (2001). Roles of phosphatidylethanolamine and of its several biosynthetic pathways in Saccharomyces cerevisiae. Mol. Biol. Cell 12, 997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birner R., Nebauer R., Schneiter R., Daum G. (2003). Synthetic lethal interaction of the mitochondrial phosphatidylethanolamine biosynthetic machinery with the prohibitin complex of Saccharomyces cerevisiae. Mol. Biol. Cell 14, 370–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codlin S., Mole S. E. (2009). S. pombe btn1, the orthologue of the Batten disease gene CLN3, is required for vacuole protein sorting of Cpy1p and Golgi exit of Vps10p. J. Cell Sci. 122, 1163–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crider B. P., Xie X. S. (2003). Characterization of the functional coupling of bovine brain vacuolar-type H(+)-translocating ATPase. Effect of divalent cations, phospholipids, and subunit H (SFD). J. Biol. Chem. 278, 44281–44288 [DOI] [PubMed] [Google Scholar]
- Croopnick J. B., Choi H. C., Mueller D. M. (1998). The subcellular location of the yeast Saccharomyces cerevisiae homologue of the protein defective in the juvenile form of Batten disease. Biochem. Biophys. Res. Commun. 250, 335–341 [DOI] [PubMed] [Google Scholar]
- Gaigg B., Simbeni R., Hrastnik C., Paltauf F., Daum G. (1995). Characterization of a microsomal subfraction associated with mitochondria of the yeast, Saccharomyces cerevisiae. Involvement in synthesis and import of phospholipids into mitochondria. Biochim. Biophys. Acta 1234, 214–220 [DOI] [PubMed] [Google Scholar]
- Glick B. S., Pon L. A. (1995). Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 260, 213–223 [DOI] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H. (1999). Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541–1553 [DOI] [PubMed] [Google Scholar]
- Gulshan K., Shahi P., Moye-Rowley W. S. (2010). Compartment-specific synthesis of phosphatidylethanolamine is required for normal heavy metal resistance. Mol. Biol. Cell 21, 443–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson P. K., Grant A. M., Nichols J. W. (2002). NBD-labeled phosphatidylcholine enters the yeast vacuole via the pre-vacuolar compartment. J. Cell Sci. 115, 2725–2733 [DOI] [PubMed] [Google Scholar]
- Hobert J. A., Dawson G. (2007). A novel role of the Batten disease gene CLN3: association with BMP synthesis. Biochem. Biophys. Res. Commun. 358, 111–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W. P., Klionsky D. J. (2002). Autophagy in yeast: a review of the molecular machinery. Cell Struct. Funct. 27, 409–420 [DOI] [PubMed] [Google Scholar]
- Jarvela I., Sainio M., Rantamaki T., Olkkonen V. M., Carpen O., Peltonen L., Jalanko A. (1998). Biosynthesis and intracellular targeting of the CLN3 protein defective in Batten disease. Hum. Mol. Genet. 7, 85–90 [DOI] [PubMed] [Google Scholar]
- Kennedy E. P., Weiss S. B. (1956). The function of cytidine coenzymes in the biosynthesis of phospholipides. J. Biol. Chem. 222, 193–214 [PubMed] [Google Scholar]
- Kim Y., Ramirez-Montealegre D., Pearce D. A. (2003). A role in vacuolar arginine transport for yeast Btn1p and for human CLN3, the protein defective in Batten disease. Proc. Natl. Acad. Sci. USA 100, 15458–15462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchler K., Daum G., Paltauf F. (1986). Subcellular and submitochondrial localization of phospholipid-synthesizing enzymes in Saccharomyces cerevisiae. J. Bacteriol. 165, 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyttala A., Ihrke G., Vesa J., Schell M. J., Luzio J. P. (2004). Two motifs target Batten disease protein CLN3 to lysosomes in transfected nonneuronal and neuronal cells. Mol. Biol. Cell 15, 1313–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Holliday L. S., Zhang L., Dunn W. A., Jr, Gluck S. L. (2001). Interaction between aldolase and vacuolar H+-ATPase: evidence for direct coupling of glycolysis to the ATP-hydrolyzing proton pump. J. Biol. Chem. 276, 30407–30413 [DOI] [PubMed] [Google Scholar]
- Lu M., Sautin Y. Y., Holliday L. S., Gluck S. L. (2004). The glycolytic enzyme aldolase mediates assembly, expression, and activity of vacuolar H+-ATPase. J. Biol. Chem. 279, 8732–8739 [DOI] [PubMed] [Google Scholar]
- Lu M., Ammar D., Ives H., Albrecht F., Gluck S. L. (2007). Physical interaction between aldolase and vacuolar H+-ATPase is essential for the assembly and activity of the proton pump. J. Biol. Chem. 282, 24495–24503 [DOI] [PubMed] [Google Scholar]
- Menon A. K., Stevens V. L. (1992). Phosphatidylethanolamine is the donor of the ethanolamine residue linking a glycosylphosphatidylinositol anchor to protein. J. Biol. Chem. 267, 15277–15280 [PubMed] [Google Scholar]
- Narayan S. B., Rakheja D., Tan L., Pastor J. V., Bennett M. J. (2006). CLN3P, the Batten’s disease protein, is a novel palmitoyl-protein Delta-9 desaturase. Ann. Neurol. 60, 570–577 [DOI] [PubMed] [Google Scholar]
- Nebauer R., Rosenberger S., Daum G. (2007). Phosphatidylethanolamine, a limiting factor of autophagy in yeast strains bearing a defect in the carboxypeptidase Y pathway of vacuolar targeting. J. Biol. Chem. 282, 16736–16743 [DOI] [PubMed] [Google Scholar]
- Ohsumi Y., Anraku Y. (1981). Active transport of basic amino acids driven by a proton motive force in vacuolar membrane vesicles of Saccharomyces cerevisiae. J. Biol. Chem. 256, 2079–2082 [PubMed] [Google Scholar]
- Opekarova M., Robl I., Tanner W. (2002). Phosphatidyl ethanolamine is essential for targeting the arginine transporter Can1p to the plasma membrane of yeast. Biochim. Biophys. Acta 1564, 9–13 [DOI] [PubMed] [Google Scholar]
- Padilla-Lopez S., Pearce D. A. (2006). Saccharomyces cerevisiae lacking Btn1p modulate vacuolar ATPase activity in order to regulate pH imbalance in the vacuole. J. Biol. Chem. 281, 10273–10280 [DOI] [PubMed] [Google Scholar]
- Pearce D. A., Sherman F. (1997). BTN1, a yeast gene corresponding to the human gene responsible for Batten’s disease, is not essential for viability, mitochondrial function, or degradation of mitochondrial ATP synthase. Yeast 13, 691–697 [DOI] [PubMed] [Google Scholar]
- Pearce D. A., Sherman F. (1998). A yeast model for the study of Batten disease. Proc. Natl. Acad. Sci. USA 95, 6915–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce D. A., Ferea T., Nosel S. A., Das B., Sherman F. (1999). Action of BTN1, the yeast orthologue of the gene mutated in Batten disease. Nat. Genet. 22, 55–58 [DOI] [PubMed] [Google Scholar]
- Phillips S. N., Benedict J. W., Weimer J. M., Pearce D. A. (2005). CLN3, the protein associated with batten disease: structure, function and localization. J. Neurosci. Res. 79, 573–583 [DOI] [PubMed] [Google Scholar]
- Rouser G., Siakotos A. N., Fleischer S. (1966). Quantitative analysis of phospholipids by thin-layer chromatography and phosphorus analysis of spots. Lipids 1, 85–86 [DOI] [PubMed] [Google Scholar]
- Sautin Y. Y., Lu M., Gaugler A., Zhang L., Gluck S. L. (2005). Phosphatidylinositol 3-kinase-mediated effects of glucose on vacuolar H+-ATPase assembly, translocation, and acidification of intracellular compartments in renal epithelial cells. Mol. Cell. Biol. 25, 575–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch S., Pohl S., Braulke T. (2004). A dileucine motif and a cluster of acidic amino acids in the second cytoplasmic domain of the batten disease-related CLN3 protein are required for efficient lysosomal targeting. J. Biol. Chem. 279, 53625–53634 [DOI] [PubMed] [Google Scholar]
- Storey M. K., Clay K. L., Kutateladze T., Murphy R. C., Overduin M., Voelker D. R. (2001). Phosphatidylethanolamine has an essential role in Saccharomyces cerevisiae that is independent of its ability to form hexagonal phase structures. J. Biol. Chem. 276, 48539–48548 [DOI] [PubMed] [Google Scholar]
- Su Y., Zhou A., Al-Lamki R. S., Karet F. E. (2003). The a-subunit of the V-type H+-ATPase interacts with phosphofructokinase-1 in humans. J. Biol. Chem. 278, 20013–20018 [DOI] [PubMed] [Google Scholar]
- The International Batten Disease Consortium (1995). Isolation of a novel gene underlying Batten disease, CLN3. The International Batten Disease Consortium. Cell 82, 949–957 [DOI] [PubMed] [Google Scholar]
- Trotter P. J., Voelker D. R. (1995). Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 270, 6062–6070 [DOI] [PubMed] [Google Scholar]
- Trotter P. J., Pedretti J., Voelker D. R. (1993). Phosphatidylserine decarboxylase from Saccharomyces cerevisiae. Isolation of mutants, cloning of the gene, and creation of a null allele. J. Biol. Chem. 268, 21416–21424 [PubMed] [Google Scholar]
- Vitiello S. P., Wolfe D. M., Pearce D. A. (2007). Absence of Btn1p in the yeast model for juvenile Batten disease may cause arginine to become toxic to yeast cells. Hum. Mol. Genet. 16, 1007–1016 [DOI] [PubMed] [Google Scholar]
- Vitiello S. P., Benedict J. W., Padilla-Lopez S., Pearce D. A. (2010). Interaction between Sdo1p and Btn1p in the Saccharomyces cerevisiae model for Batten disease. Hum. Mol. Genet. 19, 931–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. W., Klionsky D. J. (2003). The molecular mechanism of autophagy. Mol. Med. 9, 65–76 [PMC free article] [PubMed] [Google Scholar]
- Wolfe D. M., Padilla-Lopez S., Vitiello S. P., Pearce D. A. (2011). pH-dependent localization of Btn1p in the yeast model for Batten disease. Dis. Model. Mech. 4, 120–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimori T. (2004). Autophagy: a regulated bulk degradation process inside cells. Biochem. Biophys. Res. Commun. 313, 453–458 [DOI] [PubMed] [Google Scholar]
- Young B. P., Shin J. J., Orij R., Chao J. T., Li S. C., Guan X. L., Khong A., Jan E., Wenk M. R., Prinz W. A., et al. (2010). Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science 329, 1085–1088 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.