Abstract
A decade ago, RNA interference was proposed to serve as a physiologic means of regulating long-term gene expression in the mammalian brain. However, during the intervening years, this hypothesis appeared to be contradicted by both experimental data and theoretical considerations. More recently, the advent of deep sequencing technology has permitted a re-assessment of this issue. As reviewed here, a large population of small RNAs having features characteristic of endogenous siRNAs are detected within adult mouse hippocampus, which derive from genes involved in synaptic structure and signaling, and which show a significant, though modest (16–22%) up-regulation during olfactory discrimination training. Small RNAs derived from abundant cellular noncoding RNAs are also detected; in particular, a subpopulation of RNAs 25–30 nt. in length shows very large (>100 fold) up-regulation during olfactory discrimination training. Preliminary data suggest that the 25–30 nt. RNAs may associate with MIWI rather than Argonaute 1–4 homologues. I conclude that, despite their apparent low abundance, endogenous siRNAs and noncoding RNA-derived small RNAs are likely to play an important role in regulating synaptic plasticity.
Keywords: dicer, microRNA, RNA interference, siRNA, synaptic plasticity, gene expression, Argonaute, piRNA, MIWI
Introduction
The phenomenon of RNA interference (RNAi) was first noticed and analyzed in lower model organisms including plants, fungi and C. elegans (Baulcombe, 2003; Fire, 2006). Briefly, double-stranded RNA (dsRNA, which can arise from diverse sources including transgenes, viruses, transposons, and cellular genes) is processed by the RNAse III enzyme dicer to form ~22 nt. dsRNAs. One strand (called a small inhibitory RNA, or siRNA) is incorporated into RISC (a complex containing an Argonaute homolog and other proteins) which cleaves homologous RNAs in a sequence-sequence manner. This type of post-transcriptional gene silencing is extremely potent. In plants and C. elegans, at least, an individual siRNA can bind to RNAs and undergo primer extension using the enzyme RNA-dependent RNA polymerase (RdRP) to form additional long dsRNA molecules that are cleaved by dicer, thus creating a self-amplifying process. Moreover (again, at least in plants and C. elegans), gene silencing can be propagated from cell to cell, involving all of the cells of the body, including the germline (Fire et al, 1998). Finally, siRNAs can also silence genes via transcriptional interference (e.g. Grishok et al, 2005).
In 2001, we suggested that the features of RNA interference would be ideal as an endogenous mechanism for regulating long-term gene expression in the nervous system (Smalheiser et al, 2001). A scheme was proposed in which specific antisense transcripts are induced at the onset of learning, forming sense-antisense transcript hybrids that are processed to form siRNAs. In turn, the siRNAs would cause long-term silencing of the sense mRNAs encoding genes that regulate synaptic structure, function, and plasticity (e.g., NMDA and other glutamate receptors; PSD95 and other postsynaptic scaffold proteins; SynGAP1 and other signaling adaptor proteins; FMRP, HuD and other RNA-binding proteins; synapsin I, GAP43 and other presynaptic proteins; spinophilin and other actin-binding proteins; etc.). Although one might expect that learning would be associated with long-term gene activation (rather than silencing), many genes are known to be down-regulated during learning, and one can envision that genes involved in synaptic stability may need to be turned off in order for remodeling and growth to occur. At the time, it was not clear whether all of the features of RNAi seen in invertebrates should also be found in vertebrates – especially self-amplification and cell-to-cell silencing (see below). However, the RNAi machinery is clearly present within mammalian neurons, since when used as an experimental technique, both long exogenous dsRNAs and short siRNAs are effective for gene silencing (Yu et al, 2002; Krichevsky and Kosik, 2002).
So what happened to the idea that endogenous siRNAs regulate neuronal gene expression?
Our hypothesis paper appeared in April 2001. In October of the same year, a trio of papers were published in Science announcing the discovery of a large family of small RNAs, called microRNAs (miRNAs) (Lagos-Quintana et al, 2001; Lau et al, 2001; Lee and Ambros, 2001). MicroRNAs arise when primary miRNA gene transcripts (pri-miRs) are processed by a complex including drosha and DGCR8 proteins to form 70–100 nt. small miRNA hairpin precursors (pre-miRs); the pre-miRs are exported from the nucleus, processed further by dicer and its co-factors (TRBP and PACT), and incorporated into RISC. It became clear that hundreds of microRNAs are expressed in brain, and that some are brain-specific (or at least highly enriched) and developmentally regulated (Lagos-Quintana et al, 2002; Krichevsky et al, 2003). Subsequent studies have established microRNAs as major regulators of protein translation and mRNA stability with roles in cell fate, differentiation, and synaptic plasticity (Gao, 2010; Siegel et al, 2011). In contrast, numerous cloning studies that revealed abundant microRNAs failed to detect the expression of endogenous siRNAs in C. elegans or other animals.
More detailed sequencing analyses (Ambros et al, 2003; Ruby et al, 2006) suggested that endogenous siRNAs are indeed expressed in C. elegans, but had been under-detected because the usual cloning strategy added adaptors selectively to RNAs having 5’-monophosphate and 3’-hydroxyl groups (characteristic of RNAse III products), whereas many of the endogenous siRNAs had modified 5’-ends (e.g., 5’-triphosphate groups are found on secondary siRNAs, characteristic of RdRP products) and/or had modified 3’-ends (e.g., 2-O-methyl groups). The putative targets of the siRNAs (i.e., the mRNAs of the host genes giving rise to the siRNAs) comprised specific functional categories (Lee et al, 2006; Asikainen et al, 2008) and appeared to modulate expression of the target mRNAs (Lee et al, 2006). Lim DH, Oh CT, Lee L, Hong JS, Noh SH, Hwang S, Kim S, Han SJ, Lee YS. The endogenous siRNA pathway impacts development, stress resistance and lifespan (Lucchetta et al, 2009; Lim et al, 2011).
Nevertheless, there were numerous objections raised to the idea that endogenous siRNAs should regulate gene expression in a physiologic manner, especially in mammals: First, the prevailing concept was (and still is) that endogenous siRNAs have the primal function of fighting transposons and viruses (Fire, 2006). Second, the canonical RNA-dependent RNA polymerase enzyme, which is crucial for the self-amplifying nature of RNAi, has no gene homologue in mammals. Third, long double-stranded RNAs are potent stimulators of the interferon response, so apart from early embryonic cell types that lack this response (e.g., ES cells and oocytes), one would not expect long dsRNAs to be tolerated within cells. As Fire (2006) states, “We presume that once the RNAi mechanism is in place, cells would evolve very diligently to avoid producing dsRNA in amounts that would shut off important endogenous genes.”
These objections slowly evaporated, one by one, in the late 2000s: First, further cloning analyses of small RNAs, particularly using deep sequencing technology, revealed the widespread expression of endogenous siRNAs (as well as piRNAs and a variety of other small RNAs, see below) in Drosophila (Czech et al, 2008; Okamura et al, 2008; Kawamura et al, 2008; Lau et al, 2009). Endogenous siRNAs are dicer dependent, modified at their 3’-ends and associated with Argonaute proteins. They were detected as well in mouse cell types that lack an interferon response, namely, oocytes (Tam et al, 2008; Watanabe et al, 2008) and ES cells (Calabrese et al, 2007; Babiarz et al, 2008). Endogenous siRNAs do arise prominently from transposons and other repeat elements, but they also arise from regions known to transcribe both sense and antisense transcripts as well as from sequences having inverted repeat secondary structure (“hairpins”) within pseudogenes and protein-coding genes (reviewed in Golden et al, 2008; Ghildiyal and Zamore, 2009). Second, noncanonical RdRP enzymes were discovered in mammals (Maida et al, 2009). Third, numerous studies had shown that long dsRNA can be used for sequence-specific gene silencing even in cells capable of generating an interferon response (reviewed in Smalheiser et al, 2011a). Moreover, endogenously expressed dsRNAs differ from exogenous dsRNAs in many ways that might be distinguished by cells -- in length, subcellular localization, 5’-end modifications, abundance, etc. (Wang and Carmichael, 2004; Schlee et al, 2006).
The first study to analyze small RNAs within a differentiated mammalian cell type was Kawaji et al (2008), who examined the HepG2 liver cell line. They detected evidence for endogenous siRNAs that derive from loci expressing both sense and antisense transcripts (they were also the first to detect several novel classes of small RNAs derived from abundant noncoding RNAs, see below). Yi et al (2009) detected low abundance endogenous siRNAs in developing skin, and confirmed that their expression is dependent on dicer but independent of DGCR8. Thus, the stage was set for carrying out deep sequencing of mammalian brain.
Sense-antisense transcript hybrids as a source of endogenous siRNAs
As mentioned above, there is strong circumstantial evidence that endogenous siRNAs are associated with mRNA loci known to express sense and antisense transcripts. However, an entirely different line of investigation has been concerned with whether sense and antisense transcripts are co-expressed within individual cells; whether (and where) they bind to each other to form double-stranded hybrids; and whether the hybrids are indeed processed by dicer to form endogenous siRNAs. Certainly, in the past decade, it has become well established that natural antisense transcripts are specifically induced in a number of physiologic and pathologic situations and regulate the expression of their sense counterparts (Faghihi et al, 2010; Werner and Swan, 2010). It has been demonstrated in numerous cases that sense-antisense hybrids can form within cells (e.g., Soldà et al, 2005; Faghihi et al, 2008). However, the relationship between antisense and sense transcript is not necessarily negative: Often sense and antisense transcripts show a concordant, not discordant co-expression pattern, and hybrid formation sometimes has a stabilizing effect on the sense mRNA (e.g., Faghihi et al, 2008). It is likely that hybrids are not efficiently processed by dicer in the cytoplasm under resting conditions (Carlile et al, 2008), although it is conceivable that RNAi may occur if dicer is activated or mobilized (Lugli et al, 2005). Numerous sense and antisense transcripts are co-expressed within isolated synaptic fractions in adult mouse forebrain (Smalheiser et al, 2008), some of which correspond to loci that give rise to small RNAs (see below).
On the other hand, sense-antisense transcript hybrids may be expected to form naturally within the nucleus. Several studies have demonstrated nuclear transport of Argonaute and other RNAi proteins as well as miRNAs and other small RNAs (e.g., Weinmann et al, 2009); indeed, RNAi is effective in silencing targets within the nucleus (e.g., Robb et al, 2005; Carlile et al, 2008), a compartment that is thought to be protected against eliciting the interferon response (Wang and Carmichael, 2004). Dicer, drosha and Ago are also involved in processing of pre-rRNAs, which is a nuclear event (Liang and Crooke, 2011). As discussed below, many novel small RNAs have been shown to derive from intronic elements and nuclear ncRNAs as well, suggesting that the nucleus may not be excluded, and may in fact be a preferred locus of small RNA biogenesis.
In summary, there is some evidence that endogenous siRNAs may arise from sense-antisense transcript hybrids, but it is unknown where this would occur within cells and it is not clear whether formation of the hybrids would automatically lead to their processing.
Endogenous siRNAs and noncoding RNA-derived small RNAs are expressed in adult mouse hippocampus and are up-regulated in olfactory discrimination training
To examine directly whether small RNAs having the characteristics of endogenous siRNAs could be detected within adult mouse hippocampus, we utilized tissue from mice that had been subjected to an olfactory discrimination training paradigm (Larson and Sieprawska, 2002; Smalheiser et al, 2010). In this manner, we hoped to identify these and any other “interesting” small RNAs that showed large, specific changes with training (Smalheiser et al, 2011a).
Mice (male C57Bl/6 strain, 2 months of age) were first trained to execute nose-poke responses for water reinforcement in two 20-trial sessions per day. Mice were then randomly assigned to three experimental groups: The first group (“training”) received olfactory discrimination training. Mice were trained in a series of 20-trial sessions in which each trial began with the simultaneous presentation of two discriminative odors (S+ and S−) to the West sniff ports. The spatial position of the two odors on any given trial was randomly determined except that no more than three identical trials could occur in succession. A nose-poke response at the port carrying the S+ odor (L-carvone) terminated the trial, was scored as correct, and was rewarded with a drop of water; a response at the port carrying the S– odor (α-phellandrene) terminated the trial, was scored as incorrect, and was not rewarded. Each trial had a maximum duration of 60 sec and was followed by a 10-sec intertrial interval. The learning criterion was 14 or more correct trials in a 20-trial session, which is the earliest point at which learning differs significantly from random guessing at p = 0.05. The second group (“pseudo-training”) received the same two odors and trial events as in the training group, except that rewards were not contingent upon responding at the correct odor port. A nose-poke response to either odor terminated the trial and was rewarded with a drop of water. Each mouse in the pseudo-training group was yoked to a mouse in the training group in terms of number of training sessions. The third group (“nose-poke”) simply continued nose-poke training and had no odors presented. Each mouse in this experiment was yoked to a mouse in the training group in terms of number of training sessions.
Hippocampal tissue from these mice had previously been measured individually with regard to microRNA expression using high throughput RT-PCR plates (Smalheiser et al, 2010). The most striking finding was a global up-regulation of miRNA expression (by 9- 12% across the population of miRNAs) that was observed in the training vs. pseudo-training comparison. As well, there was a reorganization of miRNA expression that was detected by identifying pairs of miRNAs that were highly correlated across individuals in the training group, but un-correlated among individuals in the pseudo-training or nose-poke groups (Smalheiser et al, 2010).
Total RNA from these mice were pooled such that each pool contained RNA from 3–4 mice of the same group, and each treatment group was represented by two pools. These samples were size-selected to obtain RNAs in the range of 18–30 nt., and adaptors were added using a strategy designed to selectively amplify RNAse III products (i.e., RNAs having 5’-monophosphate groups and 3’-hydroxyl groups). cDNA libraries were made and sequenced on an Illumina Genome Analyzer II, giving 12–13 million raw sequence reads per pooled sample. We only analyzed sequences that aligned (“mapped”) exactly and uniquely to the reference mouse genome, and which mapped to exons or introns of Mouse Genome Informatics (MGI) annotated genes (consisting mostly of protein-coding genes). (Sequences that aligned to known miRNA genes were placed in a separate file and analyzed separately.)
In this fashion, we focused on putative endogenous siRNAs. However, note that this strategy may have under-detected a large proportion of endogenous siRNAs – namely, those that undergo RNA editing, those that derive from genomic repeat elements, and those which have 5’-triphosphate modifications (i.e. secondary siRNAs) or that have 3’ 2-O-methyl groups (which are characteristic of siRNAs in lower model organisms).
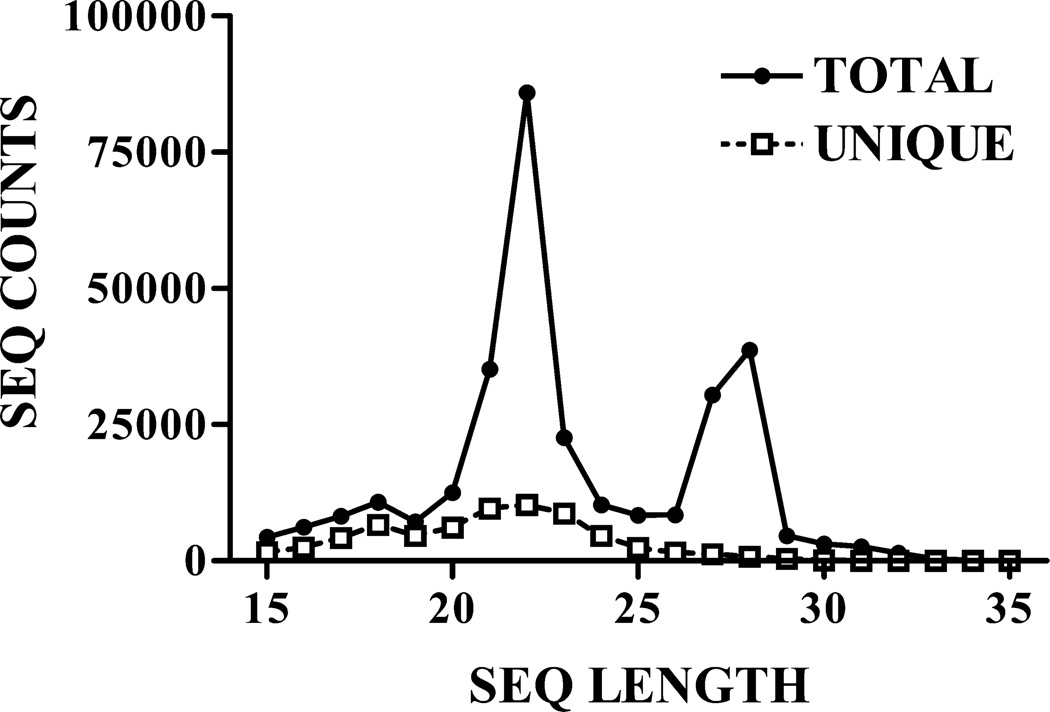
Across the filtered data set, a total of 65,516 unique RNA sequences mapped uniquely and exactly to 14,583 known genes. These sequences exhibited a sharp peak in abundance at the 21- to 22-nt size class (Fig. 1), as expected for endogenous siRNAs. A 16% global elevation of expression with training was observed across all putative siRNAs (i.e., the population of 20–23 nt. small RNAs in the dataset), which was similar to that seen for the population of miRNAs as a whole (9–12%; Smalheiser et al, 2010, 2011a).
Figure 1. Length distribution of small RNAs in the data set.
Shown are the total number of sequence reads, and the total number of unique sequences, observed in the entire data set (across all samples and treatment groups). A more detailed breakdown of the data set is presented in Table 1. All figures are reprinted from Smalheiser et al (2011a) with permission.
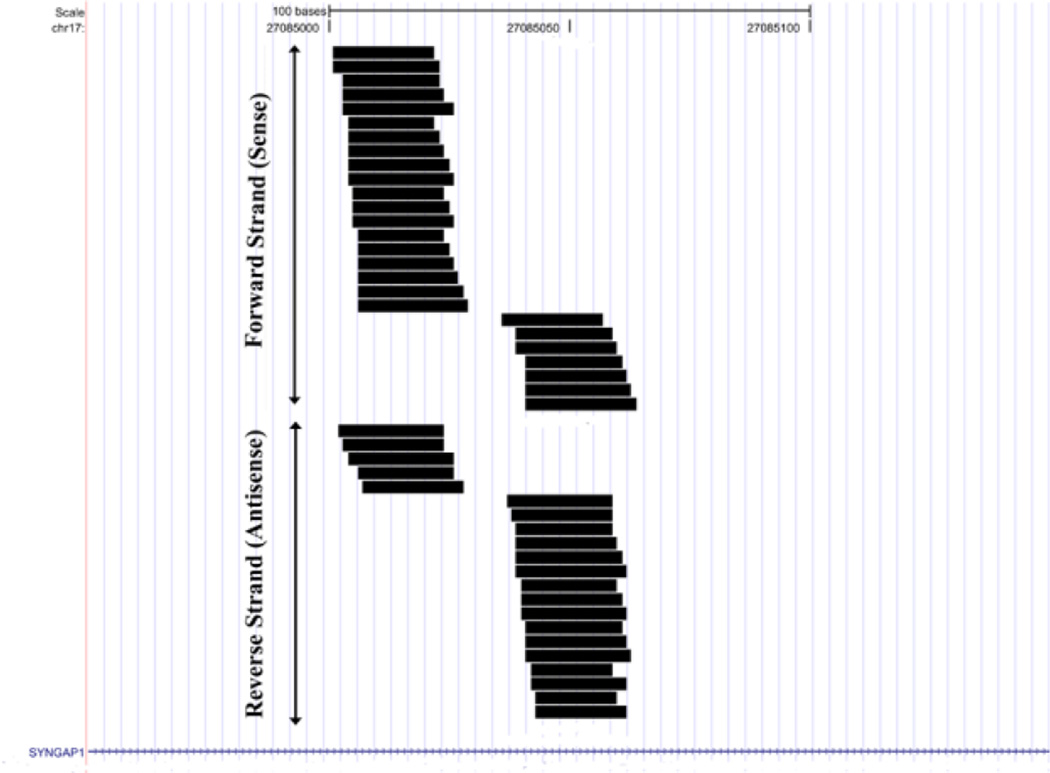
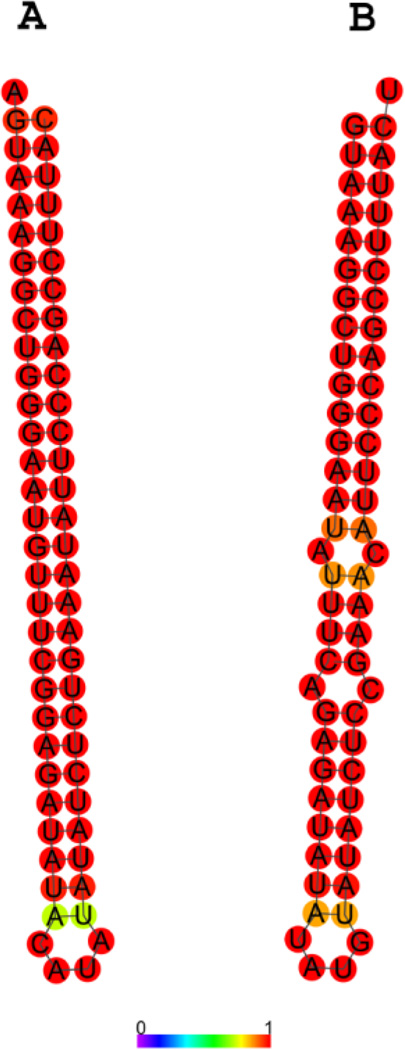
The mouse SynGAP1 locus gave rise to the most abundant set of siRNAs, about 50 unique sequences comprising several hundred counts in each sample, which mapped to both sides of an inverted repeat (“hairpin”) located within an intron (fig. 2). This locus was actually somewhat complex, since both sense and antisense transcripts were detected in this region and both sense and antisense strands are predicted to fold separately into inverted repeats (fig. 3) that are excellent substrates for dicer in vitro (Smalheiser et al, 2011a). Thus, a given small RNA processed at this locus will bind perfectly or near-perfectly to 4 different RNAs – the SynGAP1 pre-mRNA, the antisense transcript, the complementary small RNA arising from the sense hairpin, and the complementary small RNA arising from the antisense hairpin. This allows for quite sophisticated computation, since the actions of a given small RNA may differ depending on the concentration of the sense and antisense RNAs, whether the sense and antisense RNA transcripts are present as hybrids, and the extent to which the transcripts have been processed to small RNAs. It has also not been excluded that a novel ncRNA may be present as an independent transcriptional unit within the SynGAP1 intron.
Figure 2. Small RNAs aligned to the SynGAP1 locus.
Shown are all unique sequences that mapped to SynGAP1, including those that aligned to the forward or plus strand (placed on top) and to the reverse or minus strand (placed below the forward sequences).
Figure 3. Predicted secondary structure of RNA corresponding to the region within the SynGAP1 locus that aligns with small RNAs.
(A) The RNA encoded on the forward strand in the region covered by small RNAs (see Fig. 2) is predicted to form a perfect hairpin inverted repeat. (B) RNA encoded on the reverse strand forms an almost-perfect hairpin as well. Colors indicate the probability of base-pairing at each particular residue.
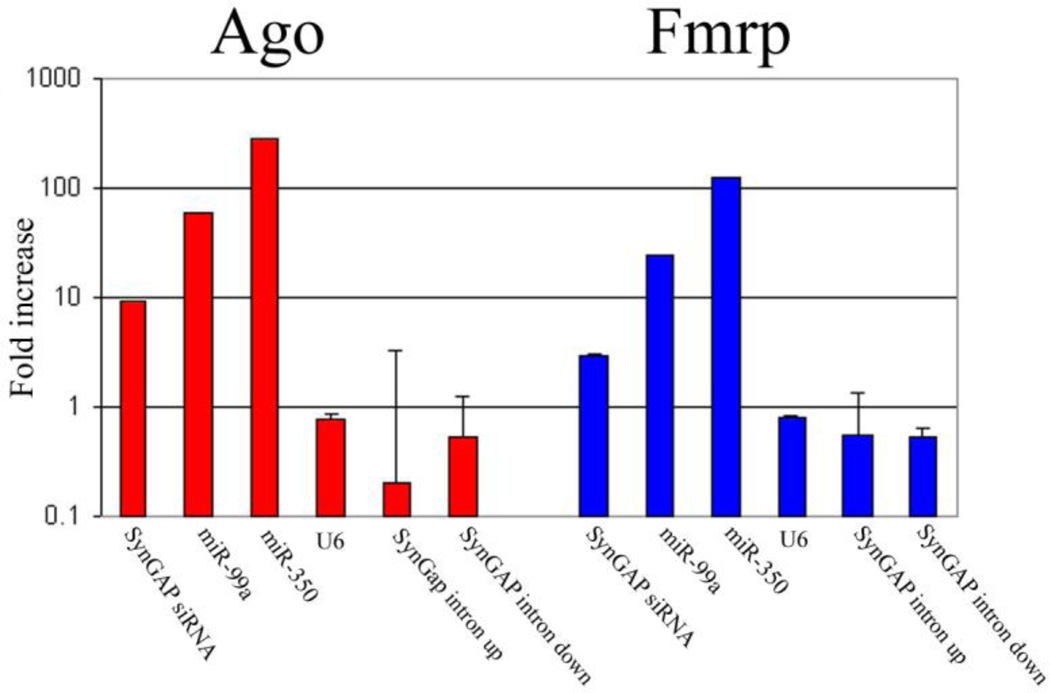
The major SynGAP1 siRNA is presumably involved in gene silencing since it is associated with Argonaute homolog proteins and FMRP (Fig. 4). The locus of siRNA biogenesis and action is still not entirely clear. Although the SynGAP1 pre-mRNA is likely to be spliced predominantly within the nucleus, we have also detected transcripts in synaptic fractions containing the intronic inverted repeat (Lugli et al, ms. in preparation), suggesting the possibility that a subset of SynGAP1 mRNA containing retained introns may be transported into dendrites (Buckley et al, 2011) and processed by dicer (Lugli et al, 2005, 2008) to form SynGAP1 siRNA locally near synapses.
Figure 4. Co-immunoprecipitation of SynGAP1 siRNA with Argonaute homolog proteins and FMRP.
The adult mouse forebrain S1 supernatants were immunoprecipitated using equal amounts of 4F9 anti-Ago antibody, anti-Fmrp antibody, or an irrelevant antibody against synapsin I. Equal fractions of the immunoprecipitates were measured for RNA content by qPCR as described in Materials and Methods. Each experiment was carried out on duplicate samples (error bars, SDs across duplicate samples), and each sample was assayed in duplicate. For each RNA, we plotted the ratio of the RNA abundance detected in the 4F9 or Fmrp immunoprecipitates relative to the synapsin I immunoprecipitates. SynGAP1 siRNA binding to 4F9 was impressively almost 10 times above baseline, although not quite as enriched as several miRNAs (mir-99a and mir-350). In contrast, U6 RNA did not show any specific binding nor did SynGAP1 intronic sequences derived from the sense strand just upstream or downstream from the intronic hairpin that gives rise to the siRNA. SynGAP1 and miRNAs also showed detectable binding to Fmrp, which in brain is a RISC-associated protein (e.g., Lugli et al. 2005; data not shown).
A genome-wide examination of small RNAs that derive from inverted repeats revealed a total of eight gene loci (Abca2, Arhgef17, Camk2a, Gap43, Rab40b, Slc17a7, Syn1, and SynGAP1) that mapped to closely adjacent sites in sense orientation which were not annotated as being contained within ncRNAs or genomic repeats.. Almost all (>99%) of the sequences mapping to these loci were 20–23 nt in length. All of these mapped within introns, but they do not appear to represent intronic miRNAs, since the small RNAs did not map to ESTs or annotated transcripts of the size of pre-miRs, did not map to genomic regions that show high cross-species conservation between mouse and man, and did not map near any known miRNAs. Interestingly, half of the hairpin endo-siRNAs are major synaptic components and/or regulators of synaptic plasticity, including SynGAP1, GAP43, CAMK2a, and synapsin I, and several are regulators of signaling (Arhgef17, Rab40b, Slc17a7). When the set of hairpin endo-siRNAs (Abca2, Arhgef17, Camk2a, Gap43, Rab40b, Slc17a7, Syn1, and SynGAP1) was pooled and tested as a group, a 22% increase was observed in the training vs. pseudo-training comparison that was highly significant (P = 0.00012); the pseudo-training and nose-poke control groups were not significantly different from each other.
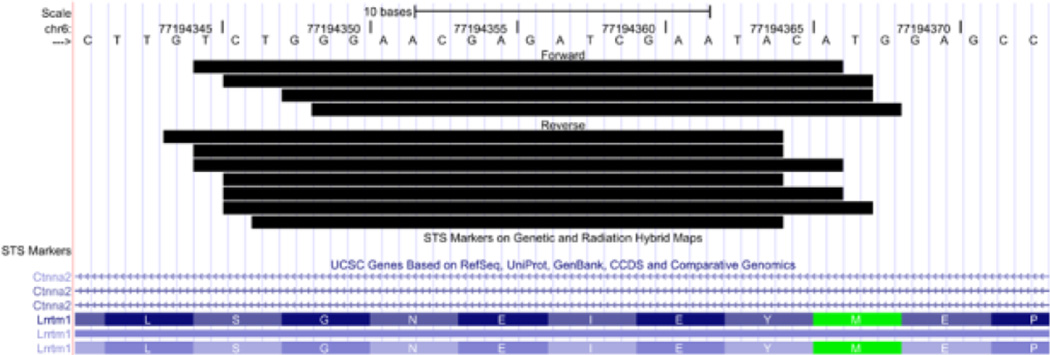
We then looked for small RNAs that mapped to overlapping regions on both sense and antisense strands. One prominent example was found: More than fifty unique sequences mapped to Ctnna2 (catenin [cadherin associated protein], α2) in both sense and antisense orientations (eight to 13 sequences were detected in each sample). The majority of these mapped to the Lrrtm1 gene, which is encoded on the plus strand and lies entirely within an intron of Ctnna2 encoded on the minus strand. Most importantly, 11 distinct sense and antisense sequences overlapped each other extensively, exhibited overhangs of 0–3 nt, and were co-expressed in three of the samples (Fig. 5). The LRRTM gene family (Laurén et al. 2003) has been identified as having roles as synaptic organizers (Linhoff et al. 2009), and Lrrtm1 is a candidate gene for schizophrenia (Francks et al. 2007). Similarly, Ctnna2 is a candidate gene for schizophrenia (Mexal et al. 2008) and α-N-catenin protein has been described as participating in the stabilization of dendritic spines in rodent hippocampal neurons (Abe et al. 2004).
Figure 5. Small RNAs aligned to the Ctnna2 locus that putatively arise from processing of sense-antisense RNA hybrids.
Multiple sequences align to a region of the Ctnna2 gene that also encodes the Lrrtm1 locus on the opposite strand. The small RNAs shown here align to both forward and reverse strands and exhibit a high degree of overlap.
Besides this “smoking gun”, many other small RNAs aligned to both strands of loci known to express both sense and antisense transcripts, including Bdnf and many of the loci reported in Smalheiser et al (2008), such as beta-site APP cleaving enzyme (Bace1), sirtuin 3, SNAP25, integrin-linked kinase, and activating transcription factor 5.
Other small RNAs expressed within hippocampus
As expected, deep sequencing of the hippocampus revealed a wide variety of microRNAs and their variants (RNA editing variants, 5’-end and 3’-end variants, miR* sequences, offset miRNAs, antisense miRNAs, etc.). We also detected many mirtrons (pre-miRs that are processed by splicing-out and trimming from short introns without the action of drosha, followed by dicer processing into small RNAs) (Berezikov et al, 2007). A few sequences were hard to classify; for example, a set of small RNAs was derived from the protein-coding region of DGCR8 in sense orientation (Smalheiser et al, 2011a). Small RNAs were also detected arising from specific sites within abundant cellular noncoding RNAs including snoRNAs (C/D box and H/ACA box), 28S rRNA, 18S rRNA, Y1 RNA, Rmrp (Smalheiser et al, 2011a) as well as certain tRNAs and a mitochondrial noncoding transcript (Smalheiser et al, 2011b) [this list would likely be even longer if we had not excluded RNAs that align to multiple sites in the genome, since many ncRNAs are represented by multiple gene copies]. Some of the noncoding RNAs (e.g., Y1 RNA) fold into classical double-stranded regions that would be expected to be good dicer substrates, whereas others (e.g., most snoRNAs) show atypical precursor secondary structures. Nonetheless, others have shown that at least some of these noncoding RNA-derived small RNAs are formed in a dicer-dependent manner (e.g., Cole et al, 2009; Taft et al, 2009) and independent of drosha or DGCR8 (e.g, Babiarz et al, 2011).
Unexpectedly, we observed that small RNAs deriving from mirtrons and cellular noncoding RNAs were expressed in a range of sizes. Sequences in the 20–23 nt. range were constitutively expressed in all treatment groups, showing only modest elevations with training as described for siRNA-like small RNAs. In contrast, longer sequences in the 25–30 nt. range were observed predominantly in the training group. These 25–30 nt. sequences shared the same precise 5’-end as the 20–23 nt. sequences, but had variable 3’-ends. Strikingly, the 25–30 nt. small RNAs as a class showed little expression in the pseudo-training or nose-poke groups and exhibited up to 100–200 fold elevations in the training group (Table 1). Neither putative siRNAs nor most microRNAs had associated sequences in the 25–30 nt size range; thus, this phenomenon was restricted to small RNAs deriving from mirtrons and noncoding RNAs (Smalheiser et al, 2011a).
Table 1.
All RNAs in the dataset, divided by size class and treatment group.
| Length | Group | Unique Seq |
Seq Counts |
p-value |
|---|---|---|---|---|
| 15–17 | nose-poke | 8244 | 2744.57 | 7.25E-08** |
| 15–17 | pseudo-training | 8244 | 2052.58 | |
| 15–17 | training | 8244 | 1778.35 | 0.0069** |
| 20–23 | nose-poke | 34801 | 14820.52 | 0.0036** |
| 20–23 | pseudo-training | 34801 | 20440.43 | |
| 20–23 | training | 34801 | 23633.06 | 4.59E-07** |
| 25–30 | nose-poke | 6410 | 703.23 | 0.436 |
| 25–30 | pseudo-training | 6410 | 666.31 | |
| 25–30 | training | 6410 | 43269.34 | 7.99E-07** |
| 31–35 | nose-poke | 219 | 19.5 | 2.20E-05** |
| 31–35 | pseudo-training | 219 | 59.62 | |
| 31–35 | training | 219 | 2153.03 | 0.017* |
Shown are the total number of unique sequences and total number of normalized sequence counts in each size class for each treatment group. Training and nose-poke groups are each compared to the pseudo-training control group.
p < 0.05 by t-test.
p < 0.01 by t-test.
Reprinted from Smalheiser et al (2011a) with permission.
Recently, Lee et al (2011) reported the existence of a subset of piRNAs that are expressed in hippocampus, which arise from unique genomic loci and which associate with the Argonaute homologue MIWI. These “piRNAs” actually derive from known noncoding RNAs and were included in our dataset – for example, the four most abundant sequences described in this study (DQ541777, DQ705026, DQ555094 and DQ719597) align to a Y1 RNA, a snoRNA, a rRNA, and a snoRNA, respectively. In our dataset, the piRNA sequences are overlapped by a variety of small RNA sequences ranging from 18 to over 30 nt., and as described above, those in the >25 nt. size range show large elevations in the training group (see Supplement 1 in Smalheiser et al, 2011a). Interestingly, Lee et al (2011) showed that DQ541777 and MIWI are expressed within dendrites, and functional blockade of the piRNA in cultured hippocampal neurons affected dendritic spine area. This suggests that the 25–30 nt. size class of training-induced small RNAs may have functional effects within dendrites, but may differ from endogenous siRNAs, miRNAs and ncRNA-derived 20–23 nt. small RNAs in that they are associated with a different type of Argonaute family homologue (MIWI instead of Ago).
There are, indeed, many similarities between endogenous siRNAs and piRNAs in terms of being associated with Argonaute family homologues, sharing 3’ modifications, and forming a silencing complex that can target both repeat elements and cellular genes. However, it is important to distinguish the population of “piRNAs” that derive from abundant ncRNAs from the better characterized population of piRNAs, identified first in testes, which also have recently been detected and characterized within many other tissues including macaque monkey cortex (Yan et al, 2011). Typical piRNAs arise mainly from intergenic loci and show a very strong tendency (~80%) to have a U residue at the 5’-end; many of the sequences form by the so-called ping-pong pathway involving both MIWI and MILI. In contrast, only 24% of the ncRNA-derived piRNAs (27–28 nt.) detected in our mouse forebrain dataset expressed 5’ U residues (Smalheiser et al, 2011a, supplementary data).
In contrast to the results of Smalheiser et al (2011a) which examined B57Bl/6 mice at P60 or greater, Babiarz et al (2011) did not detect significant expression of endogenous siRNAs in their study of P21 mouse hippocampus, although they did detect mirtrons and snoRNA-derived small RNAs and demonstrated that those RNA classes were DGCR8- independent / dicer-dependent. The reason for this discrepancy is unknown, although differences in RNA isolation methods, mouse age and strain, or different methods of sequence filtering and analysis may be involved.
Discussion. Where are we now?
Deep sequencing of adult mouse hippocampus has revealed a large population of small RNAs having characteristics of endogenous siRNAs. Moreover, the examples having the best documentation are derived from genes involved with synaptic proteins or signaling. Thus, these observations support our original hypothesis that RNA interference may regulate long-term gene expression in the brain. However, we have not yet proven that siRNAs arise from dicer processing of sense-antisense transcript hybrids and intronic hairpins in vivo.
Moreover, we did not originally anticipate that small RNAs would also be processed from abundant cellular noncoding RNAs, nor that longer 25–30 nt. RNA species would be formed specifically during training. In fact, the dramatic ~100-fold up-regulation of the 25–30 nt. small RNAs is much more striking and selective than the modest 10–20% increase that is observed in the 20–23 nt. siRNA/miRNA classes. (We are currently attempting to verify that this apparent increase in abundance is, indeed, due to an actual increase in sequence counts rather than to a selective, learning-specific loss of 3’-modifications which might cause more efficient detection in the deep sequencing pipeline.) The available evidence suggests that the siRNAs associate with Ago whereas the 25–30 nt. RNAs may associate with MIWI. Moreover, there is circumstantial evidence that at least some of the siRNAs and 25–30 nt. RNAs may be processed locally within dendrites, where they may regulate specific targets.
These findings suggest that endogenous siRNAs and ncRNA-derived small RNAs might be major regulators of synaptic plasticity and learning. The relatively low expression of siRNAs in deep sequencing studies should not be interpreted to mean that they are functionally unimportant – at least, not until further studies can be carried out to learn whether deep sequencing assays under-detect the class of siRNAs because they carry specific 5’- and/or 3’-end modifications (McCormick et al, 2011). As well, siRNAs are expected to be much more potent than miRNAs because they bind perfectly to their targets, so their levels should not (perhaps even must not) be as high as the high abundance miRNAs.
Small RNAs are currently thought to be associated with an Argonaute homologue that targets other RNAs to regulate their translation and/or stability. Unlike the case of miRNAs, which have multiple targets of relatively low affinity, siRNAs should silence one or a small number of perfect targets, though they probably show miRNA-like targeting of trans targets as well (Song et al, 2011). However, we know little about how (or where in the cell) small RNAs may function in the context of synaptic plasticity. For example, it is not clear whether siRNAs in mammalian brain utilize noncanonical RdRP enzymes to form secondary siRNAs, which may be expected to have long-lasting or self-amplifying effects. Besides the direct interactions of small RNAs with target RNAs, different classes of small RNAs might potentially compete with the entire population of miRNAs for binding to RISC (cf. Khan et al, 2009; Haussecker et al, 2010). In the nucleus, it is possible that small RNAs may have either enhancing or inhibitory effects on gene transcription in an Ago-dependent manner (Chu et al, 2010), or may control alternative splicing (Allo et al, 2009). At least in plants, yeast and C. elegans, siRNAs regulate DNA methylation of specific genes and heterochromatin domains, and there is increasing evidence that this may occur in mammals as well (Palanichamy et al, 2010).
It is also likely that small RNAs not only affect the translation of specific targets, but also have general effects on the protein synthesis machinery. For example, most of the noncoding RNAs that give rise to small RNAs are involved in protein synthesis and are induced during plasticity by stimulation of the mTOR pathway (Smalheiser et al, 2011a). Processing of pre-rRNAs has been shown to be dependent on drosha, dicer and Ago (Liang and Crooke, 2011). Thus, one function for the induction of ncRNA-derived small RNAs may be to support long-term protein synthesis and cellular growth following synaptic stimulation.
Conclusions. Where are we going?
The research agenda for the next few years is simple: Perturb individual endogenous siRNAs and related small RNAs in neural systems, and learn how they regulate the expression of their target genes, as well as the read-out of neural development, synaptic plasticity, and behavior. As is the case with many of the miRNAs, individual siRNAs are likely to confer robustness on gene networks and their functions may be most apparent when organisms are subjected to environmental stressors (cf. Lucchetta et al, 2009).
To date, gene mapping and association studies in neuropsychiatric diseases have focused largely on exons of protein-coding genes, although several studies have sought to identify SNPs that occur in miRNAs and miRNA target sites. It will be important to confirm that endogenous siRNAs are expressed in human brain and to examine whether loci that are processed to form endogenous siRNAs and other small RNAs correlate with susceptibility to neuropsychiatric diseases.
Endogenous siRNAs are only one part of a larger revolution in our understanding of genomics. The number of genes encoding noncoding RNAs is now known to be ~4 times as great as the number of protein-coding genes in the human genome. Bidirectional transcription at promoters is now appreciated as the rule rather than the exception, and natural antisense transcripts are now known to be common regulators of protein-coding genes. MiRNAs, the best understood class of small RNAs, can arise from at least three different pathways of biogenesis in mammals (drosha dependent, drosha-independent mirtrons and Ago dependent). Besides endogenous siRNAs derived from protein-coding genes and those derived from abundant cellular noncoding RNAs, there are piRNAs (of several types), promoter-associated RNAs, splice-site RNAs, tRNA-derived small RNAs, vault-derived small RNAs, 3’-UTR derived small RNAs, transposon-derived small RNAs, centrosomal repeat RNAs, and so on. The implications of this revolution for neuroscience have only begun to be explored (Mercer et al, 2008; Mattick, 2011). Finally, endogenous siRNAs may relate to three emerging new concepts that are likely to become prominent themes in neuroscience over the coming decade:
First, there is increasing recognition that genomic repeat elements (transposons and “junk” DNA) have physiologic roles in regulating normal gene expression, and that they may be major driving forces in evolution, including evolution of the human brain (Britten, 2010; Mattick, 2011). Genomic repeat elements can be a source of novel microRNA genes (Smalheiser and Torvik, 2005) as well as a source of miRNA target sites within mRNAs (Smalheiser and Torvik, 2006). Genomic repeats may act in real time within the life of an organism as well: transposition of LINE1 elements has been shown to occur within individual precursors of mammalian CNS neurons (Muotri et al, 2010), and cellular stresses cause the rapid, transient expression of Alu-containing transcripts (Liu et al, 1995). Although this review has emphasized the importance of endogenous siRNAs that target neuronal genes, in fact, the majority of endogenous siRNAs within cells are thought to target genomic repeats (Golden et al, 2008; Ghildiyal and Zamore, 2009). Thus, endogenous siRNAs may be an integral part of gene networks that involve genomic repeats. Conditional dicer knockouts in mice have produced neurodegeneration in several types of neurons, which has been assumed to reflect loss of miRNAs (e.g., Schaefer et al, 2007; Hébert et al, 2010). However, Kaneko et al (2011) have shown that dicer knockout induced degeneration in retinal pigmented epithelial cells is actually due to accumulation of toxic Alu-containing transcripts, which are normally destroyed by direct dicer cleavage and/or by siRNAs arising from such cleavage.
Second, there is increasing awareness that cross-species conservation is not the only hallmark of functional transcripts. In fact, a large number of miRNAs and long ncRNAs are specific to the human lineage. Within the human genome, intronic inverted repeats are highly enriched in genes involved in synaptic functions (Wang and Leung, 2009), suggesting that many hairpin-derived endogenous siRNAs are likely to be human specific as well.
Third, there is extensive evidence for intercellular RNA transfer not only in lower model organisms (plants and C. elegans), but also in mammals: specifically, cells in the immune system can transfer miRNAs, mRNAs and other RNAs among themselves via exosomes and microvesicles (e.g., Théry et al, 2009) both at a distance as well as by local contact (Mittelbrunn et al, 2011). Since neurons and glial cells release exosomes which can have functional effects on recipient cells (e.g., Lachenal et al, 2011; Wang et al, 2011), it is likely that intercellular RNA transfer occurs within the nervous system as well (reviewed in Smalheiser, 2007). Thus, a small RNA made in one cell may have its functional target located in a different cell. Deep sequencing of glioblastoma microvesicles has revealed the existence of many different types of RNAs besides miRNAs, including high concentrations of genomic repeat (Alu and LINE1) containing transcripts (Balaj et al, 2010), suggesting that endogenous siRNAs may also participate in the regulation of intercellular RNA transfer.
Highlights.
A decade ago, RNA interference was proposed to serve as a physiologic means of regulating long-term gene expression in the mammalian brain.
During the intervening years, this hypothesis appeared to be contradicted by both experimental data and theoretical considerations.
More recently, the advent of deep sequencing technology has permitted a reassessment of this issue.
A large population of small RNAs having features characteristic of endogenous siRNAs are detected within adult mouse hippocampus, which derive from genes involved in synaptic structure and signaling, and which showed a significant, though modest (16−22%) up-regulation during olfactory discrimination training.
Small RNAs derived from abundant cellular noncoding RNAs were also detected, especially a subpopulation of RNAs 25–30 nt. in length that showed very large (>100 fold) up-regulation during olfactory discrimination training. Preliminary data suggest that the 25–30 nt. RNAs may associate with MIWI.
Despite their apparent low abundance, endogenous siRNAs and noncoding RNA-derived small RNAs are likely to play an important role in regulating synaptic plasticity.
Highlights.
-
*
Endogenous siRNAs are detected within adult mouse hippocampus.
-
*
These derive from genes involved in synaptic structure and signaling.
-
*
Small RNAs derived from abundant cellular noncoding RNAs are also detected.
-
*
25–30 nt. RNAs showed very large (>100 fold) changes during learning.
-
*
Endo-siRNAs and ncRNA-derived small RNAs may regulate synaptic plasticity.
Acknowledgments
I thank my colleagues Ed Cook, John Davis, Yogesh Dwivedi, John Larson, Giovanni Lugli, Hari Manev, and Vetle Torvik, in alphabetical order, for their support and contributions to this project. Research was supported by NIMH, NIDA and the Stanley Medical Research Institute.
Abbreviations
- Ago
Argonaute homologue protein
- DGCR8
DiGeorge syndrome critical region gene 8
- dsRNA
double-stranded RNA
- ES cells
embryonic stem cells
- miRNA
microRNA
- MIWI
piwil1 (piwi-like homolog 1) in mouse
- ncRNA
noncoding RNA
- piRNA
piwi-interacting RNA
- RdRP
RNA-dependent RNA polymerase
- RISC
RNA induced silencing complex
- RNAi
RNA interference
- siRNA
small inhibitory RNA
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe K, Chisaka O, Van Roy F, Takeichi M. Stability of dendritic spines and synaptic contacts is controlled by αN-catenin. Nat. Neurosci. 2004;7:357–363. doi: 10.1038/nn1212. [DOI] [PubMed] [Google Scholar]
- Alló M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, Klinck R, Chabot B, Kornblihtt AR. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat. Struct. Mol. Biol. 2009;16:717–724. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans . Current Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- Asikainen S, Heikkinen L, Wong G, Storvik M. Functional characterization of endogenous siRNA target genes in Caenorhabditis elegans. BMC Genomics. 2008;9:270. doi: 10.1186/1471-2164-9-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Hsu R, Melton C, Thomas M, Ullian EM, Blelloch R. A role for noncanonical microRNAs in the mammalian brain revealed by phenotypic differences in Dgcr8 versus Dicer1 knockouts and small RNA sequencing. RNA. 2011;17:1489–1501. doi: 10.1261/rna.2442211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, Skog J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nature Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D. Overview of RNA interference and related processes. Current Protoc. Molec. Biol. 2003 doi: 10.1002/0471142727.mb2601s62. May;Chapter 26:Unit 26.1. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Molec. Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley PT, Lee MT, Sul JY, Miyashiro KY, Bell TJ, Fisher SA, Kim J, Eberwine J. Cytoplasmic intron sequence-retaining transcripts can be dendritically targeted via ID element retrotransposons. Neuron. 2011;69:877–884. doi: 10.1016/j.neuron.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer’s role in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U S A. 2007;104:18097–18102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Yue X, Younger ST, Janowski BA, Corey DR. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res. 2010;38:7736–7748. doi: 10.1093/nar/gkq648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, Hannon GJ, Brennecke J. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Kocerha J, Modarresi F, Engström PG, Chalk AM, Brothers SP, Koesema ESt, Laurent G, Wahlestedt C. RNAi screen indicates widespread biological function for human natural antisense transcripts. PLoS One. 2010;5:pii: e13177. doi: 10.1371/journal.pone.0013177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CESt, Laurent G, 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nature Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire AZ. Gene silencing by double-stranded RNA. Nobel Prize Lecture. 2006 doi: 10.1002/anie.200701979. http://nobelprize.org/nobel_prizes/medicine/laureates/2006/fire_lecture.pdf. [DOI] [PubMed]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Francks C, Maegawa S, Laurén J, Abrahams BS, Velayos-Baeza A, Medland SE, Colella S, Groszer M, McAuley EZ, Caffrey TM, et al. LRRTM1 on chromosome 2p12 is a maternally suppressed gene that is associated paternally with handedness and schizophrenia. Molec. Psychiat. 2007;12:1129–1139. doi: 10.1038/sj.mp.4002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB. Context-dependent functions of specific microRNAs in neuronal development. Neural Dev. 2010;5:25. doi: 10.1186/1749-8104-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nature Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden DE, Gerbasi VR, Sontheimer EJ. An inside job for siRNAs. Molec.Cell. 2008;31:309–312. doi: 10.1016/j.molcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Sinskey JL, Sharp PA. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans . Genes Dev. 2005;19:683–696. doi: 10.1101/gad.1247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert SS, Papadopoulou AS, Smith P, Galas MC, Planel E, Silahtaroglu AN, Sergeant N, Buée L, De Strooper B. Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum Molec. Genet. 2010;19:3959–3969. doi: 10.1093/hmg/ddq311. [DOI] [PubMed] [Google Scholar]
- Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, Karikó K, Yoo JW, Lee DK, Hadziahmetovic M, Song Y, Misra S, Chaudhuri G, Buaas FW, Braun RE, Hinton DR, Zhang Q, Grossniklaus HE, Provis JM, Madigan MC, Milam AH, Justice NL, Albuquerque RJ, Blandford AD, Bogdanovich S, Hirano Y, Witta J, Fuch E, Littman DR, Ambati BK, Rudin CM, Chong MM, Provost P, Kugel JF, Goodrich JA, Dunaief JL, Baffi JZ, Ambati J. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nature Biotechnol. 2009;27:549–555. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, Kosik KS. RNAi functions in cultured mammalian neurons. Proc. Natl. Acad. Sci. U S A. 2002;99:11926–11929. doi: 10.1073/pnas.182272699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachenal G, Pernet-Gallay K, Chivet M, Hemming FJ, Belly A, Bodon G, Blot B, Haase G, Goldberg Y, Sadoul R. Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Molec. Cell Neurosci. 2011;46:409–418. doi: 10.1016/j.mcn.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Current Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Larson J, Sieprawska D. Automated study of simultaneous-cue olfactory discrimination learning in adult mice. Behav Neurosci. 2002;116:588–599. [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans . Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lau NC, Robine N, Martin R, Chung WJ, Niki Y, Berezikov E, Lai EC. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res. 2009;19:1776–1785. doi: 10.1101/gr.094896.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurén J, Airaksinen MS, Saarma M, Timmusk T. A novel gene family encoding leucine-rich repeat transmembrane proteins differentially expressed in the nervous system. Genomics. 2003;81:411–421. doi: 10.1016/s0888-7543(03)00030-2. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans . Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee RC, Hammell CM, Ambros V. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA. 2006;12:589–597. doi: 10.1261/rna.2231506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Banerjee S, Zhou H, Jammalamadaka A, Arcila M, Manjunath BS, Kosik KS. Identification of piRNAs in the central nervous system. RNA. 2011;17:1090–1099. doi: 10.1261/rna.2565011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Crooke ST. Depletion of key protein components of the RISC pathway impairs pre-ribosomal RNA processing. Nucleic Acids Res. 2011;39:4875–4889. doi: 10.1093/nar/gkr076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DH, Oh CT, Lee L, Hong JS, Noh SH, Hwang S, Kim S, Han SJ, Lee YS. The endogenous siRNA pathway in Drosophila impacts stress resistance and lifespan by regulating metabolic homeostasis. FEBS Lett. 2011 Aug 30; doi: 10.1016/j.febslet.2011.08.034. [DOI] [PubMed] [Google Scholar]
- Linhoff MW, Laurén J, Cassidy RM, Dobie FA, Takahashi H, Nygaard HB, Airaksinen MS, Strittmatter SM, Craig AM. An unbiased expression screen for synaptogenic proteins identifies the LRRTM protein family as synaptic organizers. Neuron. 2009;61:734–749. doi: 10.1016/j.neuron.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WM, Chu WM, Choudary PV, Schmid CW. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995;23:1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchetta EM, Carthew RW, Ismagilov RF. The endo-siRNA pathway is essential for robust development of the Drosophila embryo. PLoS One. 2009;4:e7576. doi: 10.1371/journal.pone.0007576. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli G, Larson J, Martone ME, Jones Y, Smalheiser NR. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J. Neurochem. 2005;94:896–905. doi: 10.1111/j.1471-4159.2005.03224.x. [DOI] [PubMed] [Google Scholar]
- Lugli G, Torvik VI, Larson J, Smalheiser NR. Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J. Neurochem. 2008;106:650–661. doi: 10.1111/j.1471-4159.2008.05413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maida Y, Yasukawa M, Furuuchi M, Lassmann T, Possemato R, Okamoto N, Kasim V, Hayashizaki Y, Hahn WC, Masutomi K. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461:230–235. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. The central role of RNA in human development and cognition. FEBS Lett. 2011;585:1600–1616. doi: 10.1016/j.febslet.2011.05.001. [DOI] [PubMed] [Google Scholar]
- McCormick KP, Willmann MR, Meyers BC. Experimental design, preprocessing, normalization and differential expression analysis of small RNA sequencing experiments. Silence. 2011;2:2. doi: 10.1186/1758-907X-2-2. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mariani J, Kosik KS, Mehler MF, Mattick JS. Noncoding RNAs in Long-Term Memory Formation. Neuroscientist. 2008;14:434–445. doi: 10.1177/1073858408319187. [DOI] [PubMed] [Google Scholar]
- Mexal S, Berger R, Pearce L, Barton A, Logel J, Adams CE, Ross RG, Freedman R, Leonard S. Regulation of a novel αN-catenin splice variant in schizophrenic smokers. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:759–768. doi: 10.1002/ajmg.b.30679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González MÁ, Bernad A, Sánchez-Madrid F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nature Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanichamy JK, Mehndiratta M, Bhagat M, Ramalingam P, Das B, Das P, Sinha S, Chattopadhyay P. Silencing of integrated human papillomavirus-16 oncogenes by small interfering RNA-mediated heterochromatization. Mol Cancer Ther. 2010;9:2114–2122. doi: 10.1158/1535-7163.MCT-09-0977. [DOI] [PubMed] [Google Scholar]
- Robb GB, Brown KM, Khurana J, Rana TM. Specific and potent RNAi in the nucleus of human cells. Nature Struct. Molec.Biol. 2005;12:133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Schaefer A, O’Carroll D, Tan CL, Hillman D, Sugimori M, Llinas R, Greengard P. Cerebellar neurodegeneration in the absence of microRNAs. J. Exp. Med. 2007;204:1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Hornung V, Hartmann G. siRNA and isRNA: two edges of one sword. Molec. Ther. 2006;14:463–470. doi: 10.1016/j.ymthe.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel G, Saba R, Schratt G. microRNAs in neurons: manifold regulatory roles at the synapse. Current Opinion Genet. Dev. 2011 May 9; doi: 10.1016/j.gde.2011.04.008. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Smalheiser NR. Exosomal transfer of proteins and RNAs at synapses in the nervous system. Biol. Direct. 2007;2:35. doi: 10.1186/1745-6150-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Lugli G, Thimmapuram J, Cook EH, Larson J. Endogenous siRNAs and noncoding RNA-derived small RNAs are expressed in adult mouse hippocampus and are up-regulated in olfactory discrimination training. RNA. 2011a;17:166–181. doi: 10.1261/rna.2123811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Lugli G, Lenon AL, Davis JM, Torvik VI, Larson J. Olfactory discrimination training up-regulates and reorganizes expression of microRNAs in adult mouse hippocampus. ASN Neuro. 2010;2:e00028. doi: 10.1042/AN20090055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Lugli G, Thimmapuram J, Cook EH, Larson J. Mitochondrial small RNAs that are up-regulated in hippocampus during olfactory discrimination training in mice. 2011b doi: 10.1016/j.mito.2011.08.014. Mitochondrion [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Lugli G, Torvik VI, Mise N, Ikeda R, Abe K. Natural antisense transcripts are co-expressed with sense mRNAs in synaptoneurosomes of adult mouse forebrain. Neurosci. Res. 2008;62:236–239. doi: 10.1016/j.neures.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Manev H, Costa E. RNAi and brain function: was McConnell on the right track? Trends Neurosci. 2001;24:216–218. doi: 10.1016/s0166-2236(00)01739-2. [DOI] [PubMed] [Google Scholar]
- Smalheiser NR, Torvik VI. Mammalian microRNAs derived from genomic repeats. Trends in Genet. 2005;21:322–326. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Smalheiser NR, Torvik VI. Alu elements within human mRNAs are probable microRNA targets. Trends in Genet. 2006;22:532–536. doi: 10.1016/j.tig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Soldà G, Boi S, Duga S, Fornasari D, Benfante R, Malcovati M, Tenchini ML. In vivo RNA-RNA duplexes from human alpha3 and alpha5 nicotinic receptor subunit mRNAs. Gene. 2005;345:155–164. doi: 10.1016/j.gene.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Song R, Hennig GW, Wu Q, Jose C, Zheng H, Yan W. Male germ cells express abundant endogenous siRNAs. Proc Natl Acad Sci U S A. 2011;108:13159–13164. doi: 10.1073/pnas.1108567108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nature Rev. Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- Wang S, Cesca F, Loers G, Schweizer M, Buck F, Benfenati F, Schachner M, Kleene R. Synapsin I is an oligomannose-carrying glycoprotein, acts as an oligomannose-binding lectin, and promotes neurite outgrowth and neuronal survival when released via glia-derived exosomes. J. Neurosci. 2011;31:7275–7290. doi: 10.1523/JNEUROSCI.6476-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Carmichael GG. Effects of length and location on the cellular response to double-stranded RNA. Microbiol. Molec. Biol Rev. 2004;68:432–452. doi: 10.1128/MMBR.68.3.432-452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Leung FC. A study on genomic distribution and sequence features of human long inverted repeats reveals species-specific intronic inverted repeats. FEBS J. 2009;276:1986–1998. doi: 10.1111/j.1742-4658.2009.06930.x. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, Surani MA, Sakaki Y, Sasaki H. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Weinmann L, Höck J, Ivacevic T, Ohrt T, Mütze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell. 2009;136:496–507. doi: 10.1016/j.cell.2008.12.023. [DOI] [PubMed] [Google Scholar]
- Werner A, Swan D. What are natural antisense transcripts good for? Biochem. Soc. Trans. 2010;38:1144–1149. doi: 10.1042/BST0381144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Hu HY, Jiang X, Maierhofer V, Neb E, He L, Hu Y, Hu H, Li N, Chen W, Khaitovich P. Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res. 2011;39:6596–6607. doi: 10.1093/nar/gkr298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JY, DeRuiter SL, Turner DL. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl. Acad. Sci. U S A. 2002;99:6047–6052. doi: 10.1073/pnas.092143499. [DOI] [PMC free article] [PubMed] [Google Scholar]