Abstract
Purpose
Adolescent idiopathic scoliosis (AIS) is reported to be associated with the two traditional estrogen receptor genes, ESR1 and ESR2. Yet, the novel estrogen receptor G protein-coupled estrogen receptor 1 (GPER) has not been studied. To investigate the association of GPER gene polymorphisms with the onset and deterioration of AIS, we performed a case-control study.
Methods
Clinical information was recorded, blood samples were taken and genomic DNA was extracted. After resequencing the gene in 45 cases and 45 controls who were randomly selected, 16 tag single nucleotide polymorphisms (SNPs) were selected. Then the association study was extended by an additional 344 patients and 293 controls with direct sequencing and a TaqMan-based genotyping assay. The chi-square test and logistic regression were used to analyse the genotypic and allelic association. One-way analysis of variance was used to compare the mean maximum Cobb angles and ages with different genotypes in the case-only data set.
Results
No association was observed between the polymorphisms of the GPER gene and susceptibility to AIS. However, heterozygotes in three SNPs of the gene (rs3808351, rs10269151 and rs426655s3) were related significantly with the curve severity in AIS patients (P = 0.004, 0.048 and 0.028, respectively).
Conclusions
Our results demonstrate that GPER gene polymorphisms are associated with the severity of curvature in AIS; deficits of GPER may contribute to the deterioration of spine deformity.
Introduction
Adolescent idiopathic scoliosis (AIS) is a tridimensional deformity of the spine consisting of lateral deviation of the spine and rotation of the vertebrae, which arises mainly in girls during the pubertal growth spurt. Although its exact aetiology remains elusive, AIS has been accepted as a multifactorial disease with genetic predisposing factors [1, 2]. Several genes have been found to be associated with the disease, including those of estrogen receptors.
Estrogen signalling has been reported to be crucial to the onset and development of AIS [3, 4]. In spite of some reports showing the serum oestradiol levels to be significantly lower in AIS teenagers [5, 6], other studies found no difference in circulating estrogen levels between patients and controls [7, 8], and a hypothesis has been presented that not the estrogen itself but the response of bone cells to estrogen impacts scoliosis [4]. Estrogen works through two nuclear receptors, the estrogen receptor alpha (ERα) and the estrogen receptor beta (ERβ), and through a membrane receptor, the G protein-coupled estrogen receptor 1 (GPER) [9–11]. Both ERα and ERβ are traditional members of nuclear hormone receptors, and their genes have been found to be correlated with the susceptibility and curve severity of AIS[12–14], suggesting that deficits of these two receptors may induce skeletal malformation.
Despite studies of scoliosis focused on ERα and ERβ, the novel estrogen receptor GPER, or GPR30, has not been investigated yet. GPER is a member of the G protein-coupled receptors spanning the membrane seven times (7TM GPCRs), not only eliciting the rapid non-genomic responses of estrogen but also triggering more long-term transcriptional responses [9, 15]. It mediates a wide range of responses to estrogen in a large variety of cell types. Previous studies found GPER expressed in chondrocytes of human growth plate [16], and in osteoblasts, osteocytes and osteoclasts [17], modulating the bone growth on the foundation of a normal estrogenic response [18]. GPER seems to take part in the estrogen-related regulation of skeletal development, and it may also be involved in the initiation or deterioration of AIS.
In this respect, we undertook the first association study between the GPER gene and AIS. We conducted an investigation to detect single nucleotide polymorphisms (SNPs) present in the GPER genes in order to examine if they may confer a significant susceptibility or have an association with the curve severity of AIS.
Materials and methods
Subjects
A total of 389 AIS patients and 338 healthy controls were involved in our study. Tag SNP screening was performed on 45 patients and 45 controls; further genotyping consisted of an additional 344 AIS cases and 293 controls. All of the patients (53 men and 336 women, ages at onset ranging from ten to 19 years with an average of 15.62 ± 3.36 years) were recruited from the scoliosis clinic at the Sun Yat-sen Memorial Hospital and the First Affiliated Hospital of Sun Yat-sen University. Diagnoses of the patients were confirmed clinically by using the Adams forward bending test and radiographically with posteroanterior radiographic images of the whole spine, with the diagnosis criteria of existing rotational prominence and a maximum Cobb angle above 15°. Patients with scoliosis secondary to congenital vertebral malformation, neuromuscular disorders, syndromic disorders or complicated with other hereditary disorders were excluded from the study. Patients’ clinical information including age, gender, curve pattern, Cobb angle and Risser sign were recorded. Follow-up was performed until each patient reached a Risser sign grade 5, at which point the Cobb angle was recorded as the maximum Cobb angle. The maximum Cobb angle ranged from 15 to 90°.
Meanwhile, 49 healthy men and 289 healthy women were invited to participate in the study through advertisement (ages ranging from ten to 29 years with an average of 14.49 ± 2.58 years). All of the controls were examined with the forward bending test to exclude any scoliosis, and radiographs were taken for validation in case of any uncertainty, as in previous research [19, 20]. The controls were also eliminated if they had suffered from any congenital deformity of the spine or had a family history of scoliosis. All of the cases and controls were Han Chinese from south China. Informed consent to DNA analysis was signed by all subjects or their parents and witnessed by the clinician. The study was approved by the Clinical Research Ethics Committee of the two hospitals.
Blood sampling
Blood samples were collected from each subject through venipuncture. Genomic DNA was isolated with Tiangen DNA blood Mini kits (Tiangen, Beijing, China) according to the manufacturer’s instructions.
Resequencing of the GPER gene and SNP identification
In order to screen variations on exons of GPER in south Han Chinese, resequencing was performed first in 45 cases and 45 controls, who were randomly selected from the collection (a total of 18 men and 72 women, mean age 15.50 ± 2.76 years). Primers were designed for all exons and intron-exon boundaries of GPER, with 100–200 bp extensions into intronic regions (see Table 1). The polymerase chain reaction (PCR) products were sequenced in both directions by the ABI Sequence Analyzer 3730XL (Applied Biosystems, Foster City, CA, USA), and the results were then compared with sequences retrieved from the UCSC Genome Browser (http://genome.ucsc.edu/).
Table 1.
All exon sequencing primers
| Number | Forward | Reverse | Coverage region |
|---|---|---|---|
| 1 | TGTCCCCCCACCCACCAG | GCCTCACAAGCACCTCGC | Exon 1 |
| 2 | TCATCCAAATACAAAAAG | AAATCTGAACTCCGAAAC | Exon 2 |
| 3 | GGTTTGTATCTGTGGGTGAA | GTGAGGTGTCTGGTCTGTGT | Exon 3 |
| 4 | TGCTCCTGACACACCCA | AGAGGCACGAGAGGAAC | Exon 4 |
| 5 | AACAAACCCAACCCAAACCA | CCTGCCTCCTACATTCA | Exon 4 |
Linkage disequilibrium (LD) analysis, haplotype construction and htag SNP selection
LD analysis, haplotype construction and htag SNP selection of the gene region were performed with Haploview 4.2 [21]. SNPs obtained from the 45 sequenced healthy subjects with minor allele frequency (MAF) ≥1% were enrolled in the analysis.
Genotyping methods
Sixteen rare tag SNPs were identified for further analyses (see Table 2). They were then detected in all patients and controls by the TaqMan-based genotyping assay (7 SNPs: G1–G5, G15 and G16) or direct sequencing (9 SNPs: G6–G14) as described above on account of the difficulty in designing the TaqMan probes for those sites. All of the primers and probes are listed in Table 3. The TaqMan-based genotyping assay was carried out with the ABI 7500 real-time PCR System (Applied Biosystems, Foster City, CA, USA). The reaction mix contained ddH2O 3.25 μl, MgCl2 3 μl, buffer 1 μl, dNTP 0.25 μl, primers 0.2 μl, probes 0.05 μl, ROX reference dye 0.05 μl and DNA template 2 μl in a total volume of approximately 10 μl. Amplification was carried out with a ten minute cycle at 95°C, 50 cycles at 95°C for 30 seconds and 63°C for one minute. The result was analysed with the SDS v1.2 × System Software (Applied Biosystems, Foster City, CA, USA). For quality control in each plate, the sample genotypes confirmed by direct sequencing were used as positive controls and no template control as negative controls.
Table 2.
The 16 tag SNPs selected for the association study
| SNPs | Genome position (chr 7) | Reference SNP ID in NCBI | Location | Nucleotide substitutionsa | Residue change |
|---|---|---|---|---|---|
| G1 | 1121849 | rs12701969 | 5′-UTR | −9516A > G | – |
| G2 | 1121886 | – | 5′-UTR | −9479 G > A | – |
| G3 | 1126659 | rs3808351 | 5′-UTR | −4706 G > A | – |
| G4 | 1126876 | rs10269151 | 5′-UTR | −4489 G > A | – |
| G5 | 1127705 | rs33987461 | Intron | −3660 G > T | – |
| G6 | 1131356 | rs3802141 | 5′-UTR | −9 T > C | – |
| G7 | 1131378 | – | Coding region | 14 C > T | p.Ser5Phe |
| G8 | 1131394 | rs34497267 | Coding region | 30 G > A | p.Val10Val |
| G9 | 1131411 | rs11544331 | Coding region | 47 C > T | p.Pro16Leu |
| G10 | 1132153 | rs3808352 | Coding region | 789 G > A | p.Ala263Ala |
| G11 | 1132413 | – | Coding region | 1049 G > A | p.Arg350His |
| G12 | 1132671 | rs4266553 | 3′-UTR | 1307 C > G | – |
| G13 | 1132691 | rs1133041 | 3′-UTR | 1327 C > T | – |
| G14 | 1132720 | rs12702047 | 3′-UTR | 1356 G > A | – |
| G15 | 1132972 | rs3808353 | 3′-UTR | 1608 G > A | – |
| G16 | 1133171 | rs3808354 | 3′-UTR | 1807 G > T | – |
NCBI National Center for Biotechnology Information
aThe A of the translation initiation codon is denoted as +1
Table 3.
All primers and probes for each SNP in our research
| SNPs | Amplification primers | TaqMan probes or sequencing primers | ||
|---|---|---|---|---|
| Forward | Reverse | |||
| G1 | CTTCCTCTCAGTCTGACCATTGTTC | GCGGGAGCACTCTGACCTT | FAM-CATTCCATGGTACTCG-MGB | HEX-CATTCCGTGGTACTC-MGB |
| G2 | CGGGAGCACTCTGACCTTACC | TCCTCTCAGTCTGACCATTGTTCTT | FAM-CACCTGCACCCGACAGTGATGAGT-TRAMA | HEX-TCCACCTGCACCTGACAGTGATGAG-TRAMA |
| G3 | GTAGGAGTGAGATTCGCTGAAGTTC | CCCCCTACTGGCCATTTGA | FAM-CCTCGCTCTGCCCTCATGGG-TAMRA | HEX-CCTCGCTCTACCCTCATGGGGC-TAMRA |
| G4 | CCGCCTGCACGAGACTGT | CTTCCGGTCCCAAGCATTC | FAM-CAACATCTGGACGGCAGGTAAGTTCC-TAMRA | HEX-CAACATCTGGACAGCAGGTAAGTTCCG-TAMRA |
| G5 | CCCGCGAGGGAAGGTT | GGAGAAAAAGGGCCAGCAA | FAM-CCTCCGCCCCGCTCCTCT-TAMRA | HEX-CCTCCGCCCAGCTCCTCTCTG-TAMRA |
| G6–G9 | TGCTCCTGACACACCCA | AGAGGCACGAGAGGAAC | TGCTCCTGACACACCCA | |
| G10, G11 | AACAAACCCAACCCAAACCA | CCTGCCTCCTACATTCA | TACACGGCACTGCTGAA | |
| G12–G14 | AACAAACCCAACCCAAACCA | CCTGCCTCCTACATTCA | CCGTGTAGACAGCCTTGG | |
| G15 | GCAAGGTGCTGGTGGGTCT | ATTGCTGCCATCCTGGAAGA | FAM-CTGGACGTCGCGGTGTGTCCT-TAMRA | HEX-TGGACGTCGCAGTGTGTCCTCTG-TAMRA |
| G16 | TTCCAGGATGGCAGCAATG | CGGTCCCAGTCCACATGAGT | FAM-TGGAGCGCCCGCCGTC -TAMRA | HEX TCTGTGGAGCTCCCGCCGTC -TAMRA |
Statistical analyses
The Hardy-Weinberg equilibrium (HWE) test was performed in controls. The genotypic and allelic association analyses were performed by chi-square tests. Logistic regressions were used to adjust the confounding effects of age and gender, while Bonferroni adjustment was performed for multiple tests of all SNPs. One-way analysis of variance (ANOVA) was used in the comparison of mean maximum Cobb angles and ages of different genotypes in the case-only data set. Given the abnormal distribution of the Cobb angle a logarithmic transformation was employed. The analyses were performed using SPSS software (SPSS for Windows, Rel. 17.0.0. 2008, SPSS Inc., Chicago, IL, USA).
Results
LD structure and tag SNP selection
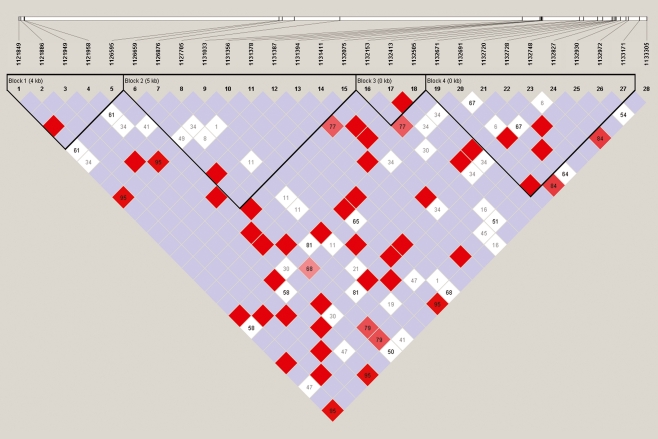
Four LD blocks in the 11 kb genomic region were revealed. The frequencies of the haplotypes in each block were also observed. We identified 16 tag SNPs: G1 and G2 in block 1, G3–G9 in block 2, G10 and G11 in block 3 and G12–G16 in block 4 (see Fig. 1 and Table 2).
Fig. 1.
LD pattern of the GPER gene from the 45 healthy subjects. There were four LD blocks in the 11 kb sequenced genomic region of the gene, which were calculated by the solid spine of the LD algorithm with a minor allele frequency ≥1%. The highlighted numbers stand for D′
Case-control association studies
The genotype distributions of the 16 SNPs, which are shown in Table 4, were all in HWE. There were no statistically significant differences of any SNPs between the patients and controls by the chi-square test, logistic regression analysis and multiple tests with Bonferroni adjustment (all SNPs Padjusted = 1). Hence, there might be no association between the polymorphism of the GPER gene and the onset of AIS.
Table 4.
Results of the case-control association studies
| SNPs | Genotypea | P value | OR (95% CI)c | |||
|---|---|---|---|---|---|---|
| Control (n = 338) | Case (n = 389) | Genotype | Genotypeb | Allele | ||
| G1 | 133/156/49 | 169/171/49 | 0.548 | 0.192 | 0.270 | 0.886 (0.715–1.098) |
| G2 | 332/6/0 | 383/6/0 | 0.810 | 0.803 | 0.815 | 0.873 (0.280–2.720) |
| G3 | 264/68/6 | 318/59/12 | 0.117 | 0.607 | 0.508 | 0.896 (0.647–1.241) |
| G4 | 308/30/0 | 355/34/0 | 0.959 | 0.823 | 0.969 | 0.990 (0.599–1.636) |
| G5 | 242/91/5 | 301/83/5 | 0.218 | 0.145 | 0.104 | 0.778 (0.575–1.053) |
| G6 | 130/162/46 | 172/168/49 | 0.296 | 0.157 | 0.188 | 0.866 (0.699–1.073) |
| G7 | 317/21/0 | 370/19/0 | 0.445 | 0.566 | 0.451 | 0.786 (0.419–1.474) |
| G8 | 305/33/0 | 360/28/1 | 0.311 | 0.450 | 0.350 | 0.786 (0.474–1.304) |
| G9 | 322/16/0 | 364/24/0 | 0.382 | 0.303 | 0.389 | 1.324 (0.698–2.515) |
| G10 | 290/45/3 | 338/49/2 | 0.798 | 0.617 | 0.611 | 0.902 (0.605–1.344) |
| G11 | 338/0/0 | 384/2/0d | 0.184 | 0.999 | 0.184 | 1.003 (0.999–1.006) |
| G12 | 306/32/0 | 352/34/0d | 0.778 | 0.875 | 0.783 | 0.933 (0.569–1.529) |
| G13 | 226/103/9 | 277/103/6d | 0.294 | 0.127 | 0.135 | 0.809 (0.612–1.069) |
| G14 | 304/34/0 | 348/41/0 | 0.811 | 0.834 | 0.816 | 1.057 (0.663–1.686) |
| G15 | 307/31/0 | 351/38/0 | 0.765 | 0.760 | 0.770 | 1.075 (0.661–1.748) |
| G16 | 293/45/0 | 347/40/2 | 0.199 | 0.597 | 0.444 | 0.846 (0.551–1.299) |
OR odds ratio, CI confidence interval
aThe three values in the “genotype” column indicate the numbers of homozygotes (major allele)/heterozygotes/homozygotes (minor allele) in each SNP, respectively
bAfter adjusting for age and gender by logistic regression
cCalculated for the alleles
dGenotype data of three patients in G11, G12 and G13 were missing, respectively; no data were missing in the control group
Genotype-phenotype studies
The differences between the maximum Cobb angles of different genotypes in G3, G4 and G12 were significant (see Table 5). Post hoc comparison for SNP G3 with the Bonferroni test found that patients with the high-risk genotype GA had larger maximum Cobb angles than with the genotype GG or AA (P = 0.018 and P = 0.019, respectively), while those with the genotype GG and AA had similar maximum Cobb angles (P = 0.316). For SNPs G4 and G12, the maximum Cobb angles in heterozygous patients were larger than in homozygous patients (P = 0.048 and P = 0.028, respectively). A haplotype test of the three loci was performed and LD was observed between G3 and G12 (D′ = 0.934), while G4 was independent of them (D′ = 0.439 and 0.012, respectively). LD pattern analysis found the three SNPs lay in different LD blocks. No differences were observed within the ages of different genotypes of these SNPs. These results imply that the heterozygous mutations in these sites of GPER are related with the curve severity of AIS. Thus, GPER may have a role in the aggravation of AIS.
Table 5.
One-way ANOVA of the maximum Cobb angles in different genotypes
| SNPs | Average of MCAa | One-way ANOVA | ||
|---|---|---|---|---|
| P (MCA) | P (MCA)b | P (age) | ||
| G1 | 25.70 ± 13°/28.12 ± 15°/28.57 ± 14° | 0.228 | 0.270 | 0.485 |
| G2 | 27.06 ± 14°/31.00 ± 17°/–c | 0.508 | 0.512 | 0.782 |
| G3 | 26.57 ± 14°/31.61 ± 15°/19.85 ± 5° | 0.009d | 0.004d | 0.428 |
| G4 | 26.61 ± 13°/32.47 ± 19°/– | 0.023d | 0.048d | 0.600 |
| G5 | 27.30 ± 14°/26.25 ± 13°/31.00 ± 14° | 0.703 | 0.734 | 0.382 |
| G6 | 26.66 ± 13°/27.52 ± 15°/27.39 ± 13° | 0.850 | 0.920 | 0.957 |
| G7 | 27.25 ± 14°/24.63 ± 13°/– | 0.441 | 0.378 | 0.409 |
| G8 | 26.68 ± 13°/32.57 ± 18°/36.00 | 0.094 | 0.128 | 0.713 |
| G9 | 27.15 ± 14°/26.71 ± 17°/– | 0.886 | 0.538 | 0.459 |
| G10 | 26.73 ± 14°/29.61 ± 15°/32.50 ± 17° | 0.371 | 0.323 | 0.726 |
| G11 | 27.22 ± 14°/15.00°/– | 0.234 | 0.135 | 0.713 |
| G12 | 26.61 ± 13°/32.79 ± 18°/– | 0.017d | 0.028d | 0.745 |
| G13 | 27.29 ± 14°/26.67 ± 13°/29.17 ± 13° | 0.881 | 0.880 | 0.328 |
| G14 | 26.94 ± 14°/28.68 ± 15°/– | 0.465 | 0.533 | 0.903 |
| G15 | 27.07 ± 14°/27.61 ± 14°/– | 0.829 | 0.791 | 0.905 |
| G16 | 26.66 ± 13°/30.90 ± 18°/32.00 ± 5° | 0.189 | 0.274 | 0.788 |
MCA maximum Cobb angle
aThe three values in this column indicate the mean MCA ± SD of homozygotes (major allele)/heterozygotes/homozygotes (minor allele) in each SNP, respectively
bAfter adjusting the distribution of Cobb angle with logarithmic transformation
cNo sample with homozygotes of minor allele had been detected in G2, G4, G7, G9, G11, G12, G14 and G15
dP < 0.05 was considered statistically significant
Discussion
Taking advantage of the history of recombination in the population, genetic association studies facilitate progress in searching for potential risk variants of complex traits such as AIS. Researchers found that the Xbal and Pvull polymorphisms in intron I of the ESR1 gene (coding the ERα) were correlated with progression of AIS [13, 14]. However, another study had a contradictory result [20]; hence, the relationship between ESR1 and AIS remains controversial. The polymorphisms of ESR2 (coding the ERβ) have been reported to associate with the susceptibility and curve severity of AIS patients [12]. In our research, we confirmed that the genomic polymorphisms of GPER, another novel receptor of estrogen, are associated with deteriorations of AIS.
This study represents the first time that alleles and genotypes of GPER SNPs have been examined in AIS patients. We analysed 16 SNPs selected by direct sequencing of all exons and intron-exon boundaries of the gene to ensure that the genotypes were representative of the local ethnic group. All cases and controls were Han Chinese from south China, thereby limiting the confounding factor of population stratification. We found no statistical difference in the distribution of genotypes and alleles in GPER between the patients and controls. However, the maximum Cobb angles with different genotypes in SNPs of G3, G4 and G12 in GPER were significantly different. Although LD was observed between G3 and G12, LD pattern analysis found they were in different LD blocks; thus, we did not perform a further haplotype study. Our data suggest that the heterozygous variants in these sites may be associated with the curve severity in AIS.
GPER was first cloned in 1996 and had been considered an orphan receptor until 2005, when it was found to bind oestradiol and act as a membrane-bound estrogen receptor. It was reported to be expressed in resting and hypertrophic chondrocytes and in all three types of bone cells [16, 17]. The staining of GPER in these cells declined with progression of puberty, suggesting that estrogen signalling via GPER may be involved in bone growth [16]. Windahl et al. reported that the estrogen-induced reduction of longitudinal bone growth was not observed in GPER-deficient mice, implying that estrogen regulates the skeletal development through GPER [18]. Although the detailed mechanism remains to be determined, GPER is considered to be a participant in estrogen-promoted closure of the growth plate and to contribute to estrogen-modulated skeletal development.
AIS progresses primarily in girls during skeletal growth, and skeletal immaturity is a crucial factor in the progression of the deformity [8, 22]. These phenomena hint that estrogen may have some relationship with the disease. Through its receptors, estrogen acts on both osteoblastic and osteoclastic cells, impacts bone remodelling and growth directly, and interacts with factors that influence the development of AIS [3, 23]. Estrogen signalling is a contributing factor in the progression of scoliosis through its role in bone formation, growth, maturation and turnover [4]. Our study suggests that heterozygotes in the SNPs of G3, G4 and G12 in the GPER gene are significantly related with the curve severity in AIS patients. The SNPs of G3 and G4 are located at the five prime untranslated region (5′-UTR), and the G12 at the 3′-UTR. These so-called regulatory SNPs (rSNPs) may impact gene regulatory sequences and finally lead to differences in gene expression and phenotypes [24]. We hypothesise that these regulatory SNPs may reduce the expression of GPER, and the deficiency of GPER may lead to delayed fusion of end plates through the estrogen signalling pathway, permitting irregular overgrowth of the spine which contributes to the curve. While it is possible that all or some of these possible risk variants could have a significant effect on the protein function, it remains for us to develop an in vitro assay to show some biological effect of them.
There are several limitations that should be considered in this study. One of them is that we only focused on the exon and intron-exon boundary regions of the gene. The relationship between AIS and the SNPs in promoter region or introns of the GPER still needs further investigation. Another restriction is due to the ethnic differences. The samples we studied are from the Han Chinese population in south China. The effect of the GPER gene on AIS needs to be clarified in other ethnic groups. The small sample size is also a weakness, and the study may be more convincing if replication is performed. Although the disease may progress with age, we did not find any difference within the age distribution of different genotypes, implying age was not a confounding factor in our study.
In summary, our study furnishes proof that GPER gene polymorphisms are associated with the severity of curvature in AIS. Our results suggest that deficits of this newly discovered estrogen receptor may contribute to the deterioration of spine deformity.
Acknowledgements
We thank the patients and their families for their cooperation. We also thank Professor Yiming Wang (Zhongshan School of Medicine and Center for Genome Research, Sun Yat-Sen University ) for providing technical guidance.
This work was supported by the National Natural Science, Foundation of China ( No. 30700456); Natural Science, Foundation of Guangdong Province (8151008901000141); Research Fund of Social Development of Guangdong Province (2010B031900023); the Fundamental Research Funds for the Central Universities (No.09ykpy39).
Footnotes
Yan Peng and Guoyan Liang contributed equally to this work.
References
- 1.Cheung KM, Wang T, Qiu GX, Luk KD. Recent advances in the aetiology of adolescent idiopathic scoliosis. Int Orthop. 2008;32:729–734. doi: 10.1007/s00264-007-0393-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lowe TG, Edgar M, Margulies JY, Miller NH, Raso VJ, Reinker KA, Rivard CH. Etiology of idiopathic scoliosis: current trends in research. J Bone Joint Surg Am. 2000;82-A:1157–1168. doi: 10.2106/00004623-200008000-00014. [DOI] [PubMed] [Google Scholar]
- 3.Letellier K, Azeddine B, Parent S, Labelle H, Rompré PH, Moreau A, Moldovan F. Estrogen cross-talk with the melatonin signaling pathway in human osteoblasts derived from adolescent idiopathic scoliosis patients. J Pineal Res. 2008;45:383–393. doi: 10.1111/j.1600-079X.2008.00603.x. [DOI] [PubMed] [Google Scholar]
- 4.Leboeuf D, Letellier K, Alos N, Edery P, Moldovan F. Do estrogens impact adolescent idiopathic scoliosis? Trends Endocrinol Metab. 2009;20:147–152. doi: 10.1016/j.tem.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Esposito T, Uccello R, Caliendo R, Martino GF, Gironi Carnevale UA, Cuomo S, Ronca D, Varriale B. Estrogen receptor polymorphism, estrogen content and idiopathic scoliosis in human: a possible genetic linkage. J Steroid Biochem Mol Biol. 2009;116:56–60. doi: 10.1016/j.jsbmb.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Kulis A, Zarzycki D, Jaśkiewicz J. Concentration of estradiol in girls with idiopathic scoliosis. Ortop Traumatol Rehabil. 2006;8:455–459. [PubMed] [Google Scholar]
- 7.Raczkowski JW. The concentrations of testosterone and estradiol in girls with adolescent idiopathic scoliosis. Neuro Endocrinol Lett. 2007;28:302–304. [PubMed] [Google Scholar]
- 8.Sanders JO, Browne RH, McConnell SJ, Margraf SA, Cooney TE, Finegold DN. Maturity assessment and curve progression in girls with idiopathic scoliosis. J Bone Joint Surg Am. 2007;89:64–73. doi: 10.2106/JBJS.F.00067. [DOI] [PubMed] [Google Scholar]
- 9.Maggiolini M, Picard D. The unfolding stories of GPR30, a new membrane-bound estrogen receptor. J Endocrinol. 2010;204:105–114. doi: 10.1677/JOE-09-0242. [DOI] [PubMed] [Google Scholar]
- 10.Prossnitz ER, Oprea TI, Sklar LA, Arterburn JB. The ins and outs of GPR30: a transmembrane estrogen receptor. J Steroid Biochem Mol Biol. 2008;109:350–353. doi: 10.1016/j.jsbmb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filardo EJ, Thomas P. GPR30: a seven-transmembrane-spanning estrogen receptor that triggers EGF release. Trends Endocrinol Metab. 2005;16:362–367. doi: 10.1016/j.tem.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Zhang HQ, Lu SJ, Tang MX, Chen LQ, Liu SH, Guo CF, Wang XY, Chen J, Xie L. Association of estrogen receptor beta gene polymorphisms with susceptibility to adolescent idiopathic scoliosis. Spine (Phila Pa 1976) 2009;34:760–764. doi: 10.1097/BRS.0b013e31818ad5ac. [DOI] [PubMed] [Google Scholar]
- 13.Wu J, Qiu Y, Zhang L, Sun Q, Qiu X, He Y. Association of estrogen receptor gene polymorphisms with susceptibility to adolescent idiopathic scoliosis. Spine (Phila Pa 1976) 2006;31:1131–1136. doi: 10.1097/01.brs.0000216603.91330.6f. [DOI] [PubMed] [Google Scholar]
- 14.Inoue M, Minami S, Nakata Y, Kitahara H, Otsuka Y, Isobe K, Takaso M, Tokunaga M, Nishikawa S, Maruta T, Moriya H. Association between estrogen receptor gene polymorphisms and curve severity of idiopathic scoliosis. Spine (Phila Pa 1976) 2002;27:2357–2362. doi: 10.1097/00007632-200211010-00009. [DOI] [PubMed] [Google Scholar]
- 15.Prossnitz ER, Maggiolini M. Mechanisms of estrogen signaling and gene expression via GPR30. Mol Cell Endocrinol. 2009;308:32–38. doi: 10.1016/j.mce.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chagin AS, Sävendahl L. GPR30 estrogen receptor expression in the growth plate declines as puberty progresses. J Clin Endocrinol Metab. 2007;92:4873–4877. doi: 10.1210/jc.2007-0814. [DOI] [PubMed] [Google Scholar]
- 17.Heino TJ, Chagin AS, Sävendahl L. The novel estrogen receptor G-protein-coupled receptor 30 is expressed in human bone. J Endocrinol. 2008;197:R1–R6. doi: 10.1677/JOE-07-0629. [DOI] [PubMed] [Google Scholar]
- 18.Windahl SH, Andersson N, Chagin AS, Mårtensson UE, Carlsten H, Olde B, Swanson C, Movérare-Skrtic S, Sävendahl L, Lagerquist MK, Leeb-Lundberg LM, Ohlsson C. The role of the G protein-coupled receptor GPR30 in the effects of estrogen in ovariectomized mice. Am J Physiol Endocrinol Metab. 2009;296:E490–E496. doi: 10.1152/ajpendo.90691.2008. [DOI] [PubMed] [Google Scholar]
- 19.Liu Z, Tang NL, Cao XB, Liu WJ, Qiu XS, Cheng JC, Qiu Y. Lack of association between the promoter polymorphisms of MMP-3 and IL-6 genes and adolescent idiopathic scoliosis: a case-control study in a Chinese Han population. Spine (Phila Pa 1976) 2010;35:1701–1705. doi: 10.1097/BRS.0b013e3181c6ba13. [DOI] [PubMed] [Google Scholar]
- 20.Tang NL, Yeung HY, Lee KM, Hung VW, Cheung CS, Ng BK, Kwok R, Guo X, Qin L, Cheng JC. A relook into the association of the estrogen receptor [alpha] gene (PvuII, XbaI) and adolescent idiopathic scoliosis: a study of 540 Chinese cases. Spine (Phila Pa 1976) 2006;31:2463–2468. doi: 10.1097/01.brs.0000239179.81596.2b. [DOI] [PubMed] [Google Scholar]
- 21.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 22.Sarwark J, Aubin CE. Growth considerations of the immature spine. J Bone Joint Surg Am. 2007;89(Suppl 1):8–13. doi: 10.2106/JBJS.F.00314. [DOI] [PubMed] [Google Scholar]
- 23.Eastell R. Role of estrogen in the regulation of bone turnover at the menarche. J Endocrinol. 2005;185:223–234. doi: 10.1677/joe.1.06059. [DOI] [PubMed] [Google Scholar]
- 24.Chorley BN, Wang X, Campbell MR, Pittman GS, Noureddine MA, Bell DA. Discovery and verification of functional single nucleotide polymorphisms in regulatory genomic regions: current and developing technologies. Mutat Res. 2008;659:147–157. doi: 10.1016/j.mrrev.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]