Summary
Identification of biomarkers contributing to disease diagnosis, classification or prognosis could be of considerable utility. For example, primary methods to diagnose multiple sclerosis include magnetic resonance imaging and detection of immunologic abnormalities in cerebrospinal fluid. We determined if gene expression differences in blood discriminated MS subjects from comparator groups and identified panels of ratios that performed with varying degrees of accuracy depending upon complexity of comparator groups. High levels of overall accuracy were achieved by comparing MS to homogeneous comparator groups. Overall accuracy was compromised when MS was compared to a heterogeneous comparator group. Results, validated in independent cohorts, indicate that gene expression differences in blood accurately exclude or include a diagnosis of MS and suggest these approaches may provide clinically useful prediction of MS.
Introduction
A cornerstone in diagnosing clinically definite multiple sclerosis (MS) is magnetic resonance imaging (MRI) detection of brain lesions disseminated in time and space1–3. Laboratory findings include cerebrospinal fluid immunologic abnormalities4, 5. Criteria, including 2001 McDonald and revised 2005 McDonald classifications are employed to standardize diagnosis of MS. However, few of the most common clinical features of MS are unique to this disease. A significant limitation in these approaches is the need for dissemination in time requiring detection of new lesions in follow-up scans, thus potentially delaying diagnosis and onset of therapies that may retard disease progression and onset of disability.
In a search for optimal diagnostics to permit effective triage of patients into specialty care, we have considered that exclusionary tests may have great utility. Since MS is relatively rare, the possibility of excluding from further consideration those individuals who do not carry specific disease markers would greatly reduce the numbers requiring further evaluation by specialists. An exclusionary test would also expedite referral to a specialist of those individuals that may require further evaluation and serve to reassure both patients and providers for those who do not. To be useful, an exclusionary test must have a high degree of accuracy so that individuals in need of further evaluation do not escape into the excluded pool. Patients who fall in the non-excluded pool would not have a specific diagnostic label attached, but would have a high likelihood that an autoimmune condition of some kind is present. At this point, the referring physician could reasonably make a decision regarding which condition is most likely and refer the patient to the appropriate specialist. By excluding patients who need no further evaluation, the medical system would deliver care efficiently to those who do. Optimization of health care delivery to improve outcomes is a major focus of the health care reform movement and paradigm-shifting exclusionary diagnostics would contribute to this goal.
With the advent of array-based technologies, the possibility that large-scale screening of DNA variants, differences in RNA expression, or differences in protein expression either at affected tissue sites or common sources (blood, plasma) could provide clinically useful information has attracted much interest. In general, it appears that analyses of DNA variants, identified thus far, either singly or combined, are limited in their ability to provide clinically useful prediction of disease6. In contrast, studies have demonstrated the potential utility of RNA or protein expression profiles to segregate subjects with a given disease from either healthy control subjects or subjects with other diseases7–18. Using this approach, we previously identified a panel of genes whose expression levels varied among different autoimmune diseases19–21. We hypothesized that expression profiles may provide a method to aid in diagnosis of autoimmune diseases, such as MS, and have performed such analyses. Limitations in our previous studies included relatively small numbers of genes included in the analysis thus reducing discriminatory power, small study size, lack of geographic heterogeneity, and lack of sufficient subjects with other inflammatory and non-inflammatory neurologic diseases and disorders in the study cohort22. These limitations have been addressed in the current study to include a significantly larger number of genes in the analysis, extended methods of analyses, and significantly larger sample cohorts drawn from various US and European sites.
Results
Gene expression patterns in distinct neurologic diseases
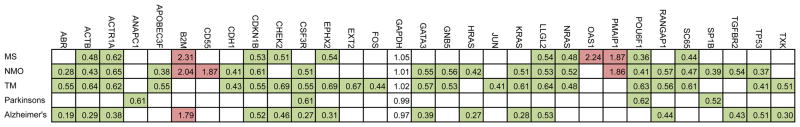
We measured expression patterns of a common set of genes assayed using a common platform in control subjects and subjects with different neurologic conditions, including autoimmune diseases. Genes for analysis were selected from prior microarray studies. Expression levels of individual genes were determined by quantitative RT-PCR by normalization to GAPDH expression levels. We employed a heatmap to depict those genes differentially expressed in individual disease cohorts relative to the control cohort, P < 0.05 (after Bonferroni correction for multiple testing) (Figure 1, red = over-expressed gene, green = under-expressed gene). Ratios of expression levels of individual genes in the indicated disease cohort relative to the control cohort were calculated and depicted within each colored box. Each disease exhibited an underlying unique pattern of gene expression. However, these profiles were sufficiently overlapping to prohibit accurate discrimination of one disease from another disease using the expression profile alone. For example, LLGL2, RANGAP1, ACTB, and POU6F1 were under-expressed in 4, 3, 4, and 4 of 5 different conditions, respectively. In contrast, other genes, e.g., ANAPC1 in Parkinson’s disease, EXT2 and FOS in TM, HRAS in NMO, were only differentially expressed in a single disease cohort. Overall, individual genes were either over-expressed, e.g. B2M, CD55, PMAIP1, or under-expressed, e.g. LLGL2, RANGAP1, ACTB, across multiple disease cohorts. Thus, each gene was differentially expressed in at least one disease cohort relative to the CTRL cohort. However, each individual disease cohort did not possess a unique expression profile distinguishing it from all other disease cohorts.
Figure 1.
Gene expression profiles across multiple autoimmune diseases. Expression levels of 44 target genes were determined by quantitative RT-PCR and normalized to expression of GAPDH. Expression levels of 31 genes are shown; expression levels of the remainder were not statistically different between CTRL and any disease cohort. Genes are identified that showed statistically significant [P < 0.05 after Bonferroni’s correction] increased [green boxes] or decreased [red boxes] expression in individual disease cohorts relative to CTRL subjects. Numerical expression ratios [disease group average/CTRL average] are displayed within the colored boxes.
Discrimination of MS from homogeneous comparator groups: identification of an optimum panel of gene expression ratios
Healthy control subjects, subjects with MS, and subjects with other inflammatory neurologic disorders (OND-I), and subjects with neurologic disorders typically considered non-inflammatory (OND-NI) were recruited from multiple U.S. and European sites (Table 1 and Supplementary Table 1). Demographic characteristics of the different disease groups, MS, OND-I, or OND-NI were matched to the CTRL cohort (Table 2). Subjects with MS included subjects with clinically isolated syndrome (CIS), newly diagnosed MS subjects who were treatment naïve and subjects with established disease (> 1 yr duration) on different therapies. Expression levels of test and control genes in blood were determined by quantitative reverse transcription polymerase chain reaction (RT-PCR) (Supplementary Table 2). We employed a search algorithm to identify those ratios of gene expression levels in which the greatest number of subjects in the test group possessed a ratio value greater than the highest ratio value in the comparator group. We employed a second algorithm to perform permutation testing of one subject group to identify the optimum set of discriminatory ratios. We reasoned that examination of expression levels of ratios of genes rather than individual genes would serve the following purposes. First, calculation of ratios normalized for differences in mRNA or cDNA template quantity and quality among different samples. Second, they obviated the need for inclusion of a ‘housekeeping’ gene in the analysis and the assumption that expression levels of ‘housekeeping’ genes did not vary among different subject populations. Third, comparisons of ratios or combinations of ratios may more accurately identify cellular phenotypes that may contribute to disease. For example, a ratio containing one gene in the numerator that is over-expressed in the test group relative to the comparator group and one gene in the denominator that is under-expressed in the test group relative to the comparator group should produce a greater ratio value difference between individuals in the two groups than a single expression value. We employed a point system to award one point to a subject if a ratio value of the test subject was greater than the ratio values of all subjects in the comparator group (Supplementary Figure 1).
Table 1.
Characteristics of Subjects
| Sites | |||
|---|---|---|---|
| Nashville | U.S.* | Europe** | |
|
| |||
| MULTIPLE SCLEROSIS (total) | 84 | 81 | 80 |
|
| |||
| CIS | 14 | 10 | |
| Treatment naïve | 30 | 4 | 55 |
| Established disease (on meds) | 40 | 77 | 15 |
|
| |||
| OND-I (total) | 1 | 85 | |
|
| |||
| Acute disseminated encephalomyelitis | 4 | ||
| Bell’s Palsy | 3 | ||
| CNS lupus | 2 | ||
| Guillaine Barre | 4 | ||
| Myasthenia Gravis | 3 | ||
| Neuromyelitis optica | 27 | ||
| Optic neuritis | 1 | 1 | |
| Transverse myelitis | 41 | ||
|
| |||
| OND-NI (total) | 1 | 128 | |
|
| |||
| Alzheimer’s | 6 | ||
| Cerebral ataxia | 2 | ||
| Cerebral bleed | 2 | ||
| Cervical radiculopathy | 6 | ||
| Drug-induced movement disorder | 1 | ||
| Dystonia | 1 | ||
| Epilepsy | 1 | 4 | |
| Essential tremor | 9 | ||
| Huntington’s disease | 1 | ||
| Hydrocephalus | 1 | ||
| Median Neuropathy | 2 | ||
| Meningioma | 1 | ||
| Migraine | 30 | ||
| Parkinson’s | 3 | 0 | |
| Peripheral Neuropathy | 1 | ||
| Pseudotumor | 3 | ||
| Restless Leg Syndrome | 1 | ||
| Seizures | 6 | ||
| Spasmodic torticollis | 1 | ||
| Stroke | 18 | ||
| Tourette’s Syndrome | 1 | ||
| Transient Ischemia | 1 | ||
|
| |||
| CONTROLS | 48 | 61 | |
six additional sites in U.S.: MA, MD, NY, SC, AZ, TX, CA, samples from sites in MS, MD, NY, AZ, and CA were obtained through the Accelerated Cure Project.
Denmark, Netherlands
Table 2.
Demographic characteristics of the different subject populations.
| AGE | P* | GENDER (% F) | P | ETHNICITY (%, C/AA/As/H)** | P | |
|---|---|---|---|---|---|---|
| MS | 43±10 | NS | 76 | NS | 80/20/0/0 | NS |
| OND-NI | 46±10 | NS | 67 | NS | 68/26/3/1 | NS |
| OND-I | 46±10 | NS | 68 | NS | 67/33/0/0 | NS |
| CTRL | 41±11 | 77 | 71/22/3/3 |
P calculated by Student’s T-test (Age) or Fisher’s exact test, NS: P > 0.05.
C, Caucasian; AA, African American; As, Asian; H, Hispanic.
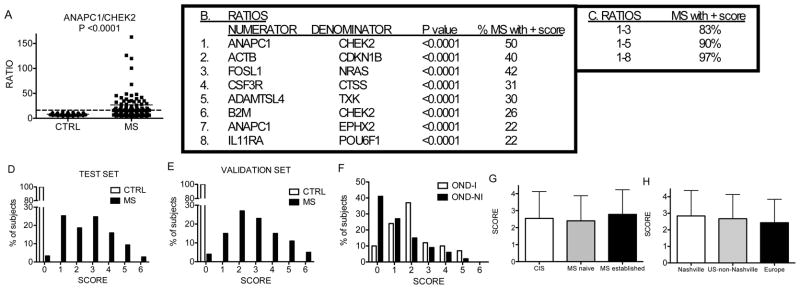
We applied this approach to determine how accurately it would distinguish subjects with MS from healthy control subjects. First, we identified ratios capable of discriminating MS subjects from control subjects. The single ratio with the greatest discriminatory power was ANAPC1/CHEK2 (Figure 2a). Fifty % of MS subjects achieved a ratio value higher than all the CTRL subjects and were awarded one point. Second, we eliminated those ratios that identified fewer than 20% of MS subjects. Third, since many ratios identified the same MS subjects, we performed another reduction to preserve only one ratio with this characteristic. A total of 8 ratios remained after this minimization process (Figure 2b). Using the point system, the combination of these 8 ratios positively identified 97% of MS subjects and eliminated 100% of CTRL subjects (Figure 2c). The score distribution was 0–6 for MS subjects and 0 for CTRL subjects (Figure 2d).
Figure 2.
Discrimination between MS and CTRL subjects with an 8 ratio scoring system. (a) Performance of the single ratio, ANAPC1/CHEK2 to discriminate MS and CTRL subjects. (b) Genes making up 8 unique discriminatory ratios. P values compare expression levels of ratios between MS and CTRL subjects. (c) Increased sensitivity with increasing numbers of ratios. (d) Score distributions among subjects using 8 ratios. (e) Validation of results by analyzing 40 new MS subjects and 40 new CTRL subjects. (f) Score distribution between OND-I and OND-NI subjects. (g) Mean scores ± std. dev. among subjects with CIS, initial diagnosis of MS, and established MS. P value is not significant among groups. (h) Mean scores ± std. dev. among MS subjects from different geographic locations. P value is not significant among groups.
Discrimination of MS from homogeneous comparator groups: validation and analysis
Our analyses depended upon determination of multiple ratios, which may create Type 1 errors. Various methods are available to correct for false discovery rates. Rather than relying upon these methods, which all make underlying assumptions, we performed a second evaluation using an independent cohort of 40 new MS subjects and 40 new CTRL subjects to validate results obtained from the initial training set. These subjects were recruited separately and the PCR analyses were performed separately. We used the same ratio values defined from the original CTRL and MS test set to award points to subjects in the validation cohort. All 40 controls were awarded a score of 0 while 4% of MS subjects received a score of 0. The remaining 96% of MS subjects achieved a score of 1–6 and the distribution of scores was similar to that observed in the training set (Figure 2e). Taken together, this demonstrates that results obtained in the training set can be replicated in an independent cohort of CTRL and MS subjects.
We applied the point system to OND-I and OND-NI subjects. In contrast to CTRL subjects, 90% of OND-I and 59% of OND-NI subjects scored ≥ 1 (Figure 2f). We compared scores among subjects with CIS, with newly diagnosed MS not yet on medications, and with established MS on different medications. Scores did not differ significantly among these three groups (Figure 2g). We also compared scores within the MS group as a function of geographic origin. Scores also did not vary significantly among MS subjects from different geographic sites (Figure 2h). Thus, subjects with CIS or subjects after their initial diagnosis of MS had a similar mean score to subjects with established MS on therapies. However, a high percentage of subjects with other neurologic conditions, especially inflammatory neurologic conditions, also scored > 0 in this analysis. Given its extremely high specificity and relatively low sensitivity, this test has greater application to exclude an individual from the diagnosis of MS rather than to establish a diagnosis of MS.
Further, we were able to obtain follow-up clinical information on 8 CIS subjects > 2 yr. after the initial consent and blood draw. Of these subjects, the 7 CIS subjects who achieved a score > 0 in the analysis now have documented MS. The 1 CIS subject who achieved a score of 0 does not have a documented case of MS.
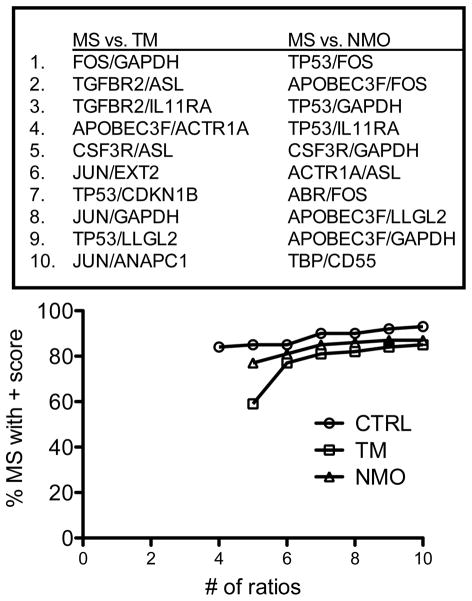
NMO and TM are inflammatory neurologic diseases that scored positive in our analysis. Therefore, we determined if we could employ a similar approach to discriminate MS from TM and MS from NMO. We identified a series of ratios that, when combined using the point system, were able to discriminate TM from MS and NMO from MS with similar overall accuracy to the MS and CTRL comparisons (Figure 3). Thus, using our approach, it was possible to distinguish MS from TM and MS from NMO with a similar degree of accuracy as obtained for the comparison of MS to CTRL. However, since each disease possessed a unique signature, it was necessary to employ separate combinations of ratios to accurately distinguish MS from NMO and MS from TM.
Figure 3.
Discrimination of MS subjects from subjects with inflammatory neurologic diseases, TM or NMO. Most discriminatory gene expression ratios were identified that segregate MS subjects from TM and NMO subjects (CTRL is included for reference). The point system was applied to combine ratio performance into a single discriminator.
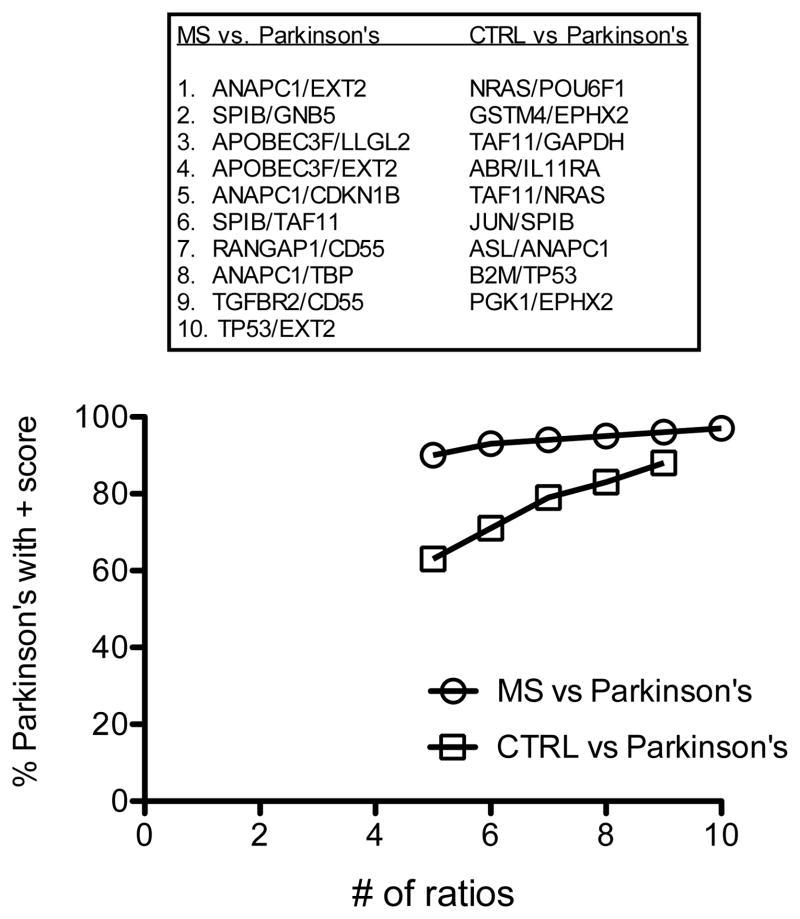
Above results demonstrate it is possible to distinguish MS from either a control cohort or even a related inflammatory disease cohort if the disease cohort is a single disease. Next, we asked if we could discriminate MS from Parkinson’s disease, a disorder typically considered non-inflammatory. To test this hypothesis in we determined if subjects with Parkinson’s disease (N = 24) segregated from MS (N = 182) and from CTRL (N = 109) using the ratio and point system. We identified 10 ratios capable of discriminating 97% of MS subjects from 100% of Parkinson’s subjects and 9 ratios capable of discriminating 88% of Parkinson’s patients from 100% of CTRL subjects (Figure 4). We interpret these results to demonstrate that subjects with Parkinson’s disease express unique gene expression signatures in blood distinguishing them from CTRL and MS subjects.
Figure 4.
Discrimination of subjects with Parkinson’s disease from MS and CTRL. Most discriminatory gene expression ratios were identified that segregate Parkinson’s disease subjects from MS subjects and CTRL subjects. Using the point system, we determined the % of Parkinson’s subjects with a score > 0, Y-axis, relative to the number of ratios, X-axis, for the different comparator groups.
Discrimination of MS from heterogeneous comparator groups
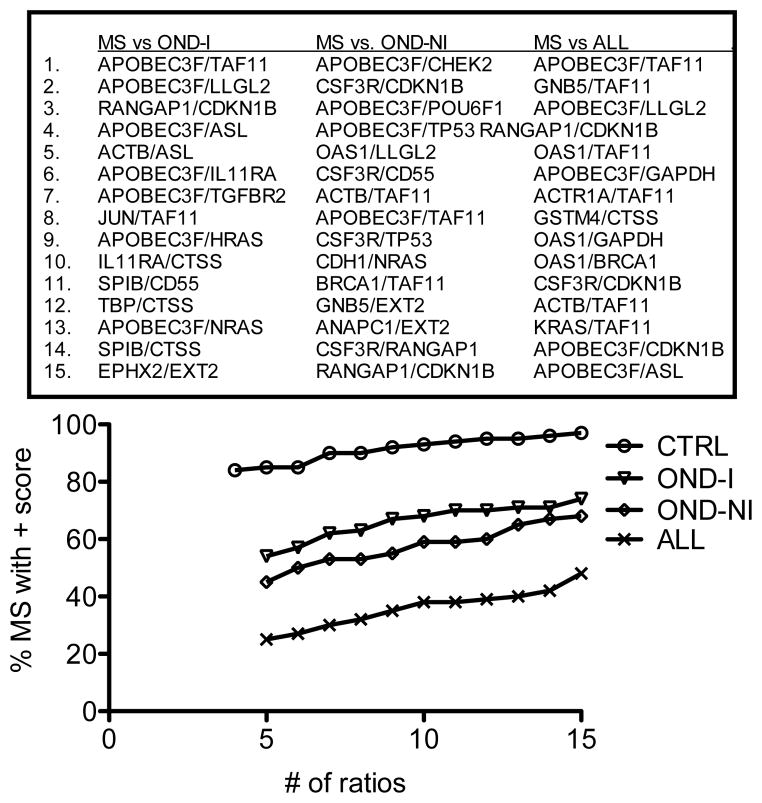
Next, we determined if we could distinguish MS from more heterogeneous groups of subjects. To do so, we combined subjects with neurologic conditions typically considered as inflammatory (other neurologic disorders-inflammatory, OND-I in Table S1) into one group. We also combined subjects with neurologic conditions typically considered non-inflammatory (other neurologic disorders-non-inflammatory, OND-NI, OND in Table S1) into a second group. We produced a third group consisting of CTRL + OND-I + OND-NI subjects (ALL). We determined the 15 best ratios using permutation testing for each comparison. Overall, comparison of MS to these heterogeneous comparator groups resulted in a marked reduction in overall discrimination ability (Figure 5). We conclude that a binary comparison such is this exhibits much reduced accuracy as the heterogeneity of the comparator group is increased.
Figure 5.
Discrimination of MS subjects from heterogeneous comparator groups. We identified the top 15 gene expression ratios with the greatest ability to discriminate MS from OND-I, OND-NI, or ALL (OND-I, OND-NI, and CTRL). Using the point system, we determined the % of MS subjects with a score > 0, Y-axis, relative to the number of ratios, X-axis, for the different comparator groups [CTRL is included for reference].
Discrimination of MS from OND-I: identification of optimum panels of gene expression ratios
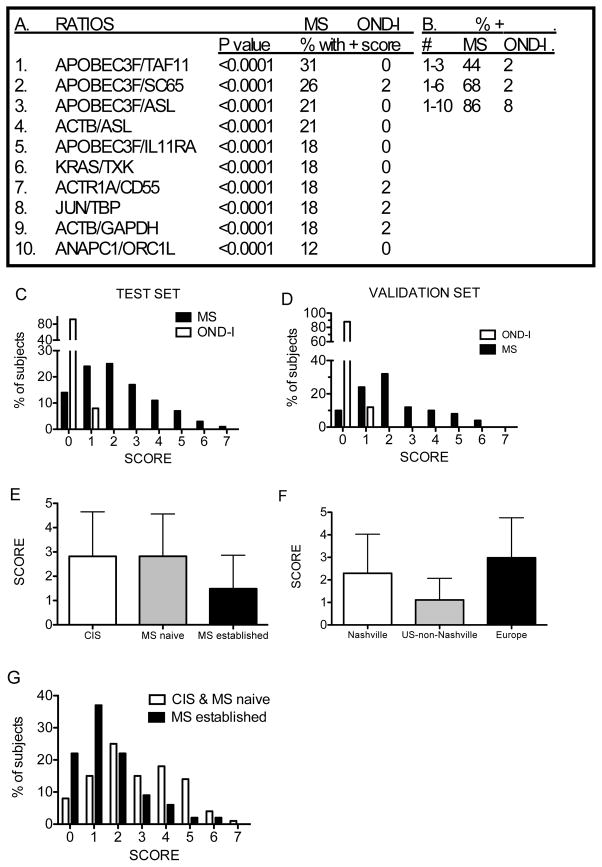
For additional analysis, we combined OND-I into one group of non-MS inflammatory neurologic disorders and investigated the ability of our approach to discriminate this combination of diseases from MS. We relaxed conditions somewhat to identify ratios with the ability to detect 0 or 1 non-MS subjects. Our best results were obtained with 10 ratios (Figure 6a). The combination of which identified 86% of MS subjects with a score > 0 and only 8% of OND-I subjects with a score > 0 (Figure 6b). Scores ranged from 0–7 for MS subjects and 0–1 for OND-I subjects (Figure 6c).
Figure 6.
Discrimination between MS and OND-I subjects using 10 gene expression ratios. (a) Genes making up 10 unique discriminatory ratios. P values compare individual ratio values between MS and OND-I subjects. (b) Increasing number of ratios increases sensitivity or ability to discriminate between MS and OND-I subjects. (c) The score distribution in MS and OND-I subjects using 10 ratios. (d) Validation of results by analyzing 40 new MS subjects and 40 new OND-I subjects (20 TM + 20 NMO). (e) Mean scores ± std. dev. among subjects with CIS, initial diagnosis of MS and established MS. P is not significant for CIS versus MS naïve, 0.03 for CIS versus established MS, and < 0.0001 for MS naïve versus established MS. (f) Mean scores ± std. dev. among subjects based upon geographic sites. P is not significant for Nashville versus Europe, < 0.0001 for Nashville versus U.S. non-Nashville, and < 0.0001 for Europe versus U.S. non-Nashville. (g) Score distributions between [CIS and MS-naïve] and established MS.
Discrimination of MS from OND-I: Validation and analysis
We performed additional analyses with 40 new MS subjects and 40 new OND-I subjects (20 NMO and 20 TM) not included in the training set. In the validation set, 88% of MS subjects achieved a score ≥ 1 and 12% of OND-I subjects achieved a score of 1 (Figure 6d), which was similar to the score distribution observed in the training set. We determined mean scores among subjects with CIS, subjects with newly diagnosed MS prior to onset of therapies, and subjects with established MS on therapies using the 10 ratios identified above. Mean scores were significantly higher in the CIS and MS-naïve groups than in the MS group with established disease (Figure 6e). We also determined mean scores based upon geographic origins of MS subjects. Subjects from Nashville and Europe had mean scores significantly greater than U.S. subjects from locations other than Nashville (Figure 6f). These results are consistent with results comparing CIS, MS-naïve, and MS-established. The majority of subjects from U.S. sites outside Nashville had established MS and were on therapies (76 of 80 subjects) while all European subjects were either CIS or newly diagnosed MS subjects not yet on therapies (N=101). The Nashville site also provided more samples with established disease (N=37) compared to CIS or treatment naïve MS (N=16) (P < 0.0001, Chi-squared test for independence among three geographic locations). The distribution of scores in the CIS and newly diagnosed MS group was also higher than that found in the established MS group. Greater than 50% of subjects with established MS achieved scores of 0 or 1 while 48% of CIS and newly diagnosed MS subjects achieved scores ≥ 3 (Figure 6g). Thus, subjects with CIS, newly diagnosed MS, and established MS from different geographic sites can be distinguished from subjects with OND-I with reasonable accuracy based upon gene expression profiles in whole blood.
Discrimination of MS from OND-NI: identification of optimum panels of gene expression ratios
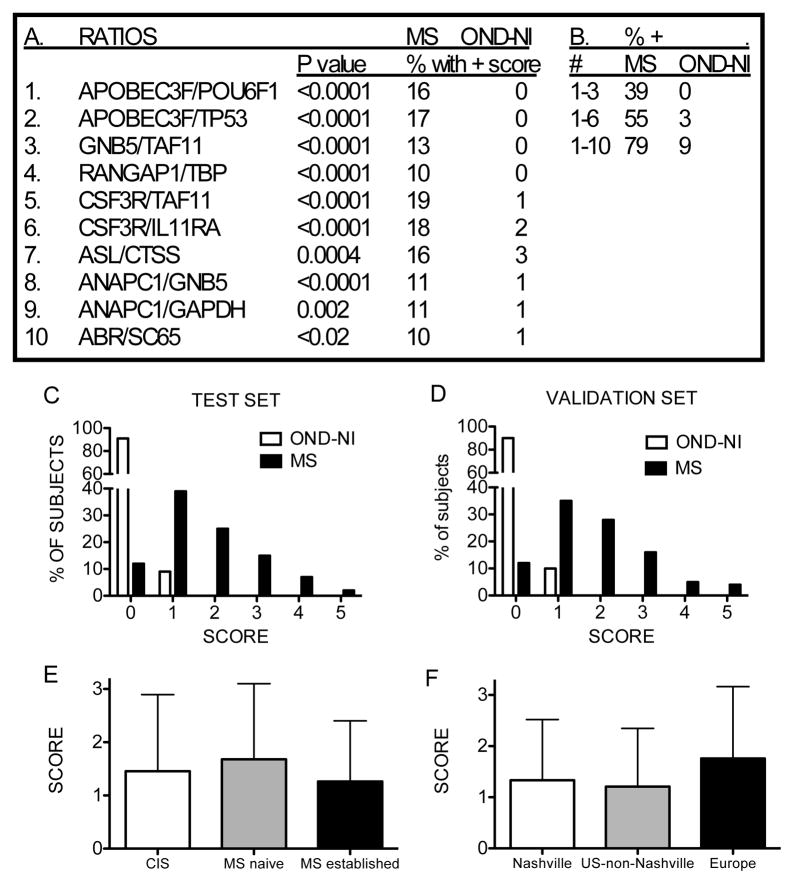
Next, we compared gene expression differences between MS and OND-NI subjects, which included Parkinson’s disease, essential tremors, migraines, and strokes. We employed the same search strategy used to compare MS and OND-I subjects and identified 10 expression ratios to construct the point system. ABOBEC3F, CSF3R, and ANAPC1 were each in the numerators of two ratios and TAF11 was in the denominator of two ratios. Each ratio alone detected > 10% of MS subjects relative to OND-NI subjects (Figure 7a). Combining ratios using the point system improved overall ability to discriminate MS subjects from OND-NI subjects (Figure 7b). Using the point system, 79% of MS subjects achieved a score ≥ 1 and 91% of OND-NI subjects achieved a score of 0, 9% achieved a score of 1 (Figure 7c).
Figure 7.
Discrimination between MS and OND-NI subjects using 10 gene expression ratios. (a) Identification of genes making up the 10 unique discriminatory ratios. P values compare individual ratio values between MS and OND-NI subjects. (b) Increasing the number of gene expression ratio increases the ability to discriminate between MS and OND-NI subjects. (c) Score distribution using 10 ratios in the training set. (d) Validation of results by analyzing 40 new MS subjects and 40 new OND-NI subjects. (e) Mean scores ± std. dev. among subjects with CIS, initial diagnosis of MS and established MS. P values were not significant among any of the comparisons. (f) Mean scores ± std. dev. among subjects based upon geographic sites. P values were not significant for any of the comparisons.
Discrimination of MS from OND-NI: Validation and analysis
We performed additional analyses with 40 new MS subjects and 40 new OND-NI subjects not included in the training set as outlined above. In the validation set, 88% of MS subjects achieved a score ≥ 1 (Figure 7d), which was a similar frequency to that observed in the training set, and 90% of OND-I subjects achieved a score of 0, 10% achieved a score of 1. As above, we determined mean scores of subjects with CIS, newly diagnosed MS and established MS and these were not statistically different among the three MS groups (Figure 7e). Similarly, mean scores of MS subjects from different geographic sites were not statistically different (Figure 7f). Using the point system, ~80% of MS subjects achieved a score ≥ 1 and 9% of OND-NI subjects achieved a score = 1 in the test set. These results demonstrate that expression in whole blood of a different set of gene ratios discriminated subjects with MS from subjects with OND-NI with reasonable accuracy.
All comparisons in these analyses were binary. Therefore, exclusion of a specific disorder by the analysis may be more accurate than inclusion of a specific disorder (see flow chart, Supplemental Figure 2). Thus, a score of 0 in the MS versus CTRL test decreased the probability that a subject had MS. A second analysis comparing MS to OND-I and MS to OND-NI would be interpreted similarly. Scores of 0 decreased the probability of MS and favored the probability of OND-I or OND-NI, respectively. Finally, specific inflammatory neurologic disorders, NMO or TM, were distinguished from MS with high degrees of accuracy. Thus, results from this single platform can be analyzed in a tiered approach to provide meaningful disease classification.
Discussion
Although our focus was on MS and other inflammatory and non-inflammatory neurologic disorders, our results support the notion that this approach could be applicable to an array of diseases. First, discrimination between MS and healthy controls or subjects with individual diseases can be achieved with a relatively high degree of accuracy. However, subjects with OND-I and OND-NI also scored positive in MS-CTRL comparisons. As such, this single comparison has greater utility as an exclusionary test rather than a test of MS inclusion. Second, it is possible to discriminate MS from groups of diseases, such as inflammatory or non-inflammatory neurologic diseases, and validate results in independent cohorts, although overall accuracy is somewhat compromised. Third, discrimination of MS from a diverse comparator group including CTRL, OND-I, and OND-NI causes a further reduction in overall accuracy. Nevertheless, a score > 0 in this analysis is highly predictive of the presence of MS. Fourth, it is possible to identify small numbers of ratios with high degrees of discriminatory power whose accuracy can be validated in independent cohorts analyzed separately.
One interpretation of our results is that many individual diseases express unique but overlapping gene expression signatures in whole blood. Given the attention paid to analyses of autoimmune diseases, it is not surprising that inflammatory neurologic diseases such as NMO and TM also express unique gene expression signatures. Perhaps somewhat surprising is that Parkinson’s disease, a disorder typically considered non-inflammatory, also possesses a unique gene expression signature distinguishing it from both CTRL and MS. Implications may be that the immune system can sense specific neurologic damage caused by Parkinson’s via responses to cytokine mediators, adhesion molecules, neurotransmitters, or other mediators read by immune cells. Alternatively, genetic risk factors associated with Parkinson’s disease may contribute to altered gene expression signatures by either direct or indirect mechanisms.
Mechanisms underlying gene expression differences among study groups or relationships to MS disease mechanism are not altogether clear. However, defects in DNA damage repair, cellular responses to DNA damage, and regulation of cell cycle progression and arrest are common properties of lymphocytes in certain autoimmune diseases, including MS, and ANAPC1, CHEK2, CDKN1B, ACTB, FOSL1, LLGL2, and NRAS encode proteins playing key roles in these fundamental cellular processes 23–27. These genes are highly represented in the ratios used to distinguish MS from comparator groups. Genes, such as ADAMTSL4, B2M, IL11RA, TXK and POU6F1, encode proteins playing key functions in regulating cells of both innate and adaptive arms of the immune system 28, 29. As such, alterations in expression of these genes may contribute to pathogenesis of MS or may represent an altered response by the immune system to MS pathogenesis.
Limitations to our study include selection of patients with pre-existing diagnoses of CIS and MS, as this may not completely represent patients in the general population in whom these tests may be performed. Our follow-up analysis of CIS patients supports the idea that initial scores > 0 will correlate with progression to MS. Future longitudinal studies are planned to better evaluate utility of these tests in this setting. Further, our binary analysis is also predicated on the fact that MS is best represented by a single set of gene expression ratios and this may not be the case. Additional analyses, such as analyses of gene expression ratios in multi-dimensional space, will address this possibility. We identified several different combinations of gene expression ratios, which performed equivalently in their ability to discriminate among subject groups. In conclusion, these minimally invasive and relatively inexpensive tests may have utility to either exclude the diagnosis of MS or to contribute to establishing a diagnosis of MS.
Materials and methods
Patients
Blood samples in PAXgene tubes were obtained from patients with a) clinically isolated syndrome (CIS), b) an initial diagnosis of MS before onset of therapy, and c) established relapsing-remitting MS on medication. Blood samples were also obtained from healthy control subjects (CTRL) and subjects with different inflammatory (OND-I) or non-inflammatory (OND-NI) neurologic conditions. MS samples were obtained from a total of 9 different sites in the U.S. and Europe. Samples from subjects with OND-I and OND-NI were obtained from 7 sites in the U.S. CTRL samples were obtained from 3 U.S. sites. Inclusion criteria for MS and other neurologic conditions were diagnosis by a neurologist using established methods and ability to provide informed consent, thus providing an un-biased study cohort. Age, race and gender were not statistically different among the different study groups. Time of the blood draw, e.g. morning/afternoon clinics, was also not statistically different among the different study groups. Relevant institutional review board approval from all participating sites was obtained.
Procedures
Total RNA, purified using Qiagen’s isolation kits by standard protocols, was reverse-transcribed using SuperScript III (Invitrogen). A TaqMan Low Density Array (TLDA) was designed to analyze expression levels of 44 genes previously identified from our microarray analysis and of 4 “housekeeping” genes in 300 ng cDNA per sample. Patient diagnosis was blinded for all experimental procedures. Relative expression levels were determined directly from the observed threshold cycle (CT), the cycle number at which fluorescence generated within reactions crosses an assigned threshold reflecting the point where sufficient amplicons have accumulated to be statistically significant above baseline. Linear expression values were determined using the formula, 2(40-CT).
Identification of discriminatory gene expression ratios
A computational algorithm was designed to identify the most discriminatory combinations of ratios22. All possible gene expression ratios were computed (e.g. ACTR1A/BRCA1, TAF11/ACTR1A, etc). To analyze individual results, we used to denote the ith ratio for the jth control and let denote the ith ratio for the kth MS patient. Here, j = 1, …, Ncontrol and k = 1, …, NMS, where Ncontrol equals the total number of controls and NMS equals the total number of MS patients in the data set. The second largest member of each data set of ratios was calculated first by { }, and designated . This was then applied to the MS data set { }. We used Ci to designate the number of MS set of ratios larger than such that 0 ≤ Ci ≤ NMS. This process was repeated for each possible ratio. The ratio that produced the largest Ci was selected as the discriminator of the two sets. This process was repeated using all possible ratios. Although more than one optimal ratio could be identified for each number of components queried, we have presented only one discriminator for each combination. Ratios were included only if > 20% of subjects within the MS group had expression values greater than all subjects in the CTRL group. A scoring system was developed to combine multiple ratios. To do so, subjects were assigned one point for each ratio in which their expression value was higher than the highest expression value within the CTRL subject group. By this approach, it was also possible to relax search criteria by setting cutoffs to the second highest expression ratio, third highest expression ratio, etc., of the comparator subject group. Using these relaxed criteria, an individual was awarded one point if the value of their expression ratio was higher than the second or third, etc., highest expression value of individuals in the comparator group, respectively. These combined ratios established a score discriminating the MS group from comparator groups.
Search algorithm for best ratios
Let D denote the set of 44 gene-expression levels associated with the disease group and C denote the set of gene-expression levels associated with the control group. For example, when D is the set of MS patients, then D is a set of 182 44-tuples; if C is associated with the Controls, then C is a set of 51 44-tuples. The algorithm that searches for the “best” set of gene ratios is the following:
80% of the control group was randomly selected and compared to the disease group in the following manner. Gene-expression level ratios were formed for elements in D and C. For each ratio, the number of elements in the disease group that were larger than the largest ratio in the control group was computed. The top 500 ratios that separate elements in D and C were saved. This calculation was repeated 200 times resulting in a set of 200 subsets of ratios (each subset having 500 ratios).
The 500 subsets were processed to identify the smallest number of ratio, R = {r1, r2, …, rn}, that produced the maximum of separation of D and C. Associated with each of the ratios in R, there were threshold values, T = {t1, t2, …, tn}, which corresponded to the highest value in the control group for each of the ratios in R.
For each member of the disease group D, the ratios in R were computed, {α1, α2, …, αn}. If αi ≥ ti, then we assigned the ratio a 1; otherwise, it was assigned a 0. In this way, we generated an n-tuple of 1’s and 0’s for each member of D. For example, if n = 6, then a typical 6-tuple would be {1,1,0,0,1,0}. This meant that this individual in the disease group would have 3 ratios that exceeded the corresponding ratios in the control group.
Lastly, the percentage of members in the disease group that had nonzero n-tuples was calculated. The larger the percentage, the better the separation of D and C.
Statistical analysis
The Welch’s corrected T-test not assuming equal variances was used to calculate P values in two-way comparisons. The Chi-squared test for independence was used to calculate P values in three-way comparisons. The Bonferroni method was employed to correct for multiple testing 30.
Supplementary Material
Acknowledgments
This work was supported by US National Institutes of Health Grant AI053984.
Footnotes
Competing Interests Statement
T.M.A. and N.J.O. are co-owners of ArthroChip.
References
- 1.Swanton JK, Rovira A, Tintore M, Altmann DR, Barkhof F, Filippi M, et al. MRI criteria for multiple sclerosis in patients presenting with clinically isolated syndromes: a multicentre retrospective study. Lancet Neurol. 2007;6(8):664–665. doi: 10.1016/S1474-4422(07)70176-X. [DOI] [PubMed] [Google Scholar]
- 2.Polman CH, Reingold SC, Edan G, Filippi M, Hartung HP, Kappos L, et al. Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria”. Ann Neurol. 2005;58(6):840–846. doi: 10.1002/ana.20703. [DOI] [PubMed] [Google Scholar]
- 3.McDonald WI, Compston A, Edan G, Goodkin D, Hartung HP, Lublin FD, et al. Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol. 2001;50(1):121–127. doi: 10.1002/ana.1032. [DOI] [PubMed] [Google Scholar]
- 4.Awad A, Hemmer B, Hartung HP, Kieseier B, Bennett JL, Stuve O. Analyses of cerebrospinal fluid in the diagnosis and monitoring of multiple sclerosis. J Neuroimmunol. 2009;219(1–2):1–7. doi: 10.1016/j.jneuroim.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 5.LInk H, Huang Y-M. Oligoclonal bands in multile sclerosis cerebrospinal fluid: An update on methodology and clinical usefulness. Journal of Neuroimmunology. 2006;180(1–2):17–28. doi: 10.1016/j.jneuroim.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Consortium TWTCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Axtell RC, de Jong BA, Boniface K, van der Voort LF, Bhat R, De Sarno P, et al. T helper type 1 and 17 cells determine efficacy of interferon-beta in multiple sclerosis and experimental encephalomyelitis. Nat Med. 2010;16(4):406–412. doi: 10.1038/nm.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keller A, Leidinger P, Lange J, Borries A, Schroers H, Scheffler M, et al. Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS One. 2009;13(10):e7440. doi: 10.1371/journal.pone.0007440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris VK, Sadiq SA. Disease biomarkers in multiple sclerosis: potential for use in therapeutic decision making. Mol Diagn Ther. 2009;13(4):225–244. doi: 10.1007/BF03256329. [DOI] [PubMed] [Google Scholar]
- 10.Quintana FJ, Farez MF, Viglietta V, Iglesias AH, Merbl Y, Izquierdo G, et al. Antigen microarrays identify unique serum autoantibody signatures in clinical and pathologic subtypes of multiple sclerosis. Proc Natl Acad Sci U S A. 2008;105(48):18889–18894. doi: 10.1073/pnas.0806310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kostka D, Spang R. Microarray Based Diagnosis Profits from Better Documentation of Gene Expression Signatures. PLoS Comput Biol. 2008;4(2):e22. doi: 10.1371/journal.pcbi.0040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ray S, Britschgi M, Herbert C, Takeda-Uchimura Y, Boxer A, Blennow K, et al. Classification and prediction of clinical Alzheimer’s diagnosis based on plasma signaling proteins. Nature Med. 2007;13(11):1359–1362. doi: 10.1038/nm1653. [DOI] [PubMed] [Google Scholar]
- 13.Quackenbush J. Microarray Analysis and Tumor Classification. N Engl J Med. 2006;354(23):2463–2472. doi: 10.1056/NEJMra042342. [DOI] [PubMed] [Google Scholar]
- 14.Hofman P. DNA Microarrays. Nephron Physiol. 2005;99(3):85–89. doi: 10.1159/000083764. [DOI] [PubMed] [Google Scholar]
- 15.Gregersen PK, Brehrens TW. Fine mapping the phenotype in autoimmune disease: the promise and pitfalls of DNA microarray technologies. Genes and Immunity. 2003;4(3):175–176. doi: 10.1038/sj.gene.6363976. [DOI] [PubMed] [Google Scholar]
- 16.Bomprezzi R, Ringnér M, Kim S, Bittner ML, Khan J, Chen Y, et al. Gene expression profile in multiple sclerosis patients and healthy controls: identifying pathways relevant to disease. Hum Mol Genet. 2003;12(17):2191–2199. doi: 10.1093/hmg/ddg221. [DOI] [PubMed] [Google Scholar]
- 17.Brynedal B, Khademi M, Wallström E, Hillert J, Olsson T, Duvefelt K. Gene expression pro ling in multiple sclerosis: A disease of the central nervous system, but with relapses triggered in the periphery? Neurobiology of Disease. 2010;37(3):613–621. doi: 10.1016/j.nbd.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 18.Harris VK, Sadiq SA. Disease biomarkers: Potential for use in therapeutic decision making. Mol Diagn Ther. 2009;13(4):225–244. doi: 10.1007/BF03256329. [DOI] [PubMed] [Google Scholar]
- 19.Maas K, Chan S, Parker J, Slater A, Moore J, Olsen N, et al. Cutting edge: molecular portrait of human autoimmune disease. J Immunol. 2002;169(1):5–9. doi: 10.4049/jimmunol.169.1.5. [DOI] [PubMed] [Google Scholar]
- 20.Liu Z, Maas K, Aune TM. Identification of gene expression signatures in autoimmune disease without the influence of familial resemblance. Hum Mol Genet. 2006;15(3):501–509. doi: 10.1093/hmg/ddi466. [DOI] [PubMed] [Google Scholar]
- 21.Maas K, Chen H, Shyr Y, Olsen NJ, Aune T. Shared gene expression profiles in individuals with autoimmune disease and unaffected first-degree relatives of individuals with autoimmune disease. Hum Mol Genet. 2005;14(10):1305–1314. doi: 10.1093/hmg/ddi141. [DOI] [PubMed] [Google Scholar]
- 22.Fossey SC, Vnencak-Jones CL, Olsen NJ, Sriram S, Garrison G, Deng X, et al. Identification of molecular biomarkers for multiple sclerosis. J Mol Diagn. 2007;9(2):197–204. doi: 10.2353/jmoldx.2007.060147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weyand CM, Fujii H, Shao L, JGJ Rejuvenating the immune system in rheumatoid arthritis. Nat Rev Rheumatol. 2009;5(10):583–588. doi: 10.1038/nrrheum.2009.180. [DOI] [PubMed] [Google Scholar]
- 24.Shao L, Fujii H, Colmegna I, Oishi H, Goronzy JJ, Weyand CM. Deficiency of the DNA repair enzyme ATM in rheumatoid arthritis. J Exp Med. 2009;206(6):1435–1449. doi: 10.1084/jem.20082251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng X, Ljunggren-Rose A, Maas K, Sriram S. Defective ATM-p53-mediated apoptotic pathway in multiple sclerosis. Ann Neurol. 2005;58(4):577–584. doi: 10.1002/ana.20600. [DOI] [PubMed] [Google Scholar]
- 26.Maas K, Westfall M, Pietenpol J, Olsen NJ, Aune T. Reduced p53 in peripheral blood mononuclear cells from patients with rheumatoid arthritis is associated with loss of radiation-induced apoptosis. Arthritis Rheum. 2005;52(4):1047–1057. doi: 10.1002/art.20931. [DOI] [PubMed] [Google Scholar]
- 27.Butowski N. Immunostimulants for malignant gliomas. Neurosurg Clin N Am. 2010;21 (1):53–65. doi: 10.1016/j.nec.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Readinger JA, Mueller KL, Venegas AM, Horai R, Schwartzberg PL. Tec kinases regulate T-lymphocyte development and function: new insights into the roles of Itk and Rlk/Txk. Immunol Rev. 2009;228(1):93–114. doi: 10.1111/j.1600-065X.2008.00757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buchner DA, Meisler MH. TSRC1, a widely expressed gene containing seven thrombospondin type I repeats. Gene. 2003;307:23–30. doi: 10.1016/s0378-1119(03)00423-2. [DOI] [PubMed] [Google Scholar]
- 30.Abdi H. The Bonferonni and Sidak corrections for multiple comparisons. Sage; Thousand Oaks, CA: 2007. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.