Abstract
Numerous loci have been found genetically associated with complex diseases, but only in a few cases has the functional variant and the molecular mechanism behind it been identified. Recently, the association of the BANK1 gene with systemic lupus erythematosus (SLE) was described. Here, we investigated the role of the associated polymorphisms on gene function and found that SNP rs17266594 located in the branch point consensus sequence has negligible effect on splicing or gene expression. The non-synonymous SNP rs10516487 located in exon 2 influenced splicing efficiency by creating an exonic splicing enhancer site for the SRp40 factor. Further, this same SNP generates protein isoforms with differential and measurable self-association properties. The full-length protein isoform containing the R61 variant forms larger protein scaffold complexes in the cell cytoplasm compared to the protective BANK1-61H variant. We also observed that, contrary to the full-length isoform, the short Δ2 isoform of BANK1 displays a homogeneous cytoplasmic distribution, underscoring the potential role of the exon 2-coded protein domain in the scaffolding function of BANK1.
We provide evidence that the non-synonymous SNP rs10516487 (G<A; R61H) shows a dual nature by first, influencing mRNA splicing and consequently the quantity of protein, and, second, by producing a risk variant-containing protein isoform with increased potential for multimerization.
Keywords: SLE, BANK1, isoforms, splicing, sub-cellular localization, cytoplasmic punctae
Introduction
Systemic lupus erythematosus (SLE) is a complex autoimmune disease primarily characterized by the elevated production of antibodies to self-antigens. The excessive production of auto-antibodies by B cells leads to the formation of immune complexes that deposit in tissues causing their damage and provoking chronic inflammation.
The basis of B-cell hyperactivation and autoantibody production is not fully understood, but it is clear that both genetic and environmental factors contribute to the loss of tolerance against self-antigens. Over the last two decades, many genes contributing to susceptibility to SLE have been identified 1. But despite this wealth of genetic information, little it is known about the mechanistic contribution of each gene to the disease, even less is known about the particular contribution of a given risk allele.
We previously reported the identification of the B-cell specific gene BANK1 as a susceptibility gene for lupus in Europeans 2. This result has been corroborated independently for European-Americans 3, 4 and Asians 5, 6. Moreover, the BANK1 gene was found in one of the loci implicated in susceptibility to lupus-like disease in dogs 7. The association signal for the human BANK1 gene is located in the region between exons 1 and 3, with two polymorphisms in high linkage disequilibrium (LD) with each other, rs17266594 and rs10516487 2. The first SNP is located in a putative branch point site (BPS) in intron 1. The T allele correlated with higher levels of the full-length transcript and lower levels of the Δ2 isoform, while the C allele correlated with lower levels of the full-length and higher levels of the Δ2 isoform. Of note, the mechanism how the splicing efficiency could be altered by the SNP rs17266594 was not fully understood. The second SNP, rs10516487 is located in exon 2 just 154 bp downstream from the first variant and causes an R (arginine) to H (histidine) substitution in amino acid position 61. The major SLE risk haplotype consists of the T allele of rs17266594 and the G allele of rs10516487 coding for arginine, while the minor protective haplotype is made of the alleles C and A, respectively. The role of the R61 risk variant on the BANK1 protein function was not investigated yet. Furthermore, apart from a full-length transcript of BANK1, we identified a short transcript with an in-frame deletion of the entire exon 2 (Δ2) that codes a short protein isoform.
The BANK1 gene codes for an adaptor/scaffold protein 8. It shares its overall structure with the Drosophila cytoplasmic molecule downstream of FGFR, Dof, also known as Heartbroken or Stumps, and with the vertebrate B-cell adaptor protein BCAP 9, 10. BANK1 lacks predicted enzymatic activity but contains a number of putative sites for tyrosine phosphorylation and proline-rich motifs that could contribute to the protein-protein interaction through SH2 and SH3 domains. BANK1 posses two ankyrin repeats and a conserved region designated as Dof-BCAP-BANK motif (DBB motif) involved in dimerization 9. BANK1 is phosphorylated upon BCR stimulation and thereafter promotes phosphorylation of the IP3 receptor type 2 by the Src tyrosine kinase Lyn, leading to Ca2+ mobilization from endoplasmic reticulum stores 8. Aiba et al. described a knock-out mouse for BANK1 that showed normal B-cell maturation, slight increase of germinal center formation and overproduction of IgM antibodies in response to T-dependent antigens 11. These authors demonstrated that BANK1 acts as a negative regulator of CD40-mediated Akt activation.
In the current study, we present an analysis of the effects of the BANK1 associated polymorphisms on gene expression and protein function. We found that the coding non-synonymous SNP in exon 2 exerts its effect at the level of RNA splicing, and also has a measurable effect on the molecular behavior of the BANK1 protein isoforms. We also show that BANK1 could form protein multimers. Thus, the SNP rs10516487 has a dual effect by increasing the gene expression and generating a protein variant more prone to self-association. This double effect could contribute to the development of SLE and other autoimmune diseases.
Results
Negligible effect of the SNP rs17266594 located in the putative branch point on splicing efficiency
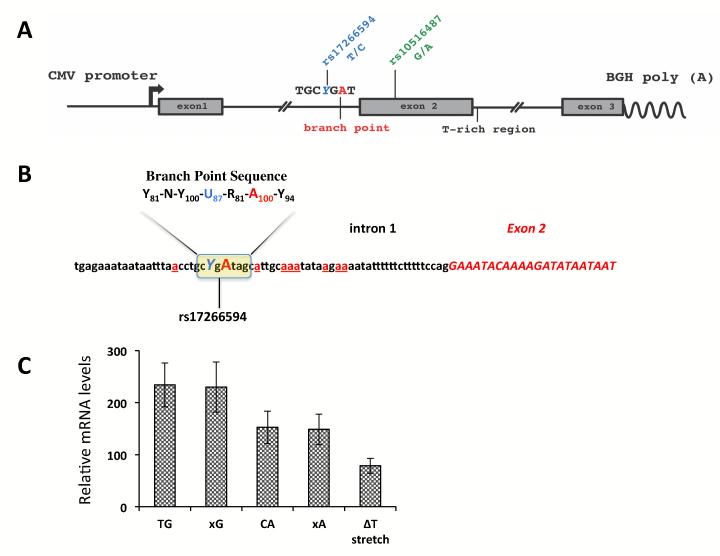
In order to investigate the role of the SNP rs17266594 on exon 2 splicing efficiency in detail, we prepared two haplotype-specific minigene constructs containing the first 3 exons and about 500 bp of the upstream and downstream intronic sequences (Figure 1A,B). The expression of the minigenes assessed 36 hours after transfection showed differential expression of the full-length isoform (P=0.0293 for TG vs. CA haplotype-specific constructs) (Figure 1C). The expression levels of the Δ2 isoform produced from the minigenes were about 100 times lower compared to the levels of the full-length isoform and did not show any significant differences between the constructs (data not shown). Thus we excluded this transcript from further analyses in the minigene assay.
Figure 1.
The minigene analysis of BANK1 exon 2 splicing.
(A) The schematic structure of the BANK1 minigene constructs cloned in the pcDNA3.1 vector.
(B) The putative branch point sequence in intron 1. The invariant adenosine is shown in capital red, the SNP rs17266594 is shown in blue. The exonic sequence in presented in italic capital letters colored in red. Alternative cryptic branch point adenosines are shown in red and underlined. The consensus branch point sequence is shown on top with frequency scores in subscript 12.
(C) mRNA expression levels for the full-length isoform produced from minigenes with particular haplotypes. Expression was normalized to the levels of the neomycin mRNA transcribed from the same plasmid. The values represent the means of 4 independent experiments +/−SD. TG vs. CA, p=0.0293. The constructs designated as xG and xC contain mutated branch point sequence, the ΔT stretch construct is similar to the TG construct but lacks the poly (T) stretch downstream of exon 2.
The genomic sequence surrounding exon 2 has only two predicted splice sites with the high strength for the 3′ acceptor site (3′AS, score 0.99), and somewhat low strength for the 5′ donor site (5′DS, score 0.5), as predicted by the NNSPLICE site predictor available at http://www.fruitfly.org/seq_tools/splice.html. The relatively weak 5′DS might be counterbalanced by a long polypyrimidine stretch downstream of the human exon 2. We then deleted this stretch from the minigene. The expression levels of the full-length isoform dropped significantly indicating an important role for this T-rich region in determining the proper 5′ donor splice site for exon 2 (Figure 1C).
Next, we mutated the branch point site in both haplotype-specific minigenes and analyzed expression of the full-length isoform (Figure 1C). The critical A residue of the BPS and the polypyrimidine tract in the 3′-end of intron 1 are conserved only among the primate species. The intronic sequence near to the acceptor site may have a number of other cryptic branch points, although their predicted strength is much weaker compared to the sequence −46 to −39 which match best to the human BPS consensus (YNYURAY) (Figure 1B) 12. Nevertheless, the complete removal of the strongest branch point sequence did not alter the levels of the full-length transcript at all, suggesting that this branch point is not essential for splicing and could be substituted by other adenosines.
The branch point SNP is in strong LD with the non-synonymous autoimmunity-associated SNP rs10516487 located 150 bp downstream in exon 2 (R61H). Although this variant is located deep in the exon and given that rs17266594 does not appear to play any significant role in splicing, this SNP could be another possible candidate to influence the splicing efficiency of the full-length transcript.
The BANK1 non-synonymous SNP rs10516487 influences splicing of exon 2
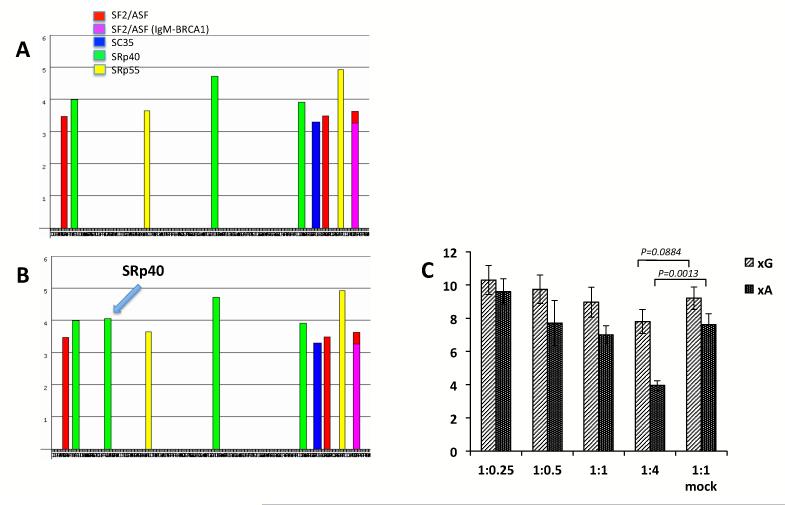
Among all exons in the BANK1 gene, exon 2 is the longest (399 bp) and the only one undergoing alternative splicing. To test whether the SNP rs10516487 could influence the splicing efficiency of the BANK1 isoforms observed in minigene expression, we performed a more detailed analysis of this SNP. The bioinformatic analysis of exon 2 with ESEfinder (http://rulai.cshl.edu/cgibin/tools/ESE3/esefinder.cgi?process=home) predicted a strong recognition site for the splicing enhancer protein SRp40 for the A allele, while the G allele completely lacks any sites at this position (Figure 2A, 2B) 13. The SRp40 factor is a member of the arginine and serine rich protein family (SR proteins) which bind to exonic splicing enhancer elements and promote splicing 14. A number of other polypeptides including SF2/ASF, Sc35, SRp55 and members of the hnRNP family together with small nuclear RNAs participate in splicing by ensuring the correct recognition of exons and excision of introns from transcribed RNA15. The first SRp40 binding site in the BANK1 exon 2 is predicted to be located in the beginning of exon next to the SF2/ASF motif and may help determine the correct 3′-exon border. The presence of a novel high-affinity binding site (the affinity score is over 4.0, compared to the SRp40 binding threshold 2.67) located further inside the exon might alter the normal assembly of the splicing factors on pre-mRNA. In order to study whether the SRp40 protein influence splicing, we co-transfected the xG and xA reporters together with a plasmid overexpressing SRp40 in HEK293T cells. We found that the gradual increase of the SRp40 from 1:0.25 to 1:4 inhibited splicing with the most significant effect on the xA construct which has a novel SRp40 binding element (P=0.013 for xA+SRp40 versus xA+mock, 1:4) (Figure 2C). Low amounts of SRp40 either slightly increased splicing of the full-length transcript produced from both minigenes or did not change it at all. Thus, we can conclude that the presence of a novel SRp40 recognition site hinder the splicing of exon 2 that carry the A allele of rs10516487.
Figure 2.
Effect of the non-synonymous SNP rs10516487 on BANK1 exon 2 splicing.
(A) The recognition sites for exonic splicing enhancers across the entire exon 2 with the G allele of rs10516487, as analyzed by ESEfinder. Only the highest score peaks for splicing factors are shown.
(B) The A allele of rs10516487 creates the strong site for the SRp40 factor.
(C) Co-transfection of the minigenes with different amounts of the plasmid expressing the SRp40 factor. The minigenes used for this experiment contained mutated branch point sequence, xG and xA constructs. The empty pcDNA vector without any insert was used as a mock control. All transfections were repeated 6 times. The values represent the means+/− SEM.
The endogenous BANK1 protein expression correlates with the mRNA levels and depends on the SNP rs10516487
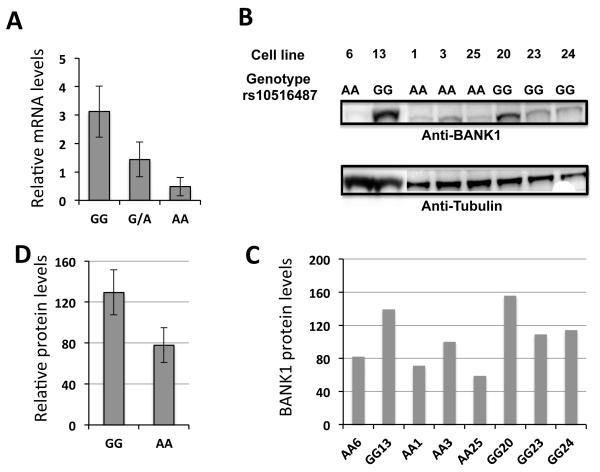
We have previously shown that the BANK1 lupus-associated polymorphism correlate with the increased expression of the full-length BANK1 transcript in peripheral blood mononuclear cells2. In order to establish if the BANK1 protein amount correlate with the mRNA levels and thereby depends on the allelic variant of rs10516487, we genotyped 33 lymphoblastoid cell lines (LCLs) and analyzed the BANK1 mRNA expression in these cells using quantitative RT-PCR (Figure 3A). We then chose eight homozygous cell lines for protein quantification by Western blot (Figure 3B,C). The results clearly show that cell lines with the GG genotype indeed produce not only more BANK1 mRNA, but also more BANK1 protein compared to the cells with the AA genotype (p-value= 0.01) (Figure 3D). The heterozygous lines show an intermediate level of expression, as expected (data for protein are not shown). These results support the direct relationship between the levels of the transcript and the protein and further support the functional importance of this SNP.
Figure 3.
Expression of endogenous BANK1 in lymphoblastoid cell lines.
(A) mRNA expression levels of the full-length BANK1 isoform in 16 lymphoblastoid cell lines homozygous for the GG genotype, 11 heterozygous and 6 homozygous lines for the AA genotype of rs10516487. GG vs AA, p=0.013. Expression was normalized to the levels of the 18S rRNA. The mean values +/− SD are shown.
(B) Protein expression in individual lymphoblastoid cell lines as determined by Western blotting with anti-human anti-BANK1 antibody (1:300) detecting the full-length protein. The blots were re-probed with anti-β-Tubulin (1:500) to ensure equal loading amounts and transfer.
(C) Normalized amounts of BANK1 protein to the arbitrarily chosen reference cell line AA3. The amount of BANK1 protein in this line was defined as 100.
(D) Averaged amounts of BANK1 protein measured on the panel C for four homozygous for GG and four homozygous for AA cell lines. Error bars represent +/− SD. GG vs AA, p=0.01.
Ectopic expression of BANK1 shows differential sub-cellular distribution of different protein isoforms
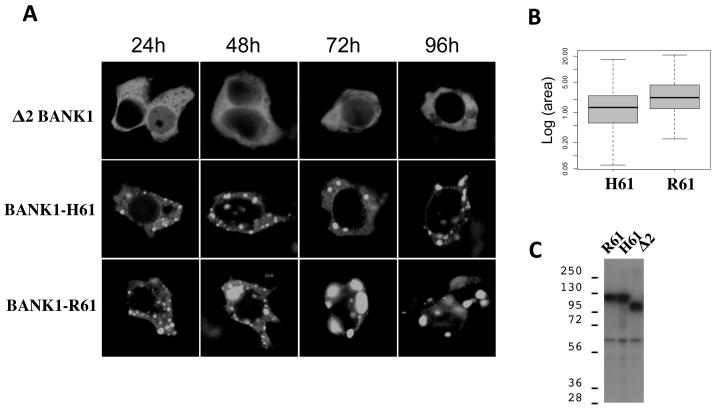
We then searched for biochemical differences between the BANK1 isoforms. We expressed the three major isoforms, namely the two full-length proteins representing the R61 and H61 variants and the Δ2 isoform fused to GFP in HEK293T cells and determined their subcellular localization. The Δ2 BANK1 isoform was homogeneously distributed within the cytoplasm whereas the FL protein showed a consistent punctate pattern (Figure 4A). Furthermore, we observed that the SLE associated isoform BANK1-R61 generates larger protein complexes compared to the BANK1-H61 and the difference became more apparent with time. We measured the area of the spots at 24 h post-transfection and found that the mean area of the spots with the BANK1-R61 was 3.4 μm2, whereas the spots with BANK1-H61 were significantly smaller - 1.8 μm2 (Figure 4B). We analyzed 377 spots with BANK1-R61 and 403 with BANK1-H61. The time course study showed that the short Δ2 isoform did not form cytoplasmic punctae over the complete period of observation (Figure 4A).
Figure 4.
Time-course analysis of protein complex formation in cells expressing different isoforms of BANK1.
(A) Confocal images of HEK293 cells transfected with plasmids expressing BANK1 isoforms tagged at the C-terminus with green fluorescent protein. Cells were analyzed 24 hours post-transfection. The both full-length Bank1 isoforms, BANK1-R61 and BANK1-H61, form discrete protein complexes in the cytoplasm while the short isoform Δ2 is homogeneously distributed in the cytoplasm. Cells expressing the BANK1-R61 isoform show larger spots compared to the cells expressing the BANK1-H61 isoform.
(B) Measurement of the cytoplasmic dots using the ImageJ program. The analysis was done blindly using coded digital pictures corresponding to 9 different transfections. Two different constructs for each isoforms were used, one tagged with CFP and the other with GFP.
(C) Western blot of cells transfected with BANK1 isoforms (BANK1-R61, BANK1-H61, and Δ2-BANK1) tagged with FLAG-epitope at the N-terminus and probed by anti-FLAG antibody. The intensity of the bands indicates equal expression of BANK1 in transfected cells. The band above 56 KDa is a nonspecific signal detected also in untransfected cells.
To exclude that the isoform-specific pattern was not due to protein degradation or cleavage, we tested the integrity of expressed proteins. In case of degradation of the N-terminus of the Δ2-fusion protein, the GFP tag would have been released and would show an equal distribution through the cytoplasm. To discard this possibility, we probed the blot with antibodies against the N-terminal FLAG tag. Western blots showed first, absence of degradation, and second, that all isoforms were expressed in comparable amounts. Thus we excluded the effect of unequal protein expression on the formation of the cytoplasmic dots (Figure 4C). In addition, Western blot analysis of the isoforms tagged at the C-terminus with V5 epitope or with fluorescent proteins did not show degradation (Figure 5A,B and Supplementary Figure 1). The expressed proteins run at 120 kDa and 95 kDa in SDS-PAGE just above their calculated molecular masses, which could be due to the acidic nature of the proteins and the presence of posttranslational modifications. We conclude that the protein domain coded by the exon 2 determines the differential cellular distribution observed between the FL and the Δ2 isoforms, and that the BANK1-R61 isoform associated with lupus is prone to form larger protein complexes.
Figure 5.
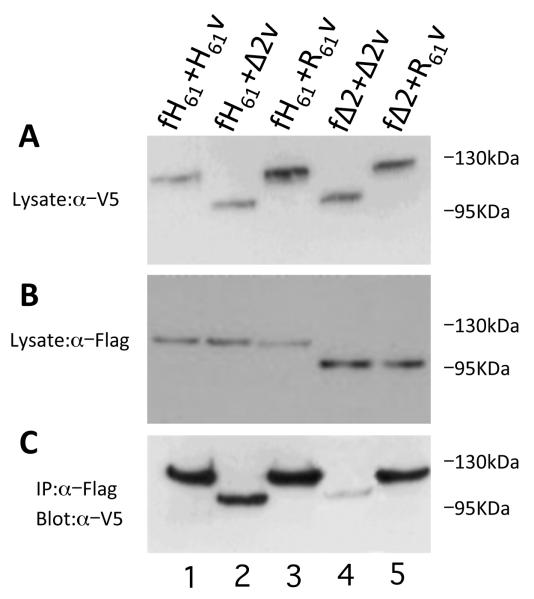
Intermolecular association of BANK1 isoforms.
(A) Protein extract of cells co-transfected with the BANK1 isoforms tagged either with FLAG-epitope at the N-term (labeled as f) or V5-epitope at the C-term (labeled as v) and probed with anti-V5 antibody.
(B) The same extracts were probed with anti-FLAG antibody.
(C) The co-immunoprecitation of different isoforms labeled differently either with FLAG-epitope or with V5-epitope indicates inter-molecular association, or dimer formation between BANK1 proteins.
BANK1 protein isoforms form dimers
The propensity of the full-length BANK1 to form large cytoplasmic structures suggests the presence of ongoing inter-molecular interactions, which can be important for providing the signaling outcome by the scaffold/adaptor protein BANK1. We studied the inter-molecular association of the isoforms in more detail. For this, we co-tranfected the constructs coding for different BANK1 isoforms tagged with either a FLAG-tag or a V5-tag and performed co-immunoprecipitation. The isoforms detected in the cell lysates by Western blot were of the predicted sizes (Figure 5A and 5B). The immunoprecipitation carried out with antibodies against the FLAG epitope and interrogated with anti-V5 antibody, showed that BANK1 isoforms form dimers (Figure 5C). Visualization of the immunoprecipitates showed that the intensity of the bands was stronger when the full-length isoforms were co-expressed (lanes 1 and 3, the results for the R61 construct are similar to that for H61 and not shown), while dimer formation between a full-length isoform and the short Δ2 isoform was weaker (lanes 2 and 5). The homodimerization of the short Δ2 isoform is even weaker, but detectable (Figure 5C, lane 4). These results imply that the sequence in exon 2 may be essential for the inter-molecular association of BANK1.
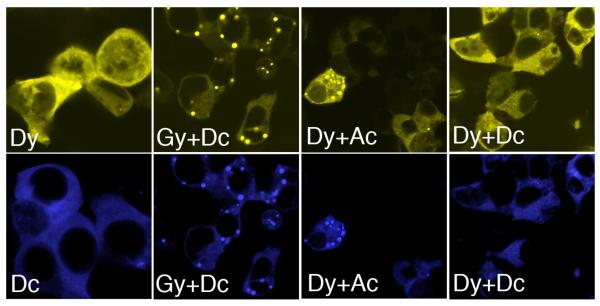
Since it is likely that the two full-length isoforms (in heterozygotes for rs10516487) and the short isoform co-exist in the same cell at the same time, we decided to study how the isoforms influence each other’s cellular distribution when co-expressed in HEK293T cells. In order to be able to discriminate each isoform, we fused them to different fluorescent proteins, yellow (y) and cyan (c). We observed, as expected, that the short isoform distributed homogeneously, in a similar pattern independently of the fluorescent tag (compare Dy versus Dc, Figure 6). Co-expression of both tagged D-BANK1 isoforms (Δ2) did not produce any dots, indicating that co-transfection and quantity of plasmid DNA used did not affect the formation of the protein complexes (see Dy+Dc). On the other hand, the short isoform was also detected in dots when co-expressed with the full-length isoform (compare the yellow and the cyan channel for Gy+Dc and Dy+Ac). This observation indicates that the full-length isoforms re-direct the short isoform to specific cellular compartments and act as dominant determinants of the distribution of the short isoform. These experiments are also in agreement with our co-immunoprecipitation results and further support the view of the existence of multimeric protein complexes containing all three of the BANK1 isoforms.
Figure 6.
Co-localization of expressed BANK1 isoforms.
Confocal images of HEK239 cells expressing different combinations of BANK1 isoforms tagged with YFP (y) or CFP (c). The first row of pictures is visualized with the yellow channel and the second with the blue channel. The short Δ2 BANK1 isoform designated as D acquires a punctate distribution when co-expressed with the full-length isoforms, G for BANK1-R61 and A for BANK1-H61.
Discussion
Despite the flood of studies reporting genetic associations with complex diseases and SLE in particular, little is known about the molecular mechanisms behind the associated variants. The majority of the associated SNPs were found in non-coding regions of genes that present certain difficulties in defining their function. SNPs located in close proximity of exon-intron boundaries can obviously influence splicing of the pre-mRNA, while non-synonymous SNPs are more likely to affect protein function due to amino-acid substitution. However, coding polymorphisms including nonsense, missense or silent substitutions could also influence the efficiency and precision of splicing by altering the regulatory elements of auxiliary splicing factors 16, 17.
Previously, we reported that the BANK1 transcript undergoes alternative splicing generating two major isoforms, a full-length isoform and a truncated Δ2 isoform 2. The short transcript lacks the entire exon 2 and codes for a protein without a putative domain for binding with the IP3 receptor type II 8. In agreement with Hertel, who suggested that alternative exons, unlike the constitutive ones could be very sensitive to minute changes in exonic and surrounding intronic sequences, we found that the BANK1 exon 2 splicing is indeed adjusted by the surrounding intronic sequence and the coding polymorphism in the exon itself 18. Moreover, the length of exon, which is longer than the majority of coding mammalian exons, could pose another obstacle for correct splicing. Subtle differences in the content and amount of splice factors present in a particular cell could alter the splicing of exon 2.
SNP rs17266594 is located in the branch point sequence in intron 1. Mutations located in exactly the same position of the branch point consensus sequence were reported to be crucial for splicing of the human lecithin:cholesterol acyltransferase gene (LCAT), the COL5A1 gene and the tyrosine hydroxylase (TH) gene 19-22. The C allele at position −2 from the branch point adenosine in intron 4 of the LCAT gene was found in patients with fish-eye disease and led to complete abolishment of splicing due to intron retention resulting in LCAT deficiency. While, for some other genes such as the low density lipoprotein (LDL)-receptor gene, the integrin β4 gene (IGTB4) and HLA-DQB1, only moderately reduced splicing efficiency was observed when polymorphisms or mutations in the branch point sequences occurred 23-25.
Our minigene experiments showed that the branch point sequence that bears rs17266594 is apparently not essential for splicing of BANK1 exon 2 and could be substituted by some other adenosines. We also observed that the level of the Δ2 isoform produced from minigenes was very low. This contrast to the previous observations reporting only 3-4 times lower levels of Δ2 compared to the full-length transcript in the fresh human PBMCs 2 and may indicate that the constructs might have missed some crucial sites that increase the efficiency of exon skipping and lead to the higher production of Δ2 from the gene. Of interest, when we analyzed the expression of the BANK1 isoforms in mice, we found that Δ2 was barely detectable in the mouse spleen cells (data not shown). Although the overall sequence homology between the human and mouse exon 2 is about 81%, the intronic regions differ between the two species. The mouse Bank1 gene has very strong splicing sites for exon 2 (score for 5′DS is 0.96 and 0.97 for 3′AS), but lacks the T-rich region. Thus, the structure of intron-exon border signals could favor to the more efficient splicing of the full-length isoform in mouse. Yet, since the Δ2 isoform is expressed in mice at low but detectable amounts, this could suggest that the splicing of the BANK1 exon 2 is rather complex and involves many other splicing signals and factors.
We further found that another SNP, the non-synonymous rs10516487 (Arg61His, R61H) located in exon 2 could play a role in splicing. It was shown before that intra-exonic mutations or polymorphisms could either create or destroy the recognition sites for splicing factors 26, 27. The published examples so far describe variants located close to the intron-exon border 26, 28-30, while the non-synonymous SNP rs10516487 in the BANK1 exon 2 is 112 bp downstream of the 3′ acceptor site. The minor protective allele A coding for His61 creates the recognition site for the SRp40 splicing enhancer factor 13, 31. Although this site is located deep in the exon, its strength is higher than that of another SRp40 element upstream (Figure 2). The binding of an additional SRp40 factor to the region located deeper in the exon might displace the other factors and thus exert a negative effect on the spliceosome assembly which would lead to a repression of splicing of exon 2 32, 33. Indeed, in the co-transfection experiment the SRp40 significantly suppressed splicing of the full-length isoform produced from the construct with the A allele. The similar function of SRp40 on inhibiting normal splicing of exon 6 in PDHA1 gene was described by Okajima et al. 34. Thus, the precise role of SR proteins in splicing could be dependent on the availability of other splicing factors, the strength of the splicing signals, the location of the binding motifs for splicing factors, the RNA folding 35-39.
Furthermore, we found that this lupus associated SNP exerts an effect on the protein function as well. The three BANK1 isoforms, two full-length with R61 and H61 and the Δ2 isoform, show different patterns of cytoplasmic localization and self-assembly. Ectopic expression of the full-length isoforms in HEK293 cells leads to the formation of protein complexes with a punctate phenotype, while the Δ2 isoform shows homogeneous distribution of the protein in the cytoplasm. The related to BANK1 protein Dof also showed a punctate pattern, and interestingly, the deletion of a region upstream of the DBB motif in Dof generated a more uniform distribution 40.
The protein domain coded by the BANK1 exon 2 constitutes a highly hydrophobic region (http://mobyle.rpbs.univ-paris-diderot.fr), which could render the protein to form cellular aggregates. It was shown that the hydrophobic patches of the DSCR1 protein (Down syndrome critical region 1, also named calcipressin 1 or regulator of calcineurin 1, RCAN1) are responsible for protein aggregation and neuropathology 41. The formation of multiprotein complexes or aggregates is in agreement with the proposed role for BANK1 as a scaffold/adaptor protein 8. Such proteins may form dimers or even more complex structures in the cell and thereby effectively bind other effector molecules to facilitate their function during intracellular signaling. Of note, the full-length isoforms of BANK1 self-associate efficiently as revealed on the Western blot after co-immunoprecipitation and the large punctate structures observed in the cytoplasm suggest the formation of complexes containing multiple molecules. The Δ2 isoform is less efficient in self-association probably because of the lack of a second dimerization domain and does not appear to form multimers, although it could engage in multimerization by the co-expressed full-length isoform. Our data may also indicate that the inter-molecular association is mediated by at least two domains of BANK1. If only one domain coded by exon 2 was involved, the dimerization of the Δ2 and hetero-dimerization between the full-length and the Δ2 protein isoforms would not occur. The second dimerization domain although not formally tested, is probably localized in the DBB region just downstream of the region coded by exon 2. It has been shown that the conserved domain DBB in two other related proteins Dof and BCAP is required for dimer formation 9.
The human Δ2 isoform could serve as a dominant negative isoform by competing with the full-length isoform for substrates thereby distorting B cell signaling. The ratio of one or the other might fine-tune the signaling. Further fine-tuning may depend on the presence of the R61 or 61H full-length protein isoform. The BANK1-R61 isoform formed significantly larger protein structures compared to BANK1-H61 suggesting a stronger ability for self-assembly. The functional importance of these BANK1 protein complexes is not known at present and awaits more studies. As the risk variant of BANK1 due to splicing efficiency leads to the production of more protein and as this protein shows increased self-assembly ability, the intracellular scaffold created is larger and more rigid in cells expressing the lupus risk allele. The outcome of this is at present unclear, and the effect of BCR signaling on self-assembly needs also to be analyzed. Nevertheless, assuming that the risk allele might relate to stronger self-assembly and a more rigid scaffold, the final consequence of this would be a less flexible modulation of the BCR response. This failure to modulate B-cell activation has been observed in SLE patients after BCR engagement 42. In this regard, modulation of the B-cell response could be mediated by adaptor/scaffold protein such as BANK1. For instance, we have shown recently that the full-length BANK1isoform can modulate trafficking of the BLK kinase to the plasma membrane (Castillejo-Lopez, submitted) and the capacity of BANK1 to modulate the trafficking of BLK or LYN may influence or distort BCR signaling.
In summary, in the current study we analyzed the effects of the associated polymorphisms on BANK1 gene function. We provide evidence that the non-synonymous SNP rs10516487 exerts dual function through the influence the mRNA splicing and by altering the behavior of the BANK1 protein. The risk allele G leads to higher production of the protein with an increased self-assembly ability, which could eventually result in the more sustained BCR-mediated signaling in patients with SLE.
Materials and Methods
Minigenes, SRp40 and BANK1 expression constructs
The BANK1 minigene constructs containing different allelic variants in intron 1 and exon 2 were prepared as follows: the human BANK1 exon 1, 2 and 3 together with the 500 bp of the intronic regions upstream and downstream of the exons were amplified by PCR and cloned in pcDNA3.1 D/V5-His -TOPO vector (Invitrogen).
The deletion of the branch point site in intron 1 was achieved by site-directed mutagenesis. Briefly, the wild-type branch point sequence was replaced with the unrelated sequence coding for a site for Msp I restriction endonuclease for easy detection of the mutant clones. The inverse PCR was performed using the plasmid DNA coding for the original minigenes, followed by digestion of the PCR products by the Dpn I endonuclease and ligation at low concentration. DNA was transformed into TOP10 competent cells (Invitrogen) and positive clones were selected by Dpn I digestion.
To prepare the expression plasmid for the SRp40 splicing factor, we cloned the SRp40 cDNA amplified by PCR with the following primers 5′-CACCATGAGTGGCTGTCGGGTATT-3′ and 5′-TTAATTGCCACTGTCAACTGATCTG-3′ in pcDNA3.1 vector. The neomycin gene present in the plasmid backbone was destroyed in order to prevent the transcription of the gene.
The BANK1 isoforms were amplified by PCR using cDNAs from individuals with known genotypes. The open reading frames with and without the stop codon for the V5-tagged protein were cloned into pcDNA3.1D/V5-His. In order to generate the fusion between the BANK1 and various fluorescent proteins, the cDNA coding for CFP, YFP and GFP were amplified with the primers 5′-GGGCGGCCGCATGGTGAGCAAGGGCGA-3′and 5′-GGTCTAGATCTTGTACAGCTCGTCCAT-3′ and subcloned in frame at the NotI/XbaI sites. The constructs coding for the N-terminal FLAG-tagged BANK1 isoforms were prepared by sequential PCR using overlapping primers for the 5′-extension and subcloned into pIRES S2-EGFP vector (Clontech) (Supplementary Table 1). All constructs were verified by sequencing.
Mouse exon 2 sequence analysis
The 1.6 kb DNA fragment encompassing the mouse exon 2 and the upstream and downstream intronic sequence was amplified with the following primers 5′-GATGTGGTGTCAGGTGGGAAAGAGA-3′ and 5′-CATGAGTGAAGACAGCAGCACTTCAG-3′ using genomic DNA from C57BL/6, BALB/cJ, NOD/Lt, DBA/1J, NZBWF1, NZW/LacJ, NZB/BINJ mouse strains. The sequences were deposited to GenBank with the following accession numbers: JF915711, JF915712, JF915713.
Cell transfection
All transfections were performed in HEK293T cells using Lipofectamine 2000 (Invitrogen). The cells were maintained in DMEM medium supplemented with 10% FCS, L-glutamine (2 mM) and 100 international units/ml of penicillin/streptomycin.
For the minigene experiments, the transfection was performed in a 24-well plate format using 400 ng of plasmid DNA per well. For the co-transfection experiments with SRp40, 800 ng of the minigenes and different amounts of the SRp40-coding plasmid were used. To ensure the equal DNA load, the total amount of DNA was adjusted to 1.3 μg by the addition of the empty pcDNA3.1 vector. The cells were collected 36 hours after transfection. All transfection experiments were performed in triplicates.
RNA extraction and expression analysis
Total RNA was extracted using Trizol reagent (Invitrogen) and treated with RQ1 DNase I (Promega). cDNA synthesis was performed using 1μg of DNase-treated RNA in a buffer containing 1 unit of MuLV reverse transcriptase (ABI), oligo(dT) primers, 1 mM dNTPs and RNase inhibitor (ABI). The quantitative real-time PCR was performed as described in 2 using SYBR green dye for detection. The expression levels of the full-length and the Δ2 transcripts generated from the minigenes were normalized to the levels of the neomycin gene expressed from the same plasmid using the comparative 2-ΔCt method. The following forward primers were used for the BANK1 full-length transcript 5′-GAGTATCATATTCAAAGATTCTGAAGAC-3′ and the Δ2 transcript 5′-CAGCGCCCCCAGATTCTGAAG-3′, and a common reverse primer 5′-CACATGGAATTTCAGTGGGAAGCAC-3′; the primers for neomycin transcript were 5′-TGGCGGACCGCTATCAGGACATA-3′ and 5′-ACCCCAGAGTCCCGCTCAGAAG-3′. Statistical analysis was performed using an unpaired 2-tailed t-test with GraphPad software (http://www.graphpad.com).
For detection of the BANK1 transcripts in lymphoblastoid cell lines, cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum and penicillin-streptomycin. RNA purification, cDNA synthesis and quantitative RT-PCR were performed as described above. The gene expression was normalized to the levels of 18S rRNA amplified with a commercial kit (Applied Biosystems).
Antibodies
The polyclonal anti-human BANK1 (ET52) antibody was generated by immunization of rabbits with a synthetic peptide ETKHSPLEVGSESSC corresponding to amino acids 467 to 480 of the full-length BANK1 protein. The serum was further affinity purified against the peptides using the SulfoLink Kit (Pierce). Additional antibodies used in this study include the following: anti-V5 (Invitogen); monoclonal anti-β-tubulin, monoclonal anti-Flag M2 and rabbit anti-Flag (Sigma); anti-rabbit and anti-mouse IgG HRP (Zymed).
Western Blotting
For detection of the native BANK1 isoforms in lymphoblastoid cell lines, 1×106 cells were lysed for 15 minutes on ice in RIPA buffer (50mM Tris-Cl pH 7.4, 150mM NaCl, 1% NP40, 0.25% Na-deoxycholate, 1mM PMSF, 1× Roche complete mini protease inhibitor cocktail) and the lysates were further pre-cleared by centrifugation at 14 000 rpm at +4°C for 5 minutes. Protein extracts corresponding to 4×105 cells were boiled in SDS sample buffer and run on 4-12% gradient SDS-PAGE gel (Invitrogene) followed by the transfer of proteins onto a nitrocelulose membrane. After blocking in PBS containing 5% non-fat dry milk and 0.1% Tween-20, the membranes were probed with the primary ET-52 antibody, secondary anti-rabbit HRP and detected with enhaced chemiluminescent reagent (Santa Cruz Biothechnology, Inc). For quantification, the membranes were stripped and reprobed with anti-β-tubulin and anti-mouse IgG HRP. The intensity of bands was analyzed using ImageJ software (http://rsb.info.nih.gov/ij/). The AA3 clone was chosen as a reference for normalization (score 100).
Immunoprecipitation
HEK293T cells were grown in 6-well plates and co-transfected with 1μg of each of the DNA constructs coding for the BANK1 isoforms tagged either with the FLAG-epitope at the N-terminus or the V5-epitope at the C-terminus. Cells were lysed in 200 μl of the Lysis Buffer A ((1% Triton X-100, 50mM HEPES pH 7.1, 150mM Nacl, 1mM EDTA, 2mM Na3VO4, 10 % Glycerol, 0.1% SDS) containing protease inhibitors (Roche) and 1mM PMSF) for 15-20 min on ice and cell extracts were pre-cleared by centrifugation at 14 000 rpm at +4°C for 5 min and subsequent incubation with 10 μl of Sepharose beads in a rotating wheel for 30 min at +4°C. About the 10% of the pre-cleared extracts were saved for Western blot analysis as control for the input. Immunoprecipitation (IP) was carried out using 150 μl of cell extracts, 1μg of Anti-Flag Ab and 20μl protein G-Sepharose beads. The IP was performed in a rotating wheel for 10 h at +4°C. The beads were washed in Lysis buffer A and resuspended in the loading buffer before SDS-PAGE and Western blotting. The proteins were detected with using anti-Flag and anti-V5 antibodies.
Confocal microscopy
Cells were grown and transfected on Lab-Tek chamber slides coated with poly-D-lysine. Twenty-four hours after transfection cells were fixed at room temperature for 20 minutes with 3,7% paraformaldehyde in PBS/0.18% Triton-X. Fluorescence fusion proteins were visualized directly after fixation, FX enhancer treatment (Invitrogen) and mounted in Vectashield (Vector Lab). Confocal microscopy was performed using a Zeiss 510 Meta confocal scanning microscope with a Zeiss plan-Apochromat 63× oil-immersion objective. Dual- or triplecolor images were acquired by consecutive scanning with only 1 laser line active per scan to avoid cross-excitation. CFP was revealed using the 405-nm line with a 405/488/543-nm excitation filter, a 515-nm dichroic and a 470-500-nm emission filter. YFP was revealed using the 514-nm line with a 458/514 nm excitation filter, a 515-nm dichroic and a 530-600 nm emission filter. Image analysis was carried out using ImageJ software.
Measurement of the cytoplasmic punctae area
Protein spots were measured using the ImageJ program Analyze Particles, version 1.42q. The following setup was used: threshold minimum and particle range between 0.2 μm2-50 μm2. The result files were inspected by eye and areas not well discriminated were corrected manually. The analysis was done double-blinded with coded digital pictures corresponding to 9 different transfections. Two different constructs for each protein isoform were used, one tagged with CFP and the other one with GFP.
Supplementary Material
ACKNOWLEDGMENTS
This work has been supported by the Swedish Research Council to MEAR, the King Gustaf Vth-80th Jubilee Fund to MEAR and SVK, Clas Groschinskys Fund and Olle Engkvist Byggmästare Fund and Marcus Borgström Fund to SVK and the Swedish Association Against Rheumatism to CCL, MEAR and SVK. We also acknowledge the NIH COBRE grant RR020143, the OCAST grant HR09-106/7569 and the Alliance for Lupus Research and the Instituto de Salud Carlos III (PS09/00129) partially co-financed with FEDER funds from the European Union to MEAR. We are grateful to Ammar Zaghlool for cloning of the ΔT minigene and SRp40 expression construct and help with initial transfection data analysis.
Footnotes
CONFLICT OF INTEREST
Authors declare no conflict of interest.
REFERENCES
- 1.Delgado-Vega A, Sanchez E, Lofgren S, Castillejo-Lopez C, Alarcon-Riquelme ME. Recent findings on genetics of systemic autoimmune diseases. Curr Opin Immunol. 2010;22(6):698–705. doi: 10.1016/j.coi.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kozyrev SV, Abelson AK, Wojcik J, Zaghlool A, Linga Reddy MV, Sanchez E, et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat Genet. 2008;40(2):211–6. doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- 3.Guo L, Deshmukh H, Lu R, Vidal GS, Kelly JA, Kaufman KM, et al. Replication of the BANK1 genetic association with systemic lupus erythematosus in a European-derived population. Genes Immun. 2009;10(5):531–8. doi: 10.1038/gene.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41(11):1228–33. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang W, Shen N, Ye DQ, Liu Q, Zhang Y, Qian XX, et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet. 2010;6(2):e1000841. doi: 10.1371/journal.pgen.1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang YK, Yang W, Zhao M, Mok CC, Chan TM, Wong RW, et al. Association of BANK1 and TNFSF4 with systemic lupus erythematosus in Hong Kong Chinese. Genes Immun. 2009;10(5):414–20. doi: 10.1038/gene.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilbe M, Jokinen P, Truve K, Seppala EH, Karlsson EK, Biagi T, et al. Genome-wide association mapping identifies multiple loci for a canine SLE-related disease complex. Nat Genet. 2010;42(3):250–4. doi: 10.1038/ng.525. [DOI] [PubMed] [Google Scholar]
- 8.Yokoyama K, Su Ih IH, Tezuka T, Yasuda T, Mikoshiba K, Tarakhovsky A, et al. BANK regulates BCR-induced calcium mobilization by promoting tyrosine phosphorylation of IP(3) receptor. Embo J. 2002;21(1-2):83–92. doi: 10.1093/emboj/21.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Battersby A, Csiszar A, Leptin M, Wilson R. Isolation of proteins that interact with the signal transduction molecule Dof and identification of a functional domain conserved between Dof and vertebrate BCAP. J Mol Biol. 2003;329(3):479–93. doi: 10.1016/s0022-2836(03)00489-3. [DOI] [PubMed] [Google Scholar]
- 10.Okada T, Maeda A, Iwamatsu A, Gotoh K, Kurosaki T. BCAP: the tyrosine kinase substrate that connects B cell receptor to phosphoinositide 3-kinase activation. Immunity. 2000;13(6):817–27. doi: 10.1016/s1074-7613(00)00079-0. [DOI] [PubMed] [Google Scholar]
- 11.Aiba Y, Yamazaki T, Okada T, Gotoh K, Sanjo H, Ogata M, et al. BANK negatively regulates Akt activation and subsequent B cell responses. Immunity. 2006;24(3):259–68. doi: 10.1016/j.immuni.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Krainer AR. Pre-mRNA splicing by complementation with purified human U1, U2, U4/U6 and U5 snRNPs. Nucleic Acids Res. 1988;16(20):9415–29. doi: 10.1093/nar/16.20.9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31(13):3568–71. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ibrahim EC, Schaal TD, Hertel KJ, Reed R, Maniatis T. Serine/arginine-rich protein-dependent suppression of exon skipping by exonic splicing enhancers. Proc Natl Acad Sci U S A. 2005;102(14):5002–7. doi: 10.1073/pnas.0500543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6(5):386–98. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 16.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3(4):285–98. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 17.Pagani F, Baralle FE. Genomic variants in exons and introns: identifying the splicing spoilers. Nat Rev Genet. 2004;5(5):389–96. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- 18.Hertel KJ. Combinatorial control of exon recognition. J Biol Chem. 2008;283(3):1211–5. doi: 10.1074/jbc.R700035200. [DOI] [PubMed] [Google Scholar]
- 19.Kuivenhoven JA, Weibusch H, Pritchard PH, Funke H, Benne R, Assmann G, et al. An intronic mutation in a lariat branchpoint sequence is a direct cause of an inherited human disorder (fish-eye disease) J Clin Invest. 1996;98(2):358–64. doi: 10.1172/JCI118800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li M, Pritchard PH. Characterization of the effects of mutations in the putative branchpoint sequence of intron 4 on the splicing within the human lecithin:cholesterol acyltransferase gene. J Biol Chem. 2000;275(24):18079–84. doi: 10.1074/jbc.M910197199. [DOI] [PubMed] [Google Scholar]
- 21.Burrows NP, Nicholls AC, Richards AJ, Luccarini C, Harrison JB, Yates JR, et al. A point mutation in an intronic branch site results in aberrant splicing of COL5A1 and in Ehlers-Danlos syndrome type II in two British families. Am J Hum Genet. 1998;63(2):390–8. doi: 10.1086/301948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janssen RJ, Wevers RA, Haussler M, Luyten JA, Steenbergen-Spanjers GC, Hoffmann GF, et al. A branch site mutation leading to aberrant splicing of the human tyrosine hydroxylase gene in a child with a severe extrapyramidal movement disorder. Annals of human genetics. 2000;64(Pt 5):375–82. doi: 10.1046/j.1469-1809.2000.6450375.x. [DOI] [PubMed] [Google Scholar]
- 23.Webb JC, Patel DD, Shoulders CC, Knight BL, Soutar AK. Genetic variation at a splicing branch point in intron 9 of the low density lipoprotein (LDL)-receptor gene: a rare mutation that disrupts mRNA splicing in a patient with familial hypercholesterolaemia and a common polymorphism. Hum Mol Genet. 1996;5(9):1325–31. doi: 10.1093/hmg/5.9.1325. [DOI] [PubMed] [Google Scholar]
- 24.Chavanas S, Gache Y, Vailly J, Kanitakis J, Pulkkinen L, Uitto J, et al. Splicing modulation of integrin beta4 pre-mRNA carrying a branch point mutation underlies epidermolysis bullosa with pyloric atresia undergoing spontaneous amelioration with ageing. Hum Mol Genet. 1999;8(11):2097–105. doi: 10.1093/hmg/8.11.2097. [DOI] [PubMed] [Google Scholar]
- 25.Kralovicova J, Houngninou-Molango S, Kramer A, Vorechovsky I. Branch site haplotypes that control alternative splicing. Hum Mol Genet. 2004;13(24):3189–202. doi: 10.1093/hmg/ddh334. [DOI] [PubMed] [Google Scholar]
- 26.Liu HX, Cartegni L, Zhang MQ, Krainer AR. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat Genet. 2001;27(1):55–8. doi: 10.1038/83762. [DOI] [PubMed] [Google Scholar]
- 27.Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet. 2002;30(4):377–84. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 28.Kashima T, Manley JL. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet. 2003;34(4):460–3. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 29.Teraoka SN, Telatar M, Becker-Catania S, Liang T, Onengut S, Tolun A, et al. Splicing defects in the ataxia-telangiectasia gene, ATM: underlying mutations and consequences. Am J Hum Genet. 1999;64(6):1617–31. doi: 10.1086/302418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pagani F, Stuani C, Tzetis M, Kanavakis E, Efthymiadou A, Doudounakis S, et al. New type of disease causing mutations: the example of the composite exonic regulatory elements of splicing in CFTR exon 12. Hum Mol Genet. 2003;12(10):1111–20. doi: 10.1093/hmg/ddg131. [DOI] [PubMed] [Google Scholar]
- 31.Liu HX, Zhang M, Krainer AR. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12(13):1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graveley BR. Sorting out the complexity of SR protein functions. Rna. 2000;6(9):1197–211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Y, Zhang Y, Zhang J. Distribution of exonic splicing enhancer elements in human genes. Genomics. 2005;86(3):329–36. doi: 10.1016/j.ygeno.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 34.Okajima K, Warman ML, Byrne LC, Kerr DS. Somatic mosaicism in a male with an exon skipping mutation in PDHA1 of the pyruvate dehydrogenase complex results in a milder phenotype. Mol Genet Metab. 2006;87(2):162–8. doi: 10.1016/j.ymgme.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 35.Ibrahim EC, Schaal TD, Hertel KJ, Reed R, Maniatis T. Serine/arginine-rich protein-dependent suppression of exon skipping by exonic splicing enhancers. Proc Natl Acad Sci U S A. 2005;102(14):5002–7. doi: 10.1073/pnas.0500543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuo BA, Uporova TM, Liang H, Bennett VD, Tuan RS, Norton PA. Alternative splicing during chondrogenesis: modulation of fibronectin exon EIIIA splicing by SR proteins. J Cell Biochem. 2002;86(1):45–55. doi: 10.1002/jcb.10188. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Smith PJ, Krainer AR, Zhang MQ. Distribution of SR protein exonic splicing enhancer motifs in human protein-coding genes. Nucleic Acids Res. 2005;33(16):5053–62. doi: 10.1093/nar/gki810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang H, Tuan RS, Norton PA. Overexpression of SR proteins and splice variants modulates chondrogenesis. Exp Cell Res. 2007;313(8):1509–17. doi: 10.1016/j.yexcr.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 39.Tyson-Capper AJ, Bailey J, Krainer AR, Robson SC, Europe-Finner GN. The switch in alternative splicing of cyclic AMP-response element modulator protein CREM{tau}2{alpha} (activator) to CREM{alpha} (repressor) in human myometrial cells is mediated by SRp40. J Biol Chem. 2005;280(41):34521–9. doi: 10.1074/jbc.M505344200. [DOI] [PubMed] [Google Scholar]
- 40.Wilson R, Battersby A, Csiszar A, Vogelsang E, Leptin M. A functional domain of Dof that is required for fibroblast growth factor signaling. Mol Cell Biol. 2004;24(6):2263–76. doi: 10.1128/MCB.24.6.2263-2276.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma H, Xiong H, Liu T, Zhang L, Godzik A, Zhang Z. Aggregate formation and synaptic abnormality induced by DSCR1. J Neurochem. 2004;88(6):1485–96. doi: 10.1046/j.1471-4159.2003.02294.x. [DOI] [PubMed] [Google Scholar]
- 42.Taher TE, Parikh K, Flores-Borja F, Mletzko S, Isenberg DA, Peppelenbosch MP, et al. Protein phosphorylation and kinome profiling reveal altered regulation of multiple signaling pathways in B lymphocytes from patients with systemic lupus erythematosus. Arthritis Rheum. 2010;62(8):2412–23. doi: 10.1002/art.27505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.