Abstract
Most of eukaryotic genes are interrupted by introns that need to be removed from pre-mRNAs before they can perform their function. This is done by complex machinery called spliceosome. Many eukaryotes possess two separate spliceosomal systems that process separate sets of introns. The major (U2) spliceosome removes majority of introns, while minute fraction of intron repertoire is processed by the minor (U12) spliceosome. These two populations of introns are called U2-type and U12-type, respectively. The latter fall into two subtypes based on the terminal dinucleotides. The minor spliceosomal system has been lost independently in some lineages, while in some others few U12-type introns persist. We investigated twenty insect genomes in order to better understand the evolutionary dynamics of U12-type introns. Our work confirms dramatic drop of U12-type introns in Diptera, leaving these genomes just with a handful cases. This is mostly the result of intron deletion, but in a number of dipteral cases, minor type introns were switched to a major type, as well. Insect genes that harbor U12-type introns belong to several functional categories among which proteins binding ions and nucleic acids are enriched and these few categories are also overrepresented among these genes that preserved minor type introns in Diptera.
Keywords: U12-type introns, minor spliceosome, insect evolution.
Introduction
Most eukaryotic protein coding genes are intervened by non-coding sequences called introns (Intervening regions) 1, which are being removed from the primary transcript in the process of splicing 2-3. There are four recognized major groups of introns, namely group I, II, III, and spliceosomal/nuclear introns. While introns from the first three groups undergo self-splicing, the latter endure splicing with the aid of complex machinery called spliceosome. The spliceosomes consist of four small nuclear ribonucleoproteins (snRNPs) and over a hundred of non-snRNP proteins that associate with snRNPs at some point during the splicing 4-6. There are two distinct types of spliceosomal introns; U2-type and U12-type introns, which are excised by the major and minor spliceosomes, respectively 7-8. Both the spliceosomes are structurally and functionally similar. The major difference lies in the ribonucleotide components, while the major spliceosome contains U1, U2, U4, and U6 snRNPs, the minor one consists of functionally equivalent U11, U12, U4atac, and U6atac snRNPs, with U5 snRNP present in both spliceosomes 9-10.
The U12-type introns were discovered in the early 1990s thanks to atypical splice site (SS) dinucleotides AT-AC 8. They contain highly conserved/prominent and consistent 5´splice site (A/G) TATCCTT (at +1 to +8 at 5' SS), a less conserved branch point site (BPS) TCCTTAACT and A(C/G) at 3' splice site 8, 11-13. However, as reported recently by Lin et al. U12-type introns can be flanked by different terminal dinucleotides indicating that the donor and acceptor sites are degenerate 14. The BPS of U2-type introns is usually located 18-40 nucleotides upstream of the 3' splice site, in contrast to the U12-type introns, where it is restricted to 12-15 nucleotides 8, 11-12, 15. Additionally, U12-type introns lack a polypyrimidine tract between the BPS and the 3′ splice site.
Although U12-type introns are highly conserved in specific lineages, they do undergo some evolutionary changes, for instance intron deletion or spliceosomal type switching 7. It has been suggested that a few mutations in the donor SS of U12-type intron may change the intron type to the major one. Because of the stronger signal constrain at the 5´ SS of U12-type introns, this process is believed to be unidirectional, as switching intron type from U2 to U12 would require too many concurrent changes 7, 14, 16. Interestingly, AT-AC U12-type introns often get converted to GT-AG U12-type intron in the process called subtype switching, which seems to be initial step in intron type switching 7, 14.
U12-type introns comprise less than half percent of all spliceosomal introns 13, 17. They are present in most eukaryotic genomes from the basic such as jellyfish to the higher chordates and plants 7, 16. Interestingly, U12-type introns are absent in some organisms such as the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, the nematode Caenorhabditis elegans and many protists. However, a recent study of phylogenetic distribution of the spliceosomal snRNA genes has shown a wider distribution of minor type intron than anticipated before 18. It is clear that U12-type introns and cognate splicing machinery were lost independently a number of times during eukaryotic evolution. In some other lineages, although not completely lost, the number of U12-type introns has been dramatically reduced. Previously, we studied the evolutionary dynamic of U12-type introns among eighteen metazoan genomes including three insects and several vertebrates 14. More insect genomes have been sequenced recently, covering 400 million years of metazoan evolution 19 and thus creating ideal resources for evolutionary studies at the genomic level. With seventeen additional insect genomes available for the analyses, we were able to study mechanism of the evolution of U12-type introns with greater details.
Here, we present a comprehensive study of the evolutionary dynamics of U12-type intron on the insect phylogeny. Our results unveiled a dramatic drop of the U12-type intron number in Diptera that occurred mostly by removal of U12-type introns from many dipteral genes and to a lesser extent by U12 to U2-type intron conversion. Interestingly, in our dataset we found evidence neither for subtype switching nor for U12 AT-AC-subtype intron conversion directly to U2-type intron.
Materials and Methods
The sequence data
The initial dataset of insect U12-type intron containing genes was downloaded from the U12-type intron database (ver. 1.0 http://genome.imim.es/cgi-bin/u12db/u12db.cgi) 20 and it consisted of seventeen U12-type genes of Drosophila melanogaster, two U12-type genes of Anopheles gambiae, and twenty-three genes from Apis mellifera. This set was complemented by forty-nine Apis mellifera genes described previously by Mount et al. 21 and two more genes in Drosophila described by Lin et al. 14. These seventy U12-type-intron bearing genes were used as a starting point to probe the U12-type intron status in twenty insect genomes D. ananassae (GenBank accession number: AAPP01019547.1), D. erecta (AAPQ01006465.1), D. grimshawi (AAPT01020220.1), D. mojavensis (AAPU01011093.1), D. melanogaster (AABU01002774.1), D. persimilis (AAIZ01002952.1), D. pseudoobscura (AAFS01001852.1), D. sechellia (AAKO01000279.1), D. simulans (AASV01031774.1), D. virilis (AANI01017120.1), D. willistoni (AAQB01008615.1), D. yakuba (AAEU02000019.1), Aedes aegypti (NZ_AAGE02004256.1), Anopheles gambiae str. PEST (NZ_AAAB02008817.1), Culex quinquefasciatus (NZ_AAWU01005511.1), Apis mellifera (NW_001253366.1), Bombyx mori (BABH01019700.1), Nasonia vitripennis (AAZX01004521.1), Pediculus humanus corporis, (AAZO01003896.1) and Tribolium castaneum (NW_001094314.1).
Orthologs identification and intron/exon structure determination
All the insect orthologs of U12-type-intron-containing genes were identified by querying protein sequences coded by the seventy genes against the twenty insect genomes using NCBI's BLAST (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi?organism=insects) 22 with the insect genomes database and default values for all other parameters. The intron-exon boundaries for the orthologs found in other insect genomes besides the query genomes were identified and marked using manual search and NCBI BLAST 22 based annotation, ORF Finder 23 , Expasy translate tool 24, and UCSC genome browser 25. In cases when the annotated gene structures were incomplete, trace archives and/or EST databases for the complete gene sequence were searched.
Since significant splicing signals are present only at the intron's termini, each intron/exon boundary was represented by forty (twenty nt upstream + twenty nt downstream of the 5´splice site) and seventy (fifty nt upstream + twenty nt downstream of the 3´splice site) nucleotides and the multiple sequence alignment of the extracted sequences was calculated using T-Coffee 26. The alignments were then inspected using BIOEDIT 27 searching for the conserved 5' and branch point signals.
Intron status determination
Intron status was determined for each of the seventy-one introns individually; one of the genes harbors two U12-type introns. The following intronic events were considered: intron type switching, i.e. U12-type to U2-type or vice versa, U12-subtype switching, i.e. AT-AC to GT-AG or vice versa, deletion and insertion of an intron in the insect lineage. In ambiguous cases, other metazoans were considered as outgroups with the human data as the first choice because of the high number of U12-type introns and accurate annotation of the human genome. We investigated the mode of evolution of U12-type introns applying parsimony principle on the species trees of the analyzed genes.
Functional annotation of genes
Gene Ontology enrichment analysis was done using DAVID (http://david.abcc.ncifcrf.gov/) 28 for seventy D. melanogaster genes for which at least one of the insect orthologous genes harbor U12-type introns in at least one insect genome. These genes were compared against the rest of the genes in D. melanogaster genome as a background data using the Functional Annotation Clustering tool in DAVID with default parameters. Classification stringency 'lowest' was used for the inclusion of all ontology terms. Similar analysis was also done for a gene set limited to Diptera lineage. The data set consisted of twenty-six genes containing a U12-type intron in any dipteran genome and was compared against all other D. melanogaster's genes.
Results and Discussion
U12-type intron number
Among the seventy-one U12-type insect introns, fourteen were of AT-AC U12-subtype, fifty-six of GT-AG subtype, and one of GC-AG intron. Number of U12-type introns in insects varies from fifteen in the C. quinquefasciatus genome to sixty-three in the A. mellifera genome (see Table 1). The parsimony analysis indicates that the ancestral insect genome contained at least seventy U12-type introns.
Table 1.
Number of U12-type introns in different insect genomes.
| Species | Number of introns available for the analysis | Number of detected U12-type introns | Genome size in Mb |
|---|---|---|---|
| Drosophila simulans | 71 | 19 | 146.7 |
| Drosophila sechellia | 71 | 19 | 166.26 |
| Drosophila melanogaster | 71 | 19 | 176.04 |
| Drosophila yakuba | 71 | 19 | 166.26 |
| Drosophila erecta | 71 | 19 | 146.7 |
| Drosophila ananassae | 71 | 17 | 185.82 |
| Drosophila pseudoobscura | 71 | 19 | 156.48 |
| Drosophila persimilis | 70 | 17 | 176.04 |
| Drosophila willistoni | 71 | 17 | 205.38 |
| Drosophila mojavensis | 71 | 19 | 166.26 |
| Drosophila virilis | 71 | 19 | 332.52 |
| Drosophila grimshawi | 71 | 19 | 234.72 |
| Aedes aegypti | 71 | 17 | 1376 |
| Culex quinquefasciatus | 71 | 15 | 579 |
| Anopheles gambiae | 70 | 17 | 278 |
| Bombyx mori | 68 | 32 | 530 |
| Tribolium castaneum | 71 | 34 | 160 |
| Apis mellifera | 71 | 63 | 264.0 |
| Nasonia vitripennis | 71 | 46 | 335 |
| Pediculus humanus | 71 | 39 | 107.58 |
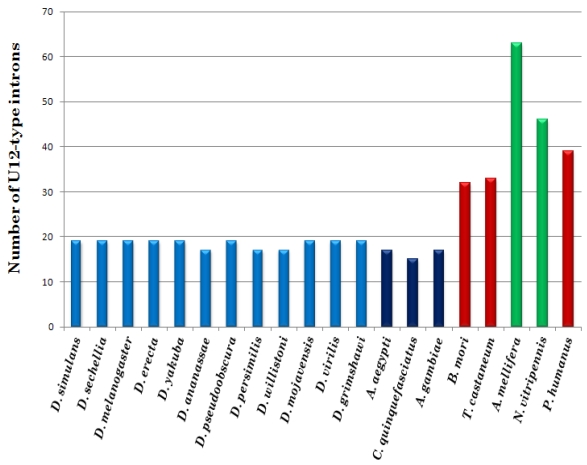
All fifteen genomes of the order Diptera have fewer minor introns than any other species analyzed in this study. Our insect genome dataset reveals a high disparity in number of U12-type introns among different genomes and even within the same genus. For instance, nine Drosophila sp. harbor nineteen U12-type introns, while D. ananassae, D. persimilis and D. willistoni contain only seventeen. Three mosquito genomes have even lower number of minor type introns, with seventeen cases in A. aegypti and A. gambiae, and only fifteen in Culex quinquefasciatus. The latter is the smallest number of U12-type introns in any investigated genome and likely, this set includes genes for which U12-type intron is essential in their functional regulation. In other insect genomes, the number of U12-type introns is two to three times higher than in dipteran genomes (see Fig. 1). The highest number of the minor type introns is observed in A. mellifera and Nasonia vitripennis of the order Hymenoptera. However, none of the investigated genomes harbor all the seventy-one U12-type introns detected in insects. Interestingly, A. mellifera's U12-type intron content is three fourth of the urochordate Ciona intestinalis, an additional hint that the genome of the last common insect ancestor accommodated at least about seventy minor-type introns 21. Nearly seventy five percent of the U12-type introns are conserved between A. mellifera and Homo sapiens. Although most of the insect genomes lost some of the U12-type intron, the process for some reason has accelerated in the Diptera order, where most of the minor type introns were lost during the last 280 million years of their evolution since divergence from the rest of insects 29.
Figure 1.
Number of U12-type introns in twenty insect genomes. Blue bars indicate Diptera order including Drosophilidae and Culicidae families, light and dark shade, respectively; green bars represent genomes of Hymenoptera order and the red bars represent neither Diptera nor Hymenoptera orders.
All sampled dipteral genomes contain less than twenty minor introns and their number in insect genomes is not correlated with the genome size (R = 0.08; see Table 1 and Supplementary Material: Fig. S1). In order to find out if this apparent loss of U12-type introns is related to the overall intron loss in Diptera or the result of selective removal of minor introns, we have compared total number of introns in each genome with the number of minor-type introns. For instance, three of dipterans (D. melanogaster, A. aegypti and A. gamabiae) have much lower number of introns than non-dipteral A. mellifera and Nasonia vitripennis (see Supplementary Material: Table S4). Although, there is a clear trend of reduction in overall number of introns in the dipteral genomes, U12-type introns disappear from those genomes in even faster rate. Interestingly, this trend is statistically significant when dipteral data are compared against the honeybee genome but not when compared against wasp data. Unfortunately, annotation and sequence quality of other insect genomes didn't allow for more extensive analysis.
Even though the genome of D. virilis is twice the size of D. melanogaster and other Drosophila sp., it harbors similar number of U12-type introns. Larger genomic size of D. virilis is due to various factors such as the presence of larger heterochromatic content 30-31, long and highly polymorphic microsatellites 31-32, and longer introns 33-34. Consequently, length of U12-type introns is also bigger in D. virilis as compared to other Drosophila species; average U12-type intron size is 1,067 nt in D. virilis in contrast to 687 nt in D. melanogaster. Interestingly, two analyzed mosquito genomes contain few U12-type introns, even though the A. aegypti genome is almost ten times larger than most Drosophila of the genomes 35. However, unlike Drosophila, half of the mosquito genome is composed of Transposable Elements rendering a large genomic size 36.
The analyzed genes contain at most one U12-type intron. The only exception is the Ca-alpha1 D gene, which codes for voltage-sensitive calcium channel and in some species, contains two GT-AG U12-type introns. The first U12-type intron, which lies between exons two and three, is absent in all the three mosquitoes and Pediculus but is conserved in the rest of the insects; whereas, the second U12-type intron is absent in all Drosophila and Pediculus, and has been converted into U2-type intron in Bombyx mori. Interestingly, many voltage channel genes contain more than one U12-type introns in vertebrates, which strongly suggest that two-U12-intron arrangement is the ancestral one 14, 37. It was also shown before that although some of the U12-type introns are being removed randomly from these genes, they usually preserve at least one of them suggesting some important role played by these introns 38. Consequently, in our dataset most of the Ca-alpha1 D genes preserved at least one of U12-type introns with the only exception being Pediculus genome. However, Pediculus has lost all but one intron in this gene and this single intron is not homologous to any insect introns, which may suggest complicated evolutionary history of the Pediculus gene that could involve retrogene activation followed by an intron gain as described recently by Szczesniak et al. 39.
Evolutionary fate of U12-type introns
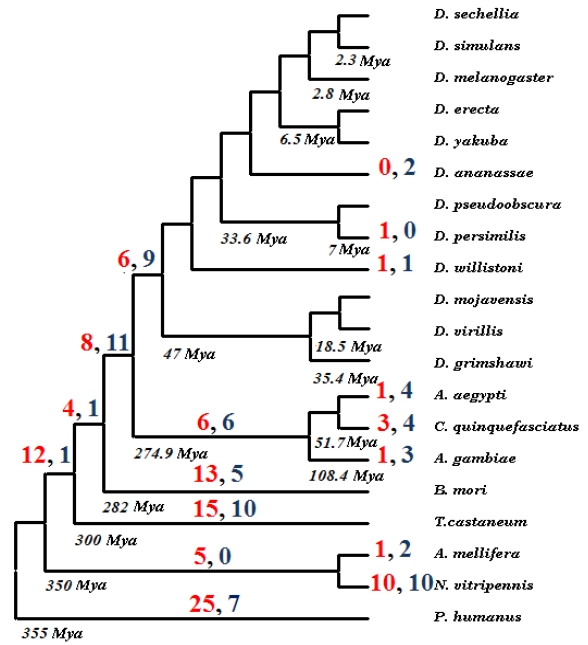
Comparative genomics analysis showed that ancestral insect genome harbored at least seventy U12-type introns. None of the analyzed extant genomes contain so many U12-type introns and relatively small number of these introns is rather norm with the Culex genome harboring fifteen U12 introns being an extreme case. So, it is a clear trend that minor type introns disappear from the insect genomes. There are two possible pathways of an U12-type intron disappearance - it can be either deleted from the host gene or converted to an U2-type intron. We took advantage of twenty complete insect genomes to understand dynamics of minor type introns shrinking repertoire.
In order to investigate which of the two pathways is more common, we applied parsimony rule on insect phylogeny for all seventy U12-type introns that were likely present in the ancestral insect genome. Supplementary Material: Figure S2 presents a matrix of the analyzed genes representing status of each intron in each of the twenty insect genomes. Each intron can be represented by one of three states: U12-type intron, U2-type intron, and intron absence. In five cases, lack of the genomic data prevents the determination of the current status of an intron. The evolution of each intron was then inferred on insect phylogeny using Dollo parsimony and number of evolutionary events on the tree, i.e. intron conversion or deletion was calculated (see Fig. 2). Overall, we have observed 112 deletions and 76 intron-type conversions. It agrees with the fact that the cooperative recognition of 5' SS and BPS and its high sensitivity to mutations, U12-type introns are highly susceptible to intron conversion to U2-type and intron loss 40. Assuming that the last common ancestor of all analyzed genomes existed 355 Mya 41 it appears that, on average, there was one intron deletion every 63 MY and one intron-type conversion every 93 MY during insect evolution. Obviously, the deletion rate of U12-type introns is much lower than overall intron loss in Drosophila branch reported by Coulombe-Huntintgton et al. 42 but this is due to much smaller total number of minor-type introns.
Figure 2.
Evolutionary fate of seventy ancestral insect U12-type introns. Red numbers indicate number of deletions in a given lineage, while numbers in blue represent U12 to U2 intron type conversions.
U12-type introns are divided into two subtypes based on the terminal dinucleotides: AT-AC or GT-AG termini. Fourteen out of seventy-one analyzed here introns are of AT-AC subtype. It's been hypothesized that the ancestor minor type introns were of AT-AC subtype and switching to GT-AG subtype might be the initial step into intron type switching from minor to major spliceosome 43. Surprisingly, in our dataset we have perfect separation of the subtype introns, i.e. in the given set of orthologous introns they are always either U2-type or U12-type of the same subtype. In other words, we haven't observed mixing the AT-AC and the GT-AG subtype introns in the same cluster of orthologous introns. This suggests that switching from minor to major intron type might be possible directly from AT-AC subtype, in contrast to previous suggestions. However, this holds only in insect dataset, because our recent analysis of chordates yielded orthologous clusters with both U12-subtypes in a single set [Janice and Makalowski, unpublished observation].
Twintron arrangement
Twintrons are referred to a special arrangement of alternatively spliced introns, in which two introns occupying the same position are processed by two different spliceosomes. In the “classical” example described in the prospero gene U2-type intron is embedded into U12-type intron. As a result of this arrangement in D. melanogaster, alternative splicing of U2-type intron leads to a twenty-nine amino acid larger protein 44-45. The twintron arrangement appeared early in evolution of insects and plays important role in embryonic development. It was shown that the two forms of the prospero transcript are unequally expressed with the “U2 form” dominating in early developmental stages, while after twelfth hour “U12 form” is taking over expression of the gene 44. Interestingly, the U12-type intron of prospero is one of only two minor type introns, which are present in all analyzed insect and vertebrate genomes. However, the twintron arrangement in this position is limited to insects only. Expression of both alternative forms has been confirmed for D. melanogaster but conservation of the splicing signals suggests that the U2-type intron appeared early in the insect evolution.
Another twintron in insect genomes was recently reported by Lin et al. in ZRSR2 gene that codes for zinc finger protein 14. Interestingly, the apparent ancestral intron in the twintron position is of U2-type, which leads to conclusion that U12-type intron appeared de novo in insect evolution. Our extensive analysis of that intron suggests that the twintron arose after Diptera separated from the rest of insects leading to Diptera specific twintron. This is also the only known case of recent U12-type intron addition to a genome. However, it should be pointed that the applied in this study methodology doesn't allow discovery of newly emerged introns in non-seed genomes (see methods section and Supplementary Material: Table S1). Although in neither case, we observed full conversion of one type of an intron to another, the twintron arrangement opens new path of intron conversion, namely instead of gradual degeneration of U12-type splicing signal by point mutation and its transition into U2-type signal, activation of a cryptic splicing site near an original one may create an alternatively spliced transcript, followed by “switching off” the original form. This would result in a new constitutive intron of different type.
Functional analysis of U12-type intron-containing genes
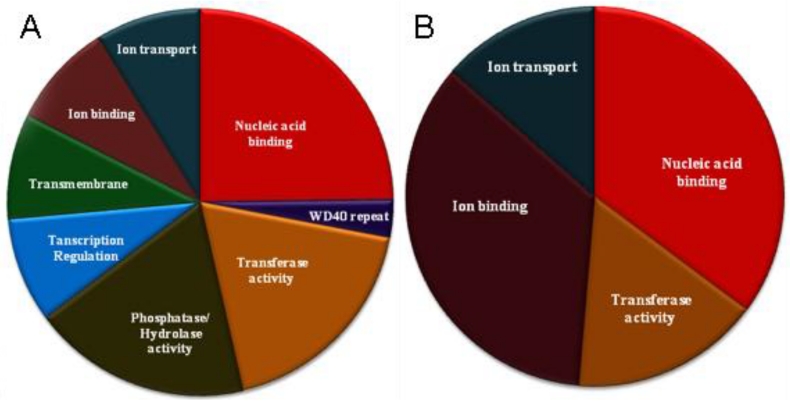
One of the most intriguing questions related to the decaying U12 spliceosomal system is why in some genomes, despite a high energetic cost of the maintaining a separate spliceosome, a minute number of U12-type introns persists. To partially answer this question, we looked if there is a functional relationship between genes harboring U12-type introns. Using DAVID system 28, we were able to assign all the insect U12-intron-containing genes into ten clusters (Fig. 3a and Supplementary Material: Table S2). Interestingly, several of these clusters group proteins of somewhat similar biological activities, e.g. proteins binding other biologically active molecules or proteins involved in signaling pathways. Not surprisingly some of the proteins belong to more than one category, for instance proteins that bind ions often are responsible for ion transport and are located in a membrane, e.g. FBGN0001991 spans these three categories. However, there is no single category that clearly dominates and most populous group, nucleic acid binding proteins, consists of sixteen genes (nineteen percent of all genes).
Figure 3.
Functional categories over-represented in genes containing the U12-type introns. A. Seventy insect genes. B. Twenty-five dipteral genes.
This picture changes a bit, when we limit our data set to genes that contain U12-type introns in Diptera only. There are twenty-five such genes and they cluster just into four groups (see Fig. 3b and Supplementary Material: Table S3). However, nucleic acid binding proteins comprise the largest group (thirteen genes) along with ion binding proteins (thirteen genes). Proteins involved in ion transport and exhibiting transferase activity complement the set. Many of the nucleic acid binding proteins are involved in transcription regulation suggesting significant influence of U12-type intron-containing genes on these processes. Interestingly, both genes that harbor twintrons (prospero and ZRSR2) belong to this category and what is even more intriguing, the latter codes for the protein related to the U2AF splicing factor 46, which functions in the proper recognition of 3' splicing site by a major spliceosome 47. This case exemplifies the delicate interplay between two spliceosomes and may partially explain why total removal of the U12 spliceosomal system from a genome is not a simple process.
Conclusions
U12-type spliceosomal system is a puzzling phenomenon. On the one hand, it has been lost repeatedly in the course of eukaryotic evolution. On the other hand, in some genomes it persists even though it is required to process just a handful of introns. Insects seems to be ideal system to track evolutionary events governing minor type introns because easily manageable number of these introns and many complete genomes with known phylogeny spanning over 350 million years of evolution.
The ancestral insect genome contained at least seventy minor type introns and none of the extant genomes harbors such a high number of these introns suggesting continuous removal of the introns with the extreme case being the genome of C. quinquefasciatus with only fifteen U12-type introns preserved. Only two minor type introns are present in all studied genomes. Our study shows that the intron deletion is more likely that its conversion to U2-type intron. Moreover, our results show that such a conversion is equally possible from both subtypes of the minor introns. Since we didn't observe any subtype switching during insect evolution, this process is not as common as previous analyses suggested.
One of the two completely preserved introns is a twintron residing in the Prospero gene. The conserved nature of the twintron strongly suggests important role of this arrangement in regulation of this gene and consequently pupa development. Interestingly, most of the genes containing minor type introns are involved in regulation of the cellular processes and/or in binding biologically active molecules. It has been suggested that U12-type introns are limiting factors of pre-mRNA processing and it is tempting to speculate that U12-type introns might prevent the over-expression of such genes. Hence, the analysis of the expression of the U12-type intron-containing genes may help in understanding the molecular mechanisms behind the down regulation of these genes and shade more light on the U12-type introns biological significance.
Supplementary materials
Figure S1. Scatter plot of correlation between genome size and number of U12-type introns.Figure S2. Status of seventy-one U12-type introns in the human and twenty insect genomes.Table S1. Insect genes harboring U12-type introns.Table S2. Functional categories over-represented in genes containing the U12-type introns in Insecta. Table S3. Functional categories over-represented in genes containing the U12-type introns in Diptera.Table S4. Total and U12-type intron number in selected genomes.
Acknowledgments
This work has been supported by the Institute of Bioinformatics funds.
References
- 1.Gilbert W. Why genes in pieces? Nature. 1978;271:501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- 2.Crick F. Split genes and RNA splicing. Science. 1979;204:264–71. doi: 10.1126/science.373120. [DOI] [PubMed] [Google Scholar]
- 3.Burge CB, Tuschl T, Sharp P.A. Splicing of precursors to mRNAs by the Spliceosome. In: Gesteland RF, Cech TR, Atkins JF, editors. RNA world. New York, USA: Cold Spring Harbor Laboratory Press; 1999. pp. 525–60. [Google Scholar]
- 4.Lamond AI. The spliceosome. Bioessays. 1993;15:595–603. doi: 10.1002/bies.950150905. doi:10.1002/bies.950150905. [DOI] [PubMed] [Google Scholar]
- 5.Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011. doi:cshperspect.a003707 [pii]10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed]
- 6.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–18. doi: 10.1016/j.cell.2009.02.009. doi:S0092-8674(09)00146-9 [pii]10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Burge CB, Padgett RA, Sharp PA. Evolutionary fates and origins of U12-type introns. Mol Cell. 1998;2:773–85. doi: 10.1016/s1097-2765(00)80292-0. doi:S1097-2765(00)80292-0 [pii] [DOI] [PubMed] [Google Scholar]
- 8.Hall SL, Padgett RA. Conserved sequences in a class of rare eukaryotic nuclear introns with non-consensus splice sites. J Mol Biol. 1994;239:357–65. doi: 10.1006/jmbi.1994.1377. doi:S0022-2836(84)71377-5 [pii]10.1006/jmbi.1994.1377. [DOI] [PubMed] [Google Scholar]
- 9.Tarn WY, Steitz JA. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell. 1996;84:801–11. doi: 10.1016/s0092-8674(00)81057-0. doi:S0092-8674(00)81057-0 [pii] [DOI] [PubMed] [Google Scholar]
- 10.Tarn WY, Steitz JA. Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science. 1996;273:1824–32. doi: 10.1126/science.273.5283.1824. [DOI] [PubMed] [Google Scholar]
- 11.Dietrich RC, Peris MJ, Seyboldt AS, Padgett RA. Role of the 3' splice site in U12-dependent intron splicing. Mol Cell Biol. 2001;21:1942–52. doi: 10.1128/MCB.21.6.1942-1952.2001. doi:10.1128/MCB.21.6.1942-1952.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hastings ML, Resta N, Traum D, Stella A, Guanti G, Krainer AR. An LKB1 AT-AC intron mutation causes Peutz-Jeghers syndrome via splicing at noncanonical cryptic splice sites. Nat Struct Mol Biol. 2005;12:54–9. doi: 10.1038/nsmb873. doi:nsmb873 [pii]10.1038/nsmb873. [DOI] [PubMed] [Google Scholar]
- 13.Levine A, Durbin R. A computational scan for U12-dependent introns in the human genome sequence. Nucleic Acids Res. 2001;29:4006–13. doi: 10.1093/nar/29.19.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin CF, Mount SM, Jarmolowski A, Makalowski W. Evolutionary dynamics of U12-type spliceosomal introns. BMC Evol Biol. 2010;10:47.. doi: 10.1186/1471-2148-10-47. doi:1471-2148-10-47 [pii]10.1186/1471-2148-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarn WY, Steitz JA. Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge. Trends Biochem Sci. 1997;22:132–7. doi: 10.1016/s0968-0004(97)01018-9. doi:S0968000497010189 [pii] [DOI] [PubMed] [Google Scholar]
- 16.Basu MK, Rogozin IB, Koonin EV. Primordial spliceosomal introns were probably U2-type. Trends Genet. 2008;24:525–8. doi: 10.1016/j.tig.2008.09.002. doi:S0168-9525(08)00230-8 [pii]10.1016/j.tig.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharp PA, Burge CB. Classification of introns: U2-type or U12-type. Cell. 1997;91:875–9. doi: 10.1016/s0092-8674(00)80479-1. doi:S0092-8674(00)80479-1 [pii] [DOI] [PubMed] [Google Scholar]
- 18.Davila Lopez M, Rosenblad MA, Samuelsson T. Computational screen for spliceosomal RNA genes aids in defining the phylogenetic distribution of major and minor spliceosomal components. Nucleic Acids Res. 2008;36:3001–10. doi: 10.1093/nar/gkn142. doi:gkn142 [pii]10.1093/nar/gkn142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimaldi D EM. Evolution of insects. Cambridge: Cambridge university press; 2005. [Google Scholar]
- 20.Alioto TS. U12DB: a database of orthologous U12-type spliceosomal introns. Nucleic Acids Res. 2007;35:D110–5. doi: 10.1093/nar/gkl796. doi:gkl796 [pii]10.1093/nar/gkl796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mount SM, Gotea V, Lin CF, Hernandez K, Makalowski W. Spliceosomal small nuclear RNA genes in 11 insect genomes. RNA. 2007;13:5–14. doi: 10.1261/rna.259207. doi:rna.259207 [pii]10.1261/rna.259207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. doi:10.1006/jmbi.1990.9999S0022283680799990 [pii] [DOI] [PubMed] [Google Scholar]
- 23.Rombel IT, Sykes KF, Rayner S, Johnston SA. ORF-FINDER: a vector for high-throughput gene identification. Gene. 2002;282:33–41. doi: 10.1016/s0378-1119(01)00819-8. doi:S0378111901008198 [pii] [DOI] [PubMed] [Google Scholar]
- 24.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–8. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT. et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–4. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–17. doi: 10.1006/jmbi.2000.4042. doi:10.1006/jmbi.2000.4042 S0022-2836(00)94042-7 [pii] [DOI] [PubMed] [Google Scholar]
- 27.Hall TA. BioEdit: a user- friendly biological sequence alignment editor and analysis program for windows 95/98 NT. Nucleic Acids Symposium Series. 1999;41:95–8. [Google Scholar]
- 28.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. doi:nprot.2008.211 [pii] 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 29.Honeybee Genome Sequencing Consortium. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–49. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gall JG, Cohen EH, Polan ML. Reptitive DNA sequences in drosophila. Chromosoma. 1971;33:319–44. doi: 10.1007/BF00284948. [DOI] [PubMed] [Google Scholar]
- 31.Schweber MS. The satellite bands of the DNA of Drosophila virilis. Chromosoma. 1974;44:371–82. doi: 10.1007/BF00284897. [DOI] [PubMed] [Google Scholar]
- 32.Schlotterer C, Harr B. Drosophila virilis has long and highly polymorphic microsatellites. Mol Biol Evol. 2000;17:1641–6. doi: 10.1093/oxfordjournals.molbev.a026263. [DOI] [PubMed] [Google Scholar]
- 33.Gregory TR, Johnston JS. Genome size diversity in the family Drosophilidae. Heredity. 2008;101:228–38. doi: 10.1038/hdy.2008.49. doi:hdy200849 [pii] 10.1038/hdy.2008.49. [DOI] [PubMed] [Google Scholar]
- 34.Moriyama EN, Petrov DA, Hartl DL. Genome size and intron size in Drosophila. Mol Biol Evol. 1998;15:770–3. doi: 10.1093/oxfordjournals.molbev.a025980. [DOI] [PubMed] [Google Scholar]
- 35.Gregory TR, Nicol JA, Tamm H, Kullman B, Kullman K, Leitch IJ. et al. Eukaryotic genome size databases. Nucleic Acids Res. 2007;35:D332–8. doi: 10.1093/nar/gkl828. doi:gkl828 [pii] 10.1093/nar/gkl828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ. et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–23. doi: 10.1126/science.1138878. doi:1138878 [pii] 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu Q, Krainer AR. AT-AC pre-mRNA splicing mechanisms and conservation of minor introns in voltage-gated ion channel genes. Mol Cell Biol. 1999;19:3225–36. doi: 10.1128/mcb.19.5.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat Rev Mol Cell Biol. 2003;4:960–70. doi: 10.1038/nrm1259. doi:10.1038/nrm1259 nrm1259 [pii] [DOI] [PubMed] [Google Scholar]
- 39.Szczesniak MW, Ciomborowska J, Nowak W, Rogozin IB, Makalowska I. Primate and rodent specific intron gains and the origin of retrogenes with splice variants. Mol Biol Evol. 2011;28:33–7. doi: 10.1093/molbev/msq260. doi:msq260 [pii] 10.1093/molbev/msq260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frilander MJ, Steitz JA. Initial recognition of U12-dependent introns requires both U11/5' splice-site and U12/branchpoint interactions. Genes Dev. 1999;13:851–63. doi: 10.1101/gad.13.7.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2. doi: 10.1093/bioinformatics/btl505. doi:btl505 [pii] 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- 42.Coulombe-Huntington J, Majewski J. Intron loss and gain in Drosophila. Mol Biol Evol. 2007;24:2842–50. doi: 10.1093/molbev/msm235. doi:msm235 [pii] 10.1093/molbev/msm235. [DOI] [PubMed] [Google Scholar]
- 43.Basu MK, Makalowski W, Rogozin IB, Koonin EV. U12 intron positions are more strongly conserved between animals and plants than U2 intron positions. Biol Direct. 2008;3:19.. doi: 10.1186/1745-6150-3-19. doi:1745-6150-3-19 [pii] 10.1186/1745-6150-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scamborova P, Wong A, Steitz JA. An intronic enhancer regulates splicing of the twintron of Drosophila melanogaster prospero pre-mRNA by two different spliceosomes. Mol Cell Biol. 2004;24:1855–69. doi: 10.1128/MCB.24.5.1855-1869.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borah S, Wong AC, Steitz JA. Drosophila hnRNP A1 homologs Hrp36/Hrp38 enhance U2-type versus U12-type splicing to regulate alternative splicing of the prospero twintron. Proc Natl Acad Sci U S A. 2009;106:2577–82. doi: 10.1073/pnas.0812826106. doi:0812826106 [pii] 10.1073/pnas.0812826106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mount SM, Salz HK. Pre-messenger RNA processing factors in the Drosophila genome. J Cell Biol. 2000;150:F37–44. doi: 10.1083/jcb.150.2.f37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3' splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–5. doi: 10.1038/45590. doi:10.1038/45590. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Scatter plot of correlation between genome size and number of U12-type introns.Figure S2. Status of seventy-one U12-type introns in the human and twenty insect genomes.Table S1. Insect genes harboring U12-type introns.Table S2. Functional categories over-represented in genes containing the U12-type introns in Insecta. Table S3. Functional categories over-represented in genes containing the U12-type introns in Diptera.Table S4. Total and U12-type intron number in selected genomes.