Abstract
Chronic inflammation and selenium deficiency are considered as risk factors for colon cancer. The protective effect of selenium might be mediated by specific selenoproteins, such as glutathione peroxidases (GPx). GPx-1 and -2 double knockout, but not single knockout mice, spontaneously develop ileocolitis and intestinal cancer. Since GPx2 is induced by the chemopreventive sulforaphane (SFN) via the nuclear factor E2-related factor 2 (Nrf2)/Keap1 system, the susceptibility of GPx2-KO and wild-type (WT) mice to azoxymethane and dextran sulfate sodium (AOM/DSS)-induced colon carcinogenesis was tested under different selenium states and SFN applications. WT and GPx2-KO mice were grown on a selenium-poor, -adequate or -supranutritional diet. SFN application started either 1 week before (SFN4) or along with (SFN3) a single AOM application followed by DSS treatment for 1 week. Mice were assessed 3 weeks after AOM for colitis and Nrf2 target gene expression and after 12 weeks for tumorigenesis. NAD(P)H:quinone oxidoreductases, thioredoxin reductases and glutathione-S-transferases were upregulated in the ileum and/or colon by SFN, as was GPx2 in WT mice. Inflammation scores were more severe in GPx2-KO mice and highest in selenium-poor groups. Inflammation was enhanced by SFN4 in both genotypes under selenium restriction but decreased in selenium adequacy. Total tumor numbers were higher in GPx2-KO mice but diminished by increasing selenium in both genotypes. SFN3 reduced inflammation and tumor multiplicity in both Se-adequate genotypes. Tumor size was smaller in Se-poor GPx2-KO mice. It is concluded that GPx2, although supporting tumor growth, inhibits inflammation-mediated tumorigenesis, but the protective effect of selenium does not strictly depend on GPx2 expression. Similarly, SFN requires selenium but not GPx2 for being protective.
Introduction
Selenium deficiency has for long been suggested to facilitate cancer development (1). Dietary Se supplementation decreased prostate, lung and colon cancer in a large clinical trial (2) with a significant effect in individuals entering the study with a low selenium status (3). However, selenium did not decrease cancer incidence when comparing a healthy cohort supplemented with selenomethionine (200 μg/day) with matched selenium placebo in the larger Selenium and Vitamin E Cancer Prevention Trial (4). It, thus, still appears conceivable that selenium deficiency, which is inevitably associated with an impaired oxidative stress response, represents a significant cancer risk. Accordingly, hydroperoxide metabolizing pathways involving selenoproteins, such as glutathione peroxidases (GPx) or thioredoxin reductases (TrxR) are most commonly discussed to mediate the protective effect of selenium. However, the relative contributions of the individual selenoproteins to cancer prevention remain unclear.
Mice deficient in both glutathione peroxidases, GPx1 and GPx2, spontaneously developed early-onset ileocolitis and intestinal cancer after 6–9 months (5,6). The cancer incidence was correlated with inflammation severity. GPx2 is predominantly expressed in proliferative zones of the intestinal mucosa (7), and its absence increases apoptosis of crypt epithelial cells (8,9). Therefore, maintenance of mucosal homeostasis has been suggested to be a physiological role of GPx2. In addition, GPx2 exerts anti-inflammatory activity and inhibits migration and invasion of tumor cells, both partially mediated by the suppression of COX-2 expression (10,11). However, GPx2 is induced by β-catenin/T-cell factor in the Wnt pathway (12) and promotes cancer cell growth (10). Therefore, the role of GPx2 in tumorigenesis appears controversial.
Gpx2 gene expression is regulated by the nuclear factor E2-related factor 2 (Nrf2) (13), which is activated by oxidizing or electrophilic agents and also by chemopreventive isothiocyanates, including the glucoraphanin-derived isothiocyanate sulforaphane (SFN) (14,15). Glucoraphanin is found in vegetables of the brassicaceae family. Epidemiological studies suggest that the intake of such vegetables may lower cancer risk, particularly at sites of the respiratory and gastrointestinal tract (16,17). SFN can inhibit phase I enzymes which activate carcinogens and activate phase II enzymes which reduce oxidative stress and detoxify carcinogens (18). Phase II enzymes include glutathione-S-transferases (GST), NAD(P)H:quinone oxidoreductase (NQO1) and, apart from GPx2, also the selenoprotein TrxR1 (19). SFN decreased azoxymethane (AOM)-induced aberrant crypt foci (ACF) formation in rats (20) and retarded tumor development in adenomatous polyposis coli mice (21). We, therefore, tested whether (i) GPx2 by itself can prevent inflammation and, thus, carcinogenesis to confirm in vitro findings in vivo, (ii) selenium exerts its anticarcinogenic effects by enhancing GPx2 synthesis, and (iii) SFN is anticarcinogenic by induction of GPx2 as suggested previously (22) in the model of inflammation-associated carcinogenesis [AOM/dextran sulfate (DSS)] (23).
Material and methods
Animals and diets
C57BL/6J wild-type (WT) and GPx2-KO mice, generated as C57BL/6J × 129SV/J hybrid, three times backcrossed to a C57BL/6J background (24) and a fourth time before the experiment started, were housed under specific pathogen free conditions. WT and GPx2-KO mice were reproduced by in-house breeding. The study was approved by local authorities (MLUV 32-2347/4+68). Different diets containing 0.086 mg Se/kg in Se-poor (−Se), 0.15 mg/kg in Se-adequate (+Se) and 0.64 mg/kg in Se-supranutritional (++Se) diet were obtained by mixing the respective amounts of selenomethionine into torula-based Se-poor diet as described (9). Pelleted diets were fed directly after weaning over the entire experiment. SFN [1-isothiocyanato-(4R,S)-methylsulfinyl)-butane] was synthesized as described (25).
Experimental design
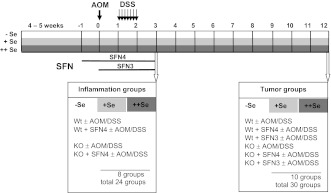
Feeding with different selenium diets started after weaning and was maintained during the entire experiment. After 4–5 weeks when the selenium state had been established, animals (weighing 24–27 g) were randomized for AOM/DSS treatment and SFN application as shown in Figure 1. Ten milligram per kilogram body wt AOM (Sigma, Steinheim, Germany) or saline (Sigma) was injected intraperitoneally. One week later, mice were given drinking water containing 1% DSS (MP Biomedicals, Illkirch, France) for 7 days. SFN (10 μmol every other day) or solvent (100 μl of 0.1 M KPi buffer) was applied by gavage either 1 week before or concomitantly with AOM. Mice were killed under isoflurane anesthesia by cervical dislocation 3 weeks (inflammation groups) or 12 weeks (tumor groups) after AOM application.
Fig. 1.
Mouse model of inflammation-triggered colon carcinogenesis. In the model, the carcinogenic DNA alkylant AOM is combined with the inflammatory DSS (23). To study the contribution of GPx2 in inflammation and/or carcinogenesis, WT and GPx2-KO mice were used. These mice were fed a selenium-poor (−Se), selenium-adequate (+Se) or selenium-supranutritional (++Se) diet to find out whether GPx2 synthesis is essential for the anticancer function of selenium. Application of SFN was included to test whether it acts via upregulation of GPx2 RNA and how much selenium is required to obtain optimal effects. Feeding of different diets started from weaning and lasting during the entire experiment. AOM (10 mg/kg body wt) was applied after 4–5 weeks when selenium status was adjusted, indicated by week 0. One week later, 1% DSS was added to the drinking water for 7 days. Mice were euthanized either 3 weeks after AOM (inflammation groups) or 12 weeks after AOM (tumor groups). Since SFN affects multiple stages of tumorigenesis, including initiation and promotion phases (18), SFN was applied either 1 week prior to AOM application for 4 weeks (SFN4) or concomitantly with AOM for 3 weeks (SFN3). Inflammation groups were treated with SFN4 and tumor groups with SFN3 or SFN4, respectively. SFN solvent groups were taken as untreated controls. Inflammation groups consisted of four animals, each representing genotype (WT and GPx2-KO), SFN4 and AOM/DSS treatment at different selenium status and respective controls, resulting in a total of 24 groups. Tumor groups consisted of 10 animals, representing the same treatments as inflammation groups plus the SFN3 groups. SFN solvent was applied for 4 weeks only, resulting in 30 tumor groups. Inflammation groups were analyzed for inflammation scores, including histological analyses, targets of Nrf2 and apoptosis. In tumor groups, numbers of ACF and tumors were counted and clinical inflammation parameters monitored during acute colitis and at the time of killing. Further details see ‘Materials and methods’, section ‘Inflammation score’.
Tissue sampling for enzyme analysis, histopathology and IHC
The proximal 2 cm of the colon and the proximal 4 cm of the ileum were freeze-clamped in liquid nitrogen and stored at −80°C until enzyme analysis. The remaining transversal and distal colon was fixed as a ‘Swiss’ roll for 24 h in 4% neutral-buffered formalin, dehydrated and embedded in paraffin. Serial sections (2 μm) of the Swiss roll were processed for inflammation scoring (hematoxylin and eosin staining), counting apoptotic cells (hematoxylin-stained) and for immunohistochemistry (IHC) as described (9). Anti-NQO1 antibody (ab34173; Abcam, Cambridge, UK), anti-human GPx2 antiserum (26) and anti-CD3 (A0452; Dako, Hamburg, Germany) were used with horseradish peroxidase-conjugated anti-rabbit antibody (AP307P; Chemicon, Hofheim, Germany) as secondary antibody. The IHC staining intensity was scored on a scale of 0–3 in a blinded fashion.
For tumor analysis, the colon was opened longitudinally, stretched on filter paper and fixed in 4% neutral-buffered formalin. Samples were blinded, stained with 0.1% methylene blue and ACF and tumor numbers counted in a stereo microscope (SZ-STU; Olympus, Japan). Tumor size was measured along the longest dimension. Since the tumors were mostly of round shape, this size was equivalent to the diameter.
Inflammation score
Scoring criteria were used as reported (27,28). Briefly, colonic inflammation was evaluated by colitis disease index (including weight loss, occult and visible blood, diarrhea) during and until 1 week after DSS application. Macroscopic abnormalities (colonic swelling and shortening) and microscopic indications (crypt and mucosa architecture, edema and infiltration) by histological investigation were monitored at the time point of tissue sampling. Histological data could only be obtained in inflammation groups, since in tumor groups entire tissues were needed for tumor counting. The maximum score was 16 when histological data were included and eight when only colitis disease index and macroscopic abnormalities were monitored.
Enzyme activities
Samples were prepared as described previously (9). NQO1 activity was measured as described (29). TrxR activity was estimated by the reduction of 5,5’-dithiobis(2-nitrobenzoic acid) to 5’ thionitrobenzoate anion (TNB) by reduced nicotinamide adenine dinucleotide phosphate (NADPH) (30). Tissue lysate, 25 μl, was mixed with 185 μl reaction mixture containing 100 mM KPi, 2 mM ethylenediaminetetraacetic acid, pH 7.4 and 15 μl 5,5’-dithiobis(2-nitrobenzoic acid) (50 mM in dimethyl sulfoxide) in a 96-well plate. The reaction was started by adding 25 μl NADPH (final concentration 2 mM). TNB production was monitored in a plate reader (Synergy 2; Biotek Instruments GmbH, Bad Friedrichshall, Germany) at 412 nm at 25°C. TrxR-independent TNB formation, determined in the absence of NADPH, was subtracted and data expressed as mU/mg protein. One unit is defined as the consumption of 1 μmol NADPH, i.e. production of 2 μmol of TNB (ϵ412 nm = 13.6 mmol/l cm) per minute. Total GST activity was determined via the conjugation of 1-chloro-2,4-dinitrobenzene and glutathione (GSH) (31).
Quantification of apoptosis
Apoptotic cells, characterized as before (9), were counted in 300 crypts/mouse of the distal colon starting 1 cm proximal to the anus in blinded Swiss rolls of untreated animals. Thus, apoptosis was quantified in the same area as described previously in 20-week-old mice and, thus, provided comparable data (9). Crypts were divided into four quarters with the fourth quarter at the crypt base.
Statistics
Significance was tested by two-way analysis of variance, Mann–Whitney U-test, Student’s t-test (GraphPad Prism® version 5.0, San Diego, CA) or Fisher’s exact test (SPSS, Version 12.0, Chicago, IL) as appropriate. A P-value < 0.05 was considered significant.
Results
Adjusting the selenium status
To verify the success of selenium feeding, plasma selenium concentration and liver GPx activity were determined (Supplementary Figure S1, available at Carcinogenesis Online). Plasma selenium increased in +Se groups 4–4.5 times compared with the −Se groups, whereas GPx activity increased 16–18 times. Under the ++Se diet, selenium levels as well as GPx activity further increased by a factor of 1.2–1.3. Although the latter increase is low, it was significant in most cases. There was no significant difference in plasma selenium content or liver GPx activity between control and AOM/DSS-treated groups at identical selenium diets (Supplementary Figure S1A and B is available at Carcinogenesis Online), verifying that all treatment groups under the same selenium diet were equally equipped with selenium.
SFN-mediated induction of Nrf2 target genes
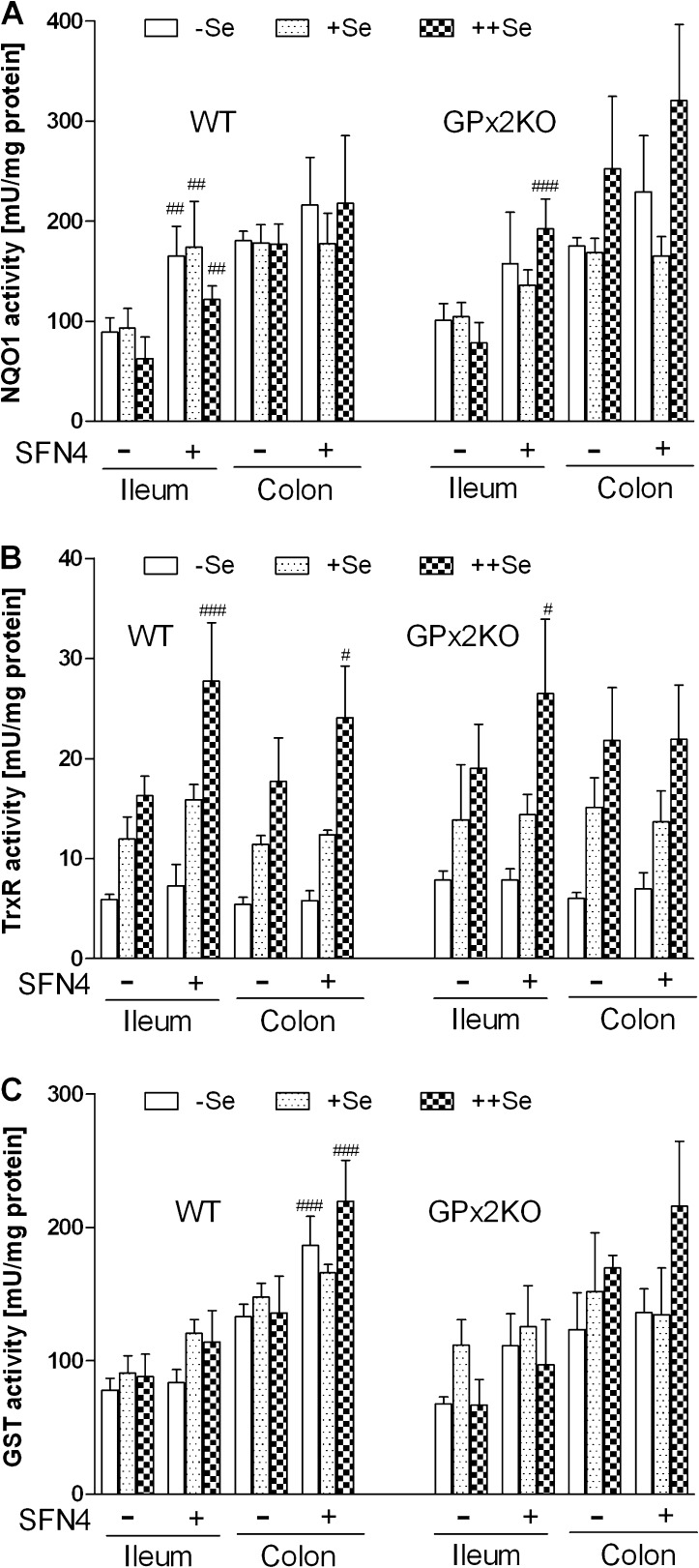
After 4 weeks of SFN application, mice without AOM/DSS treatment (Inflammation groups in Figure 1: SFN4 and respective controls) were analyzed for phase II enzyme expression to verify SFN effects. NQO1 activity was increased in ileum lysates of WT mice under all three selenium diets (Figure 2A). Basal activity was higher in the colon irrespective of the selenium state (P < 0.01 in all three states) and not enhanced by SFN. TrxR activity was increased by SFN in the ileum and colon in mice on ++Se diet only (Figure 2B). Total GST activity was enhanced by SFN in the colon of mice on −Se and ++Se diets (Figure 2C). Basal activities of all enzymes estimated were comparable in GPx2-KO mice (Figure 2A–C, right panels) to WTs, but SFN-mediated induction of NQO1 and TrxR in the ileum was only observed in the ++Se status of GPx2-KO mice, whereas induction of GST in the colon did no longer reach significance. It, thus, appears that enzyme induction by SFN in GPx2-KO mice, if any, preferentially occurs in the ++Se status.
Fig. 2.
Effects of SFN on the activity of Nrf2 target proteins. (A–C) Enzyme activities were analyzed in lysates of the ileum and colon of WT (left parts) and GPx2-KO (right parts) mice adjusted to the indicated selenium status and treated with and without SFN for 4 weeks. (A) NQO1, (B) TrxR and (C) GST. All data represent mean ± SD of four animals measured in triplicate. Not to overload the figure, significance is only shown for the SFN effect. #P < 0.05, ##P < 0.01, ###P < 0.001 versus respective controls without SFN, analyzed with two-way analysis of variance with Bonferroni’s post tests. There were no significant differences between the genotypes. The higher basal activity of NQO1 in the SFN-untreated colon compared with ileum is significant in all selenium states (P < 0.01). The enhanced activity of TrxR with increasing selenium is significant by at least P < 0.05.
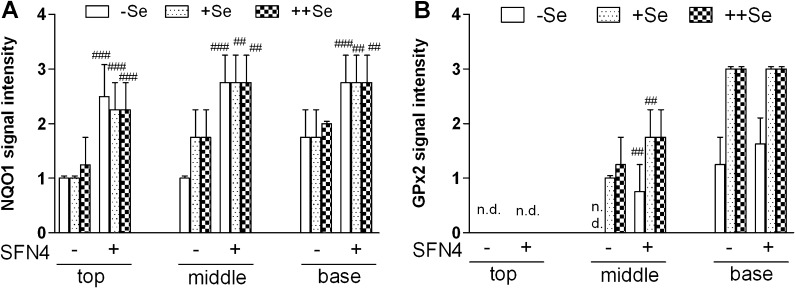
Since NQO1 induction was not detectable when measured as activity in lysates of the entire colon, NQO1 protein expression was investigated by IHC not to overlook an only localized expression. SFN4 indeed induced NQO1 expression along the villus–crypt axis in epithelial cells of the distal colon in mice on all diets (Figure 3A). Finally, GPx2 protein levels were analyzed by IHC, since lack of a specific substrate prevents discrimination of GPx1, GPx2 and GPx3 by activity measurement [Figure 3B; 32]. GPx2 protein was not detected in crypt tops and middle crypt parts of selenium-poor colon. It was increased by selenium in the middle part and bases of crypts. SFN4 significantly enhanced GPx2 in the middle of crypts in mice on −Se and +Se diet. On ++Se diet, GPx2 expression had presumably reached maximal levels and could no longer be enhanced by SFN4.
Fig. 3.
Localized upregulation of NQO1 and GPx2 by SFN. Immunoreactivity in crypts of the distal colon of WT mice was analyzed for (A) NQO1 and (B) GPx2 in Swiss rolls. Crypts were divided in top, middle and base areas. Intensity of staining was scored from 0 to 3. Data represent means ± SD of four animals measured in triplicate. Not to overload the figure, significance is only shown for the SFN effect. ##P < 0.01, ###P < 0.001 versus respective controls without SFN. The increase of GPx2 (B) in crypt middles and bases with +Se and ++Se versus −Se is significant by at least P < 0.05. n.d, not detectable.
The antiapoptotic effect of SFN in colonic crypts depends on selenium
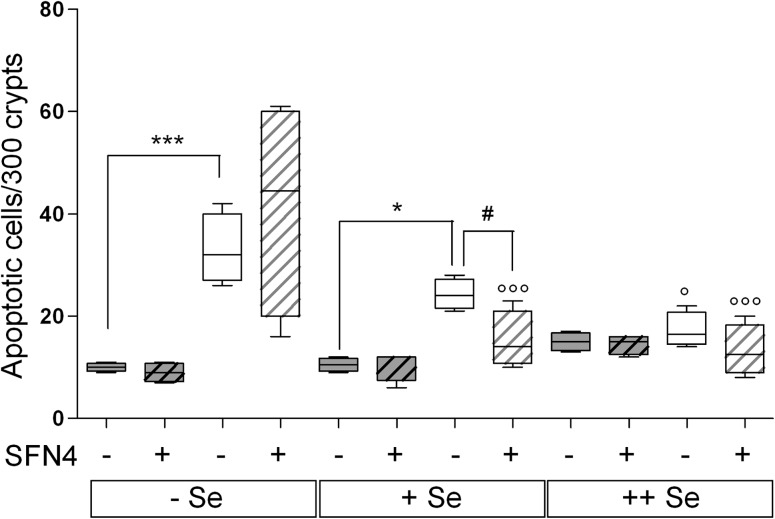
Increased apoptosis has recently been found in crypt bases in 20-week-old GPx2-KO mice under all selenium diets but most strikingly on −Se diet (9). Here, 12-week-old GPx2-KO mice (AOM/DSS-untreated controls of inflammation groups) also had drastically increased apoptosis in crypt bases on the −Se and +Se diet but not yet on the ++Se diet (Figure 3). SFN suppressed apoptosis in +Se GPx2-KO to almost the level of WT but not in −Se GPx2-KO mice.
AOM/DSS-induced colitis
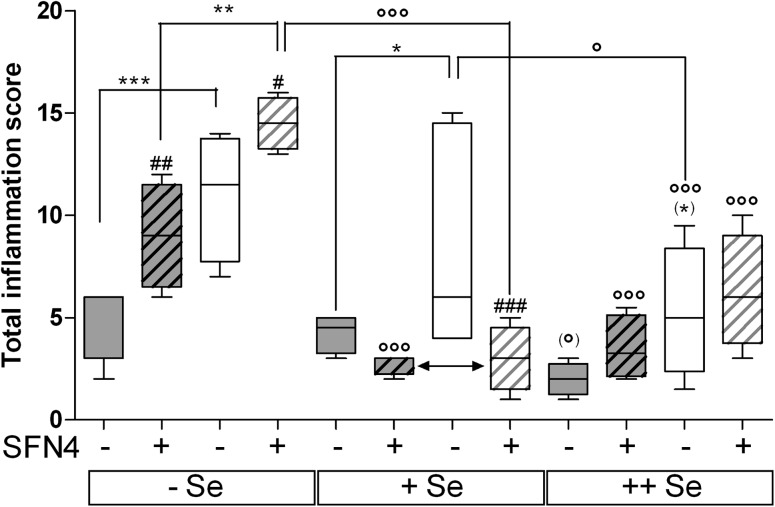
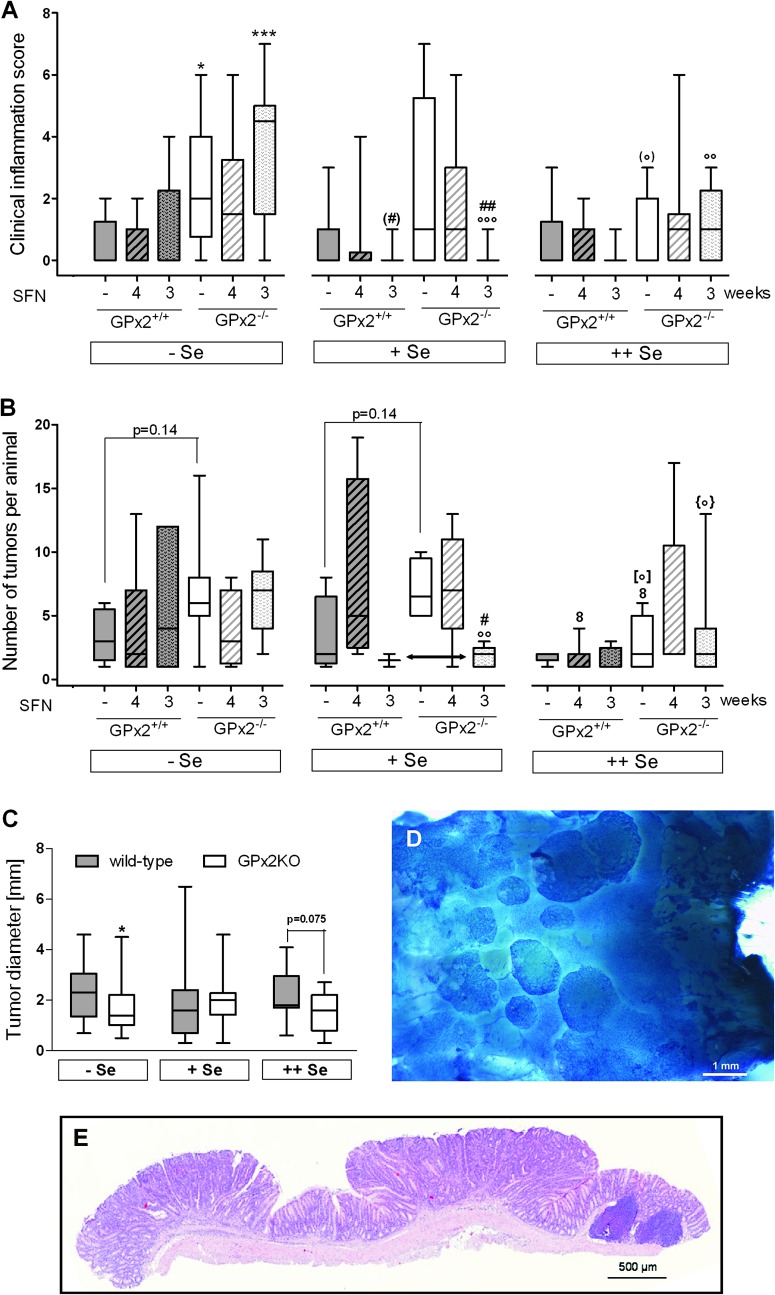
Clinical inflammation scores were determined during the acute phase after DSS application and microscopic as well as macroscopic scores 1 week after terminating DSS treatment (Figure 1), resulting in a total inflammation score. AOM/DSS generally induced inflammation (Figure 5), whereas without AOM/DSS, only one mouse in the Se-poor/SFN4 group developed a score of 0.5 (data not shown in Figure 5). GPx2-KO mice had significantly higher scores than WT mice on −Se and +Se diets, whereas only a trend (P = 0.102, indicated as *) was observed on ++Se diet. The highest inflammation scores were observed in mice on −Se diet. SFN significantly exacerbated inflammation in both GPx2-KO and WT mice at the Se-poor state but similarly alleviated inflammation in mice on +Se diet in both genotypes (Figure 5, double-headed arrow). The apparent increase of inflammation by SFN in ++Se WT mice was not significant. Representative photomicrographs of the colon as examples of different grades of inflammation are shown in the Supplementary Figure S2, available at Carcinogenesis Online.
Fig. 5.
Total inflammation scores 3 weeks after AOM application. WT and GPx2-KO mice on different selenium diet were exposed or not to AOM/DSS with and without SFN as indicated and inflammation scored 1 week after DSS treatment (see Figure 1) by clinical, macroscopical and histological signs (see Materials and methods). Only AOM/DSS-treated groups are shown. Gray background, WT; white background, GPx2-KO mice. Striped boxes indicate SFN4. Data are box plots with median and whiskers from minimum to maximum (n = 4). Significance of different treatments is marked by the same symbols used in Figure 3. * Difference between genotypes in the same selenium group. # SFN effect versus the respective untreated group and ° selenium effect as indicated above horizontal lines or versus the respective Se-poor group. One symbol, P < 0.05; two symbols, P < 0.01; three symbols, P < 0.001 estimated by two-way analysis of variance and Bonferroni’s post tests. WT +Se versus WT ++Se is significant (P < 0.05) when analyzed with Mann–Whitney U test indicated by (°). (*) indicates the trend with P = 0.102 versus WT in the same Se group, estimated with Student’s t-test.
Inflammation 12 weeks after AOM treatment
Clinical inflammation scores in the tumor groups (Figure 6A) did not exactly mirror the total scores determined in the inflammation groups (Figure 5). Whereas GPx2 deletion significantly increased the severity of inflammation in both, inflammation (Figure 5) and tumor (Figure 6A) groups on −Se diet, the difference between genotypes of tumor groups was less pronounced in mice on the +Se and ++Se diets (Figure 6A). An SFN-mediated increase in inflammation in mice on −Se diet was no longer observed. However, the SFN3 regimen clearly decreased inflammation in Se-adequate GPx2-KO mice to the level of WT mice.
Fig. 6.
Clinical inflammation scores and tumors 12 weeks after AOM application. WT and GPx2-KO mice treated as described in legend to Figure 5 were euthanized at 12th week. Not to overload the figure, only the 18 AOM/DSS-treated groups (see Figure 1) are shown in A and B. Neither inflammation nor tumors were observed in the 12 groups not treated with AOM/DSS. (A) Clinical inflammation score. Inflammation was scored by clinical signs during the acute phase and by morphological colonic changes at killing (see Materials and methods). (B) Tumor multiplicity in tumor-bearing animals. Duration of SFN is indicated by –, no application; 4, SFN4 (striped pattern); 3, SFN3 (spotted pattern). Details see Figure 1. Symbols for significance are the same as used in the other figures. * Indicates the difference between genotypes in the same selenium group; # the SFN effect versus the respective untreated group and ° the selenium effect versus the respective Se-poor and 8 versus the Se-adequate group. [°]: P = 0.051; {°}: P = 0.069 versus the respective −Se group. One symbol, P < 0.05; two symbols, P < 0.01; three symbols, P < 0.001 estimated by two-way analysis of variance and Bonferroni’s post tests in (A) and Mann–Whitney U-test in (B). Symbol in parentheses in A show significance estimated by Mann–Whitney U-test. (C) Tumor size in AOM/DSS-treated WT and GPx2-KO mice. *P < 0.05 (Mann–Whitney U-test) versus WT at the same selenium status. Gray background, WT; white background, GPx2-KO animals. All data are presented as box plots with median and whiskers (from minimum to maximum). If the median is not visible, it is zero. (A) n = 10; (B) n = number of animals with tumors in the respective groups (Table I); (C) n = number of tumors in respective animals as shown in Table I. (D) Distal colon tissue stained with methylene blue, showing various tumors accumulating close to the anus. (E) Hematoxylin & eosin stain of two resected tumors representing adenomas and a lymphoid follicle (right).
GPx2 and selenium decrease tumor numbers but SFN inhibits tumorigenesis only in mice on +Se diet and given concomitantly with AOM
The AOM/DSS dosing regime here applied is an established mouse model for inflammation-associated colon carcinogenesis (23) that is considered to closely mimic the human disease (33). By combining an established colon mutagen (AOM) with an inflammatory stimulus, adenomas develop from ACF and are ultimately transformed into adenocarcinomas after 18 weeks with strain differences in the susceptibility to AOM/DSS (34). With the medium-sensitive mice used in our study (background C57BL/6J) overwhelmingly, adenomas were still obtained 12 weeks after AOM application accumulating in the distal colon close to the anus (Figure 6D and E). A systematic differentiation into adenomas and adenocarcinomas was therefore not performed. Instead, we relied on tumor incidence and multiplicity to evaluate the anticarcinogenic influence of the GPx2 genotype and possibly interfering compounds.
Neither the incidence of tumors (Table I) nor that of ACF (Supplementary Table S1 and Figure S3 are available at Carcinogenesis Online) was significantly affected by any treatment. However, in an overall analysis, total numbers of tumors irrespective of treatment were significantly less in WT (145) compared with GPx2-KO mice (285, P < 0.05) and decreased with increasing selenium intake (P < 0.05, Table I, Fisher’s exact test). Overall effects of SFN on total tumor numbers were not significant. However, the decrease in total tumor numbers in +Se groups with SFN given together with the carcinogenic challenge (SFN3 group) is significant in both genotypes when compared pair-wise, as is the increase in −Se groups (Table I).
Table I.
Effects of GPx2, selenium and SFN on tumorigenesis in AOM/DSS-treated mice
| Diet | SFN (weeks) | GPx2+/+ |
GPx2−/− |
||
| Tumor incidence | Total tumor number* | Tumor incidence* | Total tumor number* | ||
| −Seo | 0 | 5/10 | 17 | 7/10 | 48 |
| 4 | 6/10 | 24 | 8/10 | 32 | |
| 3 | 5/10 | 30 | 9/10 | 58 | |
| +Seo | 0 | 4/10 | 13 | 4/10 | 28 |
| 4 | 4/10 | 31 | 5/10 | 37 | |
| 3 | 2/10 | 3a | 5/10 | 9a | |
| ++Seo | 0 | 5/10 | 9 | 7/10 | 19 |
| 4 | 7/10 | 12 | 5/10 | 27 | |
| 3 | 4/10 | 6 | 7/10 | 27a,b | |
Overall decrease of tumor numbers with increasing selenium supply and the presence of GPx2 was significant, o and *P < 0.05 (Fisher’s exact test) indicated at the left and lower edge. The effect of SFN in dependence of selenium, aP < 0.01 versus the respective selenium-poor group and bP < 0.01 versus the respective selenium-adequate group (Fisher’s exact test, pair-wise).
Tumor multiplicity (number of tumors per animal) only tended to be higher in GPx2-KO mice on −Se and +Se diet than in respective WT mice (Figure 6B, P = 0.14 in both dietary groups) and was significantly reduced by the ++Se diet in GPx2-KO mice compared with those fed the −Se and +Se diet (symbols ° and 8, respectively). In WTs, supranutritional selenium diminished tumors in the SFN4 group in which tumors were still high under selenium adequacy. Thus, tumor numbers were minimized in all WT groups. This correlates with the decrease in total numbers with increasing selenium intake (Table I). The slight increase in total tumor numbers in Se-poor GPx2-KO mice by the SFN3 regimen was not observed at the level of tumor multiplicity, whereas the reduction of multiplicity to the level of WT mice in Se adequacy was significant (Figure 6B, double-headed arrow). This correlates with the decrease in the clinical inflammation score by SFN3 in GPx2-KO mice to the level of WT mice (Figure 6A).
GPx2 reportedly supported tumor cell growth in a xenograft model (10). Here, tumor size was chosen as indicator for tumor growth and found significantly lowered in GPx2-KO mice on −Se diet and nearly significantly lowered in mice on the ++Se diet (Figure 6C). Accordingly, a tumor growth-promoting effect of GPx2 in vivo cannot be excluded.
Discussion
The goal of the present investigation was to elucidate the interdependence of the selenium status of the selenoprotein GPx2 and of SFN in experimental colon cancer. Tumorigenesis triggered by AOM/DSS in mouse was chosen as a revealing model, since it is considered to reflect inflammation-promoted carcinogenesis. To this end, GPx2 expression was manipulated by reverse genetics, selenium supply and SFN which, as Nrf2 activator, inter alia induces GPx2 transcription. A cooperation of Se and SFN could therefore be expected (22). The selenium states here chosen differ from those in most of the related studies. A moderate instead of a severe Se deficiency and a just supranutritional supply instead of a drastic oversupplemention were applied to reflect situations that can be expected from common selenium variations in the human diet. SFN application started either 1 week before AOM (indicated as SFN4) to optimize the status of endogenous defense systems before the carcinogenic challenge or simultaneously with AOM (indicated as SFN3) to get information about the influence of SFN on the action of the carcinogen (Figure 1). The model worked satisfactorily in yielding a substantial number of tumors 12 weeks after AOM but no tumor whatsoever in the untreated controls. The findings further disclose a clear impact of each of the variables on carcinogenesis. Their interplay and individual contribution, however, proved to be more complex than anticipated and is commented below.
The impact of the GPx2 genotype
The difference in the inflammatory response between GPx2-WT and GPx2-KO mice was most obvious in animals killed in the acute phase at all selenium states (Figure 5) and most pronounced in the −Se status. In tumor groups, only clinical signs were available for scoring during the acute phase and macroscopic colonic modifications at dissection 12 weeks after AOM. Due to the lower number of parameters and the longer recovery period after the inflammatory challenge, differences between genotypes remained significant only in the Se-poor groups (Figure 6A). Nevertheless, the overall outcome corroborates the antiinflammatory potential of GPx2, which had already been inferred from increased COX-2 expression and prostaglandin E2 production in GPx2-KO cells (11), reversal of bacteria-induced ileocolitis by one allele GPx2 in GPx1/GPx2-double KO mice (35) and inducibility of GPx2 by Nrf2-activation (13). Inhibition of intestinal inflammation might also be related to inhibition of apoptosis by GPx2, as shown in MCF7 cells (36), in the colon and ileum of mice aged 20 weeks (9) and here in the colon of 12-week-old mice of the inflammation group controls (Figure 4). Exaggerated apoptosis due to GPx2 deficiency will favor epithelial destruction, impair the barrier function and thereby predispose to inflammation-mediated neoplasia (37).
Fig. 4.
Reversal of enhanced apoptosis in colonic crypt bases of GPx2-KO mice by SFN depends on the selenium status. WT and GPx2-KO mice at different selenium states and treated with and without SFN for 4 weeks were analyzed. Apoptotic cells were counted in 300 crypt bases per animal in the transversal and distal colon in Swiss rolls. Data represent apoptosis in crypt bases and are shown as box plots with median and whiskers from minimum to maximum (n = 4). Gray background: WT, white background: GPx2-KO animals. Striped boxes indicate SFN-treated groups. Significance of the different treatments is marked by different symbols: * Represents the GPx2 effect as indicated above horizontal lines and ° the selenium effect versus the Se-poor group analyzed by two way analysis of variance with Bonferroni’s post tests. # Represents the SFN effect versus the respective untreated group analyzed by Student’s t-test. One symbol, P < 0.05; three symbols, P < 0.001.
However, how the enzymatic activity of GPx2 interferes with inflammatory processes is not entirely clear. Certainly, GPx2 contributes to H2O2 reduction; in cells where GPx2 was knocked down by small interfering RNA, total GPx activity measured with H2O2 was reduced by 35% (11). Colonic crypt cells isolated from −Se GPx2-KO mice are more susceptible to hydroperoxide stress than those from WT cells (ongoing studies). However, reduction of H2O2 is not likely the only function of GPx2 in the intestine, since this role can be taken over by GPx1, which is highly upregulated in GPx2-KO mice exactly in the areas where otherwise GPx2 is expressed (9). Recently, 12/15 lipoxygenase products have been identified as specific substrates for GPx4, which was able to inhibit apoptosis by removal of these particular substrates (38). Similar studies might help to identify a preferred or unique GPx2 substrate that is pivotal to the inflammatory process.
A difference in tumorigenesis between GPx2-WT and GPx2-KO mice could not always be verified by direct comparison of equally treated groups due to, in part scattered data and low number of animals (Figure 6B). Nevertheless, GPx2 slightly diminished tumor multiplicity in both −Se and +Se status (Figure 6B, P = 0.14) indicating that GPx2 may inhibit tumorigenesis even in the selenium-poor status. This correlates with the inhibition of inflammation (see above) and fits with its high ranking in the hierarchy (39), allowing reasonable synthesis in marginal Se deficiency (9). The small effect on tumor multiplicity, however, is supported by the significant reduction in tumor numbers evaluated by Fisher’s exact test (P < 0.05; Table I). By this approach, total tumor numbers were consistently higher in GPx2-KO mice, irrespective of the selenium status, of SFN treatment or both.
Tumor size was smaller in Se-poor GPx2-KO mice only (Figure 6C). Therefore, although still present, the tumor growth-promoting effect of GPx2 was less pronounced than recently described in a tumor xenograft model (10).
Collectively, due to its antiinflammatory potential, GPx2 tends to decrease carcinogenesis. On the other hand, GPx2 tends to support cancer cell survival and tumor growth, as has also been reported for two other selenoproteins, TrxR1 (40) and selenoprotein 15 (41). Thus, a benefit of an upregulation of GPx2 might depend on the tumor stage as discussed elsewhere (42).
Selenium effects
The plasma selenium concentration and liver GPx activity as a marker for the selenium status were comparable in all groups under the same selenium diet. The increase in GPx activity observed here in the liver (16- to 18-fold) was much higher than that observed previously in the intestine of WT mice (2- to 2.8-fold) under the same diets applied for a shorter period (9). In contrast to what has been observed in the ileum and colon of GPx2-KO mice (9), a compensatory rise in GPx activity was not observed in the liver (Supplementary Figure S1, available at Carcinogenesis Online). This is not that surprising since GPx2 is not expressed in mouse liver (43).
The changes in the levels of selenium and selenoprotein here observed in mice closely reflect those considered relevant to cancer risk in humans. It has for long been inferred from epidemiological studies that a low intake of selenium in selenium-deficient areas is associated with an increase in cancer incidence (1,44). Also, selenium supplementation reduced the cancer risk in subjects entering prospective studies with a low selenium state (1,45). Some striking parallels are observed between our mouse studies and a subgroup of the SELGEN study, where humans were given nutritionally relevant amounts of selenium (46). In humans supplemented with 100 μg sodium selenite/day for 6 weeks, plasma selenium levels increased by a factor of 1.19 or 1.23 depending on the subgroups which is in line with the difference between +Se to the ++Se groups in mice (1.2–1.3). Also, the very same selenoproteins were increased with higher selenium in human lymphocytes (46) and decreased in leukocytes (47) and colon (48) of mice fed the same Se-poor diet as fed in this study. Moreover, immune T-cell receptor signaling was upregulated by selenium in human lymphocytes, which mirrors the downregulation of the inflammatory response in mouse leukocytes under the Se-poor diet (47). Finally, the decrease in inflammation scores in WT ++Se compared with WT +Se (Figure 5) points to a better adaptation to inflammatory stimuli under the supranutritional diet. Collectively, thus, mouse studies as presented here, might well provide hints what can be expected from variable selenium intakes in humans.
A mechanistic interpretation of the selenium effects in inflammation and tumorigenesis is difficult due to the complexity of the selenoproteome (49). In the present study, selenium did not significantly affect the inflammation scores in WT mice, whereas decreased colitis with increasing selenium intake was evident in GPx2-KO mice (Figures 5 and 6A). This unexpected finding reveals that GPx2 can hardly be considered the only anti-inflammatory selenoprotein and that upregulation of another selenoprotein can compensate for the loss of GPx2. Compensation, however, appears not to be complete, since the trend for differences between genotypes persisted at ++Se diet (Figure 5(*)). In WT mice, GPx2 appears to be sufficiently expressed already in marginal selenium deficiency and an increase in the +Se to the ++Se state (Figure 3B) might not be relevant to further protection. GPx1, ranking low in the selenoprotein hierarchy, is considered the prime candidate to support GPx2 in preventing inflammation. It should rise substantially with Se supply and, in fact, is upregulated in GPx2-KO mice in Se adequacy and even more by supranutritional selenium (9). Also, the requirement of a double-KO of GPx1 and GPx2 to trigger spontaneous colitis argues for GPx1 (5). Other selenoproteins to be considered as supportive candidates would be selenoprotein W, which most sensitively responds to selenium supply (48) or a TrxR, as TrxR activity is upregulated by ++Se in WT mice in both colon and ileum and in GPx2-KO mice at least in the ileum (Figure 2B). Thus, GPx2 is certainly a major antiinflammatory selenoprotein of the intestine, but the effects of high selenium intakes are likely mediated by others.
Also, total tumor numbers were reduced by selenium (Table I). Already the moderate Se deficiency here established, facilitated tumor formation, which complies with the apparently increased cancer risk under limited selenium supply (1,3). Increasing the selenium intake further reduced tumor numbers in both genotypes. These data, however, do not conflict with previous findings showing that both, deficiency of selenoproteins in general, which is quite different from the moderate Se deficiency as well as excessive selenium supply (2.25 mg/kg diet corresponding to three times our supranutritional dose) inhibited cancer formation in a transgenic mouse model for hepatocarcinoma (50). When selenium was applied in supranutritional amounts, the difference between WT and GPx2-KO was largely reduced (see also Figure 6B), indicating that in respect to carcinogenesis selenium also works in a GPx2-independent manner, which again points to a pivotal role of other selenoproteins in cancer prevention. The candidate selenoprotein should not be maximally expressed in the Se-adequate status. A most likely candidate is again GPx1, whereas TrxR1 was reported to promote tumorigenesis (40).
SFN in Nrf2 target gene expression, inflammation and tumorigenesis
The induction of Nrf2 target genes here reported largely complies with published data, although with discrete differences, see Results and (29,51–54). However, the SFN effects on inflammation scores and tumorigenesis surprised in several respects. First of all, the SFN4 regimen meant to establish a full Nrf2-mediated adaptive response before exposing the mice to AOM/DSS proved to be largely inefficient in respect to tumorigenesis, whereas SFN3, administered simultaneously with the carcinogenic challenge, yielded significant results. Although this finding is consistent with earlier reports showing that SFN inhibits tumor development more efficiently when applied together with the carcinogen (55,56), a convincing explanation cannot be offered. The need for AOM to be metabolized by CYP2E1 for inducing DNA mutations and the inhibition thereof by SFN has been discussed (57), but it is hardly conceivable why a preexisting SFN level should not similarly interfere with AOM metabolism. The second surprise was that SFN4 increased apoptosis in colonic crypt bases (Figure 4) and triggered colitis (Figure 5) under moderate selenium deficiency, particularly in GPx2-KO mice, which at first glance appears to conflict with its protective potential as Nrf2 activator. However, the mechanisms by which SFN presumably activates the Keap1/Nrf2 system, inter alia oxidative stress and/S-alkylation, may equally trigger apoptosis and inflammation under certain circumstances. Indeed, SFN was reported to induce apoptosis in cancer cells (58) and oxidative stress has been suggested as underlying mechanism (18). Alternatively, SFN as Michael acceptor reacts with thiols including GSH. Formation of SFN-GSH adducts might temporarily decrease GSH and impair the ability to cope with oxidative stress. Thus, the toxic potential of SFN becomes unmasked under selenium restriction, where putatively counteracting enzymes, such as GPx1, selenoprotein W, H and M, are missing (48).
In Se adequacy, the protective role of SFN becomes obvious. SFN4 reduced apoptosis in colon crypt bases (Figure 4) and dampened acute inflammation (Figure 5). SFN3 reduced sustained inflammation (Figure 6A), decreased tumor multiplicity (Figure 6B) and total tumor numbers (Table I), whereas its efficacy appeared less pronounced at the supranutritional Se level. Although GPx2 induction in colon crypts by SFN was verified (Figure 3), the protective function of SFN cannot be mediated by GPx2 alone, since tumor prevention was equally observed in GPx2-KO and WT mice (indicated by double-headed arrows in Figures 5 and 6B).
Taken together, the most striking effects of SFN were observed in selenium-adequate GPx2-KO animals where apoptosis, inflammation and tumorigenesis in WT and GPx2-KO mice became equalized. Thus, SFN can have adverse effects at a low selenium status and appears to require another selenoprotein or a selenium-dependent process to act beneficially, with GPx1 being the most plausible candidate. However, GPx1 is not a target of SFN and the mutual interplay of GPx1 and GPx2 has so far remained mechanistically unclear. Nevertheless, SFN and selenium indeed may act cooperatively as suggested previously (22), although not necessarily by upregulation of GPx2.
Synopsis
In essence, our study allows the following conclusions:
(i) GPx2 clearly inhibits apoptosis and thereby inflammation-mediated carcinogenesis.
(ii) Supranutritional selenium can compensate the loss of GPx2 in respect to tumorigenesis but is less effective in respect to inflammation. Adequate selenium supply appears to provide best protection against apoptosis, inflammation and carcinogenesis, which, however, cannot be explained by optimizing GPx2 synthesis alone.
(iii) SFN inhibits carcinogenesis when given simultaneously with the mutagenic challenge. In any case, SFN requires selenium for being protective but not necessarily the synthesis of GPx2, which points to the involvement of other selenoproteins. Surprisingly and most importantly, SFN proved to be proinflammatory under selenium restriction but inhibits acute inflammation in selenium adequacy, again irrespective of GPx2 expression.
These findings corroborate the importance of GPx2 in the process of inflammation-triggered colon carcinogenesis, but also disclose GPx2-independent protective effects of adequate selenium intakes, which can be enhanced by Nrf2-activating SFN. However, the results equally highlight the risk of uncritical intakes of glucosinolates if the selenium status is low.
Supplementary material
Supplementary Table S1 and Figures S1–S3 can be found at http://carcin.oxfordjournals.org/.
Funding
German Research Foundation (Br 778/8-1); SAW-procedure of the Leibniz-Association (to R.B.-F); National Cancer Institute, National Institutes of Health, USA (R01 CA114569) (to F.-F.C).
Supplementary Material
Acknowledgments
The authors thank Stefanie Deubel and the animal facility team, especially Elke Thom, Elisabeth Meyer and Swetlana König, for excellent technical assistance and Dr Martin Osterhoff for help with statistics. Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- ACF
aberrant crypt foci
- AOM
azoxymethane
- DSS
dextran sulfate
- GPx
glutathione peroxidases
- GST
glutathione-S-transferases
- IHC
immunohistochemistry
- SFN
sulforaphane
- NADPH
reduced nicotinamide adenine dinucleotide phosphate
- NQO1
NAD(P)H:quinone oxidoreductase
- Nrf2
nuclear factor E2-related factor 2
- TrxR
thioredoxin reductases
- TNB
5′ thionitrobenzoate anion
References
- 1.Shamberger RJ, et al. Possible protective effect of selenium against human cancer. Can. Med. Assoc. J. 1969;100:682. [PMC free article] [PubMed] [Google Scholar]
- 2.Clark LC, et al. Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin. A randomized controlled trial. Nutritional Prevention of Cancer Study Group. JAMA. 1996;276:1957–1963. [PubMed] [Google Scholar]
- 3.Duffield-Lillico AJ, et al. Baseline characteristics and the effect of selenium supplementation on cancer incidence in a randomized clinical trial: a summary report of the Nutritional Prevention of Cancer Trial. Cancer Epidemiol. Biomarkers Prev. 2002;11:630–639. [PubMed] [Google Scholar]
- 4.Lippman SM, et al. Effect of selenium and vitamin E on risk of prostate cancer and other cancers: the Selenium and Vitamin E Cancer Prevention Trial (SELECT) JAMA. 2009;301:39–51. doi: 10.1001/jama.2008.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esworthy RS, et al. Mice with combined disruption of Gpx1 and Gpx2 genes have colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2001;281:G848–G855. doi: 10.1152/ajpgi.2001.281.3.G848. [DOI] [PubMed] [Google Scholar]
- 6.Chu FF, et al. Bacteria-induced intestinal cancer in mice with disrupted Gpx1 and Gpx2 genes. Cancer Res. 2004;64:962–968. doi: 10.1158/0008-5472.can-03-2272. [DOI] [PubMed] [Google Scholar]
- 7.Florian S, et al. Cellular and subcellular localization of gastrointestinal glutathione peroxidase in normal and malignant human intestinal tissue. Free Radic. Res. 2001;35:655–663. doi: 10.1080/10715760100301181. [DOI] [PubMed] [Google Scholar]
- 8.Chu FF, et al. Role of Se-dependent glutathione peroxidases in gastrointestinal inflammation and cancer. Free Radic. Biol. Med. 2004;36:1481–1495. doi: 10.1016/j.freeradbiomed.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 9.Florian S, et al. Loss of GPx2 increases apoptosis, mitosis, and GPx1 expression in the intestine of mice. Free Radic. Biol. Med. 2010;49:1694–1702. doi: 10.1016/j.freeradbiomed.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banning A, et al. Glutathione peroxidase 2 inhibits cyclooxygenase-2-mediated migration and invasion of HT-29 adenocarcinoma cells but supports their growth as tumors in nude mice. Cancer Res. 2008;68:9746–9753. doi: 10.1158/0008-5472.CAN-08-1321. [DOI] [PubMed] [Google Scholar]
- 11.Banning A, et al. GPx2 counteracts PGE2 production by dampening COX-2 and mPGES-1 expression in human colon cancer cells. Antioxid. Redox Signal. 2008;10:1491–1500. doi: 10.1089/ars.2008.2047. [DOI] [PubMed] [Google Scholar]
- 12.Kipp A, et al. Activation of the glutathione peroxidase 2 (GPx2) promoter by beta-catenin. Biol. Chem. 2007;388:1027–1033. doi: 10.1515/BC.2007.137. [DOI] [PubMed] [Google Scholar]
- 13.Banning A, et al. The GI-GPx gene is a target for Nrf2. Mol. Cell. Biol. 2005;25:4914–4923. doi: 10.1128/MCB.25.12.4914-4923.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itoh K, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 15.Surh YJ, et al. Nrf2 as a master redox switch in turning on the cellular signaling involved in the induction of cytoprotective genes by some chemopreventive phytochemicals. Planta Med. 2008;74:1526–1539. doi: 10.1055/s-0028-1088302. [DOI] [PubMed] [Google Scholar]
- 16.Hayes JD, et al. The cancer chemopreventive actions of phytochemicals derived from glucosinolates. Eur. J. Nutr. 2008;47:73–88. doi: 10.1007/s00394-008-2009-8. [DOI] [PubMed] [Google Scholar]
- 17.Herr I, et al. Dietary constituents of broccoli and other cruciferous vegetables: implications for prevention and therapy of cancer. Cancer Treat. Rev. 2010;36:377–383. doi: 10.1016/j.ctrv.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Juge N, et al. Molecular basis for chemoprevention by sulforaphane: a comprehensive review. Cell. Mol. Life Sci. 2007;64:1105–1127. doi: 10.1007/s00018-007-6484-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hintze KJ, et al. Induction of hepatic thioredoxin reductase activity by sulforaphane, both in Hepa1c1c7 cells and in male Fisher 344 rats. J. Nutr. Biochem. 2003;14:173–179. doi: 10.1016/s0955-2863(02)00282-6. [DOI] [PubMed] [Google Scholar]
- 20.Chung FL, et al. Chemoprevention of colonic aberrant crypt foci in Fischer rats by sulforaphane and phenethyl isothiocyanate. Carcinogenesis. 2000;21:2287–2291. doi: 10.1093/carcin/21.12.2287. [DOI] [PubMed] [Google Scholar]
- 21.Hu R, et al. Cancer chemoprevention of intestinal polyposis in ApcMin/+ mice by sulforaphane, a natural product derived from cruciferous vegetable. Carcinogenesis. 2006;27:2038–2046. doi: 10.1093/carcin/bgl049. [DOI] [PubMed] [Google Scholar]
- 22.Brigelius-Flohé R, et al. Sulforaphane and selenium, partners in adaptive response and prevention of cancer. Free Radic. Res. 2006;40:775–787. doi: 10.1080/10715760600722643. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka T, et al. A novel inflammation-related mouse colon carcinogenesis model induced by azoxymethane and dextran sodium sulfate. Cancer Sci. 2003;94:965–973. doi: 10.1111/j.1349-7006.2003.tb01386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esworthy RS, et al. Low glutathione peroxidase activity in Gpx1 knockout mice protects jejunum crypts from gamma-irradiation damage. Am. J. Physiol. Gastrointest. Liver Physiol. 2000;279:G426–G436. doi: 10.1152/ajpgi.2000.279.2.G426. [DOI] [PubMed] [Google Scholar]
- 25.Haack M, et al. Breakdown products of neoglucobrassicin inhibit activation of Nrf2 target genes mediated by myrosinase-derived glucoraphanin hydrolysis products. Biol. Chem. 2010;391:1281–1293. doi: 10.1515/BC.2010.134. [DOI] [PubMed] [Google Scholar]
- 26.Böcher M, et al. Synthesis of mono- and bifunctional peptide-dextran conjugates for the immobilization of peptide antigens on ELISA plates: properties and application. J. Immunol. Methods. 1997;208:191–202. doi: 10.1016/s0022-1759(97)00149-x. [DOI] [PubMed] [Google Scholar]
- 27.Kruschewski M, et al. Changes of colonic mucosal microcirculation and histology in two colitis models: an experimental study using intravital microscopy and a new histological scoring system. Dig. Dis. Sci. 2001;46:2336–2343. doi: 10.1023/a:1012334727509. [DOI] [PubMed] [Google Scholar]
- 28.Geboes K, et al. Influence of treatment on morphological features of mucosal inflammation. Gut. 2002;50(suppl. 3):III37–III42. doi: 10.1136/gut.50.suppl_3.iii37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mueller M, et al. Nrf2 target genes are induced under marginal selenium-deficiency. Genes Nutr. 2010;5:297–307. doi: 10.1007/s12263-010-0168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gromer S, et al. Human placenta thioredoxin reductase: preparation and inhibitor studies. Methods Enzymol. 2002;347:382–394. doi: 10.1016/s0076-6879(02)47038-3. [DOI] [PubMed] [Google Scholar]
- 31.Habig WH, et al. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J. Biol. Chem. 1974;249:7130–7139. [PubMed] [Google Scholar]
- 32.Brigelius-Flohé R, et al. Estimation of individual types of glutathione peroxidases. Methods Enzymol. 2002;347:101–112. doi: 10.1016/s0076-6879(02)47011-5. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka T. Colorectal carcinogenesis: review of human and experimental animal studies. J. Carcinog. 2009;8:5.. doi: 10.4103/1477-3163.49014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki R, et al. Strain differences in the susceptibility to azoxymethane and dextran sodium sulfate-induced colon carcinogenesis in mice. Carcinogenesis. 2006;27:162–169. doi: 10.1093/carcin/bgi205. [DOI] [PubMed] [Google Scholar]
- 35.Esworthy RS, et al. Epithelium-specific glutathione peroxidase, Gpx2, is involved in the prevention of intestinal inflammation in selenium-deficient mice. J. Nutr. 2005;135:740–745. doi: 10.1093/jn/135.4.740. [DOI] [PubMed] [Google Scholar]
- 36.Yan W, et al. GPX2, a direct target of p63, inhibits oxidative stress-induced apoptosis in a p53-dependent manner. J. Biol. Chem. 2006;281:7856–7862. doi: 10.1074/jbc.M512655200. [DOI] [PubMed] [Google Scholar]
- 37.Edelblum KL, et al. Regulation of apoptosis during homeostasis and disease in the intestinal epithelium. Inflamm. Bowel Dis. 2006;12:413–424. doi: 10.1097/01.MIB.0000217334.30689.3e. [DOI] [PubMed] [Google Scholar]
- 38.Seiler A, et al. Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent- and AIF-mediated cell death. Cell Metab. 2008;8:237–248. doi: 10.1016/j.cmet.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Wingler K, et al. mRNA stability and selenocysteine insertion sequence efficiency rank gastrointestinal glutathione peroxidase high in the hierarchy of selenoproteins. Eur. J. Biochem. 1999;259:149–157. doi: 10.1046/j.1432-1327.1999.00012.x. [DOI] [PubMed] [Google Scholar]
- 40.Yoo MH, et al. Thioredoxin reductase 1 deficiency reverses tumor phenotype and tumorigenicity of lung carcinoma cells. J. Biol. Chem. 2006;281:13005–13008. doi: 10.1074/jbc.C600012200. [DOI] [PubMed] [Google Scholar]
- 41.Irons R, et al. Deficiency in the 15-kDa selenoprotein inhibits tumorigenicity and metastasis of colon cancer cells. Cancer Prev. Res. 2010;3:630–639. doi: 10.1158/1940-6207.CAPR-10-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brigelius-Flohé R, et al. Glutathione peroxidases in different stages of carcinogenesis. Biochim. Biophys. Acta. 2009;1790:1555–1568. doi: 10.1016/j.bbagen.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 43.Chu FF, et al. Expression, characterization, and tissue distribution of a new cellular selenium-dependent glutathione peroxidase, GSHPx-GI. J. Biol. Chem. 1993;268:2571–2576. [PubMed] [Google Scholar]
- 44.Rayman MP. Selenium in cancer prevention: a review of the evidence and mechanism of action. Proc. Nutr. Soc. 2005;64:527–542. doi: 10.1079/pns2005467. [DOI] [PubMed] [Google Scholar]
- 45.Lee EH, et al. Effects of selenium supplements on cancer prevention: meta-analysis of randomized controlled trials. Nutr. Cancer. 2011;63:1185–1195. doi: 10.1080/01635581.2011.607544. [DOI] [PubMed] [Google Scholar]
- 46.Pagmantidis V, et al. Supplementation of healthy volunteers with nutritionally relevant amounts of selenium increases the expression of lymphocyte protein biosynthesis genes. Am. J. Clin. Nutr. 2008;87:181–189. doi: 10.1093/ajcn/87.1.181. [DOI] [PubMed] [Google Scholar]
- 47.Kipp A, et al. Marginal selenium deficiency down-regulates inflammation-related genes in splenic leukocytes of the mouse. J. Nutr. Biochem. 2011 doi: 10.1016/j.jnutbio.2011.06.011. , Nov. 30 [epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 48.Kipp A, et al. Four selenoproteins, protein biosynthesis, and Wnt signalling are particularly sensitive to limited selenium intake in mouse colon. Mol. Nutr. Food Res. 2009;53:1561–1572. doi: 10.1002/mnfr.200900105. [DOI] [PubMed] [Google Scholar]
- 49.Duntas LH. Selenium and inflammation: underlying anti-inflammatory mechanisms. Horm. Metab. Res. 2009;41:443–447. doi: 10.1055/s-0029-1220724. [DOI] [PubMed] [Google Scholar]
- 50.Novoselov SV, et al. Selenoprotein deficiency and high levels of selenium compounds can effectively inhibit hepatocarcinogenesis in transgenic mice. Oncogene. 2005;24:8003–8011. doi: 10.1038/sj.onc.1208940. [DOI] [PubMed] [Google Scholar]
- 51.Munday R, et al. Induction of phase II detoxification enzymes in rats by plant-derived isothiocyanates: comparison of allyl isothiocyanate with sulforaphane and related compounds. J. Agric. Food Chem. 2004;52:1867–1871. doi: 10.1021/jf030549s. [DOI] [PubMed] [Google Scholar]
- 52.McWalter GK, et al. Transcription factor Nrf2 is essential for induction of NAD(P)H:quinone oxidoreductase 1, glutathione S-transferases, and glutamate cysteine ligase by broccoli seeds and isothiocyanates. J. Nutr. 2004;134:3499S–3506S. doi: 10.1093/jn/134.12.3499S. [DOI] [PubMed] [Google Scholar]
- 53.Keck AS, et al. Food matrix effects on bioactivity of broccoli-derived sulforaphane in liver and colon of F344 rats. J. Agric. Food Chem. 2003;51:3320–3327. doi: 10.1021/jf026189a. [DOI] [PubMed] [Google Scholar]
- 54.Burk RF, et al. Selenium deficiency activates mouse liver Nrf2-ARE but vitamin E deficiency does not. Free Radic. Biol. Med. 2008;44:1617–1623. doi: 10.1016/j.freeradbiomed.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, et al. Anticarcinogenic activities of sulforaphane and structurally related synthetic norbornyl isothiocyanates. Proc. Natl Acad. Sci. USA. 1994;91:3147–3150. doi: 10.1073/pnas.91.8.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kassie F, et al. Chemoprevention of 2-amino-3-methylimidazo[4,5-f]quinoline (IQ)-induced colonic and hepatic preneoplastic lesions in the F344 rat by cruciferous vegetables administered simultaneously with the carcinogen. Carcinogenesis. 2003;24:255–261. doi: 10.1093/carcin/24.2.255. [DOI] [PubMed] [Google Scholar]
- 57.Barcelo S, et al. CYP2E1-mediated mechanism of anti-genotoxicity of the broccoli constituent sulforaphane. Carcinogenesis. 1996;17:277–282. doi: 10.1093/carcin/17.2.277. [DOI] [PubMed] [Google Scholar]
- 58.Rudolf E, et al. Activation of several concurrent proapoptic pathways by sulforaphane in human colon cancer cells SW620. Food Chem. Toxicol. 2009;47:2366–2373. doi: 10.1016/j.fct.2009.06.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.