Abstract
The aim of this project is to identify new therapeutic microRNAs (miRNAs) for von Hippel-Lindau (VHL)-inactivated renal cancer cells. We initially identified several potential miRNAs targeting CTNNB1 and MEK1 using several targets scan algorithms. Only miR-1826 was found to target CTNNB1 and MEK1. Therefore, we focused on miRNA-1826 and performed 3′ untranslated region (UTR) luciferase assay, functional analyses and association study between miR-1826 expression and renal cancer patient outcomes. miR-1826 expression was significantly lower in renal cancer tissues compared with non-neoplastic areas and lower expression was significantly associated with overall shorter survival and earlier recurrence after radical nephrectomy. Following miR-1826 transfection, 3′ UTR luciferase activity and protein expression of beta-catenin and MEK1 were significantly downregulated in renal cancer cells. Introduction of miR-1826 also inhibited renal cancer cell proliferation, invasion and migration. Additionally, miR-1826 promoted apoptosis and G1 arrest in VHL-inactivated renal cancer cells. Knockdowns of CTNNB1 and MEK1 by small interfering RNAs reproduced the tumor-suppressive effect of miR-1826. Our data suggest that the miR-1826 plays an important role as a tumor suppressor by downregulating beta-catenin and MEK1 in VHL-inactivated renal cancers.
Introduction
Renal cell carcinoma (RCC) is the third leading cause of death among urological tumors, accounting for 2% of adult malignancies (1). Although the rate of detection of incidental RCC has increased with improved diagnostic techniques, metastatic lesions are still found at diagnosis in ∼30% of RCC patients (2). Renal cancer patients with localized kidney cancer have a 90% 5 years survival rate. However, patients with advanced cancer (stage IV) have a significantly reduced 5 years survival rate (<30%) (1). Compared with other cancers, there are very few tumor markers for renal cancer (3). Also, renal cancer patients respond poorly to conventional chemotherapy because RCC is regarded as a multidrug-resistant cancer (4). Recently, three multikinase inhibitors, two mammalian target of rapamycin inhibitors and a vascular endothelial growth factor-neutralizing antibody have been approved for the treatment of advanced RCC but are not globally used (5). The most common histological type of renal cancer is clear cell renal cell carcinoma (cc-RCC, 70%), and the ‘von Hippel-Lindau’ (VHL) tumor-suppressor gene is associated with tumorigenesis in cc-RCC (6,7). Approximately 60% of cc-RCC patients have a mutated or inactivated VHL gene. The main function of VHL protein (p-VHL) is to inhibit beta-catenin (CTNNB1) and hypoxia inducible factor (HIF)-1 (6–8). Therefore, mutation of the VHL gene may activate oncogenic pathways, such as beta-catenin or HIF-1 (6–8). HIF-1 is known to stimulate the Ras–Raf–MEK–extracellular signal-regulated kinase (ERK) pathway (9).
Recently, a number of microRNAs (miRNAs) have been identified and reported to be important in several cancer treatments (10). miRNAs are small non-coding RNAs of ∼22 nucleotides in length that are capable of regulating gene expression at both the transcription and translation levels (11). miRNAs bind to the 3′ untranslated region (UTR) of target messenger RNA (mRNA) and repress translation from mRNA to protein or induce mRNA cleavage and thereby regulate the expression of target genes (11). Based on several target scan algorithms, we searched for miRNAs targeting both beta-catenin and HIF-1 and downstream oncogenes using microRNA Target Prediction and Functional Study Database (miRDB) (http://mirdb.org/miRDB/) (12) and other target predicting algorithms (microRNA.org and TargetScan). As a result only one, miR-1826, was found targeting both beta-catenin and MEK1. The role of MEK1 has been reported in several cancers and MEK inhibitors are also used for cancer treatment in several cancers (13,14).
Based on these results, we hypothesized that miRNA-1826 may be a new tumor suppressor for VHL mutated or inactivated RCC. To test this hypothesis, we performed 3′ UTR luciferase assays to confirm whether miR-1826 binds to the 3′ UTR of these target gene's mRNAs and affected the function (proliferation, invasion, migration, apoptosis and cell cycle) of renal cancer cells. We also knocked down CTNNB1 and MEK1 mRNAs using a small interfering RNA (siRNA) technique to examine the mechanism of miR-1826 tumor-suppressive function.
Materials and methods
The design and schematic representation of this project
A schematic representation of the role of beta-catenin and MEK1 in VHL-inactivated renal cancer cells is shown in Figure 1A. The aim of this project is to identify new therapeutic miRNAs related to VHL-inactivated renal cancer cells. VHL gene mutation and inactivation results in constitutive HIF activation and aberrant accumulation of cytoplasmic and nuclear beta-catenin (Figure 1A). In order to identify miRNAs targeting these oncogenes, we initially used miRDB and found several miRNAs targeting beta-catenin and HIF downstream genes (MEK1/2 and ERK1/2). The miRNAs identified are shown in Figure 1B.
Fig. 1.
Schematic representation of the role of beta-catenin and MEK1 in VHL-inactivated renal cancer cells and potential miRNAs targeting beta-catenin and MEK1 based on miRDB. (A) When the VHL gene is mutated or inactivated, protein VHL (p-VHL) fails to regulate HIF-1 and beta-catenin, resulting in constitutive HIF activation and aberrant accumulation of cytoplasmic and nuclear beta-catenin. (B). Based on miRDB, several miRNAs targeting beta-catenin and HIF downstream genes (MEK1/2 and ERK1/2) were identified.
Clinical samples
A total of 46 patients (31 male and 15 female) with pathologically confirmed conventional RCC were enrolled in this study (Toho University Hospital, Tokyo, Japan). The mean age of the patients was 60.8 years (range 37–77 years). They were classified according to the World Health Organization criteria and staged according to the tumor-node-metastasis (TNM) classification. Namely T refers to the size of the renal cancer and whether or not it has invaded nearby tissue, N refers to whether regional lymph nodes are involved and M whether there is distant metastasis or not. The pathology of all the patients was cc-RCC. Samples were obtained from the patients after written informed consent was obtained in Toho University hospital.
Cell culture
Renal cancer cell lines with inactivated VHL [A-498, American Type Culture Collection (ATCC) number: HTB-44; 786-O, ATCC number: CRL-1932] and with intact VHL (Caki-1, ATCC number: HTB-46) were purchased from the ATCC (Manassas, VA). The cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum.
Total RNA and protein extraction
RNA (miRNA and total RNA) was extracted from formalin-fixed paraffin-embedded human renal cancer and matched adjacent non-cancerous normal kidney tissues using a miRNeasy formalin-fixed paraffin-embedded kit (Qiagen) after microdissection. To digest DNA, the Qiagen RNase-Free DNase kit was used. Total RNA was also extracted from renal cancer cell lines using an miRNeasy mini kit (Qiagen). Cells were lysed with RIPA buffer (Pierce, Brebieres, France) containing protease inhibitors (Sigma, St Louis, MO). Protein quantification was done using a BCA protein assay kit (Pierce).
MiRNA transfection
Pre-miR™ miRNA precursors [negative control (miR-NC) (catalog#AM17110) or hsa-miR-1826 (miR-1826) (catalog#AM17100; Ambion)] were transiently transfected into renal cancer cells by Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Cell viability, cell invasion and wound healing assay
Cell viability was measured with CellTiter 96 AQueous One Solution Cell Proliferation Assay (MTS)(Promega) 3 days after transfection. Data are the mean ± SD of six independent experiments. Cell invasion assay was performed with the CytoSelect 24-well cell invasion assay kit (Cell BioLab, San Diego, CA) as described previously (15). Transfected cells (miR-NC or miR-1826 transfectant—48 h) were resuspended in culture medium and put into the upper chamber in triplicate. After 48 h incubation at 37°C (5% CO2), cells migrating through the membrane were stained. The results were expressed as invaded cells quantified at optical density 560 nm. The wound healing process begins with tissue matrix remodeling, migration and eventual closing of the wound area. Therefore, this assay is frequently used for assessment of cancer cell migration. Wound healing assay was performed with the CytoSelect 24-well wound healing assay kit as described previously (Cell BioLab)) (15). To generate a wound field, transfected cells (miR-NC or miR-1826 transfectants—48 h) were cultured until they formed a monolayer around the insert (24 h). After removing insert, a 0.9 mm open-wound field was generated and cells were allowed to migrate from either side of the gap. Wound closure was monitored and the percent closure was measured at 10 h using miR-NC and miR-1826 transfected cells [Percent closure rate (%) = migrated cell surface area/(total surface area × 100)].
Apoptosis and cell cycle analyses
Cells (miR-NC and miR-1826 precursor transfected renal cancer cells—48 h) were washed twice with 1× phosphate-buffered saline and trypsinized. After inactivating trypsin in complete medium, the cells were resuspended in ice-cold 1× binding buffer (70 μl). Annexin V-fluorescein isothiocyanate solution (10 μl) and 7-aminoactinomycin D viability dye (20 μl) were added to 70 μl of the cell suspensions. After incubation for 15 min in the dark, 400 μl of ice-cold 1× binding buffer was added. The apoptotic distribution of the cells in each sample was then determined using a fluorescence-activated cell sorting (Cell Lab QUANTA SC; Beckman Coulter, Fullerton, CA). The various phases of cells were determined using a DNA stain (4′,6-diamidino-2-phenylindole). Cell populations (G0/G1, S and G2/M) were differentiated according to 4′,6-diamidino-2-phenylindole intensity and side scatter-measured cell volume and quantified with Cell Lab QUANTA software. Data are the mean ± SD of four independent experiments.
3′ UTR luciferase assay
To identify potential miRNAs targeting beta-catenin (CTNNB1) and genes related to the Ras–Raf–MEK–ERK pathway, we used several algorithms [TargetScan (http://www.targetscan.org/), microRNA.org (http://www.microrna.org/microrna/home.do/) and miRDB (http://mirdb.org/miRDB/)] (12). Based on these algorithms, we found several miRNAs targeting CTNNB1 (11 miRNAs), MEK1 (23 miRNAs), MEK2 (two miRNAs), ERK1 (three miRNAs) and ERK2 (26 miRNAs) (Figure 1B). Among these miRNAs, miR-1826 only targets both CTNNB1 and MEK1 genes. Therefore, we focused on miR-1826 in this project. In order to perform 3′ UTR luciferase assay, PmirGLO Dual-Luciferase miRNA Target Expression Vector was used (Promega, Madison, WI). The oligonucleotides sequences (wild-type) used are shown in Supplementary Table S1, available at Carcinogenesis Online. We also constructed mutated oligonucleotides for each of the wild-type oligonucleotides [mutated-type, Supplementary Table S2 is available at Carcinogenesis Online]. In a total volume of 20 μl, 5 μl of 100 μM forward oligonucleotide, 5 μl of 100 μM reverse oligonucleotide, 2 μl of 10× annealing buffer (100 mM Tris–HCl, pH 7.5, 1 M NaCl and 10 mM ethylenediaminetetraacetic acid) and 8 μl water were added to a 200 μl polymerase chain reaction (PCR) tube and incubated at 95°C for 5 min and then placed at room temperature for 1 h. The oligonucleotides were ligated into the PmeI–XbaI site of pmirGLO Dual-Luciferase miRNA Target Expression Vector. Colony direct PCR was performed for insert recognition using REDTaq (Sigma). The primers used were as follows: forward primer, 5′-cgtgctggaacacggtaaaa-3′; reverse primer, 5′-gcagccaactcagcttcctt-3′. PCR parameters for cycling were as follows: 94°C for 3 min, 30 cycles of PCR at 94°C for 30 s, 55°C for 30 s, 72°C for 30 s, 72°C for 10 min and 4°C for 10 min. The PCR product was digested with NotI (TaKaRa/Fisher Scientific, Pittsburgh, PA). The size of vectors containing inserts were ∼200 and 100 bp by electrophoresis since the NotI recognition sequence was incorporated into the primers. Vectors were sequenced directly by an outside vendor (MCLAB, South San Francisco, CA). For 3′ UTR luciferase assay, A-498 and 786-O cells were cotransfected with miR-NC or miR-1826 and pmirGLO Dual-Luciferase miRNA Target Expression Vectors using Lipofectamine 2000 (Invitrogen). Luciferase assay was performed using the Dual-Luciferase® Reporter Assay System (Promega) at 48 h after transfection.
Knockdown of CTNNB1 and MEK1 mRNAs in A-498 cells and functional analyses
VHL gene-inactivated A-498 cells were transiently transfected with CTNNB1 and MAP2K1 siRNAs (si-CTNNB1 and si-MAP2K1, catalog#VHS50819, #VHS40795; Invitrogen, Carlsbad, CA) or negative control siRNA (si-NC; Invitrogen) according to the manufacturer’s instructions. Briefly, cells were grown in six-well plates and transfected individually with si-NC (negative control) or si-CTNNB1 or si-MAP2K1 at a concentration of 200 pmol/well. Transfection was performed with X-tremeGene siRNA transfection reagent (Roche Diagnosis, Basel, Switzerland). Then, 48 h after transfection, RNA and protein were extracted and knockdown of CTNNB1 and MAP2K1 mRNAs and proteins were confirmed by real-time PCR and western blot analysis. Cell viability (24, 48 and 72 h after transfection), invasion (48 h after transfection) and apoptosis analysis (48 h after transfection) were performed using si-NC- or si-CTNNB1- or si-MAP2K1-transfected A-498 cells.
Quantitative real-time reverse transcription–PCR
Quantitative real-time reverse transcription–PCR was performed in triplicate with an Applied Biosystems Prism 7500 Fast Sequence Detection System using TaqMan universal PCR master mix according to the manufacturer’s protocol (Applied Biosystems, Foster City, CA). The TaqMan probes and primers were purchased from Applied Biosystems. RNU48 was used as internal control. Levels of RNA expression were determined using the 7500 Fast System SDS software version 1.3.1 (Applied Biosystems).
Western analysis
Total cell protein (15 μg) was used for western blotting. Samples were resolved in 4–20% Precise Protein Gels (Pierce) and transferred to polyvinylidene difluoride membranes (Amersham Biosciences, Fairfield, CT). The membranes were immersed in 0.3% skim milk in Tris-buffered saline containing 0.1% Tween 20 for 1 h and probed with primary polyclonal and monoclonal antibody against beta-catenin (#9562; Cell signaling Technology, Beverly, MA), survivin (#2808; Cell signaling Technology), VHL (#2738; Cell signaling Technology), beta-tubulin (#2128; Cell signaling Technology) and MEK1 (#ab47422; Cambridge, UK) overnight at 4°C. Blots were washed in Tris-buffered saline containing 0.1% Tween 20 and labeled with horseradish peroxidase-conjugated secondary anti-mouse or anti-rabbit antibody (Cell signaling Technology). Proteins were enhanced by chemiluminescence (Amersham ECL plus Western Blotting detection system; Amersham Biosciences) for visualization. The protein expression levels were expressed relative to beta-tubulin levels.
Immunohistochemical study
Immunohistochemical staining was performed following the manufacturer’s instructions (UltraVision Detection System; Thermo Fisher Scientific, Fremont, CA). Namely, formalin-fixed paraffin-embedded tissue sections were incubated in xylene at room temperature (10 min, three times), rinsed in an ascending series of ethanol (100, 100, 95 and 70% for 5 min each) and rinsed in ddH2O (3 min). Antigen retrieval was performed by incubating the slides in a microwave for 5 min (high power level) in 1× citrate buffer (pH 6.0) (#21545; Chemicon international, Temecula, CA). To reduce non-specific background staining due to endogenous peroxidase, slides were incubated in ‘hydrogen peroxidase block’ (included in kit) for 10 min and then primary antibodies (CTNNB1, MEK1 and isotype controls) were added and incubated at 4°C overnight. The antibody dilution was 1:100 based on the manufacturer’s recommendation and we used the following primary antibodies: beta-catenin (#9562; Cell Signaling Technology); MEK1 (#ab47422; Abcam, Cambridge, UK). Sections were also incubated with an appropriate isotype (IgG) matched rabbit antibody as NC (#3900; Cell Signaling Technology and #ab27478; Abcam, respectively). After overnight incubation with primary antibody, slides were washed in Tris-buffered saline buffer and incubated with secondary biotinylated goat anti-rabbit antibody (10 min, room temperature) and streptavidin peroxidase was added (10 min, room temperature). Color was developed with diaminobenzidene (40 μl of diaminobenzidene Plus Chromogen + 2ml of diaminobenzidene Plus Substrate) for 5 min at room temperature. The sections were counterstained with hematoxylin (American Master Tech Scientific, Lodi, CA). Immunohistochemical staining was evaluated by visually assessing staining intensity (0–2) using a microscope at ×200. All specimens were scored blindly by two observers. We used the Human Protein Atlas (http://www.proteinatlas.org/) as a reference for immunohistochemistry assessment. The criteria of intensity are as follows: 0, negative expression; 1+, weakly positive expression; 2+, strongly positive expression.
Statistical analysis
All statistical analyses were performed using StatView (version 5; SAS Institute, NC). A P-value of <0.05 was regarded as statistically significant.
Results
Effect of miR-1826 on VHL proficient renal cancer cell (Caki-1)
We looked at the expression and function of miR-1826-transfected VHL-proficient renal cancer cells (Caki-1) (Supplementary Figure S1 is available at Carcinogenesis Online). The expression of miR-1826 was significantly higher in VHL-proficient renal cancer cell line (Caki-1) compared with VHL-inactivated renal cancer cell lines (A-498 and 786-O) (Supplementary Figure S1A is available at Carcinogenesis Online). Next, we performed functional analyses with miR-1826 overexpressing Caki-1 cells (Supplementary Figure S1B is available at Carcinogenesis Online). As expected, miR-1826-transfected Caki-1 cells did not show inhibition of proliferation (Supplementary Figure S1C is available at Carcinogenesis Online), invasion (Supplementary Figure S1D is available at Carcinogenesis Online) and migration (Supplementary Figure S1E is available at Carcinogenesis Online). miR-1826 also did not induce apoptosis in VHL-proficient renal cancer cells (Caki-1; Supplementary Figure S1F is available at Carcinogenesis Online).
3′ UTR luciferase assay and lower protein expression in miR-1826 transfectant
CTNNB1 mRNA has one potential complimentary binding site with miR-1826 within its 3′ UTR (mirSVR score: −1.2735; Figure 2A). MEK1 mRNA has two potential complimentary binding sites with miR-1826 within its 3′ UTR (mirSVR score: −0.8212 and −1.1902, respectively). Based on these results, we performed 3′ UTR luciferase assays and observed that the relative luciferase activity was significantly decreased in miR-1826-transfected VHL-inactivated renal cancer cells (A-498 and 786-O) (Figure 2B).
Fig. 2.
Binding of miR-1826 to the 3′ UTR of CTNNB1 and MEK1 mRNAs, luciferase assays and an inverse correlation between miR-1826 and CTNNB1/MEK1 protein expression in human renal cancer tissues. (A) CTNNB1 and MEK1 3′ UTR position and complementary miR-1826-binding sequence. (B) 3′ UTR luciferase assay (miR-NC and miR-1826), MT stands for mutated plasmid sequence and WT stands for wild-type plasmid sequence. (C) CTNNB1, survivin, MEK1, beta-tubulin protein expression in miR-NC or miR-1826-transfected VHL-inactivated renal cancer cells (A-498 and 786-O). (D) Inverse correlation between miR-1826 and CTNNB1/MEK1 protein expression in renal cancer tissues.
With mutated plasmids, there was no significant difference in luciferase activity between controls and miR-1826 transfectants (Figure2B). These results suggest that CTNNB1 and MEK1 mRNAs are target oncogenes of miR-1826. CTNNB1 protein expression was also significantly decreased in miR-1826-transfected cells (Figure 2C). In order to verify beta-catenin downregulation, we looked at the expression of survivin, which are T cell-factor/lymphoid-enhancer-factor downstream effectors. As expected, the protein (survivin) was significantly downregulated in miR-1826-transfected cells (Figure 2C). MEK1 protein expression was also significantly decreased in miR-1826-transfected renal cancer cells (Figure 2C).
Inverse relationship between miR-1826 and beta-catenin/MEK1 expression levels
We investigated the relationship between miR-1826 expression and beta-catenin/MEK1 protein expression levels in clinical samples. We observed an inverse correlation between miR-1826 expression and CTNNB1 or MEK1 protein expression in renal cancer tissues (Figure 2D). Representative pictures of the stainings for beta-catenin, MEK1 and isotype control in renal cancer tissues and normal kidney tissues are shown in Supplementary Figure S2, available at Carcinogenesis Online. The expression of CTNNB1 and MEK1 was weak in normal kidney tissues.
Expression of beta-catenin and MEK1 in renal cancer cells
The beta-catenin (CTNNB1) and MAP2K1 protein expression levels were significantly higher in renal cancer cells with inactivated VHL (A-498 and 786-O) compared with those with intact VHL (Caki-1) (Supplementary Figure S3A is available at Carcinogenesis Online).
Relationship between MEK1 expression level and clinical characteristics
All patient pathology was cc-RCC. MEK1 expression in the renal cancer tissues was classified into two categories based on immunohistochemistry.
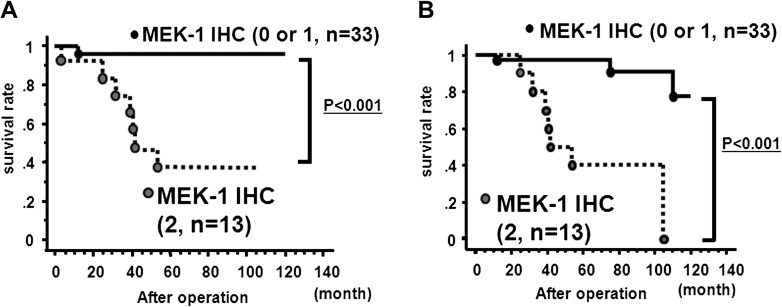
Higher MEK1 expression was associated with higher pathological stage (pTNM) and shorter overall survival and recurrence interval after radical nephrectomy (Table I). Multivariate analysis for overall survival also showed higher MEK1 expression in renal cancer tissues that was significantly associated with shorter overall survival (Table II). Furthermore, Kaplan–Meier plots showed inverse correlation between MEK1 expression and renal cancer patient outcomes (Figure 3).
Table I.
Relationship between MEK1 (MAP2K1) expression in human renal cancer tissues and clinico-pathological data
| MEK1 IHC-0 and 1 in cancer N = 33 (%) | MEK1 IHC-2 in cancer N = 13 (%) | P-value | |
| Age (mean ± SD) (years) | 60.1 ± 8.9 | 62.8 ± 9.8 | 0.37 |
| Gender | |||
| Male (n = 31) | 25 (76) | 6 (46) | 0.08 |
| Female (n = 15) | 8 (24) | 7 (54) | Reference |
| Grade | |||
| 1 (n = 13) | 11 (33) | 2 (25) | Reference |
| 2 (n = 28) | 20 (61) | 8 (35) | |
| 3 (n = 5) | 2 (6) | 3 (40) | 0.09 |
| pStage | |||
| 1 (n = 27) | 24 (73) | 3 (14) | |
| 2 (n = 10) | 5 (15) | 5 (50) | |
| 1 + 2 (n = 37) | 29 (88) | 8 (62.5) | Reference |
| 3 (n = 4) | 3 (9) | 1 (50) | |
| 4 (n = 5) | 1 (3) | 4 (80) | |
| 3 + 4 (n = 9) | 4 (12) | 5 (71) | 0.05 |
| pT | |||
| 1 (n = 27) | 24 (73) | 3 (14) | |
| 2 (n = 11) | 6 (18) | 5 (45) | |
| 1 + 2 (n = 38) | 30 (91) | 8 (24) | Reference |
| 3 (n = 7) | 3 (9) | 4 (80) | |
| 4 (n = 1) | 0 (0) | 1 (100) | |
| 3 + 4 (n = 8) | 3 (9) | 5 (83) | 0.026 |
| pN | |||
| (−) (n = 43) | 33 (100) | 10 (28) | Reference |
| (+) (n = 3) | 0 (0) | 3 (100) | 0.018 |
| pM | |||
| (−) (n = 43) | 32 (97) | 11 (31) | Reference |
| (+) (n = 3) | 1 (3) | 2 (66.7) | 0.17 |
| Overall survival | |||
| Alive (n = 38) | 32 (97) | 6 (19) | Reference |
| Dead (n = 8) | 1 (3) | 7 (87) | 0.0002 |
| Recurrence | |||
| No (n = 36) | 30 (91) | 6 (20) | Reference |
| Yes (n = 10) | 3 (9) | 7 (78) | 0.0023 |
Table II.
Association of clinical parameters, MEK1 expression and overall survival
| Coef. | Standard error | Chi-square | P-value | Exp. (Coef.) | 95% CI | |
| Univariate analysis variables | ||||||
| Grade (3 versus 1 + 2) | 1.358 | 1.014 | 1.793 | 0.1805 | 3.889 | 0.533–28.391 |
| Gender (male versus female) | −0.898 | 0.792 | 1.284 | 0.2572 | 0.407 | 0.086–1.926 |
| Age (younger versus older) | −0.671 | 0.881 | −0.762 | 0.4464 | 0.511 | 0.091–2.876 |
| pT (pT3 + pT4 versus pT1 + pT2) | 2.14 | 0.883 | 5.876 | 0.0153 | 8.5 | 1.506–47.970 |
| pN (N1 versus N0) | 2.512 | 1.301 | 3.727 | 0.0536 | 12.333 | 0.962–158.110 |
| pM (M1 versus M0) | 2.512 | 1.301 | 3.727 | 0.0536 | 12.333 | 0.962–158.110 |
| MEK1 IHC (2 versus 0 + 1) | 3.602 | 1.158 | 9.773 | 0.0018 | 37.333 | 3.858–361.255 |
| Multivariate analysis variables | ||||||
| pT (pT3 + pT4 versus pT1 + pT2) | 1.431 | 1.101 | 1.689 | 0.1938 | 4.183 | 0.483–36.219 |
| MEK1 IHC (2 versus 0 + 1) | 3.341 | 1.182 | 7.992 | 0.0047 | 28.252 | 2.786–286.504 |
CI, confidence interval; Coef, coefficient; Exp. (Coef.), e(Coefficient).
Fig. 3.
Inverse correlation between MEK1 expression and RCC patient outcomes. (A) Inverse correlation between MEK1 protein expression and overall survival after radical nephrectomy. (B). Inverse correlation between MEK1 protein expression and recurrence-free survival after radical nephrectomy.
Relationship between miR-1826 expression level and clinical characteristics
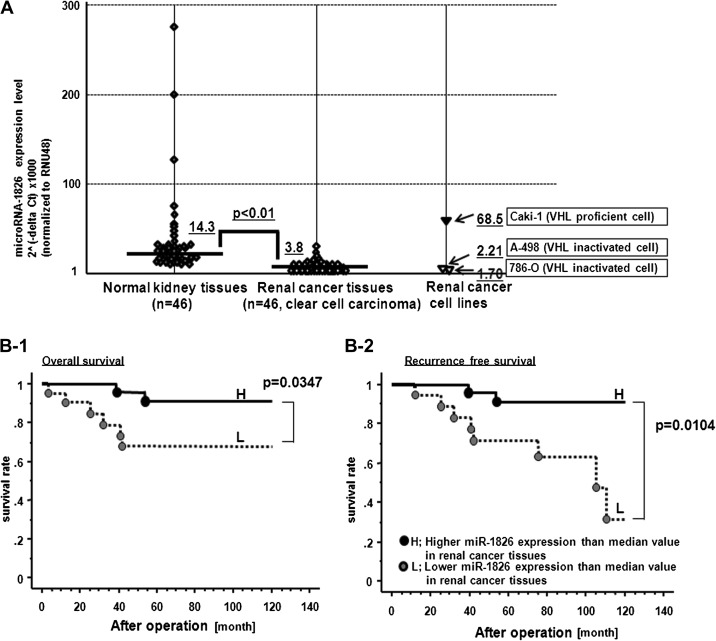
The miR-1826 expression levels were compared based on real-time PCR results. Namely, the miR-1826 expression level was significantly higher in normal kidney tissues compared with renal cancer tissues (n = 46) (Figure 4A). We also split renal cancer patients into two groups (L for low and H for high) based on median miR-1826 expression levels as a cutoff point. We investigated the relationship between miR-1826 expression level (lower or higher in cancer tissues) and clinical factors, including grade, pathologic tumor classification (pT), pathologic lymph node status (pN), pathologic metastasis status (pM) and outcomes (survival and recurrence). There was no significant association of grade and pTNM with miR-1826 expression (data not shown), but we found that patients in the lower miR-1826 category had a significantly poor outcome (Figure 4B).
Fig. 4.
miR-1826 expression level in normal kidney and matched renal cancer tissues and the relationship between miR-1826 expression and renal cancer patient outcomes. (A) miR-1826 expression level was significantly lower in renal cancer tissues compared with matched normal kidney tissues (n = 46; cc-RCC). (B-1) Inverse correlation between miR-1826 expression and overall survival. (B-2) Inverse correlation between miR-1826 and recurrence-free survival after radical nephrectomy.
Effect of miRNA-1826 on cell viability, migration, invasion, cell cycle and apoptosis in VHL-inactivated renal cancer cells
At 48 h after transfection of miR-NC or miR-1826 into renal cancer cells (A-498 and 786-O), the increased miR-1826 expression level was verified by real-time PCR (fold change: 51050 and 2513, respectively; Supplementary Figure S3B is available at Carcinogenesis Online). We observed a significant decrease in cell viability (Figure 5A), invasion (Figure 5B) and migration (Figure 5C) in miRNA-1826-transfected renal cancer cells compared with miR-NC-transfected cells. We found a significant difference in the number of apoptotic cells between miR-1826-transfected cells (A-498 and 786-O) and control cells (Figure 5D). We also found a significant difference in each of the cell cycle phases (G0/G1, S and G2/M) between miR-1826 transfectants and controls. Namely, miR-1826 induced a significant G1 arrest in A-498 renal cancer cells (Supplementary Figure S4 is available at Carcinogenesis Online).
Fig. 5.
Effect of microR-1826 overexpression on VHL-inactivated renal cancer cells (A-498 and 786-O). (A) Cell viability assay (miR-NC or miR-1826-transfected A-498 cells and 786-O cells), (B) Invasion assay, (C) Wound healing assay, (D) Flow cytometry analysis of apoptosis in miR-NC or miR-1826-transfected A-498 cells. Data are the mean ± SD of four independent experiments.
Effect of CTNNB1 and MEK1 mRNA knockdown on renal cancer cells
To look at whether miR-1826 exerts its tumor-suppressive function through CTNNB1 (beta-catenin) and MEK1 (MAP2K1), we knocked down CTNNB1 and MEK1 (MAP2K1) mRNAs using an siRNA technique. The knockdown effect was confirmed by measuring mRNA and protein expression levels. As shown in Supplementary Figure S5A, available at Carcinogenesis Online, the relative CTNNB1 and MEK1 protein expression levels were significantly decreased. Then, we examined MTS and invasion assays in si-NC, si-CTNNB1 and si-MEK1-transfected A-498 cells. As shown in Supplementary Figure S5B and C, available at Carcinogenesis Online, cell viability (days 3 and 4) and invasion (48 h) were significantly inhibited in both si-CTNNB1- and si-MEK1-transfected A-498 cells (48 h after transfection). Also, increased G1 phase arrest was inducted and apoptosis was promoted in si-CTNNB1- and si-MEK1-transfected A-498 renal cancer cells (Supplementary Figure S5D and E is available at Carcinogenesis Online).
Discussion
cc-RCC is the most common type in sporadic renal cancer and ∼60% of cc-RCC patients have mutated or inactivated VHL genes. Usually, pVHL inhibits the HIF-1 and beta-catenin signaling pathway. Thus, loss of pVHL results in the aberrant accumulation of cytoplasmic and nuclear beta-catenin and HIF, accelerating tumorigenesis (6,7).
CTNNB1 (beta-catenin) has been known as a key player in the Wnt/beta-catenin signaling pathway in several cancers including renal cancer (16). MEK1 (MAP2K1) acts as a mitogen-activated protein (MAP kinase kinase) and plays an important role as an essential component of the MAP kinase signal transduction pathway. MEK1 is also involved in several cellular processes, including proliferation and transcription regulation in several cancers including renal cancer. In the present study, we found that aberrant MEK1 expression in human renal cancer tissues was correlated with higher pathological stage (pTNM) and shorter overall survival and recurrence interval after radical nephrectomy. Kaplan–Meier plots also showed an inverse correlation between MEK1 expression and renal cancer patients’ outcomes. Several MEK1 inhibitors have been identified and have been evaluated in phases I, II and III clinical trials in several cancers (17). The effect of MEK1 inhibitors alone or in combination with others, such as mammalian target of rapamycin inhibitors or epidermal growth factor receptor inhibitors has been reported in the treatment of several cancers, including RCC (18,19). However, problems remain regarding the side effect and efficacy of these treatments. Thus, it is important to find new and safe options or approaches to achieve renal cancer remission. Therefore, we focused on miRNAs as a potential new treatment strategy. We initially searched for tumor-suppressive miRNAs inhibiting the two major cancer pathways, including beta-catenin and HIF-1 downstream genes (MEK1, MEK2, ERK1 and ERK2) with miRDB. This led to the identification of miR-1826 that only targets beta-catenin and MEK1.
Regarding the relationship between miRNA and CTNNB1, Xia et al. (20) has previously reported that miR-200a functions as a tumor suppressor by directly regulating CTNNB1 expression in nasopharyngeal carcinoma. Saydam et al. (21) found that miR-200a plays an important role as a tumor suppressor and directly targets CTNNB1 mRNA. They also showed that miR-200a blocks Wnt/beta-catenin signaling in meningioma cells (21). However, there have been no reports related to the role of miRNA and CTNNB1 in renal cancer.
Similarly, miRNAs studies involving MEK1 have found that miR-34a inhibits cell proliferation by repressing MEK1 during megakaryocytic differentiation (22). Also miR-424 regulates cell proliferation via the silencing of MEK1 and cyclin E1 in senile hemangioma (23). As far as we know, there have been no reports concerning the relationship of miRNA, MEK1 and renal cancer.
Only one miRNA-1826 was found to target both CTNNB1 and MEK1. This was also shown by 3′ UTR luciferase assays, indicating that relative luciferase levels were significantly lower in miR-1826-transfected renal cancer cells compared with miR-NC-transfected controls. Though mRNA expression of CTNNB1 and MEK1 was not changed in miR-1826-transfected cells (data not shown), the protein expression of CTNNB1 and MEK1 was significantly downregulated compared with miR-NC-transfected cells. Our results are consistent with the fact that miRNAs can bind to the 3′ UTR of target mRNA and repress translation from mRNA to protein (11). Thus, our results suggest that CTNNB1 and MEK1 are direct targets of miR-1826. We also performed an immunohistochemical study of CTNNB1 and MEK1 to examine the relationship between miR-1826 and CTNNB1 or MEK1 expression levels in human renal cancer tissues. We found an inverse correlation between miR1826 and CTNNB1 or MEK1 protein expression.
During renal cancer progression, beta-catenin (CTNNB1) is translocated into the nucleus, binds to T cell-factor/lymphoid-enhancer-factor transcriptional factors, activates target genes and thereby promotes tumorigenesis (8,24). Thus, we performed western analysis of survivin, a member of the T cell-factor/lymphoid-enhancer-factor downstream effectors, to assess whether miR-1826 inhibits downstream Wnt/beta-catenin signaling by beta-catenin downregulation. We found that expression of survivin was also downregulated by miR-1826 in transfected cells. Parker et al. (25) reported that high survivin expression is an independent predictor of cc-RCC progression and death from RCC. Moreover, several other laboratories have reported that the expression of survivin is associated with renal cancer aggressiveness and an important prognostic marker for renal cancer (26,27). Although other factors, such as insulin-like growth factor 1, interferon and nuclear factor-kappaB also regulate survivin protein expression (28,29), miR-1826 may be an additional important inhibitor of survivin in cc-RCC.
As a next step, we found that the expression of miR-1826 was significantly downregulated in renal cancer tissues compared with matched normal kidney tissues (n = 46). These results are consistent with those of miR-1826 expression in normal kidney and renal cancer cell lines, suggesting that miR-1826 may have tumor-suppressive functions in renal cancer. We did find a significantly shorter overall survival and earlier recurrence after radical nephrectomy in patients with ‘lower expression of miR-1826’. This result suggests that low miR-1826 expression in renal cancer tissues may contribute to poor patient prognosis. However, our sample number is relatively small, a larger study will be needed to look at the correlation between miR-1826 expression and clinical parameters. We also performed several functional assays using miR-1826 or miR-NC-transfected VHL-inactivated renal cancer cells (A-498 and 786-O). As expected, we found that overexpression of miR-1826 significantly inhibited renal cancer cell proliferation and also significantly inhibited renal cancer cell migration and invasion abilities. We also found that miR-1826 induced significant G1 cell cycle arrest and apoptosis in VHL-inactivated renal cancer cells. To validate whether miR-1826 plays a tumor-suppressive role by inhibiting CTNNB1 and MEK1 expression, we performed functional analyses of CTNNB1 and MEK1 in A-498 cells using a siRNA technique. The knockdown of CTNNB1 and MEK1 was confirmed at the mRNA (data not shown) and protein levels. We, then performed functional analyses (MTS and invasion analyses) and found an effect similar to that of miR-1826 overexpression. Namely, CTNNB1 or MEK1 knockdown resulted in inhibition of renal cancer cell proliferation and invasion ability. Taken together, this evidence suggests that miR-1826 exerts its tumor-suppressive effects through beta-catenin and MEK1 downregulation in renal cancer cells.
In conclusion, this is the first report documenting that miR-1826 expression is significantly decreased in human renal cancer tissues where it functions as a tumor suppressor by inhibiting CTNNB1 and MEK1 expression. Our results suggest that miR-1826 may play a therapeutically important role in renal cancer patients, especially in those with VHL-inactivated renal cancer.
Supplementary material
Supplementary Tables S1 and S2 and Figures S1–S5 can be found at http://carcin.oxfordjournals.org/.
Funding
This study was supported by grants (RO1CA130860, RO1CA160079, RO1CA138642, T32-DK07790, 1I01BX001123) from the National Institutes of Health, VA Research Enhancement Award Program (REAP), Merit Review grants and Yamada Science Foundation.
Supplementary Material
Acknowledgments
We thank Dr Roger Erickson for his support and assistance with the preparation of the manuscript.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- ATCC
American Type Culture Collection
- cc-RCC
clear cell renal cell carcinoma
- ERK
extracellular signal-regulated kinase
- HIF
hypoxia inducible factor
- miRDB
microRNA Target Prediction and Functional Study Database
- miRNA
microRNA
- mRNA
messenger RNA
- PCR
polymerase chain reaction
- RCC
renal cell carcinoma
- siRNA
small interfering RNAs
- UTR
untranslated region
- VHL
von Hippel-Lindau
References
- 1.Jemal A, et al. Cancer statistics, 2008. CA Cancer J. Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Bukowski RM. Natural history and therapy of metastatic renal cell carcinoma: the role of interleukin-2. Cancer. 1997;80:1198–1220. doi: 10.1002/(sici)1097-0142(19971001)80:7<1198::aid-cncr3>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 3.Linehan WM, et al. Molecular diagnosis and therapy of kidney cancer. Annu. Rev. Med. 2010;61:329–343. doi: 10.1146/annurev.med.042808.171650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh N, et al. Expression of multidrug resistance markers ABCB1 (MDR-1/P-gp) and ABCC1 (MRP-1) in renal cell carcinoma. BMC Urol. 2009;9:6. doi: 10.1186/1471-2490-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pirrotta MT, et al. Targeted-therapy in advanced renal cell carcinoma. Curr. Med. Chem. 2011;18:1651–1657. doi: 10.2174/092986711795471293. [DOI] [PubMed] [Google Scholar]
- 6.Linehan WM, et al. VHL loss of function and its impact on oncogenic signaling networks in clear cell renal cell carcinoma. Int. J. Biochem. Cell Biol. 2009;41:753–756. doi: 10.1016/j.biocel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linehan WM, et al. Identification of the genes for kidney cancer: opportunity for disease-specific targeted therapeutics. Clin. Cancer Res. 2007;15:671s–679s. doi: 10.1158/1078-0432.CCR-06-1870. [DOI] [PubMed] [Google Scholar]
- 8.Peruzzi B, et al. The von Hippel-Lindau tumor suppressor gene product represses oncogenic beta-catenin signaling in renal carcinoma cells. Proc. Natl Acad. Sci. USA. 2006;103:14531–14536. doi: 10.1073/pnas.0606850103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conrad PW, et al. EPAS1 trans-activation during hypoxia requires p42/p44 MAPK. J. Biol. Chem. 1999;274:33709–33713. doi: 10.1074/jbc.274.47.33709. [DOI] [PubMed] [Google Scholar]
- 10.Trang P, et al. MicroRNAs as potential cancer therapeutics. Oncogene. 2008;27:S52–S57. doi: 10.1038/onc.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inui M, et al. MicroRNA control of signal transduction. Nat. Rev. Mol. Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 12.Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–1017. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohno M, et al. Targeting the ERK signaling pathway in cancer therapy. Ann. Med. 2006;38:200–211. doi: 10.1080/07853890600551037. [DOI] [PubMed] [Google Scholar]
- 14.Yacoub A, et al. MEK1/2 inhibition promotes Taxotere lethality in mammary tumors in vivo. Cancer Biol. Ther. 2006;5:1332–1339. doi: 10.4161/cbt.5.10.3215. [DOI] [PubMed] [Google Scholar]
- 15.Hirata H, et al. DICKKOPF-4 activates the noncanonical c-Jun-NH2 kinase signaling pathway while inhibiting the Wnt-canonical pathway in human renal cell carcinoma. Cancer. 2011;117:1649–1660. doi: 10.1002/cncr.25666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barker N, et al. Mining the Wnt pathway for cancer therapeutics. Nat. Rev. Drug Discov. 2006;5:997–1014. doi: 10.1038/nrd2154. [DOI] [PubMed] [Google Scholar]
- 17.Yamaguchi T, et al. Antitumor activities of JTP-74057 (GSK1120212), a novel MEK1/2 inhibitor, on colorectal cancer cell lines in vitro and in vivo. Int. J. Oncol. 2011;39:23–31. doi: 10.3892/ijo.2011.1015. [DOI] [PubMed] [Google Scholar]
- 18.Costa LJ, et al. Upstream signaling inhibition enhances rapamycin effect on growth of kidney cancer cells. Urology. 2007;69:596–602. doi: 10.1016/j.urology.2007.01.053. [DOI] [PubMed] [Google Scholar]
- 19.Eulitt PJ, et al. Enhancing mda-7/IL-24 therapy in renal carcinoma cells by inhibiting multiple protective signaling pathways using sorafenib and by Ad.5/3 gene delivery. Cancer Biol. Ther. 2011;10:1290–1305. doi: 10.4161/cbt.10.12.13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia H, et al. miR-200a-mediated downregulation of ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma cell growth, migration and invasion. Biochem. Biophys. Res. Commun. 2010;391:535–541. doi: 10.1016/j.bbrc.2009.11.093. [DOI] [PubMed] [Google Scholar]
- 21.Saydam O, et al. Downregulated microRNA-200a in meningiomas promotes tumor growth by reducing E-cadherin and activating the Wnt/beta-catenin signaling pathway. Mol. Cell. Biol. 2009;29:5923–5940. doi: 10.1128/MCB.00332-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ichimura A, et al. MicroRNA-34a inhibits cell proliferation by repressing mitogen-activated protein kinase kinase 1 during megakaryocytic differentiation of K562 cells. Mol. Pharmacol. 2010;77:1016–1024. doi: 10.1124/mol.109.063321. [DOI] [PubMed] [Google Scholar]
- 23.Nakashima T, et al. Down-regulation of mir-424 contributes to the abnormal angiogenesis via MEK1 and cyclin E1 in senile hemangioma: its implications to therapy. PLoS One. 2010;5:e14334. doi: 10.1371/journal.pone.0014334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bilim V, et al. Altered expression of beta-catenin in renal cell cancer and transitional cell cancer with the absence of beta-catenin gene mutations. Clin. Cancer Res. 2000;6:460–466. [PubMed] [Google Scholar]
- 25.Parker AS, et al. High expression levels of survivin protein independently predict a poor outcome for patients who undergo surgery for clear cell renal cell carcinoma. Cancer. 2006;107:37–45. doi: 10.1002/cncr.21952. [DOI] [PubMed] [Google Scholar]
- 26.Crispen PL, et al. Predicting disease progression after nephrectomy for localized renal cell carcinoma: the utility of prognostic models and molecular biomarkers. Cancer. 2008;113:450–460. doi: 10.1002/cncr.23566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang GC, et al. Expression of cortactin and survivin in renal cell carcinoma associated with tumor aggressiveness. World J. Urol. 2009;27:557–563. doi: 10.1007/s00345-009-0376-2. [DOI] [PubMed] [Google Scholar]
- 28.Sato A, et al. Survivin associates with cell proliferation in renal cancer cells: regulation of survivin expression by insulin-like growth factor-1, interferon-gamma and a novel NF-kappaB inhibitor. Int. J. Oncol. 2006;28:841–846. [PubMed] [Google Scholar]
- 29.Griffith TS, et al. Induction and regulation of tumor necrosis factor-related apoptosis-inducing ligand/Apo-2 ligand-mediated apoptosis in renal cell carcinoma. Cancer Res. 2002;62:3093–3099. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.