Abstract
Monocytes in patients with systemic lupus erythematosus (SLE) are hyperstimulatory for T lymphocytes. We previously found that the normal program for expression of a negative costimulatory molecule programmed death ligand-1 (PD-L1) is defective in SLE patients with active disease. Here, we investigated the mechanism for PD-L1 dysregulation on lupus monocytes. We found that PD-L1 expression on cultured SLE monocytes correlated with TNF-α expression. Exogenous TNF-α restored PD-L1 expression on lupus monocytes. Conversely, TGF-β inversely correlated with PD-L1 in SLE and suppressed expression of PD-L1 on healthy monocytes. Therefore, PD-L1 expression in monocytes is regulated by opposing actions of TNF-α and TGF-β. As PD-L1 functions to fine tune lymphocyte activation, dysregulation of cytokines resulting in reduced expression could lead to loss of peripheral T cell tolerance.
Monocytes and dendritic cells from SLE patients display aberrant phenotypes, namely abnormal cytokine production, defective phagocytosis of apoptotic cells, and hyperstimulatory activity for allogeneic CD4+ T cells1,2,3. The mechanism which leads to dysregulated lymphocyte activation in SLE has not been elucidated, but most likely depends on differential levels of positive and negative costimulatory molecules expressed on antigen presenting cells (APCs)4,5. PD-L1 (B7-H1 or CD274) functions as a critical regulatory protein to maintain T cell self-tolerance6, and could play a major role in determining monocyte activity in SLE.
PD-L1, expressed on hematopoietic and parenchymal cells, binds to programmed death-1 (PD-1) to inhibit T cell receptor-mediated proliferation and induce T cell anergy6. Engagement of the PD-1 pathway is essential in suppressing autoimmunity, as originally demonstrated in mice lacking PD-1 expression that developed a disease similar to SLE7. Blockade of PD-1 has been shown to affect disease activity in a lupus mouse model8,9. PD-L1 deficiency does not by itself lead to SLE, but exacerbates systemic autoimmunity in lupus-prone mice6,10. The mechanistic link between PD-1 or PD-L1 expression and the pathogenesis of human SLE is not well understood. Polymorphisms in the PD-1 gene are associated with SLE susceptibility in some populations of adults and children11,12,13,14,15. However, PD-L1 gene polymorphisms were not associated with SLE16,17.
PD-L1 expression is low or absent on monocytes ex vivo18,19. When PBMC are cultured overnight in the absence of stimulation, PD-L1 expression is up-regulated on healthy monocytes and myeloid DCs. In contrast, we found a diminutive induction of PD-L1 on APCs from patients with active SLE. Cells from patients in remission regained PD-L1 expression, suggesting a reversible mechanism for regulation of PD-L119. The experimental culture system encompasses two abnormal processes that occur in SLE: the APC response to apoptotic cells and lymphocyte response to autologous antigens. The failure of APCs to up-regulate PD-L1 may occur in vivo and contribute to the breakdown of self-tolerance in SLE patients.
Cytokines abnormally expressed in SLE have been implicated in the regulation of PD-L118,20,21,22,23,24,25,26. Specifically, TNF-α has been associated with increased PD-L1 expression on synovial and peripheral macrophages derived from patients with rheumatoid arthritis24, whereas TGF-β inhibits PD-L1 expression on renal tubular cells21. Therefore, the reduced expression of PD-L1 on lupus monocytes may be the result of altered cytokine production ex vivo.
In this report we investigated the role of cytokines in regulating expression of PD-L1 in SLE. During active disease, the overexpression of TGF-β correlated with decreased levels of PD-L1 surface protein on lupus monocytes. Deficient PD-L1 expression could be restored in vitro by TNF-α, a factor required to induce PD-L1 expression on healthy monocytes. PD-L1 was not induced on isolated SLE monocytes or myeloid DCs, indicating that PD-L1 expression was not inhibited by SLE lymphocytes. TNF-α induced expression of PD-L1 mRNA in lupus cells, while TGF-β suppressed induction of the mRNA in healthy control cells, suggesting opposing transcriptional regulation by these two cytokines. These findings demonstrate that abnormal cytokine production may lead to poor PD-L1 expression on monocytes, contributing to the hyperstimulatory phenotype found in SLE.
Results
Aberrant expression of TNF-α and TGF-β correlates with PD-L1 levels on SLE monocytes
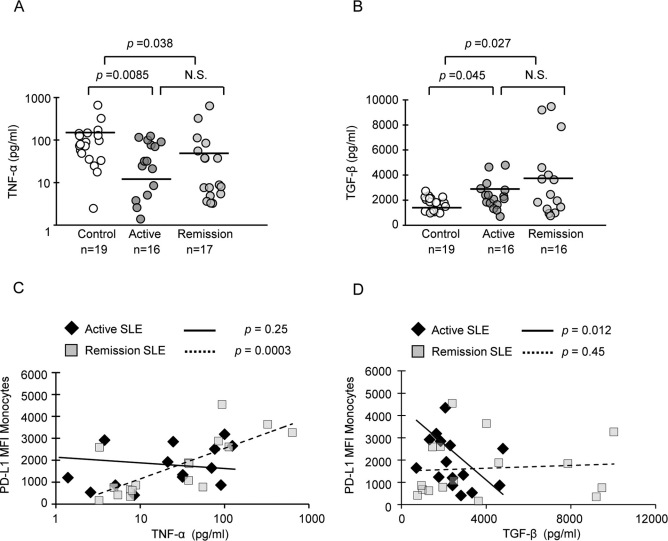
To investigate the mechanism for dysregulation of PD-L1 expression in SLE, PBMC from patients and healthy controls were cultured for 24 hours without stimulation. PD-L1 surface protein on monocytes was assayed by flow cytometry (Supplemental Figure 1). We observed no PD-L1 expression at the initiation of culture on either monocytes or myeloid DC from SLE patients or controls. PD-L1 protein was first detected by 8 hours, increased at 24 hours, and remained high until day five (Supplemental Figure 2). Deficient PD-L1 expression on cultured SLE monocytes during active disease was not related to specific medications, and is unlikely to be a gene defect, as most patients were able to restore PD-L1 protein during remission19. Hence we postulated that dysregulated cytokine production in SLE may lead to decreased expression of PD-L1 on SLE APCs. Supernatants from control and SLE PBMC cultures were assayed for expression of cytokines known to associate with the severity of SLE, including IFN-γ, IFN-α, IL-2, IL-4, IL-6, IL-8, IL-10, TNF-α, and TGF-β. The most significant differences between SLE patients and healthy controls were found in the expression of TNF-α and TGF-β. Interestingly, TNF-α, reported to be increased in SLE patient serum25, was expressed at 2.7-fold higher levels by PBMC from healthy controls compared to SLE patients in our experiments (mean, 129.4 pg ml−1 vs. 44.2 pg ml−1, Figure 1A). Though production of TNF-α was restored in some SLE patients during remission, the mean was still significantly reduced compared to controls (81.7 pg ml−1). In contrast, TGF-β was induced to higher levels in both active SLE (2393 pg ml−1) and remission (3914 pg ml−1) compared to controls (1684 pg ml−1, Figure 1B). Among other cytokines tested, IFN-γ, IL-4, IL-10, IFN-α and IL-2 were undetectable in supernatants from most subjects, an expected result considering that T lymphocytes were unstimulated. Moreover, expression of neither IL-6 nor IL-8 was significantly different in controls compared to SLE patients (data not shown).
Figure 1. PD-L1 levels correlated with TNF-α in remission and with TGF-β in patients with active disease.
(A) and (B) Levels of TNF-α and TGF-β detected in supernatants from PBMC cultured overnight without stimulation. Horizontal lines represent mean values. Cytokine levels between healthy controls and SLE patient groups were compared using the Wilcoxon-Mann-Whitney test. Significance was assigned where p<0.05; N.S., not significant. (C) PD-L1 protein levels on monocytes in the same culture was assayed by flow cytometry; TNF-α positively correlated with PD-L1 expression on monocytes in SLE patients in remission (dashed line). (D) TGF-β negatively correlated with PD-L1 protein in SLE patients with active disease (solid line). Correlations were analyzed by the Spearman's rank correlation test.
We then tested for correlations between TNF-α, TGF-β and PD-L1 expression. The expression of PD-L1 significantly correlated with TNF-α during disease remission (Figure 1C), suggesting that expression of TNF-α may be required to restore PD-L1 expression on lupus monocytes. In contrast, high expression of TGF-β was significantly correlated with low PD-L1 levels during active disease (Figure 1D). In PBMC from healthy controls, there was no correlation between cytokines and PD-L1 expression (data not shown), as most cells expressed PD-L1. Thus, correlation analyses suggested that TGF-β suppresses PD-L1 expression during active SLE, while TNF-α induces PD-L1 expression during remission and in healthy controls.
PD-L1 surface protein is induced by TNF-α and down-regulated by TGF-β
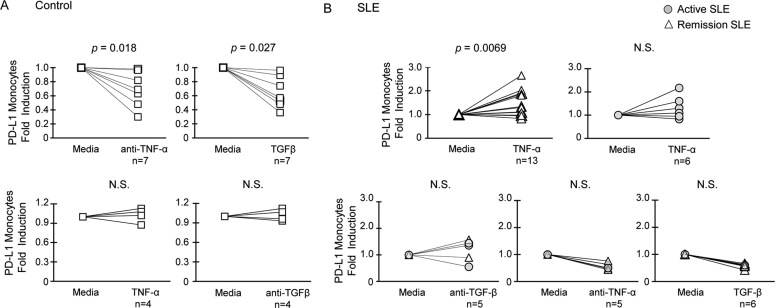
We have previously shown that cells from patients in remission restored PD-L1 expression, suggesting a reversible mechanism for regulation of PD-L1 during the disease course of SLE19. Therefore, we tested the hypothesis that expression of PD-L1 on SLE cells can be restored by TNF-α. To determine whether TNF-α is normally required for induction of PD-L1, we examined the change of PD-L1 expression on healthy cells treated with a TNF-α neutralizing antibody. Blocking TNF-α resulted in significantly decreased expression of PD-L1 surface protein on monocytes from healthy controls (Figure 2A). Likewise, treatment with TGF-β significantly suppressed PD-L1 surface protein on healthy monocytes. Addition of neither recombinant TNF-α nor TGF-β blocking antibody affected PD-L1 expression on healthy cells.
Figure 2. PD-L1 surface protein on SLE APCs is differentially regulated by TNF-α and TGF-β.
(A) Control PBMC were treated for 24 hours. Expression of PD-L1 on CD14highCD11c+ monocytes was assayed by flow cytometry, and is expressed as fold induction over untreated cells. (B) Induction of PD-L1 on monocytes in SLE PBMC treated for 24 hours or cultured in media alone. Induction of PD-L1 protein by cytokine treatments was tested for significance by the Wilcoxon signed-rank test. Results were derived from multiple independent experiments.
We found that TNF-α significantly induced PD-L1 protein expression on SLE remission monocytes (Figure 2B). However, induction of PD-L1 by TNF-α was not significant in patients with active disease. Of note, TNF-α treatment resulted in little induction of PD-L1 on myeloid DCs (data not shown), indicating that expression of PD-L1 is differentially regulated on monocytes and myeloid DCs. Blocking TGF-β with a mAb was not sufficient to restore PD-L1 on SLE monocytes (Figure 2B). The data suggests that restoration of PD-L1 expression on SLE monocytes may require TNF-α and other positive regulatory factors in addition to blocking inhibitory TGF-β. TNF-α is normally required for PD-L1 induction, whereas TGF-β may oppose the induction in active SLE. As SLE monocytes express low levels of PD-L1, inhibition of TNF-α by mAb or addition of recombinant TGF-β was not able to reduce the protein expression further (Figure 2B).
PD-L1 expression is induced by monocyte-derived factors
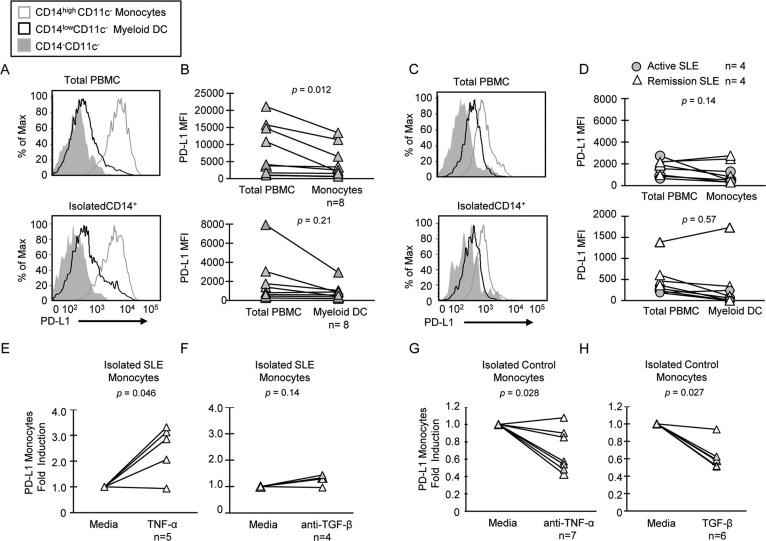
Cytokines that dysregulate expression of PD-L1 can be derived from either aberrant SLE myeloid or lymphoid cells2,27. To identify other cell populations affecting PD-L1 expression on monocytes, we compared the expression of PD-L1 on isolated CD14+ cells cultured alone with that on monocytes in total PBMC cultures. No expression of PD-L1 was detected on isolated monocytes prior to culture at time 0 (Supplemental Figure 2). PD-L1 was induced on isolated healthy CD14+ monocytes cultured overnight without stimulation, indicating that monocyte-intrinsic factors are sufficient for induction of surface PD-L1 (Figure 3A and B). However, the depletion of CD14− cells decreased expression of PD-L1 on myeloid DCs and monocytes from healthy donors that had higher PD-L1 expression (Figure 3B), suggesting that induction of PD-L1 could be enhanced by lymphocytes. Both regulatory T cells and NKT cells have been previously reported to induce PD-L1 expression, and may play roles in defective PD-L1 induction in SLE28,29. There was no change in the level of PD-L1 expression on either isolated SLE monocytes or myeloid DCs as compared to total PBMCs (Figure 3C and D), demonstrating that PD-L1 expression is not suppressed by SLE lymphocytes. Rather, SLE APCs lack intrinsic factors required for PD-L1 induction.
Figure 3. Expression of PD-L1 on isolated CD14+ cells is not entirely dependent on lymphocytes.
(A) and (C) Representative histograms demonstrate PD-L1 induction on total PBMC or isolated CD14+ cells from a control subject and an active SLE patient. Protein expression was assayed by flow cytometry after culturing cells for 24 hours without stimulation. (B) and (D) PD-L1 expression on monocytes gated from total PBMC compared to isolated monocytes and myeloid DCs in eight healthy controls and eight SLE patients. (E) and (F) Fold induction of PD-L1 MFI on isolated CD14+ SLE monocytes treated with TNF-α or with anti-TGF-β mAb. (G) and (H) Fold induction of PD-L1 MFI on isolated CD14+ monocytes from healthy donors treated with anti-TNF-α mAb or with recombinant TGF-β. Induction was quantified by dividing the MFI of cytokine-treated samples by that of untreated cells, and tested for significance by the Wilcoxon signed-rank test.
We next tested the direct effect of cytokines on isolated CD14+ monocytes and myeloid DCs. The induction of PD-L1 expression on monocytes after treatment of total PBMCs with TNF-α that we showed in Figure 2 was similarly demonstrated in isolated SLE monocytes, where expression of PD-L1 was significantly induced in four out of five patients (Figure 3E). Blocking TGF-β did not changed expression of PD-L1 protein on isolated SLE monocytes (Figure 3F), whereas blocking TNF-α significantly decreased PD-L1 expression on isolated healthy monocytes (Figure 3G). Moreover, TGF-β treatment of isolated monocytes resulted in significant repression of PD-L1 in healthy donors (Figure 3H). These findings support the hypothesis that intrinsic defects in SLE monocyte cytokine expression result in poor PD-L1 induction.
Expression of PD-L1 correlated with TNF-α in monocyte culture supernatants
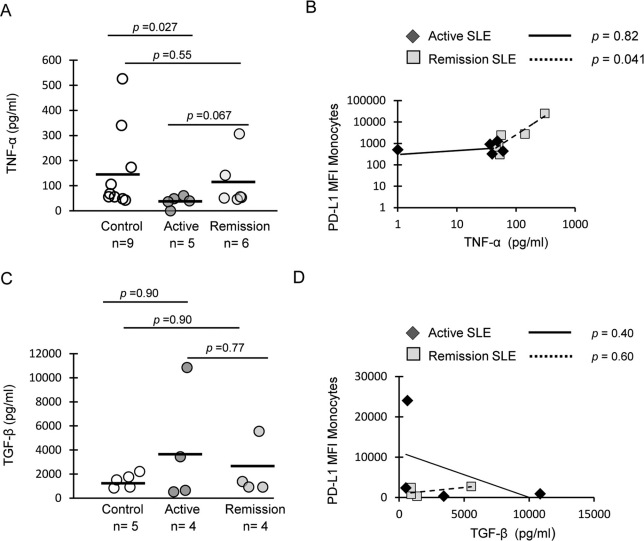
In order to determine whether or not PD-L1 is induced by monocyte-intrinsic factors, we assayed levels of TNF-α and TGF-β in purified monocyte culture supernatants. Increased TNF-α was detected in supernatants from healthy monocytes (mean, 157 pg/ml) compared to those from patients with active disease (Figure 4A, p = 0.027). TNF-α expression was higher in SLE remission (109 pg/ml) compared to active disease (37 pg/ml), but the difference was not statistically significant (p = 0.067). Expression of PD-L1 on isolated monocytes from remission patients also significantly correlated with TNF-α (Figure 4B). These data provide evidence that monocytes produce TNF-α required to induce PD-L1. Conversely, substantial expression of TGF-β was detected in monocyte supernatants (Figure 4C). Isolated monocytes from SLE patients produce higher level of TGF-β compared to healthy controls (active disease = 3527 pg/ml; remission = 2189 pg/ml; healthy controls 5 1446 pg/ml), but the difference was insignificant. No correlation was found between PD-L1 protein level and TGF-β (Figure 4D). These data suggest that cell types other than monocytes can also produce TGF-β to counter-regulate level of PD-L1 on monocytes.
Figure 4. Monocytes produce TNF-α to induce expression of PD-L1.
(A) TNF-α was assayed in supernatants from isolated CD14+ monocytes cultured overnight without stimulation. Horizontal lines represent mean values. (B) PD-L1 protein levels on monocytes in the same culture was assayed by flow cytometry; TNF-α positively correlated with PD-L1 expression on monocytes in SLE patients in remission. (C) TGF-β protein levels in monocyte supernatants. (D) Correlation between TGF-β and PD-L1 protein levels on monocytes. Cytokine levels between healthy controls and SLE patient groups were compared using the Wilcoxon-Mann-Whitney test. Correlations were determined by the Spearman's rank correlation test.
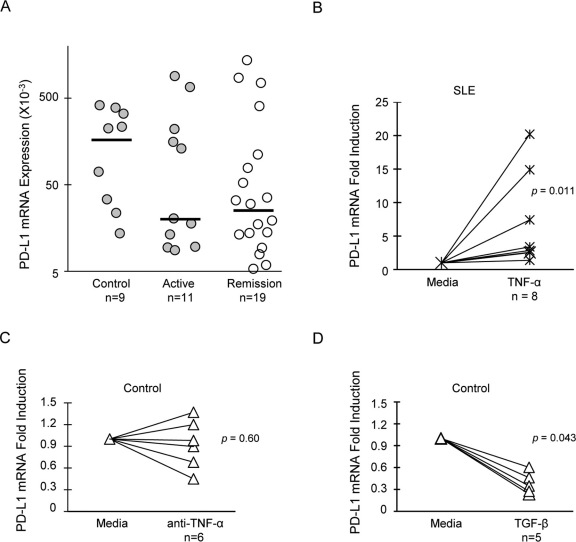
Opposing effects of TNF-α and TGF-β on PD-L1 mRNA expression
PD-L1 expression has been reported to be regulated at both transcriptional and translational levels in different model systems30,31,32. PD-L1 mRNA expression was assayed in cultured PBMC from SLE patients and healthy donors. We found that PD-L1 mRNA was expressed in cells from all control subjects (median ± S.E.M. = 0.22 ± 0.16, Figure 5A), and was on average ten-fold higher than in SLE patients (0.018 ± 0.09 for active disease, 0.024 ± 0.08 for remission), although the differences were not significant. Parallel flow cytometry assays demonstrated that some patients with active disease and high PD-L1 mRNA expression had little to no PD-L1 protein on the surface of monocytes, myeloid DCs, or lymphocytes. Moreover, there was no correlation between PD-L1 mRNA and protein expression in any subject group (data not shown), indicating that restoration of PD-L1 surface protein was not entirely dependent upon mRNA expression, but may require additional translational signals, as demonstrated in other biological models30,31.
Figure 5. PD-L1 mRNA is induced by TNF-α and suppressed by TGF-β.
Total PBMC were cultured for 24 hours. PD-L1 mRNA expression was assayed by RT-qPCR for each subject in triplicate. (A) Relative PD-L1 mRNA expression in controls and lupus patients was represented by the scatter plot. Horizontal lines represent medians. Baseline mRNA expression was set at 5×10−3. Expression of PD-L1 mRNA in control subjects and SLE patients were compared using the Wilcoxon-Mann-Whitney test. P>0.05 between all groups. (B) PD-L1 mRNA in SLE PBMC treated with recombinant TNF-α. (C) and (D) PD-L1 mRNA levels in healthy donor PBMC treated with anti-TNF-α or TGF-β. Induction of PD-L1 mRNA by cytokines was tested for significance by the Wilcoxon signed-rank test.
The signaling pathways mediated by TNF-α and TGF-β are well documented33,34. However, it is unclear how these distinct pathways converge to counter-regulate PD-L1 expression. We next demonstrated that PD-L1 mRNA is differentially regulated by TGF-β and TNF-α. PD-L1 mRNA was significantly induced in SLE cells treated with TNF-α (Figure 5B), whereas blocking TNF-α prevented induction of PD-L1 mRNA in three of six healthy donors (Figure 5C). Similarly, TGF-β significantly inhibited expression of PD-L1 mRNA in control PBMC (Figure 5D). Overall, these data support a model in which PD-L1 gene expression is tightly regulated by the action of two opposing cytokines, TNF-α and TGF-β.
Discussion
Although functional defects in monocytes and DCs are well known in SLE, the underlying molecular mechanisms are not fully understood. The present study addresses the mechanism for regulating PD-L1, a critical immunoregulatory protein that determines the pro- versus anti-inflammatory activity of APCs. PD-L1 expression is regulated by a balance of cytokines with known importance in SLE21,35. In other inflammatory diseases including rheumatoid arthritis, infection, and malignancies, PD-L1 expression has been reported to be up-regulated by inflammatory cytokines36,37. In contrast, we previously demonstrated deficient PD-L1 surface expression on monocytes and myeloid DCs with active SLE19. We have shown here that the defect on SLE monocytes can be attributed partially to overexpression of TGF-β, which inhibits PD-L1 expression, and partly to decreased production of TNF-α required for induction of PD-L1 mRNA and surface protein expression.
In the current study, some patients with active SLE produced less TNF-α, a trait reported to correlate with SLE-associated HLA-DR alleles38,39,40. Whether children with SLE expressing less TNF-α in correlation with decreased PD-L1 expression on monocytes carry the polymorphisms leading to low TNF-α expression is not known. Expression of PD-L1 may also be influenced by the availability of receptors and inducible signaling molecules and transcription factors in the TNF receptor signaling pathway. TNF receptor expression is reportedly normal in SLE, though decreased expression of TNF-α signaling proteins in SLE patients has been reported41,42, and could account for the decreased PD-L1 induction observed in unresponsive SLE monocytes from some patients, although most patient monocytes respond well to TNF-α. Anti-TNF-α therapy is effective in treating rheumatoid arthritis, auto-inflammatory diseases, and some SLE patients. However, TNF-α blockers have been shown to trigger lupus in some patients, and induce lupus-related autoantibody production. The onset of autoimmunity triggered by TNF-α blockers may be in part through activation of plasmacytoid dendritic cells, which lead to enhanced production of IFN-α43. Our data suggests that lack of normal TNF-α induction of PD-L1 expression could also contribute to disease. These studies underline the complex role of TNF-α in autoimmunity, with different effects at distinct cellular targets.
The role of TGF-β in the pathogenesis of SLE remains unclear. Previous studies have shown decreased TGF-β production by SLE lymphocytes44, in contrast to our increase in TGF-β production by SLE PBMC with concurrent suppression of PD-L1. However, TGF-β therapies in lupus-prone mice have yielded mixed results45, perhaps because TGF-β contributes to the development of both proinflammatory Th17 cells and regulatory T cells (Treg)46. TGF-β is essential for differentiation of murine Treg, but its role in human Treg is less well defined. PD-L1 has been shown to induce Treg in mice and humans28,47, suggesting defective induction of PD-L1 may lead to reduced number of functional Treg observed in SLE patients48. TGF-β receptor mRNA expression has been reported to be increased in SLE49, though whether or not this leads to increased surface protein expression and higher sensitivity to TGF-β signaling is not known.
TGF-β inhibition of PD-L1 expression on monocytes is consistent with a previous finding that TGF-β suppresses PD-L1 expression on murine renal proximal tubular epithelial cells, allowing the activation of CD8+ cytotoxic T cells21. Our data suggest that overexpression of TGF-β during active SLE can suppress PD-L1 by inhibiting protein and mRNA expression. It has been shown that TGF-β can inhibit production of TNF-α in mouse macrophages50, but further work is required to determine whether TGF-β suppresses PD-L1 expression directly, or through inhibition of TNF-α. TGF-β and TNF-α could directly counter-regulate gene expression, as occurs in mucosal epithelial cells, where TGF-β inhibits recruitment of the transcription factor NFκB to the IL-6 promoter51.
Our results differed from previous findings, which have shown increased levels of TNF-α and diminished expression of TGF-β in SLE patients53. In our experimental system, levels of cytokines were measured in culture supernatants of unstimulated cells, whereas others reported cytokine levels in serum or using a reporter assay44,52. Another group measured expression of TNF-α and TGF-β in culture supernatants stimulated by adding apoptotic cells53. Our model system may measure in part response to apoptotic cells; more apoptotic cells were found in SLE PBMC cultures compared to healthy cells. However, the number of apoptotic cells did not correlate with PD-L1 expression, and preliminary experiments showed no effect of additional apoptotic cells on PD-L1 expression on healthy monocytes. Thus, SLE monocytes may be resistant to apoptotic cell signals. Subject selection could also influence reported cytokine profiles, though neither age nor immunosuppressive medication significantly affected cytokine or PD-L1 expression in pediatric SLE patients in our cohort19.
While control monocytes expressed PD-L1 in the absence of other cell types, our results did not rule out the possibility that lymphocytes normally contribute to expression of PD-L1 on APCs. We noted decreased PD-L1 expression on cultured isolated myeloid DCs and monocytes in some control subjects and patients. Interestingly, we found that PD-L1 expression on SLE monocytes and myeloid DCs could be partially restored by co-culturing with allogenic CD4+ T cells from healthy donors (preliminary data). SLE T cells present multiple signaling aberrations27, and are reduced in number of functional Treg48, which could be required for induction of PD-L1 on APCs28. It remains to be determined whether correcting SLE T cell defects can alter expression of PD-L1 on APCs.
Although most SLE patients with active disease expressed PD-L1 mRNA, surface protein expression was generally low or undetectable. The lack of correlation between mRNA and protein suggests that additional translational mechanisms may be required for effective induction of PD-L1 protein on APCs. We hypothesized that PD-L1 could be cleaved from the cell surface. However, soluble PD-L1 was undetectable in culture supernatants and in SLE plasma when assayed with a sensitivity of 60 pg/ml. Post-transcriptional and translational regulation of PD-L1 has been shown in different biological models. In human trophoblast cells, epidermal growth factor increases recruitment of PD-L1 transcripts to active ribosomes31, and IFN-γ induces PD-L1 protein expression by inhibiting miRNA in cholangiocytes30. We are currently investigating whether additional translational regulatory mechanisms are involved in determining expression of PD-L1 surface protein on APCs.
We cannot rule out a role for additional cytokines in PD-L1 regulation in SLE. Type I and II IFNs are critical mediators of SLE1, and known inducers of PD-L1 expression20,22. However, neither IFN-γ nor IFN-α were detected in unstimulated PBMC cultures in our experiments or by others54, and thus are unlikely to be involved in the regulation of PD-L1 in our model system. IL-10 signaling through STAT3 has been shown to up-regulate PD-L1 protein in dendritic cells55,56, but in our experiments IL-10 was only detected at minimal levels in some unstimulated PBMC cultures (data not shown). IL-6 also signals through STAT3 to induce PD-L1 expression56. While IL-6 was detected in SLE culture supernatants (data not shown), the direct role of IL-6 in the regulation of PD-L1 in SLE has yet to be explored. Deficient PD-L1 expression on SLE APCs may result from the cytokine environment in vivo. Therefore, we assayed levels of 16 plasma cytokines in 66 SLE patients in remission, 48 patients with active disease, and 65 healthy donors. We found no correlation between plasma cytokine levels and PD-L1 expression. These data do not exclude the possibility that cytokines may have an effect on phenotypes of APCs in lymphoid tissues, which are inaccessible for human studies.
In the current study, we identified TNF-α and TGF-β as counter-regulators of PD-L1 expression. Monocytes are programmed to respond in autologous culture to generate intrinsic factors required to stimulate expression of PD-L1, and the reduced expression of PD-L1 on SLE monocytes was not dependent on other cell types. The lack of proper induction of PD-L1 may contribute to the hyperstimulatory phenotype of SLE APCs, resulting in reduced peripheral T cell tolerance. Better characterization of mediators regulating PD-L1 may lead to promising new therapeutic targets aimed at restoring PD-L1 expression in SLE patients.
Methods
Human subjects and blood samples
The research protocol was approved by the Institutional Review Board of Seattle Children's Hospital. Pediatric SLE patients were recruited in the Seattle Children's Hospital Rheumatology Clinic; age-matched healthy pediatric volunteers were recruited through an ongoing project to study pediatric autoimmune diseases. Informed assent and consent were obtained. All lupus patients fulfilled the current American College of Rheumatology (ACR) classification criteria for SLE diagnosis57. Subjects were excluded for infections, malignancy, or other autoimmune diseases that may affect PD-L1 expression. Peripheral venous blood was collected into Cell Preparation Tubes (CPT, BD Biosciences, San Jose, CA). PBMC were isolated and frozen in 7% DMSO (Sigma-Aldrich, St-Louis, MO). SLE scores were determined by the SLE disease activity index (SLEDAI)58. Active disease was defined by a SLEDAI >4 to exclude patients with only laboratory abnormalities.
Cell culture and flow cytometry
PBMC were thawed, washed and diluted to 106 cells ml−1 in culture medium consisting of RPMI 1640 supplemented with L-glutamine (CellGro, Manassas, VA), and 10% heat-inactivated human AB serum (Valley Biomedical, Winchester, VA), 1% penicillin/streptomycin (CellGro) and 0.1% β-mercaptoethanol. All assays were performed using frozen PBMC samples, as we previously determined that freezing does not influence PD-L1 expression. Cells were plated in round-bottom 96-well plates, and cultured for 24 hours without stimulation. Between 60–85% of total PBMC were recovered after overnight culturing. PBMC were surface-stained using fluorochrome-conjugated mAb, including: anti-CD3 (UCHT1), anti-PD-L1 (MIH1) (eBioscience, San Diego, CA), anti-CD11c (B-ly6), and anti-CD14 (MjP9) (BD Biosciences), with isotype-matched, fluorochrome-labeled antibodies as controls. All samples were blocked using 0.5% human AB serum and anti-FcR antibody (Miltenyi, Bergisch GladbachGermany) prior and during staining. Flow cytometry was performed using an LSR II cytometer (BD Biosciences), and the data analyzed using FlowJo software (Tree Star, Ashland, OR). Examination of surface markers on normal PBMC determined that the PD-L1+ cells segregated into monocyte (CD14highCD11c+) and immature myeloid DC (CD14lowCD11c+) populations (Supplemental Figure 1)19,59. A similar expression profile of PD-L1 was obtained when cells were grown on standard tissue culture plates compared to polystyrene-treated plates. For cytokine experiments, PBMC were cultured for 24 hours in the culture medium, or with 10 ng ml-1 of recombinant cytokines TNF-α (eBioscience) or TGF-β (R&D systems). In some experiments, cells were treated with 10 μg ml−1 of mAb neutralizing TNF-α or TGF-β (R&D systems). CD14+ monocytes and myeloid DCs were sorted by negative selection using the EasySep human monocyte enrichment kit (StemCell Technologies, Inc., Vancouver, BC, Canada), with purity >90%. Endotoxin levels in PBMC and isolated monocytes were determined to be <1 EU ml−1 by Limulus amoebocyte lysate clot assay (Associates of Cape Cod, East Falmouth, MA). Expression of PD-L1 on monocytes and myeloid DC was determined by subtracting the background PD-L1 mean fluorescence intensity (MFI) of the CD3−CD14−CD11c− subset, which was consistently negative for PD-L1 expression, similar to the isotype controls.
Cytokine analysis
All cytokine assays were performed on culture supernatants (24 hours PBMC culture) by the Cytokine Analysis Laboratory at the Fred Hutchinson Cancer Research Center. Cytokines were measured by the Luminex assay system with a latex bead-based multianalyte system (Luminex, Austin, TX). TGF-β was assayed by ELISA (R&D Systems).
Real-time reverse transcriptase-qPCR (RT-qPCR)
Total RNA was extracted from PBMC using the RNeasy Plus Micro Kit (Qiagen, Germantown, MD). Real-time RT-qPCR analysis was performed using a Bio-Rad iCycler, in 25-μl reaction mixtures with SYBR green PCR master mix (SABiosciences, Frederick, MD). The primers used for amplifying PD-L1 mRNA (Gene Bank accession number: NM_014143) were sense: 5′-GGCATTTGCTGAACGCAT-3′; antisense: 5′-CAATTAGTGCAGCCAGGT-3′. For GAPDH, the primers were sense: 5′-TGCACCACCAACTGCTTA-3′; antisense: 5′-GGATGCAGGGATGATGTTC-3′. Standard curves were determined for each primer set by serial dilution. Expression of each cDNA was calculated from the cycle threshold (CT). The relative PD-L1 mRNA expression was determined by normalizing to GAPDH mRNA, and dividing by the percentage of monocytes in each sample, where PD-L1 is predominantly expressed.
Statistical analyses
STATA statistical software was used to perform all analyses (Stata Corporation, College Station, TX).
Author Contributions
JNO and AMS performed study design, contributed to data analyses and wrote the manuscript. JNO performed experiments; AEW participated in study design and reviewed the manuscript.
Supplementary Material
Supplementary materials
Acknowledgments
This project was supported by grants from the Lupus Research Institute, and Lupus Foundation of America to A.M.S., and the Thrasher Research Foundation (award number: NR-0109) to J-N.O. Support was derived in part by Clinical and Translational Science Award (CTSA) (Grant Number: I ULI RR025014-02) from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH. We thank Dr. Veronika Groh-Spies, Dr. Keith Elkon and Dr. Jeffrey Ledbetter for helpful discussion and critical review of the manuscript, Kristy Seidel and Do Peterson for statistical analyses, Matthew Crabtree and Michelle Stanley for technical assistance, Elizabeth Ocheltree and Dr. Rebecca Howsmon for proofreading of the manuscript and Gretchen Henstorf for collection of clinical samples.
References
- Blanco P., Palucka A. K., Gill M., Pascual V. & Banchereau J. Induction of dendritic cell differentiation by IFN-alpha in systemic lupus erythematosus. Science 294, 1540–1543 (2001). [DOI] [PubMed] [Google Scholar]
- Katsiari C. G., Liossis S. N. & Sfikakis P. P. The Pathophysiologic Role of Monocytes and Macrophages in Systemic Lupus Erythematosus: A Reappraisal. Semin Arthritis Rheum 39, 491–503 (2009). [DOI] [PubMed] [Google Scholar]
- Zhu J. et al. T cell hyperactivity in lupus as a consequence of hyperstimulatory antigen-presenting cells. J Clin Invest 115, 1869–1878 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D., Mehta H., McCune W. J. & Kaplan M. J. Aberrant phenotype and function of myeloid dendritic cells in systemic lupus erythematosus. J Immunol 177, 5878–5889 (2006). [DOI] [PubMed] [Google Scholar]
- Monrad S. & Kaplan M. J. Dendritic cells and the immunopathogenesis of systemic lupus erythematosus. Immunol Res 37, 135–145 (2007). [DOI] [PubMed] [Google Scholar]
- Keir M. E., Butte M. J., Freeman G. J. & Sharpe A. H. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol 26, 677–704 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura H., Nose M., Hiai H., Minato N. & Honjo T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity 11, 141–151 (1999). [DOI] [PubMed] [Google Scholar]
- Kasagi S. et al. Anti-programmed cell death 1 antibody reduces CD4+PD-1+ T cells and relieves the lupus-like nephritis of NZB/W F1 mice. J Immunol 184, 2337–2347 (2010). [DOI] [PubMed] [Google Scholar]
- Wong M., La Cava A., Singh R. P. & Hahn B. H. Blockade of programmed death-1 in young (New Zealand black x New Zealand white)F1 mice promotes the activity of suppressive CD8+ T cells that protect from lupus-like disease. J Immunol 185, 6563–6571 (2010). [DOI] [PubMed] [Google Scholar]
- Lucas J. A. et al. Programmed death ligand 1 regulates a critical checkpoint for autoimmune myocarditis and pneumonitis in MRL mice. J Immunol 181, 2513–2521 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertsias G. K. et al. Genetic, immunologic, and immunohistochemical analysis of the programmed death 1/programmed death ligand 1 pathway in human systemic lupus erythematosus. Arthritis Rheum 60, 207–218 (2009). [DOI] [PubMed] [Google Scholar]
- Thorburn C. M. et al. Association of PDCD1 genetic variation with risk and clinical manifestations of systemic lupus erythematosus in a multiethnic cohort. Genes Immun 8, 279–287 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Cruz R. et al. Association of PDCD1 polymorphisms with childhood-onset systemic lupus erythematosus. Eur J Hum Genet 15, 336–341 (2007). [DOI] [PubMed] [Google Scholar]
- Lee Y. H., Woo J. H., Choi S. J., Ji J. D. & Song G. G. Association of programmed cell death 1 polymorphisms and systemic lupus erythematosus: a meta-analysis. Lupus 18, 9–15 (2009). [DOI] [PubMed] [Google Scholar]
- Suarez-Gestal M., Ferreiros-Vidal I., Ortiz J. A., Gomez-Reino J. J. & Gonzalez A. Analysis of the functional relevance of a putative regulatory SNP of PDCD1, PD1.3, associated with systemic lupus erythematosus. Genes Immun 9, 309–315 (2008). [DOI] [PubMed] [Google Scholar]
- Abelson A. K. et al. No evidence of association between genetic variants of the PDCD1 ligands and SLE. Genes Immun 8, 69–74 (2007). [DOI] [PubMed] [Google Scholar]
- Wang S. C. et al. Ligands for programmed cell death 1 gene in patients with systemic lupus erythematosus. J Rheumatol 34, 721–725 (2007). [PubMed] [Google Scholar]
- Brown J. A. et al. Blockade of programmed death-1 ligands on dendritic cells enhances T cell activation and cytokine production. J Immunol 170, 1257–1266 (2003). [DOI] [PubMed] [Google Scholar]
- Mozaffarian N., Wiedeman A. E. & Stevens A. M. Active systemic lupus erythematosus is associated with failure of antigen-presenting cells to express programmed death ligand-1. Rheumatology (Oxford) 47, 1335–1341 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. et al. Expression of B7-H1 in inflammatory renal tubular epithelial cells. Nephron Exp Nephrol 102, e81–92 (2006). [DOI] [PubMed] [Google Scholar]
- Starke A., Wuthrich R. P. & Waeckerle-Men Y. TGF-beta treatment modulates PD-L1 and CD40 expression in proximal renal tubular epithelial cells and enhances CD8 cytotoxic T-cell responses. Nephron Exp Nephrol 107, e22–29 (2007). [DOI] [PubMed] [Google Scholar]
- Waeckerle-Men Y., Starke A. & Wuthrich R. P. PD-L1 partially protects renal tubular epithelial cells from the attack of CD8+ cytotoxic T cells. Nephrol Dial Transplant 22, 1527–1536 (2007). [DOI] [PubMed] [Google Scholar]
- Schreiner B. et al. Interferon-beta enhances monocyte and dendritic cell expression of B7-H1 (PD-L1), a strong inhibitor of autologous T-cell activation: relevance for the immune modulatory effect in multiple sclerosis. J Neuroimmunol 155, 172–182 (2004). [DOI] [PubMed] [Google Scholar]
- Wan B. et al. Aberrant regulation of synovial T cell activation by soluble costimulatory molecules in rheumatoid arthritis. J Immunol 177, 8844–8850 (2006). [DOI] [PubMed] [Google Scholar]
- O'Shea J. J., Ma A. & Lipsky P. Cytokines and autoimmunity. Nat Rev Immunol 2, 37–45 (2002). [DOI] [PubMed] [Google Scholar]
- Kinter A. L. et al. The common gamma-chain cytokines IL-2, IL-7, IL-15, and IL-21 induce the expression of programmed death-1 and its ligands. J Immunol 181, 6738–6746 (2008). [DOI] [PubMed] [Google Scholar]
- Crispin J. C., Kyttaris V. C., Terhorst C. & Tsokos G. C. T cells as therapeutic targets in SLE. Nat Rev Rheumatol 6, 317–325 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarnath S. et al. Regulatory T cells and human myeloid dendritic cells promote tolerance via programmed death ligand-1. PLoS Biol 8, e1000302 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde S. et al. Human NKT cells direct the differentiation of myeloid APCs that regulate T cell responses via expression of programmed cell death ligands. J Autoimmun 37, 28–38 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong A. Y. et al. MicroRNA-513 regulates B7-H1 translation and is involved in IFN-gamma-induced B7-H1 expression in cholangiocytes. J Immunol 182, 1325–1333 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holets L. M., Carletti M. Z., Kshirsagar S. K., Christenson L. K. & Petroff M. G. Differentiation-induced post-transcriptional control of B7-H1 in human trophoblast cells. Placenta 30, 48–55 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzec M. et al. Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD-L1, B7-H1). Proc Natl Acad Sci U S A 105, 20852–20857 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parameswaran N. & Patial S. Tumor necrosis factor-alpha signaling in macrophages. Crit Rev Eukaryot Gene Expr 20, 87–103 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. & Gomis R. R. The logic of TGFbeta signaling. FEBS Lett 580, 2811–2820 (2006). [DOI] [PubMed] [Google Scholar]
- Ronnblom L. & Elkon K. B. Cytokines as therapeutic targets in SLE. Nat Rev Rheumatol 6, 339–347 (2010). [DOI] [PubMed] [Google Scholar]
- Brown K. E., Freeman G. J., Wherry E. J. & Sharpe A. H. Role of PD-1 in regulating acute infections. Curr Opin Immunol 22, 397–401 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H. & Chen X. Immunoregulatory role of B7-H1 in chronicity of inflammatory responses. Cell Mol Immunol 3, 179–187 (2006). [PMC free article] [PubMed] [Google Scholar]
- Atsumi T. [Tumor necrosis factor alpha in systemic lupus erythematosus: evaluation by restriction fragment length polymorphism and production by peripheral blood mononuclear cells]. Hokkaido Igaku Zasshi 67, 408–419 (1992). [PubMed] [Google Scholar]
- Jacob C. O. et al. Heritable major histocompatibility complex class II-associated differences in production of tumor necrosis factor alpha: relevance to genetic predisposition to systemic lupus erythematosus. Proc Natl Acad Sci U S A 87, 1233–1237 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh N. J., Owen P., Cox B., Dunphy J. & Welsh K. MHC class II, tumour necrosis factor alpha, and lymphotoxin alpha gene haplotype associations with serological subsets of systemic lupus erythematosus. Ann Rheum Dis 65, 488–494 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L. et al. Decreased expressions of the TNF-alpha signaling adapters in peripheral blood mononuclear cells (PBMCs) are correlated with disease activity in patients with systemic lupus erythematosus. Clin Rheumatol 26, 1481–1489 (2007). [DOI] [PubMed] [Google Scholar]
- Zhu L. J. et al. Altered expression of TNF-alpha signaling pathway proteins in systemic lupus erythematosus. J Rheumatol 37, 1658–1666 (2010). [DOI] [PubMed] [Google Scholar]
- Palucka A. K., Blanck J. P., Bennett L., Pascual V. & Banchereau J. Cross-regulation of TNF and IFN-alpha in autoimmune diseases. Proc Natl Acad Sci U S A 102, 3372–3377 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka K., Gray J. D., Stimmler M. M., Toro B. & Horwitz D. A. Decreased production of TGF-beta by lymphocytes from patients with systemic lupus erythematosus. J Immunol 160, 2539–2545 (1998). [PubMed] [Google Scholar]
- Saxena V. et al. Dual roles of immunoregulatory cytokine TGF-beta in the pathogenesis of autoimmunity-mediated organ damage. J Immunol 180, 1903–1912 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoreschi K., Laurence A., Yang X. P., Hirahara K. & O'Shea J. J. T helper 17 cell heterogeneity and pathogenicity in autoimmune disease. Trends Immunol, 32, 395–401 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco L. M. et al. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J Exp Med 206, 3015–3029 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Cava A. T-regulatory cells in systemic lupus erythematosus. Lupus 17, 421–425 (2008). [DOI] [PubMed] [Google Scholar]
- Hrycek A., Kusmierz D., Dybala T. & Swiatkowska L. Expression of messenger RNA for transforming growth factor-beta1 and for transforming growth factor-beta receptors in peripheral blood of systemic lupus erythematosus patients treated with low doses of quinagolide. Autoimmunity 40, 23–30 (2007). [DOI] [PubMed] [Google Scholar]
- Khera T. K., Dick A. D. & Nicholson L. B. Fragile X-related protein FXR1 controls post-transcriptional suppression of lipopolysaccharide-induced tumour necrosis factor-alpha production by transforming growth factor-beta1. Febs J 277, 2754–2765 (2010). [DOI] [PubMed] [Google Scholar]
- Haller D. et al. Transforming growth factor-beta 1 inhibits non-pathogenic Gram negative bacteria-induced NF-kappa B recruitment to the interleukin-6 gene promoter in intestinal epithelial cells through modulation of histone acetylation. J Biol Chem 278, 23851–23860 (2003). [DOI] [PubMed] [Google Scholar]
- Studnicka-Benke A., Steiner G., Petera P. & Smolen J. S. Tumour necrosis factor alpha and its soluble receptors parallel clinical disease and autoimmune activity in systemic lupus erythematosus. Br J Rheumatol 35, 1067–1074 (1996). [DOI] [PubMed] [Google Scholar]
- Sule S., Rosen A., Petri M., Akhter E. & Andrade F. Abnormal production of pro- and anti-inflammatory cytokines by lupus monocytes in response to apoptotic cells. PLoS One 6, e17495 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santer D. M., Yoshio T., Minota S., Moller T. & Elkon K. B. Potent induction of IFN-alpha and chemokines by autoantibodies in the cerebrospinal fluid of patients with neuropsychiatric lupus. J Immunol 182, 1192–1201 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M. et al. Defective IL-10 signaling in hyper-IgE syndrome results in impaired generation of tolerogenic dendritic cells and induced regulatory T cells. J Exp Med 208, 235–249 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfle S. J. et al. PD-L1 expression on tolerogenic APCs is controlled by STAT-3. Eur J Immunol 41, 413–424 (2011). [DOI] [PubMed] [Google Scholar]
- Hochberg M. C. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 40, 1725 (1997). [DOI] [PubMed] [Google Scholar]
- Bombardier C., Gladman D. D., Urowitz M. B., Caron D. & Chang C. H. Derivation of the SLEDAI. A disease activity index for lupus patients. The Committee on Prognosis Studies in SLE. Arthritis Rheum 35, 630–640 (1992). [DOI] [PubMed] [Google Scholar]
- O'Doherty U. et al. Human blood contains two subsets of dendritic cells, one immunologically mature and the other immature. Immunology 82, 487–493 (1994). [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials