Abstract
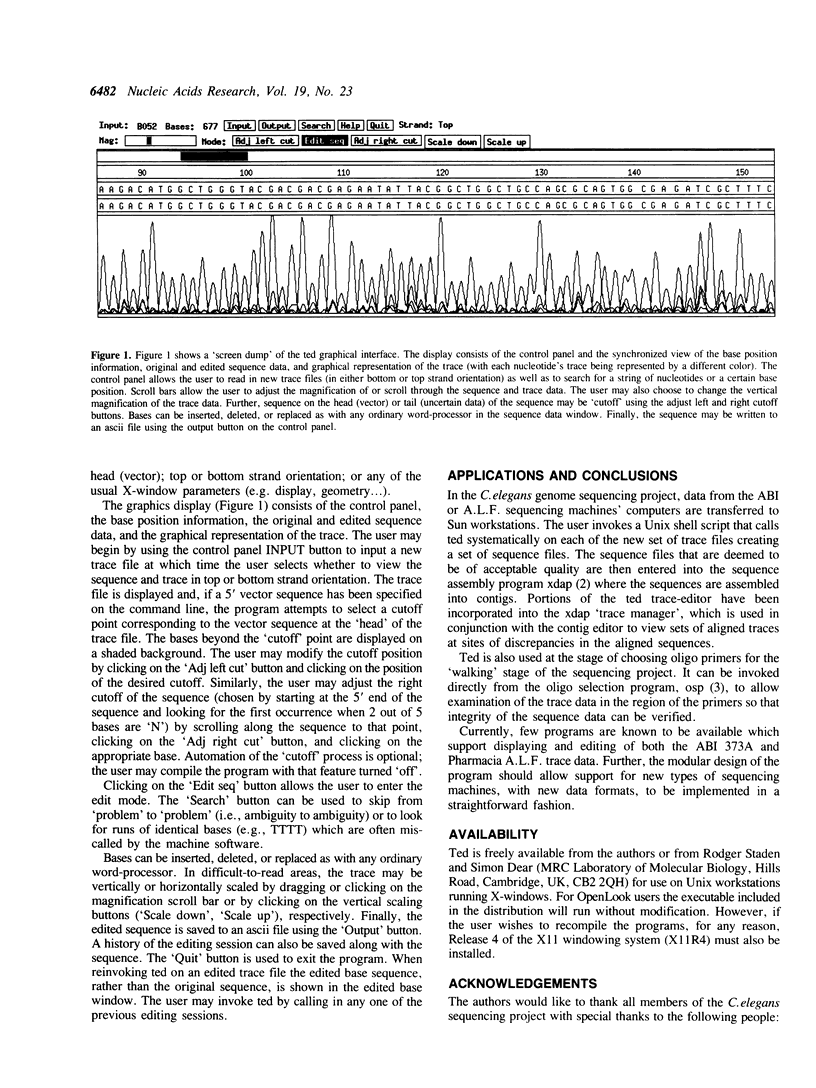
'Ted' (Trace editor) is a graphical editor for sequence and trace data from automated fluorescence sequencing machines. It provides facilities for viewing sequence and trace data (in top or bottom strand orientation), for editing the base sequence, for automated or manual trimming of the head (vector) and tail (uncertain data) from the sequence, for vertical and horizontal trace scaling, for keeping a history of sequence editing, and for output of the edited sequence. Ted has been used extensively in the C.elegans genome sequencing project, both as a stand-alone program and integrated into the Staden sequence assembly package, and has greatly aided in the efficiency and accuracy of sequence editing. It runs in the X windows environment on Sun workstations and is available from the authors. Ted currently supports sequence and trace data from the ABI 373A and Pharmacia A.L.F. sequencers.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dear S., Staden R. A sequence assembly and editing program for efficient management of large projects. Nucleic Acids Res. 1991 Jul 25;19(14):3907–3911. doi: 10.1093/nar/19.14.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier L., Green P. OSP: a computer program for choosing PCR and DNA sequencing primers. PCR Methods Appl. 1991 Nov;1(2):124–128. doi: 10.1101/gr.1.2.124. [DOI] [PubMed] [Google Scholar]


