Abstract
We have identified a novel gene, dwwA, which is required for cytokinesis of Dictyostelium cells on solid surfaces. Its product, Dd WW domain containing protein A (DWWA), contains several motifs, including two WW domains, an IQ motif, a C2 domain, and a proline-rich region. On substrates, cells lacking dwwA were multinucleated and larger and flatter than wild-type cells due to their frequent inability to sever the cytoplasmic bridge connecting daughter cells after mitosis. When cultured in suspension, however, dwwA-null cells seemed to carry out cytokinesis normally via a process not driven by the shearing force arising from agitation of the culture. GFP-DWWA localized to the cell cortex and nucleus; analysis of the distributions of various truncation mutants revealed that the N-terminal half of the protein, which contains the C2 domain, is required for the cortical localization of DWWA. The IQ motif of DWWA binds calmodulin in vitro. Given that the scission process is also defective in calmodulin knockdown cells cultured on substrates (Liu et al., 1992), we propose that DWWA's multiple binding domains enable it to function as an adaptor protein, facilitating the scission process through the regulation of Ca2+/calmodulin-mediated remodeling of the actin cytoskeleton and/or modulation of membrane dynamics.
INTRODUCTION
During cytokinesis, cells are pinched into two parts by constriction of contractile rings. Experimental evidence from a variety of eukaryotic cells lacking cell walls suggests that the contractile force produced by the active interaction of parallel filaments of actin and nonmuscle myosin II making up the rings is essential for cytokinesis. Called the purse-string model, this scenario has been thought to be the principal mechanism of cytokinesis in animal cells (Mabuchi and Okuno, 1977; Glotzer, 1997; Robinson and Spudich, 2000). On the other hand, when grown on substrates, amebic cells of the cellular slime mold Dictyostelium discoideum efficiently carry out cytokinesis without myosin II (Neujahr et al., 1997; Zang et al., 1997). Indeed, detailed microscopic observation of myosin II-null cells revealed that Dictyostelium makes use of at least three distinct types of cytokinesis. 1) Cytokinesis A, or the purse-string model, is the classic cytokinesis and is dependent on myosin II. 2) Cytokinesis B (Zang et al., 1997; Nagasaki et al., 2002), or attachment-assisted mitotic cleavage (Neujahr et al., 1997), does not depend on myosin II but does require adhesion to a solid surface. 3) Cytokinesis C, or cytofission (Spudich, 1989), is a cell cycle-uncoupled cytokinesis of adherent cells (Nagasaki et al., 2002). We recently reported morphological and genetic evidence indicating that cytokinesis B is driven by opposing traction forces generated along both polar peripheries and that cytokinesis A and B are redundant, but distinct, cell cycle-coupled pathways leading to furrow formation in the equatorial region of the cell (Nagasaki et al., 2001, 2002). Although distinct from each other, cytokinesis A and B may nonetheless share the spatial and temporal regulatory mechanisms responsible for furrow formation, as well as for the final scission of the ingression. These processes, however, remain poorly understood.
Dictyostelium is a model organism, which, for a number of reasons, is particularly suitable for molecular genetic analysis of the mechanism of cytokinesis. First, it has a relatively small haploid genome, and an efficient method of insertional mutagenesis is available. This makes it straightforward to construct a library of randomly mutagenized cells and to identify clones with recessive mutations. Second, they are simple amebic cells, yet are morphologically and biochemically similar to higher animal cells, leukocytes in particular; and large numbers of homogenous cells can be readily made available for biochemical analysis. Finally, Dictyostelium has an advantage to screen cytokinesis mutants in that cytokinetic defects are not lethal in this organism. One drawback to Dictyostelium is that it makes use of at least two parallel pathways leading to formation and constriction of the furrow. This means that special consideration must be taken when designing a screening strategy for isolating mutants in which furrow formation is defective (Nagasaki et al., 2002). Still, mutants in which processes shared by cytokinesis A and B are defective, i.e., mechanisms regulating the furrowing and final scission processes should show clear phenotypes under standard culture conditions, and these should be easy targets for genetic screening. In fact, four of the cytokinesis mutants isolated thus far exhibit a defect affecting scission of the cytoplasmic bridge between daughter cells (Adachi et al., 1997; Weber et al., 1999; Wienke et al., 1999; Palmieri et al., 2000). Despite the availability of these mutants, our understanding of the scission process is far from complete, and our understanding of the spatial and temporal regulation of the placement of the furrow is even less well understood. Therefore, to gain additional insight into the mechanisms underlying these processes, we set out to isolate mutants that show cytokinetic defects when cultured on substrates after random insertional mutagenesis of wild-type Dictyostelium cells. Using that approach, we were able to identify a cytokinesis mutant in which a novel gene, dwwA, encoding a protein containing an IQ motif, a C2 domain, and two WW domains, is disrupted. Unexpectedly, cells lacking dwwA became multinucleate when cultured on solid surfaces, but not when cultured in suspension. This was because its product, Dd WW domain containing protein A (DWWA), is critical for the final scission of cells undergoing cytokinesis on a solid surface, but not in suspension.
MATERIALS AND METHODS
Cell Culture
Parental D. discoideum wild-type AX2 cells were grown axenically in HL-5 medium (Sussman, 1987) supplemented with penicillin and streptomycin (Wako Pure Chemicals, Tokyo, Japan) at 22°C. DwwA-null cells were cultured in HL-5 in the presence of penicillin, streptomycin, and 10 μg/ml blasticidin S (Funakoshi, Tokyo, Japan). Cells carrying derivatives of the Dictyostelium expression vector pBIG (Ruppel et al., 1994) were grown in medium containing penicillin, streptomycin, and 10 μg/ml G418. The cells were usually grown on 9-cm plain plastic Petri dishes; in some experiments, however, they were grown in suspension, in conical Teflon flasks on a shaker rotating at ∼140 rpm.
Restriction Enzyme-mediated Insertional (REMI) Mutagenesis
Bsr-REMI mutagenesis was carried out as described previously (Kuspa and Loomis, 1992; Adachi et al., 1994), except that we used pmBsr, a miniaturized tagging vector with improved mutagenesis efficiency (Hibi et al., 2004). AX2 cells were grown in suspension to a density of 5 × 106 cells/ml in HL-5 medium supplemented with 5 μg/ml vitamin B12 and 200 μg/ml folate. Approximately 1.5 × 108 cells were transformed with 10 μg of pmBsr linearized with BamHI, along with 4 U of DpnII (New England Biolabs, Beverly, MA) by electroporation. The resultant transformants were cultured on plastic dishes in HL-5 medium containing 10 μg/ml blasticidin S. After 5-6 d, the colonies formed by the transformed cells were observed under a microscope; those showing large numbers of multinucleated cells were isolated from the dish, recloned on bacterial lawns, and used for further analyses.
Inverse Polymerase Chain Reaction (PCR)
Genomic DNA was isolated from mutant cells according to the method of Bain and Tsang (1991). Samples (1 μg) of genomic DNA were digested with one of the restriction enzymes that do not cut inside of the Bsr tag, i.e., Acc65I, BclI, ClaI, EcoRV, HpaI, MluI, NcoI, NdeI, SalI, and SphI (Fermentas, Hanover, MD), after which the fragments were circularized with DNA ligase (Fermentas). A pair of primers, SEB IVP (5′-TTTTTTTTATCTAGAGGATCTGTACGATAC-3′) and HEB IVP (5′-AAAAGATAAAGCTGACCCGAAAGCTAGCAT-3′), which were designed to extend outward from the Bsr tag, were then used for PCR to amplify the sequences flanking the tag. To obtain the 5′-untranslated region sequence of this gene, we performed another round of inverse PCR. In this case, the inverse PCR products were amplified using three pairs of primers: 5′-ACGATCTTTATATCACCCTCTACAA-3′ and 5′-AGCAACACCAGAAGAGGCAGCAGTT-3′ for the IVP(Bcl)2 clone, 5′-TTTTAACCTCTCTCTTTTCTGAT-3′ and 5′-GTATCGAAATTCAATTTAATAATATACCA-3′ for the IVP(Cla) and IVP(Xba) clones, and 5′-TTTTAACCTCTCTCTTTTCTGAT-3′ and 5′-CATGGTTCTTGTATCTATAATG for the IVP(Mun) clone. All of the inverse PCR products were subsequently cloned into the pGEM-T easy vector (Promega, Madison, WI).
Molecular Cloning of the dwwA Gene
Full-length dwwA cDNA was cloned from a cDNA library of vegetative Dictyostelium cells by using the rapid amplification of a cDNA ends (RACE) method. Components of the reverse transcription synthesis of the cDNA library used for amplification of the 3′ ends included 1 μg of poly(A) RNA, 1× reverse transcription buffer, 2 μM dNTP, 10 pmol of QT primer, poly T primer containing the sequence of the Q0 primer (5′-CCAGTGAGCAGAGTGACGAGGACTCGAGCTCAAGCTTTTTTTTTTTTTTTTT-3′), and 100 U of MMLV reverse transcriptase RNaseH- (Toyobo, Osaka, Japan). The reaction mixture was incubated for 1 h at 42°C. The 3′-RACE products of the dwwA cDNA were generated using a dwwA primer (5′-AGCAACACCAGAAGAGGCAGCAGTT-3) and a Q0 primer (5′-CCAGTGAGCAGAGTGACG-3′). Amplification of the 5′ end was carried out using a 5′-full RACE core set (Takara, Tokyo, Japan) according to manufacturer's instructions. Fragments of the dwwA gene were cloned into pGEM-T easy vector. Full-length cDNA and genomic DNA were amplified by PCR by using primers 5′-GGATCCAATGTCAAATAAAAATCCATTAAATAATAGTA-3′ and 5′-GAGCTCTTATTTTCTTTGTTGTACAAGTGT-3′, which added BamHI and SacI recognition sites at either end of the PCR products, enabling them to be subcloned into GFP/pBIG such that dwwA was fused to the 3′ end of GFP cDNA, the expression of which is driven by the actin 15 promoter (Nagasaki et al., 2002).
Reverse Transcription-Polymerase Chain Reaction (RT-PCR) Analysis
AX2 cells were allowed to develop on agarose plates containing 17 mM phosphate buffer (Fukui, 1990). Using ISOGEN (Nippon Gene, Tokyo, Japan), total RNA was purified from these developing AX2 cells at the indicated times, after which RT-PCR was performed for 25 cycles with primers specific to dwwA.
Generation of dwwA-Null Cells
DwwA-null cells were generated by homologous recombination. Genomic DNA encoding dwwA was cloned into pGEM-T (Promega), and the blasticidin S resistance cassette from pUCBsrΔBam was inserted at the unique SwaI site of dwwA, yielding the targeting vector. Ten micrograms of dwwA targeting vector was then linearized with PvuII and introduced into wild-type AX2 cells. Transformants were selected for blasticidin S resistance, and colonies formed on plastic dishes were isolated. Genomic PCR and Southern blotting analysis was then carried out to verify disruption of dwwA.
Fluorescence Microscopy
AX2 and dwwA-null cells were transfected with green fluorescent protein (GFP)-myosin II/pTIKL (Liu et al., 2000) or GFP-dwwA/pBIG by electroporation, after which the resultant transfectants were transferred to plastic Petri dishes with thin glass bottoms (Iwaki, Funabashi, Japan), and the culture medium was replaced with medium modified to minimize background fluorescence (Nagasaki et al., 2002). The cells were then observed using a confocal laser scanning microscope (CSU10; Yokogawa, Tokyo, Japan).
Expression and Purification of Recombinant Glutathione S-Transferase-Calmodulin (GST-CaM) Fusion Protein and In Vitro Binding Assay
The Dictyostelium CaM gene, calA, was cloned from a Dictyostelium vegetative cDNA library. A cDNA construct encoding a full-length GST-CaM fusion protein was then created by subcloning CaM cDNA into pGEX (Amersham Biosciences, Piscataway, NJ), the GST fusion protein vector, after which the GST-CaM and GST constructs were transformed into BL21 (DE3) Escherichia coli cells (Stratagene, La Jolla, CA). Cells expressing GST-CaM and GST were induced with isopropyl-1-thio-d-galactopyranoside (1 mM) for 12 h at 22°C and then lysed by sonication in phosphate-buffered saline (PBS) containing 1 mM dithiothreitol, 2 μM leupeptin, and 1 mM phenylmethylsulfonyl fluoride. GST-CaM was purified from the extract using a glutathione-Sepharose 4B (Amersham Biosciences) column.
Dictyostelium cells expressing GFP-DWWA were sonicated in PBS buffer containing protease inhibitors, after which the cell lysate was centrifuged at 25,000 × g for 20 min. The resultant supernatant was mixed with 200 μl of a 50% slurry of GST-CaM beads or control GST beads and then agitated for 2 h at 4°C in the presence of 1 mM CaCl2 or 5 mM EGTA. After incubation, the beads were washed five times with PBS containing 1 mM CaCl2 or 5 mM EGTA, and bound proteins were extracted with SDS sample buffer. The samples were then analyzed by SDS-PAGE and Western blotting by using an anti-GFP antibody.
RESULTS
REMI Mutants Exhibiting Defective Cytokinesis on a Solid Surface and Identification of a Gene Responsible for the Defect
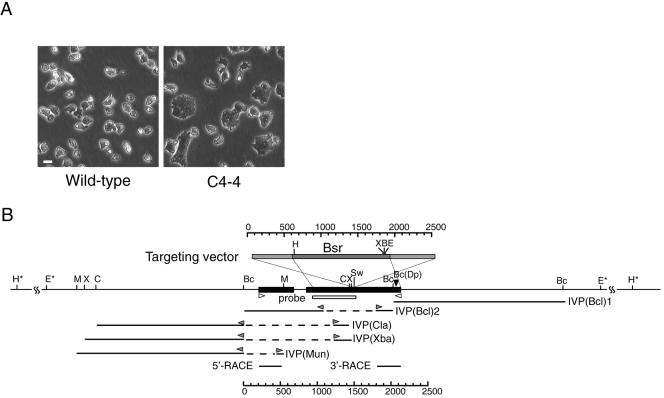
The Bsr-insertional tag was randomly integrated into the Dictyostelium genome by using a modified REMI method. The resultant mutagenized Dictyostelium cells were then maintained on plastic dishes, and cytokinesis mutants were isolated from the library by means of microscopic screening. Eight mutants that showed a high incidence of large, multinucleated cells were obtained from ∼6000 independent transformants. One clone, C4-4 (Figure 1A), exhibited a particularly severe phenotype and was therefore used for further analysis.
Figure 1.
C4-4 REMI-mutant (A) and schematic representation of the gene disrupted in C4-4 (B), which we named dwwA. (A) Phase contrast micrographs of clone C4--4 and wild-type cells grown on plastic dishes. (B) Restriction map around dwwA and the targeting vector. The targeting vector (top box) used to knockout dwwA has an insertion of the Bsr cassette (dark gray box) at the unique SwaI site, and extends to both sides of the SwaI site (light gray boxes) in exon 2. The two exons are shown as black boxes. The underlines indicate five genomic DNA fragments cloned from the C4-4 cells by inverse PCR. Bc(Dp) indicates the insertion site of the original REMI-mutant. White box indicates a region used as a probe for Southern blotting. Two white arrowheads indicate the primers used for RT-PCR. Gray arrowheads indicate the inverse PCR primers used for genome walking. Restriction enzyme sites: B, BamHI; Bc, BclII; C, ClaI; E, EcoRI; H, HindIII; M, MunI; Sw, SwaI; X, XbaI; and Dp, DpnII. Asterisks indicate restriction enzyme sites that were predicted by Southern blotting.
To identify the gene causing this phenotype, the genomic sequences flanking the inserted tag were determined. As shown in Figure 1B, one such fragment, IVP(Bcl)1 was cloned from the C4-4 genomic DNA by inverse PCR by using SEB IVP and HEB IVP primers. The tag was integrated into a BclI site within the open reading frame (Figure 1B), and the 3′ flanking sequence was contained within the fragments of IVP(Bcl)1. Starting with IVP(Bcl)1, we assembled the full-length sequences of the cDNA and genomic DNA by using fragmentary sequences found in the GenBank and the database of the Dictyostelium discoideum cDNA Project. This enabled us to design a pair of oligonucleotide primers that would amplify the full sequence from genomic DNA or a cDNA library. Sequence analysis of the complete cDNA and genomic DNA revealed this gene to have two exons (Figure 1B). To determine whether there were more exons situated upstream, we carried out genome walking by using inverse PCR. The resultant cloned fragments, which were designated IVP(Bcl)2, IVP(Cla), IVP(Xba), and IVP(Mun), respectively, contained the sequence upstream of the first exon, but no additional open reading frame was found. The positions of 5′ and 3′ ends were further confirmed by 5′-RACE and 3′-RACE.
Amino Acid Sequence of DWWA
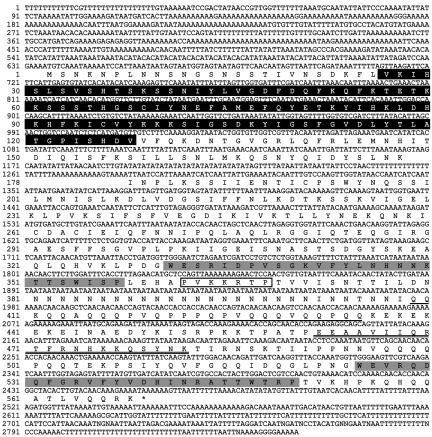
Sequence analysis of the complete cDNA revealed a polypeptide of 568 amino acid residues (Figure 2) with a calculated molecular mass of 65 kDa. The protein contained two WW domains (Espanel and Sudol, 1999), respectively spanning amino acid residues 331-357 and 525-550, so we named it DWWA. In addition, DWWA contained one IQ (CaM binding) motif spanning amino acid residues 462-482, and an N-terminal C2 domain, which is a Ca2+ and phospholipid binding domain spanning ∼130 amino acid residues. A putative nuclear localization signal was situated between amino acid residues 362 and 368, and there was a proline- and glutamine-rich sequence in the region between amino acid residues 409 and 435, which was notable because proline-rich regions are known to be protein-protein interaction motifs (Kay et al., 2000).
Figure 2.
DNA sequence of dwwA and the deduced amino acid sequence of its product: the black and white boxes indicate a putative C2 domain and putative nuclear localization signal, respectively; the two gray boxes indicate sequences corresponding to WW domains; double underlines indicated an IQ motif; and a single underline indicates a glutamine- and proline-rich sequence.
Expression of dwwA and Subcellular Localization of GFP-DWWA
To determine when during the life cycle of Dictyostelium dwwA is expressed, samples of total RNA were isolated at various stages of the developmental program leading to formation of fruiting bodies and analyzed by RT-PCR. As shown in Figure 3A, the dwwA transcript was expressed mainly during the growth phase, with expression declining during the early developmental phases. IG7 is expressed at a constant level during development and served as an internal control (Chang et al., 1996).
Figure 3.
Expression of dwwA during the life cycle of Dictyostelium and subcellular localization of GFP-DWWA. (A) RT-PCR. Wild-type cells were developed on nonnutrition agar for the times indicated, after which the DNA was amplified using the same amount of total RNA as the template in each case. Amplification of IG7 served as an internal control (Chang et al., 1996). (B) Localization of the GFP-DWWA in living wild-type cells (left) and in cells fixed with 3.7% formaldehyde (right). Full-length DWWA was concentrated along the cell cortex. (C) Localization of GFP-DWWA in fixed cells during the cell cycle. Bar, 10 μm.
To observe DWWA dynamics in living Dictyostelium cells, wild-type cells were transformed with an extrachromosomal vector harboring a gfp-dwwA fusion gene under the control of the constitutive actin 15 promoter. In living cells, the product, GFP-DWWA, localized to the cell cortex and nuclei (Figure 3B, left); when cells were fixed with 3.6% formaldehyde, the nuclear localization of GFP-DWWA was found to be limited to the nucleolus (Figure 3B, right). Figure 3C shows the localization of GFP-DWWA in fixed cells at each stage of the cell cycle.
DwwA-Null Cells Exhibit Defective Cytokinesis on a Solid Surface but Grow Normally in Suspension
After disrupting dwwA by homologous recombination with a targeting vector (Figure 1B), transformed clones were cultured on solid surfaces in HL-5 medium containing blasticidin S. The selective disruption of dwwA was confirmed by Southern blot analysis (Figure 4A). EcoRI and HindIII digestion of wild-type genomic DNA yielded 7.0- and 17-kbp bands, respectively. In dwwA-null cells, however, the bands were shifted to 4.5- and 7.0 kbp, respectively, as a result of tag insertion. One representative clone, dwwA-null cell 10-1, was selected and used for further analysis. Cells lacking dwwA showed the same phenotype as the original REMI mutants, confirming that the phenotype of the original C4-4 clone reflected the disruption of the dwwA gene.
Figure 4.
Generation of dwwA-null cells and their phenotype. (A) Genomic DNA digested with either ClaI, EcoRI, or HindIII was separated by agarose gel electrophoresis, transferred to a membrane, and probed with dwwA cDNA fragments. (B) Development. Wild-type and dwwA-null cells were allowed to form fruiting bodies on nonnutrient agar for 36 h at 22°C. Bar, 100 μm. (C) Cell size. Wild-type and mutant cells were maintained on plastic plates for 3 d (dish) or were grown in suspension for 3 d and then transferred to plastic plates (suspension) before observation under a phase contrast microscope. Bar, 10 μm.
DwwA-null cells became larger and flatter than wild-type cells when cultured on a solid surface but not when cultured in suspension (Figure 4C). On solid surfaces, 50% of dwwA-null cells were multinucleate, whereas >95% of wild-type cells were mononucleate (Figure 5, A and C). When cultured in suspension with continuous agitation, dwwA-null cells grew at a rate similar to that of wild-type cells and were less frequently multinucleate than on substrates (Figure 5, B and D). Wild-type cells, by contrast, were more frequently multinucleate in suspension than on substrates. As a result, the fractions comprised of multinucleate dwwA and wild-type cells were very similar in suspension.
Figure 5.
Phenotypic characterization of the cytokinesis defect in dwwA-null cells. Cells of each strain were transferred to a dish with a thin glass bottom for microscopy. Top and bottom panels show phase contrast and 4,6-diamidino-2-phenylindole fluorescence images of cells fixed with 3.7% formaldehyde, respectively. (A) Cells that had been continuously cultured on plastic dishes and then transferred to glass-bottomed dishes. (B) Cells cultured in suspension for three days before being transferred to glass-bottomed dishes. The dwwA O/E (overexpression) cells are the “rescued” dwwA-null cells carrying the plasmid harboring the gfp-dwwA gene. Bar, 10 μm. (C and D) Histograms of the number of nuclei per cell.
We also examined cytokinesis of dwwA-null cells in the absence of both solid substrate adhesion and agitation. For this, the cells were embedded and cultured in a gel comprised of HL-5 medium and 1% ultralow gelling temperature agarose (Cosmo Bio, Tokyo, Japan), or at the interface between HL-5 medium and 80% Percoll (Amersham Biosciences). Under either of these conditions, myosin II-null cells, which require adhesion to solid substrates for cytokinesis, became large and multinucleate within 2-3 d. By contrast, cytokinesis proceeded to completion normally in both dwwA-null and wild-type cells (our unpublished data).
We confirmed that the mutant phenotype reflected disruption of dwwA by genetic complementation with GFP-DWWA. DwwA cDNA was fused to the constitutive actin 15 promoter and GFP, inserted into a multicopy vector carrying the G418 resistance gene, and introduced into dwwA-null cells. The resultant transformants showed the normal wild-type phenotype (Figure 5, A and C), indicating the gene product was capable of reversing the defect of dwwA-null cells.
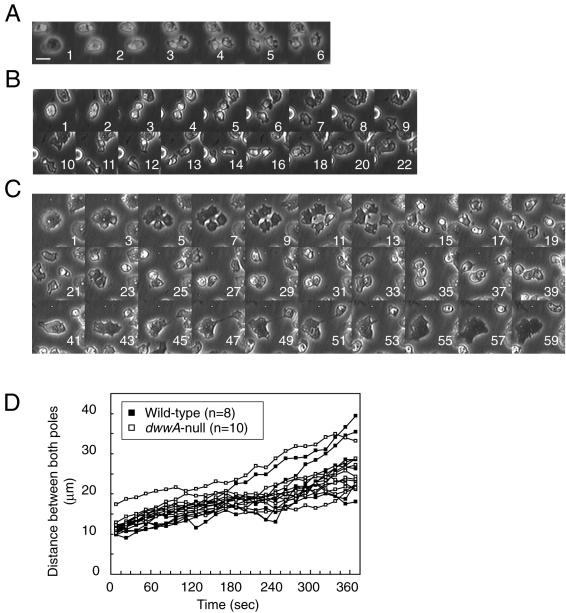
We next carried out a detailed microscopic analysis of cytokinesis in dwwA-null cells. In wild-type cells, cell division was completed within 3-4 min after initiation of furrowing (Figure 6A). Early stages of cytokinesis of dwwA-null cells seemed to proceed normally (Figure 6B), in that dwwA-null cells formed a cleavage furrow and daughter cells seemed to separate from one another at the normal rate. The separation of the daughter cells proceeded almost to completion in most cases (Figure 6B, 7-18 min); however, cleavage of the narrow cytoplasmic bridge connecting the daughter cells often failed (55% of 40 mitotic mononucleate dwwA-null cells studied), and the process subsequently reversed, yielding a single binucleate cell within 20 min. Cytokinesis of binucleate dwwA-null cells on a plastic dish proceeded in analogous manner (Figure 6C). Again, the early mitotic stages proceeded normally (Figure 6C, 3-7 min), and four daughter cells seemed to nearly complete the cytokinesis (11-35 min). In this case, however, the process reversed in three of the cells, yielding a single trinucleate cell and a single mononucleate cell. It was this abortive cytokinesis that caused the accumulation of large multinucleate cells on solid surfaces.
Figure 6.
Cytokinetic sequences in wild-type and dwwA-null cells cultured on solid surfaces. Each panel shows a series of phase contrast images recorded at the times indicated at the bottom right (minutes). (A) Mitotic wild-type cells complete cytokinesis within 2-3 min after the onset of furrowing. (B and C) Abortive cytokinesis of mitotic dwwA-null cells. Mitotic dwwA-null cells containing one (B) or two nuclei (C) seemed to have separated completely into two or four daughter cells, respectively, but in fact remained connected by thin cytoplasmic bridges. The connected cells subsequently merged to form a multinucleate cell. Bar, 10 μm. (D) Changes of distances between the poles of two daughter cells of wild-type and dwwA-null cells undergoing cytokinesis. Time zero was chosen when the mitotic cell started to elongate and division axis became apparent. Cells were placed in plastic dishes and images of cell division were analyzed using ImageJ software.
DwwA-null cells did not exhibit noticeable phenotypic defects in our other assays. Rates of separation between two daughter cells did not differ significantly between wild-type and dwwA-null cells (Figure 6D). Furthermore, speeds of interphase vegetative cells were also the same between the two cell lines (3.67 ± 0.63 μm/min for wild-type and 3.60 ± 0.97 μm/min for dwwA-null cells). These data demonstrate that the faulty cytokinesis of dwwA-null cells was not caused by general motility defects. They grew normally on bacterial lawns (our unpublished data), and formed normal fruiting bodies on agar plates (Figure 4B).
DWWA Is Required for Normal Actin Distribution in Dictyostelium Cells
The large, flat shape of dwwA-null cells grown on substrates (Figure 4C) was suggestive of cytoskeletal abnormalities. Investigation of the distribution of myosin II in wild-type and dwwA-null cells expressing GFP-myosin II showed that, at interphase, GFP-myosin II localized to the cell cortex in both cell types, and we detected no differences between the two (Figure 8A). In mitotic dwwA-null cells, moreover, GFP-myosin II localized normally to the cleavage furrow at anaphase, just as in wild-type cells (Figure 7B, left), and remained concentrated in the cytoplasmic bridge connecting the daughter cells (Figure 7B, middle and right).
Figure 8.
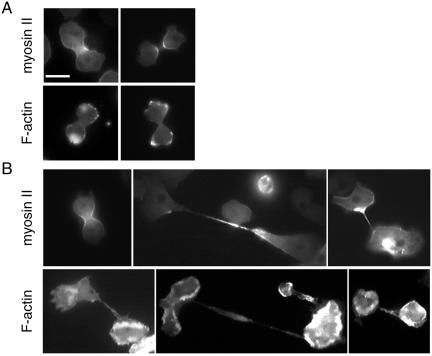
Localization of myosin II and F-actin in wild-type and dwwA-null cells. (A) Interphase wild-type and dwwA-null cells expressing GFP-myosin II. There is no difference in the distribution of GFP-myosin II in the two cell lines. (B) Cells were fixed with 3.7% formaldehyde and stained with rhodamine-phalloidin for F-actin. In wild-type cells, F-actin was localized in the crowns (arrowheads) and along the cell cortices. The pattern of F-actin distribution in dwwA-null cells differs from that in wild-type cells. Bar, 10 μm.
Figure 7.
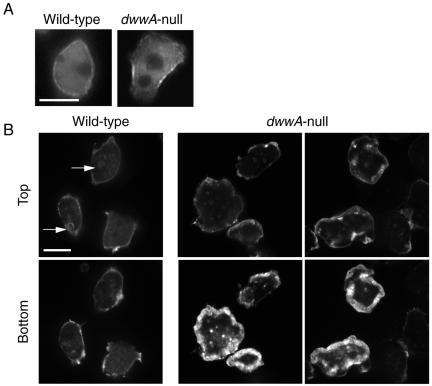
Distribution of actin and myosin II in mitotic wild-type (A) and dwwA-null (B) cells. (A) Wild-type cells were transformed with plasmid carrying the GFP-myosin II gene (top). GFP-myosin II localized to cleavage furrows in mitotic wild-type cells. F-actin was visualized by fixing cells with 3.7% formaldehyde and staining with rhodamine-phalloidin (bottom). F-actin was enriched along both poles of the two daughter cells. (B) In dwwA-null cell, GFP-myosin II in a dumbbell-shaped cell was localized in the cleavage furrow (top), as in wild-type cells. Middle and right pictures in the top panel show cells at the final step of cytokinesis. Cells that proceed to the final step of cytokinesis accumulated GFP-myosin II in the thin cytoplasmic bridge connecting the two daughter cells, as well as in the tapered region of the cell body connected to the cytoplasmic bridge. Bottom, distribution of F-actin at the final step of cytokinesis with long and thin cytoplasmic bridges. Note the excessive accumulation of F-Actin around the cell periphery. Bar, 10 μm.
When wild-type cells on substrates were fixed with formaldehyde, stained with rhodamine-phalloidin, and observed by confocal microscopy, F-actin was found to localize to crowns on the ventral surface of the cell and to thin peripheral bands surrounding the ventral surface (Figure 8B). This probably represented the cortical localization. In contrast, excessive accumulation of F-actin was observed as a thick band around the periphery of the ventral surface of dwwA-null cells (Figure 8B), and a similar abnormal distribution of F-actin was often observed in ∼50% of dwwA-null cells during cytokinesis (Figure 7B).
The N-Terminal Region of DWWA Is Required for Localization to Cortex
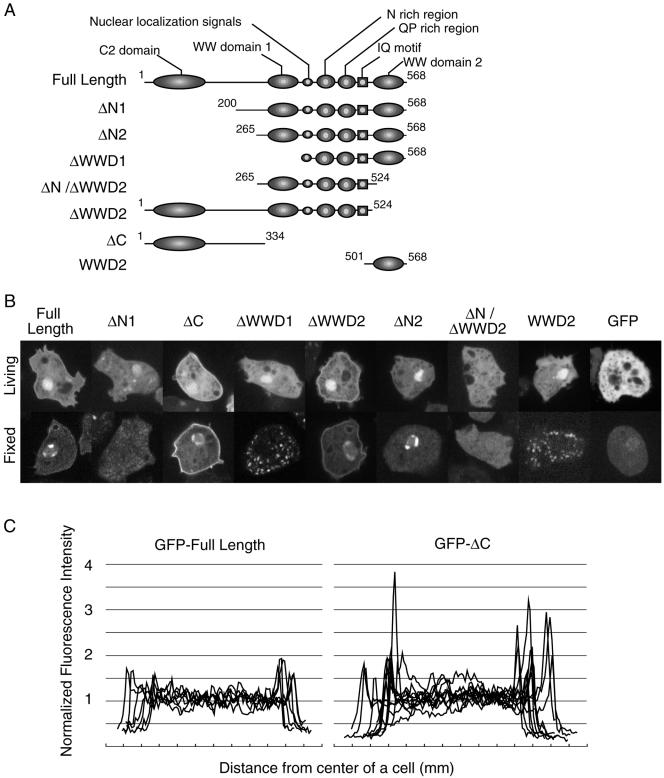
DWWA contains several motifs within its amino acid sequence. We therefore constructed seven deletion mutants, each lacking one of the motifs, to determine their respective roles in subcellular localization (Figure 9A). The mutant genes were constructed by PCR, fused with the actin 15 promoter and GFP, and subcloned into the pBIG extrachromosomal vector. GFP-DWWA served as a control and localized to the nuclei and cell cortex (Figure 9B). Four mutants lacking the N-terminal end of DWWA (GFP-ΔN1, GFP-ΔN2, GFP-ΔWWD1, and GFP-ΔN/ΔWWD2) failed to localize to the cell cortex. Mutants truncated at the C-terminal end (GFP-ΔC and GFP-ΔWWD2) were not expressed at detectable levels in the majority of transformed cells, but in cells where they were, they were clearly localized to the cortex. Indeed, GFP-ΔC, which lacked the entire C-terminal half of DWWA, localized to the cortex even more intensely than the intact GFP-DWWA (Figure 9C).
Figure 9.
Localization of a series of DWWA deletion mutants. (A) Schematic representation of the seven constructs. Each box and ellipse shows the indicated domain; the numbers indicate amino acids residues. (B) Fluorescence images of cells expressing GFP-fusion proteins. The top and bottom panels show living cells and cells fixed in 3.7% formaldehyde, respectively. GFP-full length DWWA, GFP-ΔC, and GFP-ΔWWD2 were localized in the cell cortex and nucleus, whereas GFP-ΔN1 and GFP-ΔN/ΔWWD2 were diffusely distributed within the cells, like GFP alone. (C) Profiles of the relative fluorescence intensities of GFP-DWWA and GFP-ΔC in wild-type cells. Fluorescence intensity of the GFP fusion proteins was measured by scanning across the cells, avoiding nuclei, by using NIH Image software. The intensities were then normalized to the average intensity within each cell.
DWWA Interacts with CaM In Vitro
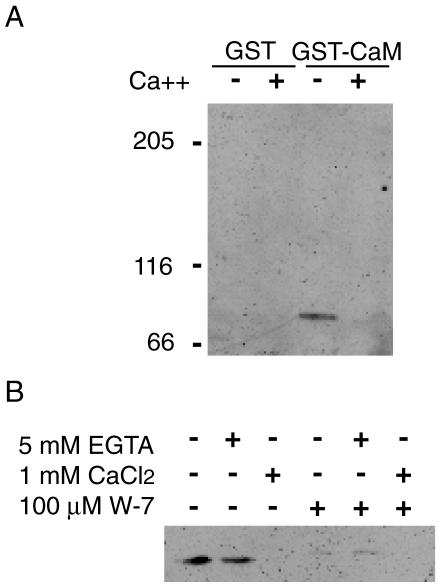
DWWA also contains an IQ motif, which is a potential CaM binding site (Figure 2). We therefore examined the interaction between recombinant GST-CaM and GFP-DWWA. To do this, GST-CaM was expressed in E. coli and then purified using GST-affinity beads. Purified GST-CaM-bound beads or control GST-bound beads were incubated with Dictyostelium cell lysates containing GFP-DWWA in the presence of 5 mM CaCl2 or 10 mM EGTA. In the absence of Ca2+, but not in its presence, GFP-DWWA coprecipitated with GST-CaM (Figure 10A), suggesting the IQ motif of GFP-DWWA exclusively binds CaM without Ca2+, i.e., apocalmodulin. The interaction of DWWA and CaM in the absence of Ca2+ was blocked by addition of W-7, an inhibitor of Ca2+/CaM (Figure 10B).
Figure 10.
(A) Interaction of DWWA and CaM. Dictyostelium CaM fused with GST and bound to glutathione-Sepharose beads and extracts from cells expressing GFP-DWWA were mixed. GST-CaM-beads were washed, extracted, and subjected to SDS-PAGE. GFP-DWWA was detected using an anti-GFP antibody on Western blots. (B) Effect of W-7 on DWWA binding to CaM. GST-CaM-beads and cell extracts containing GFP-DWWA were incubated in the absence or the presence of 100 μM W-7 in the Ca2+-free buffer.
DISCUSSION
In Dictyostelium, random insertional mutagenesis, especially REMI (Kuspa and Loomis, 1992; Adachi et al., 1994), is a powerful tool with which to generate a library of randomly mutagenized cells, and it has enabled a number of cytokinesis-related genes to be identified. We recently developed an improved REMI system, involving a miniaturized tag and inverse PCR, that enables more efficient integration and rapid identification of the disrupted locus (Hibi et al., 2004). With this improved REMI system, we were able to isolate eight cytokinesis mutants from ∼6000 transformants. One of them, C4-4, exhibited particularly severe cytokinetic defects when cultured on substrates, reflecting disruption of a novel locus, dwwA.
Interestingly, dwwA-null cells cultured in suspension were mostly mono- or binucleate, and the distributions of cell sizes and growth rates were normal. This seems consistent with an earlier proposal by ourselves and others that Dictyostelium uses two distinct, cell cycle-coupled mechanisms of cytokinesis: the purse-string method, or cytokinesis A, and attachment-assisted mitotic cleavage, or cytokinesis B (Neujahr et al., 1997; Zang et al., 1997; Nagasaki et al., 2001, 2002). The former depends on formation of a contractile ring and is driven by active interaction of actin and myosin II, but does not require substrate adhesion; the latter depends on adhesion to substrates but not on myosin II. This hypothesis is based on detailed microscopic examination of Dictyostelium cells lacking the myosin II gene. In those studies, myosin II-null cells were unable to form cleavage furrows and divide in suspension via cytokinesis A, but they did undergo cell division efficiently on solid surfaces via cytokinesis B (Neujahr et al., 1997; Zang et al., 1997). In sharp contrast, dwwA-null cells became multinucleate on solid surfaces but not in suspension. This phenotype is similar to that of cells lacking amiA (Nagasaki et al., 1998) or coronin (de Hostos et al., 1993), which have been implicated in cytokinesis B (Nagasaki et al., 2002), and suggests that DWWA may also be essential for cytokinesis B.
Detailed microscopic analysis of mitotic dwwA-null cells, however, revealed that this is not the case. Instead, Figure 6, B and C, shows a typical instance in which the residual bridge between dwwA-null daughter cells failed to be cleaved. During telophase, dwwA-null cells formed cleavage furrows like those formed by wild-type cells, with similar localization of myosin II. They then went on to constrict the furrow to a point where the division is almost complete, so that mitotic mononucleate dwwA-null cells seemed to have divided into two daughter cells (Figure 6B, 6 min), or a binucleate cell to have divided into four (Figure 6C, 11 min). But thin cytoplasmic bridges, often undetectable using conventional phase contrast microscopy, remained between the daughter cells, prompting the cells to refuse in most cases (Figure 6B, 20 min; Figure 6C, 53 min). It thus seems that DWWA is essential for scission of cytoplasmic bridges in the final stage of cytokinesis, but not for the preceding furrowing process that requires myosin II (cytokinesis A) or AmiA and coronin (cytokinesis B).
How then do dwwA-null cells divide normally in suspension? One obvious possibility is that in shaking cultures shear forces generated by the agitation severs the thin cytoplasmic bridges. To test this possibility, we embedded cells in an ultralow gelling temperature agarose gel containing HL-5 medium. Under those conditions, myosin II-null cells were unable to carry out cytokinesis, but both dwwA-null and wild-type cells divided and grew normally. DwwA-null and wild-type cells were also able to divide and grow when submerged in HL-5 on a cushion of Percoll. This makes it unlikely that division of dwwA-null cells in normal suspension cultures is supported by shear forces arising from agitation, and indicates that the requirement of DWWA for scission of the residual cytoplasmic bridge between daughter cells is adhesion dependent. This finding was somewhat unexpected, because we had assumed that a common scission mechanism would be used, regardless of whether the cells are adherent. On the other hand, substrate adhesion is known to profoundly affect the actin cytoskeleton (Gerald et al., 1998), and this may be related to the differential requirement of DWWA for scission in Dictyostelium cells.
GFP-DWWA localized to the cell cortex during both interphase and the mitotic phases, as well as to the cortex of the cleavage furrow during the scission process. This is consistent with the idea that DWWA functions within cytoplasmic bridges and that the absence of this activity inhibits cytokinesis in mutants lacking DWWA. Observation of GFP-DWWA deletion mutants revealed that the N-terminal region of DWWA is essential for localization to the cell cortex. In that regard, we found a weak homology to the C2 domain in the N-terminal of DWWA. The C2 domain was first identified as one of four conserved functional domains in protein kinase C (Newton, 1995), where it serves as a Ca2+ binding motif; Ca2+ binding leads the association of protein kinase C with the plasma membrane. Since then, C2 domains have been identified in numerous other proteins, where it was also involved in Ca2+-dependent lipid binding (Rizo and Sudhof, 1998). The fact that in the present study a deletion mutant containing only the C2 domain-like region (GFP-ΔC) was able to localize to the cell cortex suggests that the N-terminal region of DWWA plays a key role in determining its subcellular localization within Dictyostelium cells.
DWWA also contains two WW domains, which are small modules comprised of ∼40 amino acid residues (Sudol, 1996; Sudol et al., 2001). The ability of this domain to bind a variety of proline-rich sequences provides it with a great deal of functional diversity. Based on the pattern of amino acid sequences or their ligand specificities, WW domains have been classified into four groups (Bedford et al., 2000). Both of the WW domains in DWWA are similar to those previously identified in YAP65 or Nedd4, which have been classified to group I, i.e., they bind polypeptides with the minimal core consensus of PPXY. GFP-ΔWWD2, which lacks the WW domain 2, localized to the cell cortex normally, whereas GFP-WWD2, which lacks everything but domain 2, was diffusely distributed throughout the cytoplasm. This suggests that whatever the WW domain 2 of DWWA is binding, it is not directly involved in recruiting DWWA to the cortex; rather, it perhaps recruits a binding partner to the cortex.
Finally, the C-terminal region of DWWA contains an IQ motif, which is a CaM binding motif found in a number of molecules (Bahler and Rhoads, 2002). Some IQ motifs bind CaM in a Ca2+-dependent manner, whereas others promote Ca2+-independent retention of CaM (Jurado et al., 1999). The IQ motif of DWWA bound apocalmodulin exclusively. Ca2+ signaling involving CaM has been implicated in a variety of cell functions (Hinrichsen, 1993; Hoeflich and Ikura, 2002), including cytokinesis. For instance, CaM and a CaM binding protein colocalized to the division furrow during cytokinesis in Tetrahymena, and the CaM antagonist W-7 blocked cytokinesis of synchronous Tetrahymena cells (Gonda et al., 1999). In addition, GFP-CaM has been observed on the septum in fission yeast (Moser et al., 1997) and in the cleavage furrow in HeLa cells; moreover, cytokinesis in HeLa cells was blocked or delayed by injection of a CaM-specific inhibitory peptide during early anaphase (Li et al., 1999). Dictyostelium has one calmodulin gene, calA (Liu et al., 1992), and one calmodulin-like gene, calB (Rosel et al., 2000). CalA mRNA is expressed during both the vegetative and development stages, whereas calB mRNA is expressed mainly during the development stage. Liu et al. (1992) constructed a strain in which calA expression could be conditionally suppressed through inducible expression of calA antisense RNA, and they found that cytokinesis was disrupted when cells expressing calA antisense RNA were cultured on substrates. The cytokinetic defect in these calA knockdown cells is similar to that of dwwA-null cells in that both cell types are unable to cleave the cytoplasmic bridges at the final step of cytokinesis. This striking similarity between dwwA-null cells and CaM knockdown cells, together with the demonstrated ability of DWWA's IQ motif to bind CaM, strongly suggests that DWWA and CaM cooperate to efficiently carry out the scission process. This hypothesis is further supported by two unpublished observations: that DWWA lacking the IQ motif (ΔC) was unable to genetically complement the defects of dwwA-null cells and that wild-type cells treated with 125 μM W-7, which inhibited CaM-DWWA binding in vitro, failed to cleave the cytoplasmic bridge between daughter cells (our unpublished data). Looked at another way, the phenotypic similarity between dwwA-null cells and CaM knockdown cells suggests that DWWA is a major effector of CaM in Dictyostelium. At the final step of cytokinesis, the plasma membrane must undergo a major rearrangement so that fusion occurs between the two opposing membrane bilayers, resealing the separated daughter cells. We propose that DWWA and Ca2+/CaM cooperate in these processes, but it remains unclear whether and how abnormalities of the actin cytoskeleton are related to defective membrane dynamics in dwwA-null cells.
It is noteworthy that a novel protein similar to DWWA was recently identified in human (Kremerskothen et al., 2003). Using yeast two hybrid screening, this protein, named KIBRA, was isolated as a Dendrin binding protein and found to possess two WW domains and a C2 domain. It is tempting to speculate that DWWA-like adaptor proteins are conserved in eukaryotic cells and carry out functions related to those described here.
Acknowledgments
We thank the Dictyostelium cDNA project in Japan and Dictyostelium genome project for allowing access the sequence information. We also thank the Japan Society for the Promotion of Science for the fellowship to A.N.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-05-0329. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-05-0329.
References
- Adachi, H., Hasebe, T., Yoshinaga, K., Ohta, T., and Sutoh, K. (1994). Isolation of Dictyostelium discoideum cytokinesis mutants by restriction enzyme-mediated integration of the blasticidin S resistance marker. Biochem. Biophys. Res. Commun. 205, 1808-1814. [DOI] [PubMed] [Google Scholar]
- Adachi, H., Takahashi, Y., Hasebe, T., Shirouzu, M., Yokoyama, S., and Sutoh, K. (1997). Dictyostelium IQGAP-related protein specifically involved in the completion of cytokinesis. J. Cell Biol. 137, 891-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler, M., and Rhoads, A. (2002). Calmodulin signaling via the IQ motif. FEBS Lett. 513, 107-113. [DOI] [PubMed] [Google Scholar]
- Bain, G., and Tsang, A. (1991). Disruption of the gene encoding the p34/31 polypeptides affects growth and development of Dictyostelium discoideum. Mol. Gen. Genet. 226, 59-64. [DOI] [PubMed] [Google Scholar]
- Bedford, M., Sarbassova, D., Xu, J., Leder, P., and Yaffe, M.B. (2000). A novel pro-Arg motif recognized by WW domains. J. Biol. Chem. 275, 10359-10369. [DOI] [PubMed] [Google Scholar]
- Chang, W.T., Newell, P.C., and Gross, J.D. (1996). Identification of the cell fate gene stalky in Dictyostelium. Cell 87, 471-481. [DOI] [PubMed] [Google Scholar]
- de Hostos, E.L., Rehfuess, C., Bradtke, B., Waddell, D.R., Albrecht, R., Murphy, J., and Gerisch, G. (1993). Dictyostelium mutants lacking the cytoskeletal protein coronin are defective in cytokinesis and cell motility. J. Cell Biol. 120, 163-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espanel, X., and Sudol, M. (1999). A single point mutation in a group I WW domain shifts its specificity to that of group II WW domains. J. Biol. Chem. 274, 17284-17289. [DOI] [PubMed] [Google Scholar]
- Fukui, Y. (1990). Actomyosin organization in mitotic Dictyostelium amoebae. Ann. N.Y. Acad. Sci. 582, 156-165. [DOI] [PubMed] [Google Scholar]
- Gerald, N., Dai, J., Ting-Beall, H., and De Lozanne, A. (1998). A role for Dictyostelium racE in cortical tension and cleavage furrow progression. J. Cell Biol. 141, 483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer, M. (1997). The mechanism and control of cytokinesis. Curr. Opin. Cell Biol. 9, 815-823. [DOI] [PubMed] [Google Scholar]
- Gonda, K., Katoh, M., Hanyu, K., Watanabe, Y., and Numata, O. (1999). Ca2+/calmodulin and p85 cooperatively regulate an initiation of cytokinesis in Tetrahymena. J. Cell Sci. 112, 3619-3626. [DOI] [PubMed] [Google Scholar]
- Hibi, N., Nagasaki, A., Takahashi, M., Yamagishi, A., and Uyeda, T.Q.P. (2004). Dictyostelium discoideum talin A is crucial for myosin II-independent and adhesion-dependent cytokinesis. J. Muscle Res. Cell. Motil. (in press). [DOI] [PubMed]
- Hinrichsen, R.D. (1993). Calcium and calmodulin in the control of cellular behavior and motility. Biochim. Biophys. Acta 1155, 277-293. [DOI] [PubMed] [Google Scholar]
- Hoeflich, K.P., and Ikura, M. (2002). Calmodulin in action: diversity in target recognition and activation mechanisms. Cell 108, 739-742. [DOI] [PubMed] [Google Scholar]
- Jurado, L.A., Chockalingam, P.S., and Jarrett, H.W. (1999). Apocalmodulin. Physiol. Rev. 79, 661-682. [DOI] [PubMed] [Google Scholar]
- Kay, B., Williamson, M., and Sudol, M. (2000). The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 14, 231-241. [PubMed] [Google Scholar]
- Kremerskothen, J., Plaas, C., Buther, K., Finger, I., Veltel, S., Matanis, T., Liedtke, T., and Barnekow, A. (2003). Characterization of KIBRA, a novel WW domain-containing protein. Biochem. Biophys. Res. Commun. 300, 862-867. [DOI] [PubMed] [Google Scholar]
- Kuspa, A., and Loomis, W. (1992). Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc. Natl. Acad. Sci. USA 89, 8803-8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C.J., Heim, R., Lu, P., Pu, Y., Tsien, R.Y., and Chang, D.C. (1999). Dynamic redistribution of calmodulin in HeLa cells during cell division as revealed by a GFP-calmodulin fusion protein technique. J. Cell Sci. 112, 1567-1577. [DOI] [PubMed] [Google Scholar]
- Liu, T., Williams, J.G., and Clarke, M. (1992). Inducible expression of calmodulin antisense RNA in Dictyostelium cells inhibits the completion of cytokinesis. Mol. Biol. Cell 3, 1403-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., Ito, K., Lee, R., and Uyeda, T.Q.P. (2000). Involvement of tail domains in regulation of Dictyostelium myosin II. Biochem. Biophys. Res. Commun. 271, 75-81. [DOI] [PubMed] [Google Scholar]
- Mabuchi, I., and Okuno, M. (1977). The effect of myosin antibody on the division of starfish blastomeres. J. Cell Biol. 74, 251-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser, M.J., Flory, M.R., and Davis, T.N. (1997). Calmodulin localizes to the spindle pole body of Schizosaccharomyces pombe and performs an essential function in chromosome segregation. J. Cell Sci. 110, 1805-1812. [DOI] [PubMed] [Google Scholar]
- Nagasaki, A., de Hostos, E.L., and Uyeda, T.Q.P. (2002). Genetic and morphological evidence for two parallel pathways of cell-cycle-coupled cytokinesis in Dictyostelium. J. Cell Sci. 115, 2241-2251. [DOI] [PubMed] [Google Scholar]
- Nagasaki, A., Hibi, M., Asano, Y., and Uyeda, T.Q.P. (2001). Genetic approaches to dissect the mechanisms of two distinct pathways of cell cycle-coupled cytokinesis in Dictyostelium. Cell Struct. Funct. 26, 585-5891. [DOI] [PubMed] [Google Scholar]
- Nagasaki, A., Sutoh, K., Adachi, H., and Sutoh, K. (1998). A novel Dictyostelium discoideum gene required for cAMP-dependent cell aggregation. Biochem. Biophys. Res. Commun. 244, 505-513. [DOI] [PubMed] [Google Scholar]
- Neujahr, R., Heizer, C., and Gerisch, G. (1997). Myosin II-independent processes in mitotic cells of Dictyostelium discoideum: redistribution of the nuclei, re-arrangement of the actin system and formation of the cleavage furrow. J. Cell Sci. 110 (Pt 2), 123-137. [DOI] [PubMed] [Google Scholar]
- Newton, A.C. (1995). Protein kinase C. Seeing two domains. Curr. Biol. 5, 973-976. [DOI] [PubMed] [Google Scholar]
- Palmieri, S.J., Nebl, T., Pope, R.K., Seastone, D.J., Lee, E., Hinchcliffe, E.H., Sluder, G., Knecht, D., Cardelli, J., and Luna, E.J. (2000). Mutant Rac1B expression in Dictyostelium: effects on morphology, growth, endocytosis, development, and the actin cytoskeleton. Cell Motil. Cytoskeleton 46, 285-304. [DOI] [PubMed] [Google Scholar]
- Rizo, J., and Sudhof, T.C. (1998). C2-domains, structure and function of a universal Ca2+-binding domain. J. Biol. Chem. 273, 15879-15882. [DOI] [PubMed] [Google Scholar]
- Robinson, D., and Spudich, J. (2000). Towards a molecular understanding of cytokinesis. Trends Cell Biol. 10, 228-237. [DOI] [PubMed] [Google Scholar]
- Rosel, D., Puta, F., Blahuskova, A., Smykal, P., and Folk, P. (2000). Molecular characterization of a calmodulin-like Dictyostelium protein CalB. FEBS Lett. 473, 323-327. [DOI] [PubMed] [Google Scholar]
- Ruppel, K.M., Uyeda, T.Q.P., and Spudich, J.A. (1994). Role of highly conserved lysine 130 of myosin motor domain. In vivo and in vitro characterization of site specifically mutated myosin. J. Biol. Chem. 269, 18773-18780. [PubMed] [Google Scholar]
- Spudich, J.A. (1989). In pursuit of myosin function. Cell Regul. 1, 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudol, M. (1996). Structure and function of the WW domain. Prog. Biophys. Mol. Biol. 65, 113-132. [DOI] [PubMed] [Google Scholar]
- Sudol, M., Sliwa, K., and Russo, T. (2001). Functions of WW domains in the nucleus. FEBS Lett. 490, 190-195. [DOI] [PubMed] [Google Scholar]
- Sussman, M. (1987). Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. In: Dictyostelium discoideum: Molecular Approaches to Cell Biology, vol. 28, ed. J.A. Spudich, Orlando: Academic Press, 9-29. [DOI] [PubMed] [Google Scholar]
- Weber, I., Gerisch, G., Heizer, C., Murphy, J., Badelt, K., Stock, A., Schwartz, J.M., and Faix, J. (1999). Cytokinesis mediated through the recruitment of cortexillins into the cleavage furrow. EMBO J. 18, 586-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienke, D.C., Knetsch, M.L., Neuhaus, E.M., Reedy, M.C., and Manstein, D.J. (1999). Disruption of a dynamin homologue affects endocytosis, organelle morphology, and cytokinesis in Dictyostelium discoideum. Mol. Biol. Cell 10, 225-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang, J.H., Cavet, G., Sabry, J.H., Wagner, P., Moores, S.L., and Spudich, J.A. (1997). On the role of myosin-II in cytokinesis: division of Dictyostelium cells under adhesive and nonadhesive conditions. Mol. Biol. Cell 8, 2617-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]