Abstract
Undergraduate biology education provides students with a number of learning challenges. Subject areas that are particularly difficult to understand include protein conformational change and stability, diffusion and random molecular motion, and molecular crowding. In this study, we examined the relative effectiveness of three-dimensional visualization techniques for learning about protein conformation and molecular motion in association with a ligand–receptor binding event. Increasingly complex versions of the same binding event were depicted in each of four animated treatments. Students (n = 131) were recruited from the undergraduate biology program at University of Toronto, Mississauga. Visualization media were developed in the Center for Molecular and Cellular Dynamics at Harvard Medical School. Stem cell factor ligand and cKit receptor tyrosine kinase were used as a classical example of a ligand-induced receptor dimerization and activation event. Each group completed a pretest, viewed one of four variants of the animation, and completed a posttest and, at 2 wk following the assessment, a delayed posttest. Overall, the most complex animation was the most effective at fostering students' understanding of the events depicted. These results suggest that, in select learning contexts, increasingly complex representations may be more desirable for conveying the dynamic nature of cell binding events.
INTRODUCTION
An understanding of molecular biology at the undergraduate level is dependent on a student's ability to assimilate dynamic and increasingly complex cellular and molecular processes. In addition, many of the difficulties associated with teaching and learning molecular biology are linked to the emerging interdisciplinarity of the field, which demands an understanding of complex systems at several different levels of organization. Unfortunately, undergraduate learning environments, which are often characterized by lecture-based courses with high enrollment, are not always conducive to higher-order learning of complex subject matter. Rather, greater emphasis is placed on factual recall than on depth of understanding (Momsen et al., 2010). Moreover, the tools available to instructors to supplement lecture and lab are not necessarily well suited to the task at hand. Confusion and misconception on the part of the student may arise from misuse of tools that are not designed with the learning objectives in mind or from tools where the idealized or simplified representation of concepts is interpreted literally.
Most biology instructors would agree that visual representations are essential to learning molecular biology. Textbook illustrations, diagrams, animations, and interactive learning tools are commonly used to make sense of molecular and cellular phenomena. The ubiquity of the Web and increasing adoption of portable devices (PDAs, tablet computers, iPads, etc.) have resulted in a tremendous growth in the availability of visual media for teaching, learning, and research. Yet, surprisingly little is known about the efficacy of visual learning tools. As the field of molecular biology continues to evolve, driven by scientific and technical innovation, it becomes critical to understand not only the impact of such visuals on students but also to develop a visual language that maximizes pedagogical outcomes and helps animators and media designers develop educationally impactful pieces. In this study, we examine the relative effectiveness of three-dimensional visualization techniques for learning about molecular biology. Specifically, the study addresses the concepts of protein conformational change, molecular crowding, and random molecular motion in association with a membrane-receptor binding event.
Challenges Related to Undergraduate Molecular Biology Education
Molecular biology is a rapidly growing field that poses a number of challenges for both teachers and students. Contemporary biology has experienced a great shift away from reductionist thinking toward a much more integrated study of complex and interconnected systems. Where biology was once more focused on isolated evidence directly related to the field, it now draws upon findings in disciplines such as chemistry, mathematics, and computer science. A recent report issued by the National Research Council (2009) called for reform to undergraduate biology education (see Woodin et al., 2010, for a discussion and summary of this report). Its authors described the new biology as more integrative, reflecting real-world research and practice and adopting knowledge from a number of related disciplines (including, e.g., chemical and computational sciences) in hopes of fostering deeper understanding of biological systems. Addressing this challenge demands a collective effort to change undergraduate biology education, an effort focused on interdisciplinarity and providing students and teachers with the skills to understand the connections within these scientific disciplines (Labov et al., 2010).
In a discussion of teaching and research challenges related to the field, Tibell and Rundgren (2010) identify four areas of key importance in addressing the future of education in molecular biology: 1) challenges relating to content selection, 2) challenges relating to understanding at multiple levels of abstraction, 3) challenges relating to domain-specific language, and 4) challenges relating to the use of visualization in molecular biology education. Content selection poses a great challenge to instructors, who are expected to keep pace with the rapid rate at which new information is generated in this field. For the novice student, the choice of content to support learning is even more difficult, given the student's inability to distinguish between key and peripheral learning material. Moreover, students experience difficulty understanding biological concepts, as these concepts involve a number of interacting processes occurring at multiple scales of time and space. To make sense of these processes, “learners must be able to sift through complex information spaces, discriminate between important and unimportant information, and recognize critical patterns and relationships” (Tibell and Rundgren, 2010). In practice, students tend to approach biology as a series of unconnected ideas or theories, rarely integrating their knowledge and making connections with real-life phenomena (Tanner and Allen, 2005; Momsen et al., 2010). Subject areas of particular difficulty for students include protein conformational change and stability (Robic, 2010), diffusion and random molecular motion (Garvin-Doxas and Klymkowsky, 2008), and molecular crowding (Ellis, 2001), all of which are poorly understood, because they are often neglected in undergraduate curriculum. Students have trouble reconciling the perceived efficiency of biological systems with the concept of randomness. In addition to the challenge of trying to come to terms with the full complexity of the molecular world, students entering biology are faced with learning a new “visual” language for which they are not prepared.
The Visual Language of Science
Biology is an inherently visual domain. Much of what we know about cell and molecular structure is derived from imaging technologies, such as x-ray crystallography and electron microscopy. These techniques provide us with a glimpse of the great complexity of the molecular world. In addition, we rely upon a range of visual depiction conventions and strategies to describe different aspects of these structures. Molecular structures may be represented in a number of ways, from detailed models showing individual atoms to coarse molecular surface representations. Understanding these depictions requires students to familiarize themselves with the visual language used to describe a world operating at multiple levels of organization. Indeed, the visual conventions used to represent molecular structure and function can be very challenging for students to understand and can lead to a number of conceptual and reasoning difficulties (Schönborn and Anderson, 2006). In part, this is owing to a lack of necessary visualization skills on the part of the student, who is newly exposed to this material. The undergraduate biology curriculum does not include training in visual literacy. Rather, students are expected to “catch on” and acquire these skills as they learn (Flannery, 2006; Schönborn and Anderson, 2006). The design and presentation of the visual learning tool also greatly impacts upon students' interpretation of a scientific phenomenon. For example, difficulties may arise from visual explanations in which phenomena are represented with “deceptive clarity” (Tasker and Dalton, 2006; Harris et al., 2009; Linn et al., 2010). This is certainly true of visual representations that offer an oversimplified explanation of scientific concepts for the sake of clarity. In this scenario, students may recall a sequence of events but retain only a superficial understanding of the concept overall. Conversely, visual explanations that introduce extraneous complexity not relevant to the learning goal may be equally misleading. Visually rich materials are often borrowed from sources that do not “contextualize” them for classroom use. Since teachers find it challenging to remain up-to-date in the rapidly evolving areas of the life sciences, the value of such visuals is diluted, and their impact is lowered. However, when carefully designed, with a clear learning objective in mind, scientific visualizations are powerful tools for describing the intricacies of cellular and molecular systems. Perhaps more than in any other area of science, visualization helps biologists grasp the complexity of events that are both too small to see with the naked eye (or microscope, in the case of biomolecules) or too rapid to observe. Aside from being the “only option” for representing such complex events in space and time, visual explanations are often more engaging and memorable than other forms of communication.
The Role of Three-Dimensional Visualization in Science Education
Visualization as an emergent field has tremendous potential to contribute to advancing science curricula, and it is a discipline that is rapidly expanding. Three-dimensional visualizations can be powerful tools of intuition, playing a critical role in transforming the way we think about the cellular and molecular world. In particular, they are expected to have an impact on the ability of students to assimilate complex spatial and temporal events, and studies suggest that teachers are increasingly positive in their use, acceptance, and attitudes toward these kinds of visually rich media (Schönborn and Anderson, 2006; Harris et al., 2009). To date, however, research examining the educational impact of visualizations is both contradictory and inconclusive (Tversky et al., 2002; Linn, 2003; Linn et al., 2008; Lowe, 2003; O'Day, 2007). In addition, a significant portion of the existing professionally produced three-dimensional cell and molecular visualizations used in the classroom lack the requisite scientific accuracy and mechanism-based design approaches that we believe are critical in the context of education. This is, in part, due to an acknowledged gap between what is known by practicing scientists and what is taught at postsecondary institutions (Howitt et al., 2008; Tibell and Rundgren, 2010). However, as discussed, there are difficulties associated with the design of communication tools themselves.
The benefits of three-dimensional visualization require a software platform that combines both the level of control and computing power demanded by the gaming and entertainment industries, while maintaining the accuracy and data import capabilities of dedicated molecular graphics software. This hybrid platform has been realized with the recent development of several plug-ins for three-dimensional animation packages from the entertainment industry—namely the Embedded Python Molecular Viewer (ePMV; Johnson et al., 2011), BioBlender (Andrei et al., 2010), and our own software toolkit Molecular Maya (mMaya; McGill, in preparation).The goal of visualization developed in this context is to merge data sets from otherwise nonintersecting fields and more accurately represent dynamic cellular landscapes (McGill, 2008; Iwasa, 2010). We feel this goal of “data integration through visualization” has positive implications both for scientists and for students, who will come to appreciate the importance of integrating knowledge from various sources.
Overview of the Experiment
The experiment reported here was structured as a repeated-measures design with a between-subject factor. We examined at three time points the relative effectiveness of three-dimensional visualization techniques for learning about molecular biology, specifically protein conformation and molecular motion in association with a membrane-receptor binding event. Student participants were randomly assigned to one of four animated treatments. Increasingly complex versions of the same receptor–ligand binding event were depicted in each of these treatments. We hypothesized that, as a result of viewing simpler versions, students would perform better on straightforward questions, such as “Does A bind to B?” We expected the opposite to be true when we asked students about more difficult, abstract concepts relating to the random nature of molecular binding events (where the simpler versions do not provide any visual cues of such behavior, while the more complex ones do). We were interested in understanding how different visual variables map to the students' performance on test questions ranging from more straightforward, fact-based understanding to more abstract intuitions of protein behavior at the molecular scale.
METHODS
Participants
Students (n = 131; year 1: 19; year 2: 52; year 3: 33; year 4: 27; age range = 18–24) were recruited from the undergraduate biology program at University of Toronto, Mississauga. Participants each received a $20 gift certificate for the University of Toronto Bookstore upon finishing the study. A condition of enrollment in the study was the completion of a first-year introductory cell biology course. We felt it was important that students have a basic understanding of cell biology in order to benefit from viewing the visualizations.
Materials
Stimulus Materials.
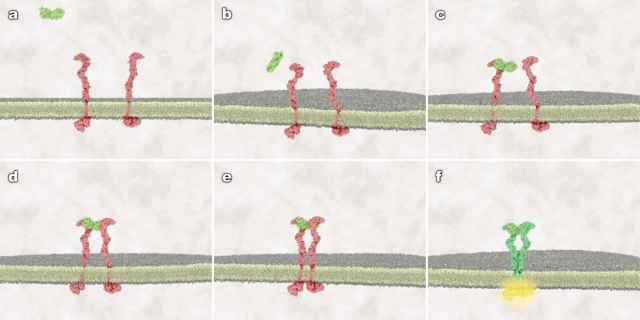
Visualization media were developed at the Center for Molecular and Cellular Dynamics at Harvard Medical School in collaboration with Digizyme (Brookline, MA). Stem cell factor (SCF) ligand and cKit receptor tyrosine kinase were used as a classical example of a ligand-induced receptor dimerization and activation event (Figure 1).
Figure 1.
Static frames from treatment 2 (random protein motion, receptor conformational flexibility, and membrane fluidity) that show the different stages of binding and receptor activation. (A) ligand in the extracellular space, receptor as monomers; (B) ligand approaches membrane, conformationally flexible receptors diffuse in the plane of membrane; (C) ligand randomly encounters and binds a receptor monomer, no receptor activation occurs; (D) ligand-bound and unbound receptors continue to diffuse, trimer forms, no immediate activation occurs; (E) ligand-induced tether of receptor extracellular domains allows cytoplasmic tails to make contact; and (F) contact between cytoplasmic tails leads to cross-phosphorylation and receptor activation.
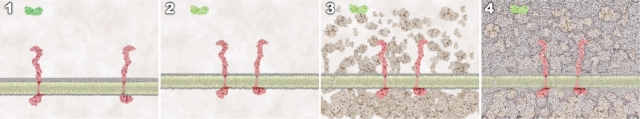
The timing and framing of the event were treated consistently across the four animations. With each successive treatment, additive layers of visual complexity were integrated (shown in Figure 2).
Figure 2.
Identical opening frames from four animations representing the same receptor–ligand binding event: 1) treatment 1 depicts the event using smooth, directed motion of the ligand toward the receptors; 2) treatment 2 introduces random, nondirected (Brownian) motion of the ligand and the receptors; 3) treatment 3 builds upon treatment 2 and introduces molecular crowding; 4) treatment 4 builds upon treatment 3 and introduces molecular water.
Not only are the overall shapes of cKit and SCF proteins relatively simple (in comparison with other, more complex receptors [e.g., epidermal growth factor receptor], a recent crystallographic study provided us with the accurate conformation of both the ligand-bound and unbound states of cKit (Yuzawa et al., 2007). Using the three-dimensional software Autodesk Maya in combination with our custom mMaya toolkit, we were able to: 1) import the Protein Data Bank crystallographic data sets for both unbound and ligand-bound states of the receptor; 2) create a surface-mesh representation of SCF and cKit that highlighted the overall secondary structure domains (namely D1D2D3, D4, D5, transmembrane [TM], and cytoplasmic [cyto] domains); 3) rig the receptor according to this secondary structure organization (D1D2D3•D4•D5•TM•cyto, where • represents a joint between domains); and 4) animate not only the proteins moving relative to one another in the scene, but also the motion of individual domains within cKit (as it undergoes ligand-induced conformational changes). The four three-dimensional animations are available for viewing through the following website: www.molecularmovies.com/bindingstudy/index.html.
Test Materials and Measures.
The evaluation instrument and ethics application (IRB) were completed at the University of Toronto concurrent with the development of materials. The study was held at the University of Toronto in a computer lab equipped with 40 iMac computers (21.5-inch display).
The learning and testing materials used in this study were developed based on a review of popular North American textbooks, and in particular, those texts used at University of Toronto (Alberts et al., 2002, 2009; Lodish et al., 2007). Moreover, questions were reviewed by the second author of the study, who holds a PhD in cell and molecular biology. Each of three test instruments (pretest, posttest, and delayed posttest) used in this study was composed of 10 short-answer questions. This testing format was used to discourage students from guessing at the right answer. The test instruments were piloted on four students, and questions were modified based upon student feedback. The tests assessed students' understanding in three areas: 1) protein conformation, 2) molecular motion, and 3) molecular crowding. Each test included questions to measure both students' surface-level understanding and their deep-level understanding. A subset of questions across the instruments was isomorphic, while other questions were designed to begin assessing students' abilities to infer broader concepts from the content of the animations. Indeed, both the posttest and delayed posttest included questions more predictive in nature, which were intended to measure students near transfer of knowledge. Examples of each question type are included in Table 1.
Table 1.
Examples of assessment questions included in each of three test instruments
| Level of measurement | Question | Test instrument |
|---|---|---|
| Surface | Proteins are inherently rigid structures. Is this statement true or false? Please explain. | Pretest |
| How many functional units or domains would you say [KIT] has? | Posttest | |
| Would you describe the movement of molecules through the extracellular space as random or directed? Please explain. | Delayed posttest | |
| Deep | What are the forces that contribute to the conformation of a protein? | Pretest |
| How is the binding of the ligand SCF to the receptor KIT mediated? Please explain. | Posttest | |
| How does SCF drive the dimerization of KIT? Please explain. | Delayed posttest | |
| Transfer | What do you think would happen if the temperature were increased in this environment? Please explain. | Posttest |
| Knowing that temperature impacts upon water vibration, what do you think would have happened in the animation you viewed if the temperature had been decreased? Please explain. | Delayed |
Procedure
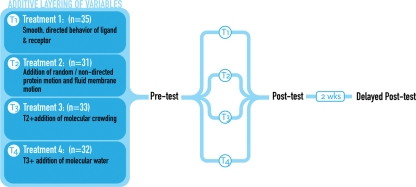
The design comprised four instructional conditions, each described in Figure 3. Students were randomly assigned to four experimental groups (G1 = 35; G2 = 31; G3 = 33; G4 = 32). Each participant was assigned a random number and received an instructional package explaining the study, a request for written consent, and a background information form. Students were assessed individually, and each assessment took approximately 40 min. Each participant completed a pretest, for which no feedback was provided. The pretest served as a baseline measure for ensuring equivalent prior knowledge in each of the four groups.
Figure 3.
Experimental protocol for assessing the impact of visual variables upon students' understanding of a receptor-ligand binding event.
Participants then viewed one of four variants of the animation. Students were given a brief description of the subject matter and were informed that the animation did not include any identifying labels or audio and that this was deliberate. Students were instructed to view the animation as many times as they desired. Upon viewing the animation, students completed a posttest assessing their factual and conceptual understanding of molecular binding events. At 2 wk following the assessment, students were asked to complete an online delayed posttest (in Blackboard), also made up of 10 short-answer questions. Students' answers at all three time points were scored as correct (1 point) or incorrect (0 points) out of a possible 10 points for each test.
Analyses
A repeated-measures analysis of variance (ANOVA), with time as the within-subjects factor, test scores as the dependent variable, and group assignment as the between-subjects factor, was conducted to measure the impact of treatment over time. To fulfill the assumptions of the repeated-measures ANOVA, we also conducted Levene's test for homogeneity of variance and Mauchly's test for the compound symmetry of the variance–covariance matrix. In addition, follow-up pairwise comparisons were conducted to determine which of the means differed significantly from one another.
RESULTS
The result of Levene's test at pretest (F(3,127) = 1.13, p = 0.339), posttest (F(3,127) = 1.24, p = 0.296), and delayed posttest (F(3,127) = 2.58, p = 0.056) indicate that homogeneity of variance was not violated. As well, Mauchly's criterion for our data (0.0085, p = 0.096) indicates that the assumption of sphericity was not violated. The results of the repeated-measures ANOVA, summarized in Table 2, show that test scores varied significantly between pretest, posttest, and delayed-posttest assessment (Wilk's Λ = 0.665, F(2,126) = 31.74, p < 0.001, multivariate η2 = 0.33).
Table 2.
Summary of results
| Mean test scores (and SD) | |||
|---|---|---|---|
| Group assignment | Pretest | Posttest | Delayed posttest |
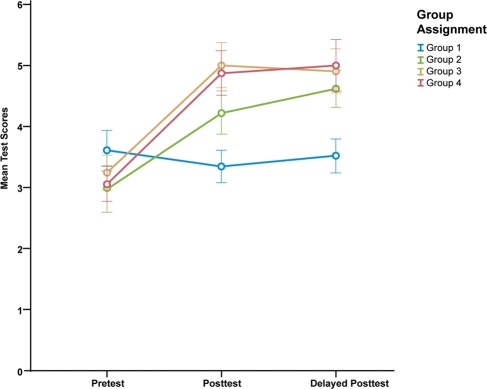
| Group 1 (n = 35) | 3.60 (1.97) | 3.34 (1.57) | 3.51 (1.67) |
| Group 2 (n = 31) | 2.97 (2.10) | 4.23 (1.98) | 4.61 (1.69) |
| Group 3 (n = 33) | 3.24 (1.64) | 5.00 (2.12) | 4.91 (2.08) |
| Group 4 (n = 32) | 3.06 (1.66) | 4.88 (2.07) | 5.00 (2.38) |
The impact of the treatment (group assignment) was also significant (Wilk's Λ = 0.795, F(6,252) = 5.09, p < 0.001, multivariate η2 = 0.11), accounting for 11% of the difference in test scores.
Post hoc analyses were conducted to evaluate pairwise differences among means at each of three time points. The Bonferroni procedure was used to control for type 1 error across comparisons. The results showed no significant differences between groups at pretest. There were significant differences between groups at posttest (F(3,127) = 5.19, p = 0.002, partial η2 = 0.11) and delayed posttest (F(3,127) = 4.10, p = 0.008, partial η2 = 0.10). There were notable differences between treatments 1 and 2 at both posttest and delayed posttest; however, these results failed to reach significance. Smaller differences were also found when comparing groups 2, 3, and 4. There were significant differences between group 1 and groups 3 and 4 at both posttest (G1 and G3 p < 0.01; G1 and G4 p < 0.05) and delayed posttest (G1 and G3 p < 0.05; G1 and G4 p < 0.05). The results are summarized in Table 3 and Figure 4.
Table 3.
Post hoc comparisons of posttest and delayed posttest results
| Posttest | Delayed posttest | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Group (I) | Group (J) | Mean (I–J) | SE | Significance | Group (I) | Group (J) | Mean (I–J) | SE | Significance |
| 1 | 2 | −0.88 | 0.479 | 0.405 | 1 | 2 | −1.10 | 0.487 | 0.154 |
| 3 | −1.66 | 0.471 | 0.004 | 3 | −1.39 | 0.479 | 0.025 | ||
| 4 | −1.53 | 0.475 | 0.010 | 4 | −1.49 | 0.483 | 0.015 | ||
Figure 4.
Plot showing mean test scores at three time intervals.
A one-way ANOVA of posttest results by question type revealed insignificant differences in student performance on surface-level (basic) questions (F(3, 127) = 0.539, p = 0.657). Student performance on questions measuring depth of understanding (advanced), however, differed significantly (F(3, 127) = 10.935, p > 0.001, partial η2 = 0.21). Further post hoc analysis (Bonferroni) identified significant differences in scores on advanced questions between groups 1 and 3 (p < 0.001) and groups 1 and 4 (p < 0.001). A more detailed breakdown of these results is reported in Table 4.
Table 4.
Post hoc comparisons of posttest results by question type
| Posttest basic questions | Posttest advanced questions | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Group (I) | Group (J) | Mean (I–J) | SE | Significance | Group (I) | Group (J) | Mean (I–J) | SE | Significance |
| 1 | 2 | −0.17 | 0.218 | 1.00 | 1 | 2 | −0.76 | 0.350 | 0.191 |
| 3 | 0.10 | 0.214 | 1.00 | 3 | −1.80 | 0.344 | 0.000 | ||
| 4 | −0.06 | 0.216 | 1.00 | 4 | −1.51 | 0.347 | 0.000 | ||
A comparison of delayed posttest results by question type showed significant differences in group performance on basic questions (F(3, 127) = 10.199, p < 0.001, partial η2 = 0.19). Further post hoc analysis (Bonferroni) revealed significant differences between groups 1 and 2 (p = .025) and groups 1 and 3 (p < 0.001), and groups 1 and 4 (p < 0.001). However, differences in group performance on questions testing depth of understanding (advanced) were not significant. A more detailed breakdown of these results is reported in Table 5.
Table 5.
Post hoc comparisons of delayed posttest results by question type
| Delayed posttest basic questions | Delayed posttest advanced questions | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Group (I) | Group (J) | Mean (I–J) | SE | Significance | Group (I) | Group (J) | Mean (I–J) | SE | Significance |
| 1 | 2 | −0.61 | 0.210 | 0.025 | 1 | 2 | −0.62 | 0.363 | 0.545 |
| 3 | 0.10 | 0.207 | 0.000 | 3 | −0.29 | 0.358 | 1.000 | ||
| 4 | −0.94 | 0.208 | 0.000 | 4 | −0.45 | 0.360 | 1.000 | ||
DISCUSSION
The present study aimed at identifying the impact of visual complexity upon students' understanding of dynamic molecular events. The primary focus of this experiment was to examine the role of key visual variables in supporting undergraduate students' understanding of corresponding features of the molecular environment that are not well understood at this level of study. These include: protein conformational changes, random molecular movement, and molecular crowding. Our data show that students' overall performance improved significantly with increasing levels of visual complexity.
In assessing student understanding of surface-level information, while we hypothesized that the simpler animations might be more effective at conveying basic concepts, this was not the case. Regardless of whether students were exposed to the most simple or the most complex visual treatment, their ability to explain basic concepts on the posttest was comparable. Furthermore, participants assigned to the more complex treatments (groups 3 and 4) scored significantly higher on basic questions in the delayed posttest than did students in group 1.
In accordance with our second hypothesis, we observed that increasingly complex animations (3 and 4) fostered greater understanding of abstract concepts related to molecular binding events. Students assigned to these groups scored significantly higher on advanced questions in the posttest than their counterparts in group 1. However, students' scores on more advanced questions in the delayed posttest were comparable across all four groups. In other words, the learning effects of the more complex visualizations were lasting, but only with regard to the more basic concepts. The results of the present study prompt us to ask how we can better design visualizations to support depth of understanding over the long term.
Finally, there are certain limitations to the study that need to be addressed in future studies. In designing this experiment, we were focused solely on the impact of visual variables in fostering understanding. Given these parameters, we were careful not to introduce confounding factors, such as narration in the form of audio or text. While we felt this was a necessary omission in the design of the stimuli, it detracted from the overall enjoyment and educational benefit of the animations for the participants. Although there was improvement in students' scores, the mean test scores in the higher-achieving groups was still a modest five out of a possible 10 points. In this way, the visualizations were not ideally suited to teaching, nor were they representative of animated media that students typically encounter in an educational setting or online environment. For visualizations to be maximally effective, according to the theory of dual processing (Paivio, 1986), they should leverage both the viewer's visual and verbal cognitive-processing skills.
A second potential limitation of the study concerns a possible lack of equivalency across the three test instruments. While we attempted to isomorphically match questions in each pretest, posttest, and delayed posttest set, this was not always possible. This was due to the fact that we wanted to ask students questions pertaining specifically to the events depicted in the animated treatments, that is, abstract, predictive questions that would test near transfer of knowledge. The largely unchanging scores of group 1 (the least educationally impactful treatment) across three time points suggest that the tests were indeed equivalent. However, the lack of a negative control in this study limits our ability to generalize from the results.
A third possible limitation of the study relates to the performance of students assigned to the more complex visual treatment. That students in groups 3 and 4 significantly outperformed group 1 participants begs the question: Did the more complex representations actually foster deeper understanding or merely provide these students with more information? Without more in-depth analysis of student responses, this is a difficult question to answer. However, it is worth noting here that the performance level of group 2 was much closer to that of groups 3 and 4 than it was to group 1, even though treatment 2 contained the same level of detail as the one shown to group 1. The single feature that distinguished treatment 2 from treatment 1 was the addition of random motion and conformational flexibility of the membrane receptors. Those small changes did not provide entirely new information, but rather encouraged students to 1) question the behavior and motion of the ligand in relation to the membrane receptors and 2) observe the structural flexibility of the membrane receptors in relation to ligand-induced activation. Our data also show that performance in group 1 decreased slightly between pretest and posttest, suggesting that this treatment might actually have been harmful to students. It should be noted that the majority of students performed similarly in this group across each of the three time points. There was no correlation between level of expertise (year, number of courses completed) and a decrease in test score between the first and second tests.
Despite the limitations of the present study, it suggests some interesting implications for the use of animated visualizations in undergraduate molecular biology. We set out to examine whether a more complex representation of a membrane-receptor binding event would impact positively or negatively upon students' understanding of molecular environments. It would appear that students were able to focus on the more perceptually salient aspects of the animation regardless of the level of detail. Both the ligand and protein receptor made use of explicit color cues as a means of helping the viewer focus on the main narrative. This finding is consistent with Lowe's (1999) study of the extraction of information from complex animation.
With this study, we have attempted to demonstrate that complexity may well be what is needed in some learning contexts to convey the truly dynamic nature of the molecular realm. As educators, we tend to rely largely on readily available static and highly schematized representations of proteins outside their cellular context—representations that lack critical structural and kinetic information and convey misconceptions about the nature of the molecular world. The insights emerging from recent advances in the study of structural cell and molecular biology call for more sophisticated visual representations. This is true not only for purposes of scientific communication among scientists, but also for undergraduate and graduate students, who are expected to develop an understanding of the cellular/molecular realms as they proceed with their studies. By studying the impact of visual variables in effectively representing protein structure and function, we hope we have drawn attention to the importance of studying the representational features that best foster understanding of these complex and dynamic processes.
Acknowledgments
The authors thank Eric Keller for work on the three-dimensional visualizations, Campbell Strong for his work on Molecular Maya, Stephen Harrison for his guidance with the study, and Jeannie Park for support with the study website development. This research was supported by a Gordon Research Conference Minigrant from the National Science Foundation.
REFERENCES
- Alberts B, Bray D, Hopkin K, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Essential Cell Biology. 3rd ed. New York: Garland Science; 2009. p. 860. [Google Scholar]
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 4th ed. New York: Garland Science; 2002. p. 1616. [Google Scholar]
- Andrei RM, Callieri M, Zini MF, Loni T, Maraziti G, Pan MC, Zoppè M. BioBlender: A Software for Intuitive Representation of Surface Properties of Biomolecules. 2010. http://arxiv.org/ftp/arxiv/papers/1009/1009.4674.pdf (accessed 28 October 2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ. Macromolecular crowding: obvious but underappreciated. Trends Biochem Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- Flannery MC. Thinking in pictures. Am Biol Teach. 2006;68:299–303. [Google Scholar]
- Garvin-Doxas K, Klymkowsky MW. Understanding randomness and its impact on student learning: lessons learned from building the Biology Concept Inventory (BCI) CBE Life Sci Educ. 2008;7:227–33. doi: 10.1187/cbe.07-08-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Peck RF, Colton S, Morris J, Chaibub Neto E, Kallio J. A combination of hand-held models and computer imaging programs helps students answer oral questions about molecular structure and function: a controlled investigation of student learning. CBE Life Sci Educ. 2009;8:29–43. doi: 10.1187/cbe.08-07-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt S, Anderson T, Costa M, Hamilton S, Wright T. A concept inventory for molecular life sciences: how will it help your teaching practice? Australian Biochemist. 2008;39:1–4. [Google Scholar]
- Iwasa JH. Animating the model figure. Trends Cell Biol. 2010;20:699–704. doi: 10.1016/j.tcb.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Johnson GT, Autin L, Goodsell DS, Sanner MF, Olson AJ. ePMV embeds molecular modeling into professional animation software environments. Structure. 2011;19:293–303. doi: 10.1016/j.str.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labov JB, Reid AH, Yamamoto KR. Integrated biology and undergraduate science education: a new biology education for the twenty-first century? CBE Life Sci Educ. 2010;9:10–16. doi: 10.1187/cbe.09-12-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linn MC. Technology and science education: starting points, research programs, and trends. Int J Sci Educ. 2003;25:727–758. [Google Scholar]
- Linn MC, Chang HY, Chiu JL, Zhang HZ, McElhaney K. Can desirable difficulties overcome deceptive clarity in scientific visualizations? In: Benjamin A., editor. Successful Remembering and Successful Forgetting: A Festchrift in Honor of Robert A. Bjork. New York: Routledge; 2010. pp. 239–262. [Google Scholar]
- Linn MC, et al. Proceedings of the 8th International Conference of the Learning Sciences, part 3. Utrecht, The Netherlands: International Society of the Learning Sciences; 2008. Improving the design and impact of interactive, dynamic visualizations for science learning; pp. 221–228. www.isls.org/icls2008. [Google Scholar]
- Lodish H, Berk A, Kaiser CA, Krieger M, Scott MP, Bretscher A, Ploegh H, Matsudaira P. Molecular Cell Biology. 6th ed. New York: W. H. Freeman; 2007. p. 1296. [Google Scholar]
- Lowe RK. Extracting information from an animation during complex visual learning. Eur J Psychol Educ. 1999;14:225–244. [Google Scholar]
- Lowe RK. Animation and learning: selective processing of information in dynamic graphics. Learn Instruct. 2003;13:157–176. [Google Scholar]
- McGill G. Molecular movies… coming to a lecture near you. Cell. 2008;133:1127–1132. doi: 10.1016/j.cell.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Momsen JL, Long TM, Wyse SA, Ebert-May D. Just the facts? Introductory undergraduate biology courses focus on low-level cognitive skills. CBE Life Sci Educ. 2010;9:435–440. doi: 10.1187/cbe.10-01-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council. A New Biology for the 21st Century: Ensuring the United States Leads the Coming Biology Revolution. Washington, DC: National Academies Press; 2009. p. 112. [PubMed] [Google Scholar]
- O'Day DH. The value of animations in biology teaching: a study of long-term memory retention. CBE Life Sci Educ. 2007;6:217–223. doi: 10.1187/cbe.07-01-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paivio A. Mental Representations: A Dual Coding Approach, Oxford, England: Oxford University Press. 1986 [Google Scholar]
- Robic S. Ten common misconceptions about protein structure, folding, and stability. CBE Life Sci Educ. 2010;9:189–195. doi: 10.1187/cbe.10-03-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönborn KJ, Anderson TR. The importance of visual literacy in the education of biochemists. Biochem Mol Biol Educ. 2006;34:94–102. doi: 10.1002/bmb.2006.49403402094. [DOI] [PubMed] [Google Scholar]
- Tanner K, Allen D. Approaches to biology teaching and learning: understanding the wrong answers—teaching toward conceptual change. Cell Biol Educ. 2005;4:112–117. doi: 10.1187/cbe.05-02-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasker R, Dalton R. Research into practice: visualisation of the molecular world using animations. Educ Res. 2006;7:141–159. [Google Scholar]
- Tibell LAE, Rundgren C-J. Educational challenges of molecular life science: characteristics and implications for education and research. CBE Life Sci Educ. 2010;9:25–33. doi: 10.1187/cbe.08-09-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tversky B, Morrison JB, Betrancourt M. Animation: can it facilitate? Int J Hum Comput Stud. 2002;57:247–262. [Google Scholar]
- Woodin T, Carter VC, Fletcher L. Vision and change in biology undergraduate education, a call for action—initial responses. CBE Life Sci Educ. 2010;9:71–73. doi: 10.1187/cbe.10-03-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzawa S, Opatowsky Y, Zhang Z, Mandiyan V, Lax I, Schlessinger J. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell. 2007;130:323–34. doi: 10.1016/j.cell.2007.05.055. [DOI] [PubMed] [Google Scholar]