Abstract
Superoxide dismutases (SOD) convert superoxide radicals into less damaging hydrogen peroxide. The opportunistic human pathogen Candida albicans is known to express CuZnSOD (SOD1) and MnSOD (SOD3) in the cytosol and MnSOD (SOD2) in the mitochondria. We identified three additional CuZn-containing superoxide dismutases, SOD4, SOD5, and SOD6, within the sequence of the C. albicans genome. The transcription of SOD5 was up-regulated during the yeast to hyphal transition of C. albicans, and SOD5 was induced when C. albicans cells were challenged with osmotic or with oxidative stresses. SOD5 transcription was also increased when cells were grown on nonfermentable substrates as the only carbon source. The Rim101p transcription factor was required for all inductions observed, whereas the Efg1p transcription factor was specifically needed for serum-modulated expression. Deletion of SOD5 produced a viable mutant strain that showed sensitivity to hydrogen peroxide when cells were grown in nutrient-limited conditions. Sod5p was found to be necessary for the virulence of C. albicans in a mouse model of infection. However, the sod5 mutant strain showed the same resistance to macrophage attack as its parental strain, suggesting that the loss of virulence in not due to an increased sensitivity to macrophage attack.
INTRODUCTION
Aerobic eukaryotic pathogens can encounter superoxide radicals (O2-) generated from several sources. These sources can be internal or external. An important internal source is the mitochondrial respiratory chain (Boveris, 1978; Casteilla et al., 2001; Lenaz, 2001), and thus the rate of respiration can have a significant impact on reactive oxygen species (ROS) production. A key external source of ROS encountered by pathogens is from phagocytes. The superoxide radical is the first intermediate in the oxidative burst generated in the phagosome, and this burst is thought to be involved in pathogen killing (Reeves et al., 2002). The superoxide radicals are known to inactivate [4Fe-4S] cluster-containing enzymes by oxidizing one iron and releasing it from the cluster (Liochev and Fridovich, 1994; Fridovich, 1995). Free iron can react with hydrogen peroxide to generate toxic hydroxyl radicals (OH-) by Fenton chemistry (Fridovich, 1978; Meneghini, 1997). The hydroxyl and superoxide radicals react with cellular components, resulting in oxidation of proteins and nucleic acids as well as lipid peroxidation. These effects can lead to inactivation of enzymes, to disruption of membranes, to mutations, and ultimately to cell death (Halliwell and Gutteridge, 1990, 1999).
To reduce the harmful effects of superoxide radicals, cells express detoxifying enzymes. Superoxide dismutase (SOD) is an antioxidant enzyme involved in elimination of superoxide anions; it catalyzes the reaction: O2- + O2- + 2H+ → H2O2 + O2. Normally, H2O2 is still toxic to the cell; therefore, another enzyme, catalase, converts it to water. Superoxide dismutases can be classified according to metal cofactor(s) bound to them: there are iron (FeSOD), manganese (MnSOD), copper and zinc (CuZnSOD), and nickel (NiSOD) versions of the enzymes. The CuZnSOD family is thought to have evolved separately, and thus these enzymes share no sequence similarity with other SOD families (Kanematsu and Asada, 1991; Smith and Doolittle, 1992).
An important aerobic eukaryotic pathogen, Candida albicans, causes the majority of human fungal infections. These infections range from thrush in immunocompetent colonized hosts, to life-threatening systemic infections in immunocompromised individuals such as patients with cancer (Corner and Magee, 1997). C. albicans can grow in several morphological forms: it can proliferate as a budding yeast, and it can also form filaments, such as true hyphae and pseudohyphae. Hyphal formation of this fungus can be induced by high temperature, basic pH, and by the presence of serum, and this morphological transition is implicated in pathogenesis of C. albicans (Brown and Gow, 1999).
Typically, eukaryotes express cytosolic CuZnSOD and mitochondrial MnSOD (Fridovich, 1995). C. albicans possesses both cytosolic CuZnSOD and mitochondrial MnSOD encoded by the SOD1 and SOD2 genes, respectively (Hwang et al., 1999; Rhie et al., 1999). In addition to these proteins, a C. albicans cytoplasmic MnSOD (the SOD3 gene product) was recently identified (Lamarre et al., 2001). SOD1 was shown to form homodimers (Lamarre et al., 2001), whereas SOD2 and SOD3 were shown to form homotetramers (Rhie et al., 1999; Lamarre et al., 2001). C. albicans Sod1p was shown to protect cells against superoxide radicals produced by macrophage, and it was shown to be important for the virulence of C. albicans in mouse model (Hwang et al., 2002). C. albicans catalase was also shown to be involved in virulence of this pathogen. C. albicans Sod2p was shown to have no effect on the virulence aspect of this fungus but rather protects the cell against intracellularly produced superoxides (Hwang et al., 2003). C. albicans seems to coordinate the production of CuZnSOD and MnSOD in a reciprocal manner: CuZnSOD shows the highest activity in the midexponential phase, whereas MnSOD has the highest activity upon the entry to stationary phase (Rhie et al., 1999; Lamarre et al., 2001). The morphology of C. albicans is coupled to CuZnSOD production because cells generate increased amounts of CuZnSOD during the switch from yeast to hyphal forms (Gunasekaran et al., 1998). In particular, the conditions of elevated production of CuZnSOD coincide with increased levels of ROS produced by C. albicans as cells produce the highest levels of ROS, including superoxide, during hyphal growth and in exponential phase (Schroter et al., 2000). In addition, C. albicans generates higher amounts of ROS when grown in the presence of salt (Schroter et al., 2000; Sander et al., 2002).
We have identified three new potential C. albicans superoxide dismutase genes. We studied the expression, regulation, and function of one of these genes, SOD5, and show that this gene is under a complex regulation of expression. Functional tests show no involvement of SOD5 in defense against macrophage-induced killing. However, the effects of the deletion of SOD5 on virulence may suggest the involvement of Sod5p in removal of superoxides produced by the host.
MATERIALS AND METHODS
C. albicans and Bacterial Strains
The C. albicans strains are listed in Table 1; plasmids and oligonucleotides used in this study are listed in Table 2. The C. albicans strains SC5314 (Fonzi and Irwin, 1993), HLC52 (Lo et al., 1997), JKC19 (Liu et al., 1994), DAY25 and DAY185 (Davis et al., 2000a), BWP17 (Wilson et al., 1999), and GKO6 (Davis et al., 2002) were used for transcription analysis; RM1000 (Negredo et al., 1997) was used as a parental strain to generate the RM1000-HU, H10, UH16, UH16-1, UH16-2, and UH16-3 strains. The Escherichia coli strain DH5α was used for all plasmid constructions and maintenance. The plasmid pVEC (Magee and Magee, 1997) was used for the generation of plasmids p37, p37H, p37U, and pSOD5-110; plasmids p5921 (Fonzi and Irwin, 1993) and pRM100 (Pla et al., 1995) were used for the amplification of hisG-CaURA3-hisG and CaHIS1 markers, respectively.
Table 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| SC5314 | Wild-type isolate | Fonzi and Irwin (1993) |
| RM1000 | URA3Δ :: λimm434/URA3Δ :: λimm434 HIS1 :: hisG/HIS1 :: hisG | Negredo et al. (1997) |
| RM1000-HU | URA3Δ :: λimm434/URA3Δ :: λimm434 HIS1 :: hisG/HIS1 :: hisG SOD5/SOD5 :: [p37H] :: [p37U] | This study |
| H10 | URA3Δ :: λimm434/URA3Δ :: λimm434 HIS1 :: hisG/HIS1 :: hisG sod5Δ :: HIS1/SOD5 | This study |
| UH16 | URA3Δ :: λimm434/URA3Δ :: λimm434 HIS1 :: hisG/HIS1 :: hisG sod5Δ :: hisG-URA3-hisG/sod5Δ :: HIS1 | This study |
| UH16-1 | URA3Δ :: λimm434/URA3Δ :: λimm434 HIS1 :: hisG/HIS1 :: hisG sod5Δ :: hisG/sod5Δ :: HIS1 | This study |
| UH16-2 | URA3Δ :: λimm434/URA3Δ :: λimm434 HIS1 :: hisG/HIS1 :: hisG sod5Δ :: hisG/sod5Δ :: HIS1 :: [p37U] | This study |
| UH16-3 | URA3Δ :: λimm434/URA3Δ :: λimm434 HIS1 :: hisG/HIS1 :: hisG sod5Δ :: HIS1/sod5Δ :: hisG :: [pSOD5-110] | This study |
| HLC52 | URA3Δ :: λimm434/URA3Δ :: λimm434 efg1Δ :: hisG/efg1Δ :: hisG-URA3-hisG | Lo et al. (1997) |
| JKC19 | URA3Δ :: λimm434/URA3Δ :: λimm434 cph1Δ :: hisG/cph1Δ :: hisG-URA3-hisG | Liu et al. (1994) |
| DAY25 | URA3Δ :: λimm434/URA3Δ :: λimm434 HIS1 :: hisG/HIS1 :: hisG arg4 :: hisG/arg4 :: hisG rim101: ARG4/rim101 :: URA3 | Davis et al. (2000a) |
| DAY185 | URA3Δ :: λimm434/URA3Δ :: λimm434 HIS1 :: hisG/HIS1 :: hisG ARG4 :: URA3 :: arg4 :: hisG/arg4 :: hisG | Davis et al. (2000a) |
| BWP17 | URA3Δ :: λimm434/URA3Δ :: λimm434 HIS1 :: hisG/HIS1 :: hisG arg4 :: hisG/arg4 :: hisG | Wilson et al. (1999) |
| GKO6 | URA3Δ :: λimm434/URA3Δ :: λimm434 HIS1 :: hisG/HIS1 :: hisG arg4 :: hisG/arg4 :: hisG mds3 :: Tn7-UAU1/mds3 :: Tn7-URA3 | Davis et al. (2002) |
Table 2.
Plasmids and oligos used in this study
| A. Plasmids | Description | Source |
|---|---|---|
| pVEC | CaARS CaURA3 | Magee and Magee (1997) |
| p37 | CaARS upstream flank 1kb CaSOD5 downstream flank 1kb | This study |
| p37H | CaARS upstream flank 1kb CaHIS1 downstream flank 1kb | This study |
| p37U | CaARS upstream flank 1kb hisG-CaURA3-hisG downstream flank 1 kb | This study |
| pSOD5-110 | CaARS CaURA3 upstream flank 1kb CaSOD5 downstream flank 1kb | This study |
| p5921 | hisG-CaURA3-hisG | Fonzi and Irwin (1993) |
| pRM100 | CaARS CaHIS1 | Pla et al. (1995) |
| B. Oligos | Sequence | Used for |
|---|---|---|
| ODH157(EcoRI) | GGAATTCAAACCTGGTCTACCAGAAGTT | SOD5 + 1kb flank F |
| ODH158 (EcoRI) | GGAATTCCGGTTATGTTAACGGTTAGAG | SOD5 + 1kb flank R |
| OMM1 (BamHI) | CGGGATCCGATGAATGGTAAGTTAGATTGATAT | Diverge SOD5 F |
| OMM2 (BamHI) | CGGGATCCGAAAAATAAAATAGATGAGCCATTT | Diverge SOD5 R |
| OMM3 (XbaI) | GCTCTAGAAAAGAAAAATAAAATAGATGAGCC | Diverge SOD5 R |
| OMM4(XbaI) | GCTCTAGAGGTACCTGGAGGATGAGGAG | CaHIS1 F |
| CB20R(BamHI) | CGGGATCCAATATTTATGAGAAACTATCACTTC | CaHIS1 R |
| OMM8 | GGAGCAGTAGAAGCCATACTA | SOD5 R Northern |
| OMM9 | ATGGGCGAGTCCTACAAAACT | SOD5 F Northern |
| OMM10 | ACCAGTGAATCATTTGAAGTT | SOD4 F Northern |
| OMM11 | AGAAGCACTAGTTGATGAACC | SOD4 R Northern |
| OMM14 | TCCAGAAGATGATGAAAGACATG | SOD1 F Northern |
| OMM15 | GTCTAGCACCAGCATGACCAG | SOD1 R Northern |
| OMM16 | AACACAACTTGCTTTGAGTTGAAA | SOD6 F Northern |
| OMM17 | ACAATCACTAGAGATTCCGTGGAT | SOD6 R Northern |
| OMM18(BamHI) | CGGGATCCCGGTTATGTTAACGGTTAGAG | SOD5 + 1kb flank R |
| OMM19 (XbaI) | GCTCTAGAAAACCTGGTCTACCAGAAGTT | SOD5 + 1kb flank F |
| UO106 | AAGCCGGTTTTGCCGGTG | ACT1 probe F |
| UO107 | TGGTGAACAATGGATGGAC | ACT1 probe R |
Media and Culture Growth Conditions
C. albicans strains were routinely cultured on YPD (1% yeast extract, 2% peptone, 2% dextrose, pH 6) medium at 30°C. Cells with integrated selection markers or disrupted genes were cultured in synthetic glucose medium containing 0.67% yeast nitrogen base, 0.15% amino acids, 2% dextrose, and appropriate supplements. All media were supplemented with uridine (25 μg/ml) for the growth of C. albicans Ura auxotrophic strains.
Because under certain conditions, C. albicans cells form hyphae at 37°C, it is difficult to accurately measure the growth of cells in the culture media. We therefore prepared a standard growth-curves YPD media at 37°C. We observed that it takes 300 min for SC5314 strain to reach an OD600 of 1.0 at 30°C, and it takes 225 min for SC5314 strain to reach an OD600 of 1.0 at 37°C. It takes 325 min for RM1000-HU and UH16-2 strains to reach an OD600 of 1.0 at 30°C, and it takes 250 min for RM1000-HU and UH16-2 strains to reach an OD600 of 1.0 at 37°C. These standard curves were used to approximate the time when the conditions should have changed.
For the Northern analyses performed with the yeast and mycelial forms of C. albicans, blastospores were first grown overnight at 30°C in the following media: mammalian cell culture media IMDM (pH 6.5) (Invitrogen, Carlsbad, CA), YPD, YP, or YP supplemented with either 2% galactose (YPGal), 3% glycerol (YPGlyc), or 1.5% methanol (YPMeth). The precultures were then diluted to an OD600 of 0.1 and incubated in the same media at either 30°C until the OD600 reached 1.0, or at 37°C until the OD600 reached 2.5. In other cases, the preculture grown in YPD medium was diluted to an OD600 of 0.1 and then incubated until the OD600 reached 1.0 at either 30 or 37°C in the following media: YPD, YPD supplemented with 10% serum, YPD adjusted to pH 8, YPD with 0.3 M NaCl, and YPD with 0.3 M NaCl at pH 8.
For some samples, the preculture grown in YPD medium was diluted to an OD600 of 0.1 and then incubated at either 30 or 37°C in YPD. In those cultures the pH, the NaCl concentration or both of those factors were increased 10, 30, or 60 min before the OD600 reached 1.0. In those conditions, the pH of the media was increased to 8.0 with 10 N NaOH; the salt concentration was increased to 0.3 M by adding appropriate amount of 5 M NaCl solution into the media.
In other cases, the preculture grown in YPD medium was diluted to an OD600 of 0.1. The culture was then supplemented with reactive oxygen species generators 1 h before the OD600 reached 1.0 at 30°C, or 1 h before the OD600 reached 2.5 at 37°C. In this case, cells were grown in YPD with 0.4 mM H2O2, YPD with 0.5 mM menadione (Sigma-Aldrich, St. Louis, MO), or YPD with 0.5 mM riboflavin (Sigma-Aldrich) (illuminated with 100-W table-lamp). All liquid cultures were rotated at 250 rotations per minute. Spot-assays were done on media solidified with 2% agar. 5′-Fluoroorotic acid (Diagnostic Chemicals, Charlottetown, PEI, Canada) containing medium was prepared as described previously (Boeke et al., 1984), except that uridine was substituted for uracil. C. albicans cells were transformed by the previously described method (Chen et al., 1992).
RNA Preparation and Northern Blot Analyses
Total RNA was extracted by the hot phenol extraction method (Carlson and Botstein, 1982). The Northern blot analyses were performed by the previously described method (Alarco et al., 1997) with the following modifications. RNA samples (80 μg) were electrophoresed on a 7.5% formaldehyde, 1% agarose gel and transferred by capillarity to a Zeta-Probe nylon membrane (Bio-Rad, Mississauga, ON, Canada). Detection of specific RNAs was performed by hybridization at 65°C in 0.5 M NaPO4, pH 7.2, 1 mM EDTA, 7% SDS, 1% bovine serum albumin with 50 ng of 32P-labeled DNA probes (single-stranded DNA was omitted from this solution). Probes were generated by using the random prime labeling system (Amersham Biosciences, Piscataway, NJ) with the following modifications: 50 ng (rather than 25 ng) of DNA was added to the reaction, the reaction was incubated at 37°C for 45 min (rather than for 10 min), and all of labeled probe (rather than 1.25 ng) was used for hybridization. The unincorporated labeled nucleotides were removed by G-50 microcolumns (Amersham Biosciences). Membranes were washed once at 65°C with a solution containing 40 mM NaPO4, pH 7.2, 5% SDS, 1 mM EDTA, 0.5% bovine serum albumin for 10 min (rather than twice for 20 min), and once with a solution containing 40 mM NaPO4, pH 7.2, 1% SDS, and 1 mM EDTA for 20 min (rather than four times for 20 min) and were visualized using a PhosphorImager (model 4255; PerkinElmer Life Sciences, Boston, MA).
All Northern probes were made by polymerase chain reaction (PCR): the 0.24-kb-long SOD5 probe used primers OMM8 and OMM9, the 0.24-kb-long SOD4 probe used primers OMM10 and OMM11, the 0.21-kb-long SOD1 probe used primers OMM14 and OMM15, and the 0.5-kb-long SOD6 probe used primers OMM16 and OMM17. A CaACT1 probe was generated with UO106 and UO107 oligos and was used as an internal control to monitor RNA loading and transfer (a gift from Dr. Ursula Oberholzer, Biotechnology Research Institute, Montreal, PQ, Canada).
The dot-blot was performed the same way as the Northern, with the difference that 80 μg of SOD1, SOD4, SOD5, and SOD6 unlabeled probes (rather than 80 μg of RNA) were spotted on the membranes.
Deletion and Reversion of SOD5
To delete the SOD5 gene, the SOD5 open reading frame (ORF) and its 1-kb flanking regions were PCR amplified from genomic DNA of the CAI4 strain with oligos ODH157 and ODH158 that contained EcoRI sites. The PCR product (2.6 kb) and plasmid pVEC were digested with EcoRI, and a 4.7-kb EcoRI fragment of pVEC was gel-purified and ligated with the EcoRI-digested 2.6-kb SOD5 PCR product. Clone p37 contained an insert with the SOD5 ORF running in the opposite orientation with respect to that of the Amp marker. The SOD5 ORF was deleted from p37 by the use of inverse PCR by using either oligonucleotides OMM1 and OMM2 or OMM1 and OMM3. The hisGURA3-hisG marker was cut from p5921 with BglII and BamHI; OMM1 and OMM2 create BamHI restriction sites in p37 into which the hisG-URA3-hisG marker was inserted to yield p37U. The HIS1 marker was PCR amplified from pRM100 with OMM4 (containing an XbaI site) and CB20R (containing a BamHI site, a gift from Dr. Catherine Bachewich, Biotechnology Research Institute, Montreal, PQ, Canada); OMM1 and OMM3 created BamHI and XbaI restriction sites, respectively, in p37 into which the HIS1 marker was ligated to yield p37H. The HIS1 and SOD5-1-kb-flanking regions were PCR amplified from p37H with oligos ODH157 and ODH158, and this PCR product was transformed into RM1000 to yield the H10 strain, in which the sod5::HIS1 fragment replaces the chromosomal SOD5 ORF. The hisG-URA3-hisG and SOD5-1-kb-flanking regions were cut out of p37U with XmnI and SpeI, and this fragment was transformed into H10 to yield the UH16 strain, in which the sod5::hisG-URA3-hisG fragment replaces the second chromosomal SOD5 ORF. Strain UH16 was plated on 5′-fluoroorotic acid-containing plates to select for Ura-auxotrophs. As a result, the UH16-1 strain was created in which one SOD5 ORF was replaced with HIS1 marker, and the second SOD5 ORF was replaced with hisG. To create the prototrophic strain UH16-2, UH16-1 was transformed with the p37U construct, which was targeted to the chromosomal SOD5-promoter region by cutting the construct in the SOD5-promoter region with NsiI. The HIS1 URA3 auxotrophic wild-type strain was transformed with the p37U and p37H constructs in a sequential manner; these were targeted to chromosomal SOD5-promoter region by cutting the constructs in the SOD5-promoter region with NsiI, creating strain RM1000-HU. For SOD5 reversion studies, the SOD5 ORF and its 1-kb-flanking regions were PCR amplified from CAI4 genomic DNA with the OMM18 and OMM19 oligos that contained BamHI and XbaI sites, respectively. The resulting PCR product (2.6 kb) and plasmid pVEC were double digested with BamHI and XbaI restriction enzymes and ligated together to yield pSOD5-110. pSOD5-110 was transformed into UH16-1 to yield revertant strain UH16-3.
Genomic DNA was prepared from all of the obtained strains, digested with NheI and XhoI, and analyzed for the correct integration events by Southern blotting with a digoxigenin system (Roche Diagnostics, Indianapolis, IN). The digoxigenin probe was generated by PCR with ODH157 and OMM1 oligos that amplified the 1-kb SOD5 promoter sequence from p37. To confirm the absence of the SOD5 ORF, genomic DNA from UH16, UH16-1, and UH16-2 strains were cut with DraI, and analyzed by Southern blotting with a probe that binds inside the SOD5 ORF. This probe was also used for Northern analysis to confirm the absence of SOD5 mRNA in the SOD5 inducing conditions (see MATERIALS AND METHODS).
Protein Preparation and In-Gel SOD Activity Assay
Protein preparation was done as described previously (Ito-Kuwa et al., 1999). The protein concentration was determined by the Bradford method using an assay kit (Bio-Rad), as described by the provider. SOD activity in polyacrylamide gels was detected using nitroblue tetrazolium (Beauchamp and Fridovich, 1971), with the modification that 5 mg of cell extracts was electrophoresed on 10% nondenaturing polyacrylamide gels in triplicates. After the electrophoresis, one gel was used for Coomassie Blue staining (Malloy et al., 1984). The other two gels were used for determining SOD activity with or without 4 mM KCN (Ito-Kuwa et al., 1999).
Spot Assay and Growth Assay
Overnight, cultures of C. albicans cells were concentrated to OD600 of 20.0. Serial twofold dilutions were done, and 5 μl of cells was spotted onto a plate and incubated for 48 h at either 30 or 37°C. Spot assays were done on the types of media described in RESULTS. The growth assay of cells in liquid media was done by diluting overnight cells in those media to OD600 of 0.1. The growth rate was monitored at OD600 for 24 h at 30 and 37°C.
Macrophage and Virulence Assays and Fungal Burden
The end point dilution survival assay was performed as described previously (Marcil et al., 2002). Dilutions of C. albicans without macrophages where colonies could be distinctively visualized were counted toward the lower limit and compared with the same dilutions with macrophages. Colonies from a total of at least 72 wells per condition were used to provide data.
For virulence assays, C. albicans strains were passaged overnight at 30°C in YPD before infection. Cells were harvested, washed in cold phosphate-buffered saline (Wisent Inc., St.-Bruno, Canada), and counted using a hemacytometer. Then cells were resuspended at a density of 5.75 × 106 cells/ml. B6 mice (8-wk-old females) (Charles River Laboratories, Wilmington, MA) were infected through the lateral tail vein with 200 μl (1.15 × 106 organisms). Mouse survival was monitored at least three times daily, and moribund mice were killed by CO2 gas. We used C. albicans strain RM1000-HU to infect 13 mice, and UH16-2 and UH16-3 to infect 12 mice. C. albicans colony-forming units (cfu) in kidney were counted from live animals killed at day 5 after the infection. The kidneys were removed aseptically from four mice per strain; the organs were weighed, homogenized, diluted in sterile saline, and plated on YPD plates. Colonies were counted after incubation of the plates at 30°C for 48 h, and the results were expressed as cfu per gram of infected organ.
RESULTS
Identification of SOD4, SOD5, and SOD6
The mammalian pathogen C. albicans is known to possess both a copper-zinc-dependent superoxide dismutase (CuZnSOD encoded by SOD1) (Hwang et al., 1999) and a manganese-dependent superoxide dismutase (MnSOD, encoded by SOD3) (Lamarre et al., 2001) in the cytosol and to encode a second MnSOD (SOD2) that is localized to mitochondria (Rhie et al., 1999). The C. albicans genome sequence revealed three additional open reading frames with high sequence homology to the copper-zinc family of superoxide dismutases: orf 19.2062, orf 19.2060, and orf 19.2108 in version 19 of Stanford's C. albicans genome sequence assembly (Stanford Genome Technology Center, http://wwwsequence.stanford.edu/group/candida/). We have designated these new genes as SOD4, SOD5, and SOD6 respectively. All three of these genes share sequence homology to the previously described SOD1 and to each other, but share no homology to the manganese-containing superoxide dismutases (Figure 1A). All three predicted proteins (Sod4p, Sod5p, and Sod6p) contain characteristic residues involved in copper and zinc binding, and amino acids involved in dimer contact (Fink and Scandalios, 2002) (Figure 1B). Northern analysis (described below) showed that the mRNAs for each of the three genes were ∼700 nucleotides in length. However, the predicted length of Sod6p of 316 amino acids was too large to be easily accommodated within this size of message. This discrepancy could be resolved if SOD6 contained an intron. Nucleotide sequence analysis shows that SOD6 contains a well-characterized 3′ splicing site “CTAGG” motif starting at nucleotide 752, a signal for lariat formation “AACTGAC” starting at nucleotide 723, and a possible 5′ splicing site “GTCGGT” starting at nucleotide 462, which closely resembles the previously reported “GTANGT” motif (Ballance, 1986). Splicing of this potential intron would generate a predicted ORF of 218 amino acids, close in size to the predicted 232 amino acids and 228 amino acids of Sod4p and Sod5p. All these proteins are likely to be cytosolic, because they lack mitochondria-targeting sequences. Intriguingly, SOD4 and SOD5 are close together in the genome, with SOD4 located 2 kb upstream of SOD5 (Nantel et al., 2002).
Figure 1.
Similarity of the CuZn-containing and Mn-containing superoxide dismutases in C. albicans. (A) Phylogenetic tree shows the similarity between all C. albicans' superoxide dismutases: Sod4p, Sod5p, and Sod6p are homologous to Sod1p, and the CuZnSODs share no sequence similarity with the MnSODs. The analysis was produced by the Multiple Sequence Alignment program of Florence Corpet (http://prodes.toulouse.inra.fr/multalin/multalin.html). (B) Amino acid sequence alignment of CuZn Superoxide dismutases (SOD1, SOD4, SOD5, and SOD6) and Mn superoxide dismutases (SOD2 and SOD3). Deduced amino acid sequences were aligned with the CLUSTALW program. Identical amino acids are boxed in black, and conservative changes are boxed in gray. Hyphens indicate gaps that were introduced to maximize the alignment. +, oligos used for Northern and Southern analysis for Sod4p, Sod5p, and Sod6p. #, oligos used for Northern and Southern analysis for Sod1p. The Sod4p, Sod5p, and Sod6p contain CuZn-SOD characteristic residues. Asterisks below Cu and Zn stand for copper and zinc metals bindings, respectively; asterisks below DC stand for amino acids involved in dimer contact (Fink and Scandalios, 2002). Numbers at the ends of the sequences represent the amino acid lengths of the proteins.
SOD5 Is Transcriptionally Induced during Yeast to Hyphal Transitions
Hyphal formation in C. albicans can be induced by incubation of the cells in the presence of serum and high temperature. Recent transcriptional profiling experiments established that the SOD5 gene was induced during the yeast to hyphal switch (Nantel et al., 2002). To study the expression pattern of the copper-zinc SOD genes, we generated probes to unique sequences within each gene (Figure 1B). Dot-blot analysis confirmed the specificity of each probe; there was an insignificant cross-hybridization between the SOD4 and SOD5 probes, and no cross-hybridization for any of the other combinations (our unpublished data). These probes were used to investigate transcriptional regulation of the SOD5 gene under a variety of conditions. Northern blot analysis of the actin (ACT1) mRNA in C. albicans strains was used as an internal loading control, because the transcription of this gene was shown to be unaffected under most of the growth conditions tested in our study (Nantel et al., 2002). In agreement with microarray data, Northern analysis demonstrated that the SOD5 transcription increases at 37°C in the presence of serum, compared with the control condition of no serum and 30°C (Figure 2A, lanes 1 and 4). We investigated whether serum, temperature or both of these factors regulated the expression of SOD5. In the absence of serum, a temperature increase from 30 to 37°C reduced the expression of SOD5 from a relative intensity of 1.0-0.3 (Figure 2A, lines 1 and 2). In contrast, the presence of serum caused high levels of SOD5 expression (Figure 2A, compare line 1 with lines 3 and 4). This suggests that serum, rather than temperature, controls the induction of SOD5 expression.
Figure 2.
Hyphal-inducing conditions induce SOD5 expression. (A) Serum, rather than temperature, induces SOD5 transcription. Northern blot analysis of the SOD5 transcripts in C. albicans strain SC5314 grown in the log phase in indicated media (YPD and YPD containing 10% serum) at indicated temperatures. (B) SOD5 transcription is up-regulated in IMDM medium. Northern blot analysis of the SOD5 transcripts in C. albicans strain SC5314 grown in the log phase in IMDM medium at 30 and 37°C. Northern blot analysis of the actin (ACT1) transcripts in C. albicans strain SC5314 was used as an internal loading control. Relative intensities are shown below each of the genes analyzed. Relative intensities of cells grown at 37°C are underlined and quantified independently from SOD5 intensities obtained at 30°C. Quantifications were performed by NIH Image software (RSB-NIMH-NIH, http://rsb.info.nih.gov/nih-image/).
The mammalian cell culture medium IMDM lacks serum but allows C. albicans to grow exclusively in a hyphal form at both 30 and 37°C. SOD5 was highly expressed in this medium at both temperatures (Figure 2B). As in Lamarre et al., 2001, we observed the accumulation of new, 3.4-kb mRNA species hybridizing to the SOD5 probe (lane 4), which does not correspond to the specific SOD1, SOD4, or SOD6 mRNAs (our unpublished data). The nature of this mRNA is unclear, but because the distance between the start codon of SOD5 and the stop codon of SOD4 on the chromosome is 3.3 kb, it is possible that the SOD5 probe hybridized to a mRNA species that contains both SOD4 and SOD5. This idea is supported by the detection of the same message by a SOD4 probe (our unpublished data).
Another way to induce the yeast-to-hyphal transition is to increase the pH and the temperature of the medium. At both temperatures, 30 and 37°C, an increase of pH from 6.0-8.0 increases the expression of SOD5 (Figure 3A, lanes 6-9 and 18-21). At both temperatures, the rise in SOD5 mRNA expression persists up to 300 min for 30°C or 225 min for 37°C of growth in the media with an increased pH. The induction peaks at 30 min for cells grown at 30°C and at 10 min for cells grown at 37°C.
Figure 3.
Alkaline conditions, osmotic and oxidative stresses affect SOD5 transcription. (A) SOD5 transcription is up-regulated in alkaline conditions and in the presence of increased amounts of salt. Northern blot analysis of the SOD5 transcripts in C. albicans strain SC5314 diluted to an OD600 of 0.1 and then incubated at either 30 or 37°C in YPD. In those cultures, the pH, NaCl concentration, or both of those factors were increased 10, 30, 60, or 300/225 min before the OD600 reached 1.0. (B) Effects of oxidative shock on SOD5 and SOD1 of C. albicans. Northern blot analysis of the SOD5 transcripts in C. albicans strain SC5314 grown in log-phase, from OD600 of 0.1 to OD600 of 1.0 at 30°C or from OD600 of 0.1 to OD600 of 2.5 at 37°C; the hydrogen peroxide/menadione/riboflavin were supplemented 1 h before reaching the final OD600 of 1.0 or 2.5. Lanes 1 and 2, no oxidative shock was applied. Lanes 3 and 4, hydrogen peroxide was added 1 h before OD600 reached 1.0. Lanes 5 and 6, menadione was added 1 h before OD600 reached 1.0. Lanes 7 and 8, riboflavin and light were added 1 h before OD600 reached 1.0. Northern blot analysis of the actin (ACT1) transcripts in C. albicans strain SC5314 was used as an internal loading control. Analysis as described in Figure 2.
Osmotic and Oxidative Stresses Affect SOD5 Transcription
C. albicans cells grown in the presence of salt were shown to generate higher amounts of ROS (Schroter et al., 2000; Sander et al., 2002). Intriguingly, previous transcription profiling established that osmotic stress also induced the expression of SOD5 (Enjalbert et al., 2003). We confirmed these results by Northern analysis, because cells grown in the presence of high salt concentrations showed an increased transcription of SOD5 at 30°C (Figure 3A). The levels of SOD5 mRNA increase as early as 10 and 30 min after the addition of salt to the media at both 30 and 37°C but stay up after 60 and 300 min only at 30°C (Figure 3A, lanes 1-5 and 13-17).
When pathogens are engulfed by neutrophils, there is an increase in both pH and salt concentration inside the phagosome (Reeves et al., 2002). Intriguingly, high salt and high pH have a cumulative effect on SOD5 transcription at 30°C; at 37°C high salt and high pH yield to a higher SOD5 response compared with the levels of SOD5 transcripts induced separately by high pH and high salt (Figure 3A, lanes 10-12 and 22-23). The fact that high salt and high pH together lead to increased SOD5 expression was expected for a model that the expression increase permits the mobilization of superoxide dismutase activity to defend against phagosome-mediated killing.
In some organisms such as S. cerevisiae, the transcription of the gene encoding the cytoplasmic superoxide dismutase is induced by treatment with various oxidative agents like hydrogen peroxide and menadione (Yoo et al., 1999; Lamarre et al., 2001; Lee et al., 2002). We investigated the transcription profile of the C. albicans CuZn superoxide dismutases in cells treated either with hydrogen peroxide or superoxide radicals. We used two types of superoxide generators: riboflavin, which generates superoxides outside of the cell (Ito-Kuwa et al., 1999), and menadione, which creates superoxides inside the cell (Lee et al., 2002). The presence of hydrogen peroxide caused increased levels of SOD1 expression (Figure 3B, compare lane 1 with lanes 3 and 4) and moderate increases in transcription of SOD5. Menadione leads to moderate increases in transcription of SOD1 at both temperatures and that of SOD5 (Figure 3B, lanes 5 and 6). Riboflavin treatment results in elevated levels of SOD1 at 37°C and that of SOD5 at both temperatures (Figure 3B, lanes 7 and 8). The SOD1 induction was weak, as described previously (Lamarre et al., 2001). The fact that the transcription of SOD5 was increased upon the induction of oxidative stress is also expected if the gene is involved in defense against a phagosome-generated oxidative burst. Because SOD5 transcription was induced by both intracellular and extracellular superoxide generators, it provides us with the first evidence that Sod5p is needed for elimination of superoxides whose source is from within the cell.
Nonfermentable Carbon Sources Affect SOD5 Expression
Because we saw that the intracellular superoxide generator menadione induced SOD5 levels, we further investigated the effect of intracellularly generated superoxide on SOD5 transcription. Studies of isolated mitochondria showed that up to 80% of the superoxide in eukaryotic cells was produced in the mitochondria by electron leakage at the QH2: cytochrome c segment (complex III) during respiration (Chance et al., 1979). Therefore, the effect of respiration on SOD5 transcription was investigated. One of the ways to increase respiration is to grow the cells in media with a nonfermentable carbon source, such as glycerol or methanol, while providing glucose or galactose as the sole carbon source allows cells to grow by fermentation (Longo et al., 1996; De Freitas et al., 2000).
At 30°C, we grew cells until they reached an OD600 of 1.0. We observed that the growth on glucose or galactose as the only carbon source leads to relatively low levels of SOD5 transcription (Figure 4, lanes 1 and 3). When cells were grown in the presence of glycerol or methanol as the only carbon source at 30°C SOD5 expression was moderately increased (Figure 4, lanes 5 and 7), compared with fermentable carbon sources at the same temperature (compare with lanes 1 and 3). At 30°C, the highest SOD5 transcript levels were detected in cells grown in the absence of any carbon source (YP) (Figure 4, lane 9).
Figure 4.
SOD5 transcription is affected by nonfermentable carbon source. Northern blot analysis of the SOD5 transcripts in C. albicans strain SC5314 grown in log-phase, from OD600 of 0.1 to OD600 of 1.0 at 30°C or from OD600 of 0.1 to OD600 of 2.5 at 37°C in YPD, YPGal, YPGlyc, YPMeth, and YP. Northern blot analysis of the actin (ACT1) transcripts in C. albicans strain SC5314 was used as an internal loading control. Analysis as described in Figure 2.
At 37°C, we grew cells until they reached an OD600 of 2.5. We observed that the growth on glucose or galactose as the only carbon source leads to very low levels of SOD5 transcription (Figure 4, lanes 2 and 4). When cells were grown in the presence of glycerol or methanol as the only carbon source at 37°C, SOD5 expression was highly increased (Figure 4, lanes 6 and 8), compared with fermentable carbon sources at the same temperature (compare with lanes 2 and 4). At 37°C, the highest SOD5 transcript levels were detected in cells grown in the absence of any carbon source (YP) (Figure 4, lane 10). These results show that growth on nonfermentable carbon sources increase SOD5 transcription, which might suggest that one of the roles of Sod5p is to eliminate intracellularly generated superoxide radicals. SOD1 and SOD4 were also up-regulated by increased temperature when grown on nonfermentable carbon sources, and SOD1, SOD4, and SOD6 were also up-regulated in the absence of carbon sources (YP) (our unpublished data).
Transcription Factors Involved in the Regulation of SOD5 Expression
Previous transcription profiling established that the nonhyphal efg1 cph1 double mutant failed to induce SOD5 expression under hyphal-inducing conditions (Nantel et al., 2002). We used Northern analysis to determine the relative impact of each transcription factor on the expression of the SOD5 gene. For each of the individual null mutants, we attempted to induce SOD5 transcription by one of the following conditions: serum and high temperature, osmotic stress (0.3 mM NaCl), and basic pH (pH8). In all of these conditions, the cph1 mutant showed the same pattern of SOD5 expression as the wild-type strain: SOD5 was induced in all three conditions as compared with the noninduced condition (YPD 30°C) (Figure 5, lanes 1-8). On the other hand, the efg1 mutant failed to increase the transcription of SOD5 specifically under serum-inducing conditions (Figure 5, lanes 9-12, compare lanes 2 and 10). In addition, under noninducing conditions the efg1 mutant showed generally elevated levels of SOD5 message (Figure 5, compare lanes 1 and 9). Interestingly, the addition of salt to the media does not increase the SOD5 mRNA levels, compared with SOD5 levels during the growth of efg1 mutant strain under noninducing conditions (Figure 5, lanes 9 and 11). The SOD5 mRNA levels were increased in the efg1 mutant in alkaline-inducing conditions, although not to the same extent as the corresponding increase in the wild-type strain (Figure 5, lane 12). To investigate the factors responsible for the pH-mediated induction of SOD5 expression, we looked at cells defective in either the Rim101p transcription factor or in Mds3p; these factors represent independent branches of the pH response (Davis et al., 2002). In contrast to both the efg1 and cph1 mutants, the rim101 mutant blocked SOD5 induction under all of the conditions tested compared with SOD5 expression in its parental strain (Figure 5, lanes 13-20). Just as in the wild-type and the cph1 mutant, the mds3 mutant showed that SOD5 was induced in all three conditions (YPD 30°C) (Figure 5, lanes 21-28). Therefore, the Cph1p transcription factor and Mds3p have essentially no effect on SOD5 transcription under the conditions tested, Efg1p is required for SOD5 transcription in the presence of serum and perhaps for suppressing the SOD5 expression under the noninducing condition, and Rim101p is important for the normal SOD5 transcription under all of the conditions tested.
Figure 5.
Transcription factors involved in the transcription of SOD5. Northern blot analysis of the SOD5 transcripts in C. albicans strains lacking indicated transcription factors (strains SC5314, JKC19, HLC52, DAY25, DAY185, BWP17, and GKO6 [mds3]; Table 1). The potential transcription of SOD5 was induced by one of the three inducing conditions: presence of serum (10%) at 37°C, presence of salt (0.3M NaCl) for 1 h at 30°C, or switch to alkaline conditions (pH8) for 1 h at 30°C. Analysis as described in Figure 2. The intensities of SOD5 in HLC52 were compared with SOD5 intensities of JKC19. The intensities of SOD5 in DAY25 were compared with SOD5 intensities of DAY185. The intensities of SOD5 in mds3 were compared with SOD5 intensities of BWP17. These Northern blot analyses were reproduced twice. The general drop in SOD5 levels in rim101 was reproduced, but the difference in relative intensities of SOD5 in the rim101 strain was not significant.
SOD5 Is Not Essential for Viability in C. albicans
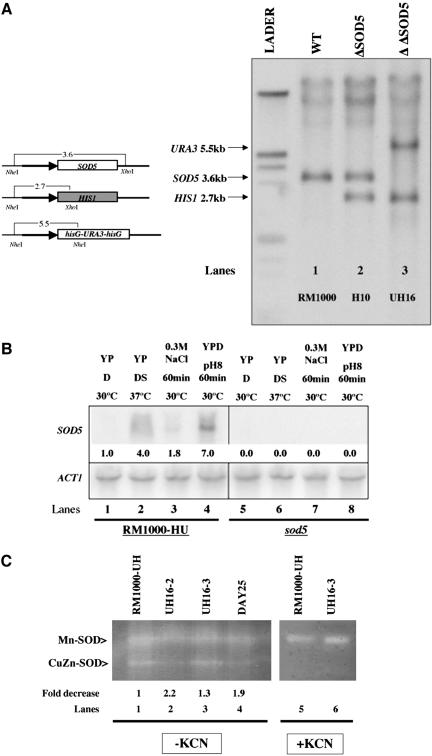
To study the function of Sod5p in C. albicans, we created a strain in which both copies of SOD5 were deleted. We generated a strain UH16 with the sod5::HIS1 sod5::hisG-URA3-hisG constructs replacing both copies of the SOD5 gene (see MATERIALS AND METHODS for details). The correct position of insertions during these steps was analyzed by Southern blotting (Figure 6A). The absence of the SOD5 gene was also confirmed with a Southern blot probed with a SOD5 internal fragment probe. This probe binds inside the SOD5 open reading frame exclusively, and it fails to generate a SOD5-corresponding band in the SOD5 double-mutant (our unpublished data). When the DNA was restricted with DraI, the SOD5 probe hybridized to an unexpected 7-kb DNA fragment, which could be derived from the accumulation of a 7-kb DNA fragment seen with ethidium bromide staining. The nature of this piece of DNA is unclear, but it might be derived from the multiple repeat sequence whose DraI restriction digest yields a 7-kb DNA fragment that has an affinity to the SOD5 probe (our unpublished data). In addition, the absence of SOD5 mRNA under either SOD5-inducing conditions (the presence of either serum or alkaline conditions or high pH) or noninducing conditions (YPD) was confirmed by Northern blotting by using the same internal probe (Figure 6B). No increase in SOD1, SOD4, and SOD6 transcript levels was observed in the sod5 mutant strain (UH16-2) (our unpublished data). Finally, we performed an in-gel activity assay that detects the activity of both types of superoxide dismutases, CuZnSOD and MnSOD. RM1000-HU cells grown in Iscove's modified Dulbecco's medium (IMDM) at 37°C express both MnSOD (Figure 6C, lane 1, top band) and high amounts of CuZnSOD (Figure 6C, lane 1, bottom band). On the other hand, UH16-2 cells showed reduced levels of the CuZnSOD activity (Figure 6D, lane 2, bottom band is missing) under the conditions tested. The CuZnSOD activity is restored in the UH16-3-revertant strain (Figure 6C, lane 3, bottom band). The rim101 mutant strain also showed reduced levels of the CuZn superoxide dismutase activity band (Figure 6C, lane 4). This observation is consistent with the results in Figure 5 and shows that the RIM101 mutant cells possess low levels of both SOD5 transcripts and Sod5p enzyme activity. The activity of the CuZnSOD can be inhibited by KCN, which blocked the CuZnSOD activity of RM1000-HU and UH16-3 cells grown in IMDM medium at 37°C (Figure 6C, lanes 5 and 6, bottom bands are missing). This shows that in IMDM media SOD5 seems to be a major contributor of overall CuZnSOD activity in C. albicans. The ability to generate the viable double-mutant strain shows that SOD5 is not an essential gene in C. albicans.
Figure 6.
SOD5 gene is deleted from UH16 strain. (A) Restriction endo-nuclease map of SOD5, of HIS1 inserted in place of SOD5 ORF and of hisG-URA3-hisG inserted in place of SOD5 ORF digested with NheI and XhoI. The arrows indicate 1 kb of SOD5 promoter region. Southern blot analysis with the use of a 1-kb probe generated with ODH157 and OMM1 oligos recognizes 1 kb of the SOD5 promoter region. The genomic DNA samples, digested with NheI and XhoI, were prepared from strains RM1000 (SOD5/SOD5; lane 1), H10 (Δsod5::HIS1/SOD5, lane 2), UH16 (Δsod5::hisG-URA3-hisG/Δsod5::HIS1, lane 3). The wild-type SOD5 band is 3.6kb, the band corresponding to the URA3 cassette is 5.5 kb, and the band corresponding to the HIS1 cassette is 2.7 kb. (B) Northern blot analysis of the SOD5 transcripts in the UH16-2 mutant and its parental strain, RM1000-HU. The transcription of SOD5 was induced by one of the inducing conditions: presence of serum (10%) at 37°C, or switch to alkaline conditions (pH 8) for 1 h at 30°C, or presence of 0.3 M NaCl for 1 h at 30°C for UH16-2 and RM1000-HU strains. Northern blot analysis of the actin (ACT1) transcripts in indicated C. albicans strains was used as an internal loading control. Analysis as described in Figure 2. (C) Superoxide dismutase activity staining revealed SOD activity by staining the 10% native polyacrylamide gel with nitro blue tetrazolium and riboflavin. A sample (5 mg) of the crude extract from exponentially growing cells in IMDM 37°C of RM1000-HU (lanes 1and 5), UH16-2 (lane 2), UH16-3 (lane 3 and 6), and DAY25 (lanes 4) was loaded into three gels. One gel (lanes 1-4) was stained for SOD activity in the absence of 4 mM KCN, the second gel (lanes 5-6) was stained for SOD activity in the presence of KCN, and the third gel was stained with Coomassie Blue as an internal loading control (our unpublished data). Relative intensities are shown below each of the genes analyzed. Quantifications were performed by NIH Image software (RSB-NIMH-NIH http://rsb.info.nih.gov/nih-image/).
Hydrogen Peroxide Together with Limited Nutrients Impairs Growth of the SOD5 Mutant Strain
We examined the growth properties of the sod5 mutant strain under conditions that were shown to increase the transcription of the SOD5 gene. The spot assay showed that there is no growth difference between the UH16-2 strain and the wild-type strain in medium that contained serum, NaCl, pH 8, or NaCl and pH 8. The sod5 mutant strain showed no heat sensitivity at 42°C, and no growth defect on Lee and Spider medium (our unpublished data). In addition, the sod5 mutant strain showed no defect with respect to hyphal development triggered by serum, IMDM media, or high pH (pH 8.0) at 37°C as shown by spot assays and growth in liquid media (our unpublished data).
In Saccharomyces cerevisiae, both of the single genes encoding CuZnSOD and MnSOD were found to be up-regulated during the metabolic transition from fermentation to respiration (Maris et al., 2001), and the S. cerevisiae CuZnSOD mutant possesses a reduced ability to grow by respiration in glycerol-rich medium (Longo et al., 1996; De Freitas et al., 2000). We therefore investigated the ability of the sod5 mutant strain UH16-2 to grow on media rich with nonfermentable carbons. We found that there was no growth difference in the liquid or solid medium that contained either glucose, galactose, glycerol, or methanol as a carbon source; in the absence of any of above-mentioned carbon source molecules, the SOD5 mutant grew as fast as its parental strain (our unpublished data). CuZnSOD deficient S. cerevisiae require lysine and methionine for aerobic growth (Bilinski et al., 1985), whereas superoxide dismutase defective E. coli require branched chain amino acids (Imlay and Fridovich, 1991). However, the C. albicans SOD5 mutant showed neither lysine, methionine, nor branched amino acid auxotrophies (our unpublished data).
The E. coli and in Helicobacter pylori SOD mutants were found to be more sensitive to the effect of hydrogen peroxide than its parental isogenic strain (Carlioz and Touati, 1986; Keyer et al., 1995; Seyler et al., 2001). We compared the growth rate of the SOD5 mutant with that of its parental isogenic strain. We found no growth difference between the SOD5 mutant and the wild-type strains grown on nutrient-rich media (YPD) supplemented with 1.0 mM hydrogen peroxide (Figure 7). However, the SOD5 mutant (UH16-2) grew poorly on SC (synthetic complete) media supplemented with 1.0 mM hydrogen peroxide, compared with its parental strain (Figure 7). The growth of UH16-3 strain showed an intermediate growth on SC media supplemented with hydrogen peroxide, consistent with the reintroduction of SOD5 in one copy into the UH16-1 background. The SOD5-phenotype suppression is observed in UH16-2 cells after incubation in SC media, which contains hydrogen peroxide and slight excess of fermentable glucose (2.5% glucose, rather than 2.0% of glucose) or in YPD media which contains hydrogen peroxide (Figure 7).
Figure 7.
Hydrogen peroxide and limited nutrients inhibit growth of UH16-2 strain. Spot assay of RM1000-HU, UH16-2, UH16-3 strains on SC or YPD, SC supplemented with hydrogen peroxide, and SC supplemented with 2,5% dextrose (SDD) and hydrogen peroxide media at 30°C for 48 h.
SOD5 Is Involved in Virulence of C. albicans but Is Not Involved in Defense of C. albicans against the Fungicidal Activity of Macrophages
The sensitivity to hydrogen peroxide is often linked to a decrease in virulence. To test directly the SOD5 involvement in macrophage-defense, the UH216-2 strain was incubated with RAW 264.7 mouse macrophage cells, and the percentage of survival (28.9 ± 5.7%) was not different from the survival of its parental RM1000-HU strain (27.6 ± 7.0%) and from the revertant UH16-3 strain (28.9 ± 7.0%). These results suggest that Sod5p is not required for C. albicans survival when cells are subjected to the fungicidal activity of macrophages.
To test directly the SOD5 involvement in an organism defense, we tested the virulence of the sod5 mutant strain in a mouse systemic infection model by intravenous tail infection (Figure 8). The results showed that 90% of the mice infected with the wild-type (RM1000-HU) died within 8 d of being inoculated. However, the homozygous sod5 mutant strain (UH16-2) had markedly diminished virulence compared with the wild-type parental strain (RM1000-HU). Mice infected with UH16-3 strain showed an intermediate survival rate consistent with the reintroduction of one copy of SOD5 gene into the UH16-1 background.
Figure 8.
Sod5p activity is required for virulence in animal model. Survival of mice infected with C. albicans sod5 mutant UH16-2 (squares), sod5 revertant UH16-3 (triangles), and wild-type parental RM1000-HU (circles) strains. Mice were inoculated by tail vein injection and survival was measured over a 30-d period.
The cfu in mice kidneys were determined on the fifth day after infection. One hundred percent of the kidneys recovered from mice infected with the wild-type parental strain (RM1000-HU) revealed an average of 1.0 × 106 cfu g-1 kidney at day of sacrifice. Seventy-five percent of the kidneys recovered from mice infected with the sod5 revertant strain (UH16-3) revealed an average of 7.0 × 105 cfu g-1 kidney at the day of sacrifice and 2.0 × 104 cfu g-1 kidney in the remaining 25%. Seventy-five percent of the kidneys recovered from mice infected with the sod5 strain (UH16-2) revealed an average of 5.0 × 104 cfu g-1 kidney on the day of sacrifice and 5.0 × 105 cfu g-1 kidney of the remaining 25%, indicating an effective clearance of the sod5 knockout strain. These results demonstrate that a functional Sod5p plays an important role in the virulence of C. albicans.
DISCUSSION
C. albicans is known to express a CuZnSOD (SOD1) and an MnSOD (SOD3) in the cytosol and a second MnSOD (SOD2) in the mitochondria (Hwang et al., 1999; Rhie et al., 1999; Lamarre et al., 2001). In this study, we report the existence of three additional candidate CuZn-containing superoxide dismutases, the products of the SOD4, SOD5, and SOD6 genes. We have characterized the SOD5 gene that seems to be important factor in the virulence of this fungus. Virulence of C. albicans is often attributed to its ability to switch from yeast to hyphal forms. Consistent with this, SOD5 expression increases with several hyphal-inducing conditions, SOD5 expression is dependent on known components of hyphal-inducing signaling pathways, and the absence of this gene leads to a defect in the virulence of this organism. As expected for superoxide dismutases, SOD5 has some roles in responding to certain stresses.
Superoxide dismutases serve to convert damaging superoxide radicals, a key form of ROS, to less damaging hydrogen peroxide that can be converted into water by catalase action. SOD5 clearly encodes a CuZn-containing superoxide dismutase: it has the highest sequence similarity to the CuZn superoxide dismutase family, and deletion of the SOD5 gene eliminates the major source of CuZn dependent (cyanide sensitive) superoxide dismutase activity detected in IMDM-grown cells.
What is the significance of C. albicans having multiple CuZn superoxide dismutase genes, most of which are potentially expressed in the cytosol? For pathogens such as C. albicans, there are both internal and external sources of superoxides. An attractive model would be to have high levels of SOD activity to protect the fungal cell against the oxidative burst inside the phagosome. The fact that SOD5 is highly expressed in the hyphal state of C. albicans (this study; Nantel et al., 2002), and hyphal cells are typically the form that interacts with the host phagocytic cells (reviewed in Gow et al., 2002) would support such a model. In addition, increases in pH, salt concentration, or presence of oxidative species lead to increases in SOD5 transcription, the same events happen inside the phagosome (Reeves et al., 2002), further strengthening a model that SOD5 could have a defense-related function. Most significantly, the strongest evidence that supports this model is the fact that the sod5 knockout strain is defective in mouse killing. It was previously shown that the C. albicans sod1 mutant strain is defective in virulence and in macrophage killing. Our results have shown that the sod5 mutant behaves similarly to sod1 with respect to the virulence in mice, but surprisingly Sod5p does not affect macrophage killing. This would suggest that there are other phagocytic mammalian cells, such as neutrophils or dendritic cells, to which Sod5p is critical for a wild-type response.
We investigated the possibility that a further function of SOD5 is to protect C. albicans cells against intracellularly generated superoxide radicals. C. albicans produces the highest levels of CuZnSOD during the midexponential phase (Rhie et al., 1999; Lamarre et al., 2001), and it generates increased amounts of CuZnSOD during the switch from yeast to hyphal forms (Gunasekaran et al., 1998). During the same conditions, hyphal growth and in exponential phase of the growth, C. albicans is known to produce more ROS, including superoxide radicals (Schroter et al., 2000). These two parameters, superoxide generation and superoxide detoxification, were correlated to respiration of C. albicans (Ito-Kuwa et al., 1999). Mitochondrial respiration was shown to be a powerful source of superoxide radicals in yeast (Chance et al., 1979), and factors such as nonfermentable carbon sources were linked to increased superoxide production by electron transport chain in C. albicans (Rikhvanov et al., 2002), S. cerevisiae (Davidson et al., 1996; Longo et al., 1996; Maris et al., 2001), and in Schizosaccharomyces pombe (Jeong et al., 2001). In our study, these conditions that increase ROS production also increase SOD5 transcription. SOD5 expression levels were increased when cells grew in the presence of salt or on nonfermentable carbon as the only carbon source. In addition, an increased amount of fermentable glucose restored the growth ability of the SOD5 mutant cells grown in nutrient limiting media in the presence of hydrogen peroxide. We also observed that SOD5 transcription was induced by both menadione and riboflavin, which are intracellular and extracellular superoxide generators, respectively. This also provides us with the evidence that Sod5p is needed for elimination of superoxides found on both sides of the cell membrane. This might suggest that Sod5p shares the redundancy with other superoxide dismutases in eliminating extracellularly and intracellularly generated superoxide radicals in C. albicans.
All the increases in SOD5 transcript levels were observed in C. albicans grown in exponential phase, the same phase in which C. albicans produces more ROS. All hyphal-inducing conditions induced SOD5 transcription, and SOD5 transcription was found to be regulated by transcription factors regulating the yeast-to-hyphal switch of C. albicans. All these lines of evidence might suggest that hyphal growth and log-phase growth processes increase mitochondrial respiration in aerobically growing C. albicans, which subsequently increase mitochondrially generated superoxide radicals, leading to increased levels of detoxification enzymes. It is possible that these detoxification enzymes include proteins such as SOD5.
Our results also show that SOD5 possesses a complex regulation of expression. Intriguingly, Efg1p was found to positively regulate SOD5 transcription in the presence of serum as seen in Nantel et al., 2002, and negatively regulate SOD5 transcription under noninducing conditions. In addition, SOD5 transcription was found to be dependent on the Rim101 transcription factor, which is required for alkaline-induced filamentation (Davis et al., 2000b). This implicates the Rim101p pathway in the response of the C. albicans cell to oxidative stress. Rim101p was shown to be required for SOD5 induction in response to a variety of external signals, not just pH. Interestingly enough, the growth of the S. cerevisiae rim101 mutant was found to be inhibited at basic pH and in the presence of NaCl (Lamb et al., 2001), the same conditions in which SOD5 transcription was found to be up-regulated. The 1-kb sequence upstream of the SOD5 start codon revealed four potential Rim101p binding sites, which closely resemble the previously described CCAAGAAA-Rim101p binding site (Ramon and Fonzi, 2003), and four potential Efg1p binding sites (CAWWTG) (Leng et al., 2001).
All recent evidence suggests that C. albicans possesses a complicated cytosolic detoxification system. During the log-phase growth of C. albicans, SOD1 and SOD5 (CuZn-containing superoxide dismutases) are expressed (this study; Lamarre et al., 2001). SOD1 seems to be primarily responsible for removal of extracellular, macrophage-generated superoxide radicals (Hwang et al., 2002). SOD5 seems to be responsible for detoxification of extracellularly and intracellularly, possibly mitochondrially, generated superoxide radicals during log phase and hyphal growth. As C. albicans cells enter the stationary phase, the expression of CuZn-containing superoxide dismutases is repressed and the expression of Mn-containing cytosolic superoxide dismutase (SOD3) is increased (Lamarre et al., 2001).
The search for sequences similar to CuZnSOD in genomes of Neurospora crassa, Aspergillus fumigatus, S. cerevisiae, and S. pombe revealed that all of these organisms have SOD1 homologs, but only filamentous fungi (N. crassa and A. fumigatus) have additional sequences homologous to SOD5 [NCU03013.1 (161682-160794) and contig:69: a fumigatus (74890-75198), respectively]. Perhaps filamentous fungi have more sources of production of harmful superoxide radicals, which applies more pressure on these fungi to possess extra isoforms of superoxide detoxificators, such SOD5. The role of SOD4 and SOD6 remain to be established, although SOD4 was recently shown to be regulated in White-Opaque phenotype switching (Lan et al., 2002).
Acknowledgments
We thank Dr. A. Mitchell for the rim101 and mds3 mutants and their wild-type parental strains, Anne Marcil for assistance with the end point dilution survival assay, Dr. U. Oberholzer for donating the ACT1 primers, Dr. C. Bachewich for the CB20R primer, and Daniel Dignard for bioinformatics assistance. This work was supported by Canadian Institutes of Health Research grant MOP-42516 (to M.W.). M.M. gratefully acknowledges a FRSQFCAR-Sante Scholarship and National Research Council Graduate Student Scholarship Supplement. This is National Research Council publication 46148.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-03-0179. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-03-0179.
References
- Alarco, A.M., Balan, I., Talibi, D., Mainville, N., and Raymond, M. (1997). AP1-mediated multidrug resistance in Saccharomyces cerevisiae requires FLR1 encoding a transporter of the major facilitator superfamily. J. Biol. Chem. 272, 19304-19313. [DOI] [PubMed] [Google Scholar]
- Ballance, D.J. (1986). Sequences important for gene expression in filamentous fungi. Yeast 2, 229-236. [DOI] [PubMed] [Google Scholar]
- Beauchamp, C., and Fridovich, I. (1971). Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44, 276-287. [DOI] [PubMed] [Google Scholar]
- Bilinski, T., Krawiec, Z., Liczmanski, A., and Litwinska, J. (1985). Is hydroxyl radical generated by the Fenton reaction in vivo? Biochem. Biophys. Res. Commun. 130, 533-539. [DOI] [PubMed] [Google Scholar]
- Boeke, J.D., LaCroute, F., and Fink, G.R. (1984). A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197, 345-346. [DOI] [PubMed] [Google Scholar]
- Boveris, A. (1978). Production of superoxide anion and hydrogen peroxide in yeast mitochondria. In: Biochemistry and Genetics of Yeast, ed. M. Bacila, B.L. Horecker, and A.O.M. Stoppane, New York: Academic Press, 66-79.
- Brown, A.J., and Gow, N.A. (1999). Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol. 7, 333-338. [DOI] [PubMed] [Google Scholar]
- Carlioz, A., and Touati, D. (1986). Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 5, 623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, M., and Botstein, D. (1982). Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell 28, 145-154. [DOI] [PubMed] [Google Scholar]
- Casteilla, L., Rigoulet, M., and Penicaud, L. (2001). Mitochondrial ROS metabolism: modulation by uncoupling proteins. IUBMB Life 52, 181-188. [DOI] [PubMed] [Google Scholar]
- Chance, B., Sies, H., and Boveris, A. (1979). Hydroperoxide metabolism in mammalian organs. Physiol. Rev. 59, 527-605. [DOI] [PubMed] [Google Scholar]
- Chen, D.C., Yang, B.C., and Kuo, T.T. (1992). One-step transformation of yeast in stationary phase. Curr. Genet. 21, 83-84. [DOI] [PubMed] [Google Scholar]
- Corner, B.E., and Magee, P.T. (1997). Candida pathogenesis: unravelling the threads of infection. Curr. Biol. 7, R691-R694. [DOI] [PubMed] [Google Scholar]
- Davidson, J.F., Whyte, B., Bissinger, P.H., and Schiestl, R.H. (1996). Oxidative stress is involved in heat-induced cell death in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93, 5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, D., Edwards, J.E., Jr., Mitchell, A.P., and Ibrahim, A.S. (2000a). Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect. Immun. 68, 5953-5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, D., Wilson, R.B., and Mitchell, A.P. (2000b). RIM101-dependent and- independent pathways govern pH responses in Candida albicans. Mol. Cell. Biol. 20, 971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, D.A., Bruno, V.M., Loza, L., Filler, S.G., and Mitchell, A.P. (2002). Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics 162, 1573-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Freitas, J.M., Liba, A., Meneghini, R., Valentine, J.S., and Gralla, E.B. (2000). Yeast lacking Cu-Zn superoxide dismutase show altered iron homeostasis. Role of oxidative stress in iron metabolism. J. Biol. Chem. 275, 11645-11649. [DOI] [PubMed] [Google Scholar]
- Enjalbert, B., Nantel, A., and Whiteway, M. (2003). Stress induced gene expression in Candida albicans: absence of a general stress response. Mol. Biol. Cell 14, 1460-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink, R.C., and Scandalios, J.G. (2002). Molecular evolution and structure-function relationships of the superoxide dismutase gene families in angiosperms and their relationship to other eukaryotic and prokaryotic superoxide dismutases. Arch. Biochem. Biophys. 399, 19-36. [DOI] [PubMed] [Google Scholar]
- Fonzi, W.A., and Irwin, M.Y. (1993). Isogenic strain construction and gene mapping in Candida albicans. Genetics 134, 717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridovich, I. (1978). The biology of oxygen radicals. Science 201, 875-880. [DOI] [PubMed] [Google Scholar]
- Fridovich, I. (1995). Superoxide radical and superoxide dismutases. Annu. Rev. Biochem. 64, 97-112. [DOI] [PubMed] [Google Scholar]
- Gow, N.A., Brown, A.J., and Odds, F.C. (2002). Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 5, 366-371. [DOI] [PubMed] [Google Scholar]
- Gunasekaran, U., Yang, R., and Gunasekaran, M. (1998). Regulation of superoxide dismutase synthesis in Candida albicans. Mycopathologia 141, 59-63. [DOI] [PubMed] [Google Scholar]
- Halliwell, B., and Gutteridge, J.M. (1990). Role of free radicals and catalytic metal ions in human disease: an overview. Methods Enzymol. 186, 1-85. [DOI] [PubMed] [Google Scholar]
- Halliwell, B., and Gutteridge, J.M. (1999). Free Radicals in Biology and Medicine, Oxford: Oxford Science Publications, 105-245.
- Hwang, C.S., Baek, Y.U., Yim, H.S., and Kang, S.O. (2003). Protective roles of mitochondrial manganese-containing superoxide dismutase against various stresses in Candida albicans. Yeast 20, 929-941. [DOI] [PubMed] [Google Scholar]
- Hwang, C.S., Rhie, G., Kim, S.T., Kim, Y.R., Huh, W.K., Baek, Y.U., and Kang, S.O. (1999). Copper- and zinc-containing superoxide dismutase and its gene from Candida albicans. Biochim. Biophys. Acta 1427, 245-255. [DOI] [PubMed] [Google Scholar]
- Hwang, C.S., Rhie, G.E., Oh, J.H., Huh, W.K., Yim, H.S., and Kang, S.O. (2002). Copper- and zinc-containing superoxide dismutase (Cu/ZnSOD) is required for the protection of Candida albicans against oxidative stresses and the expression of its full virulence. Microbiology 148, 3705-3713. [DOI] [PubMed] [Google Scholar]
- Imlay, J.A., and Fridovich, I. (1991). Isolation and genetic analysis of a mutation that suppresses the auxotrophies of superoxide dismutase-deficient Escherichia coli K12. Mol. Gen. Genet. 228, 410-416. [DOI] [PubMed] [Google Scholar]
- Ito-Kuwa, S., Nakamura, K., Aoki, S., Osafune, T., Vidotto, V., and Pienthaweechai, K. (1999). Oxidative stress sensitivity and superoxide dismutase of a wild-type parent strain and a respiratory mutant of Candida albicans. Med. Mycol. 37, 307-314. [DOI] [PubMed] [Google Scholar]
- Jeong, J.H., Kwon, E.S., and Roe, J.H. (2001). Characterization of the manganese-containing superoxide dismutase and its gene regulation in stress response of Schizosaccharomyces pombe. Biochem. Biophys. Res. Commun. 283, 908-914. [DOI] [PubMed] [Google Scholar]
- Kanematsu, S., and Asada, K. (1991). Chloroplast and cytosol isozymes of CuZn-superoxide dismutase: their characteristic amino acid sequences. Free Radic. Res. Commun. 12-13, 383-390. [DOI] [PubMed] [Google Scholar]
- Keyer, K., Gort, A.S., and Imlay, J.A. (1995). Superoxide and the production of oxidative DNA damage. J. Bacteriol. 177, 6782-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarre, C., LeMay, J.D., Deslauriers, N., and Bourbonnais, Y. (2001). Candida albicans expresses an unusual cytoplasmic manganese-containing superoxide dismutase (SOD3 gene product) upon the entry and during the stationary phase. J. Biol. Chem. 276, 43784-43791. [DOI] [PubMed] [Google Scholar]
- Lamb, T.M., Xu, W., Diamond, A., and Mitchell, A.P. (2001). Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J. Biol. Chem. 276, 1850-1856. [DOI] [PubMed] [Google Scholar]
- Lan, C.Y., Newport, G., Murillo, L.A., Jones, T., Scherer, S., Davis, R.W., and Agabian, N. (2002). Metabolic specialization associated with phenotypic switching in Candida albicans. Proc. Natl. Acad. Sci. USA 99, 14907-14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J., Kwon, E.S., Kim, D.W., Cha, J., and Roe, J.H. (2002). Regulation and the role of Cu, Zn-containing superoxide dismutase in cell cycle progression of Schizosaccharomyces pombe. Biochem. Biophys. Res. Commun. 297, 854-862. [DOI] [PubMed] [Google Scholar]
- Lenaz, G. (2001). The mitochondrial production of reactive oxygen species: mechanisms and implications in human pathology. IUBMB Life 52, 159-164. [DOI] [PubMed] [Google Scholar]
- Leng, P., Lee, P.R., Wu, H., and Brown, A.J. (2001). Efg1, a morphogenetic regulator in Candida albicans, is a sequence-specific DNA binding protein. J. Bacteriol. 183, 4090-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liochev, S.I., and Fridovich, I. (1994). The role of O2- in the production of HO.: in vitro and in vivo. Free Radic. Biol. Med. 16, 29-33. [DOI] [PubMed] [Google Scholar]
- Liu, H., Kohler, J., and Fink, G.R. (1994). Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266, 1723-1726. [DOI] [PubMed] [Google Scholar]
- Lo, H.J., Kohler, J.R., DiDomenico, B., Loebenberg, D., Cacciapuoti, A., and Fink, G.R. (1997). Nonfilamentous C. albicans mutants are avirulent. Cell 90, 939-949. [DOI] [PubMed] [Google Scholar]
- Longo, V.D., Gralla, E.B., and Valentine, J.S. (1996). Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Mitochondrial production of toxic oxygen species in vivo. J. Biol. Chem. 271, 12275-12280. [DOI] [PubMed] [Google Scholar]
- Magee, B.B., and Magee, P.T. (1997). WO-2, a stable aneuploid derivative of Candida albicans strain WO-1, can switch from white to opaque and form hyphae. Microbiology 143, 289-295. [DOI] [PubMed] [Google Scholar]
- Malloy, J.M., Rieker, J.P., and Rizzo, C.F. (1984). Quantitation of proteins on Coomassie blue-stained polyacrylamide gels based on spectrophotometric determination of electroeluted dye. Anal. Biochem. 141, 503-509. [DOI] [PubMed] [Google Scholar]
- Marcil, A., Harcus, D., Thomas, D.Y., and Whiteway, M. (2002). Candida albicans killing by RAW 264.7 mouse macrophage cells: effects of Candida genotype, infection ratios, and gamma interferon treatment. Infect. Immun. 70, 6319-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris, A.F., Assumpcao, A.L., Bonatto, D., Brendel, M., and Henriques, J.A. (2001). Diauxic shift-induced stress resistance against hydroperoxides in Saccharomyces cerevisiae is not an adaptive stress response and does not depend on functional mitochondria. Curr. Genet. 39, 137-149. [DOI] [PubMed] [Google Scholar]
- Meneghini, R. (1997). Iron homeostasis, oxidative stress, and DNA damage. Free Radic. Biol. Med. 23, 783-792. [DOI] [PubMed] [Google Scholar]
- Nantel, A., et al. (2002). Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 13, 3452-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negredo, A., Monteoliva, L., Gil, C., Pla, J., and Nombela, C. (1997). Cloning, analysis and one-step disruption of the ARG5, 6 gene of Candida albicans. Microbiology 143, 297-302. [DOI] [PubMed] [Google Scholar]
- Pla, J., Perez-Diaz, R.M., Navarro-Garcia, F., Sanchez, M., and Nombela, C. (1995). Cloning of the Candida albicans HIS1 gene by direct complementation of a C. albicans histidine auxotroph using an improved double-ARS shuttle vector. Gene 165, 115-120. [DOI] [PubMed] [Google Scholar]
- Ramon, A.M., and Fonzi, W.A. (2003). Diverged binding specificity of Rim101p, the Candida albicans ortholog of PacC. Eukaryot. Cell 2, 718-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves, E.P., Lu, H., Jacobs, H.L., Messina, C.G., Bolsover, S., Gabella, G., Potma, E.O., Warley, A., Roes, J., and Segal, A.W. (2002). Killing activity of neutrophils is mediated through activation of proteases by K+ flux. Nature 416, 291-297. [DOI] [PubMed] [Google Scholar]
- Rhie, G.E., Hwang, C.S., Brady, M.J., Kim, S.T., Kim, Y.R., Huh, W.K., Baek, Y.U., Lee, B.H., Lee, J.S., and Kang, S.O. (1999). Manganese-containing superoxide dismutase and its gene from Candida albicans. Biochim. Biophys. Acta 1426, 409-419. [DOI] [PubMed] [Google Scholar]
- Rikhvanov, E.G., Varakina, N.N., Rusaleva, T.M., Rachenko, E.I., and Voinikov, V.K. (2002). Sodium azide reduces the thermotolerance of respiratively grown yeasts. Curr. Microbiol. 45, 394-399. [DOI] [PubMed] [Google Scholar]
- Sander, C.S., Hipler, U.C., Wollina, U., and Elsner, P. (2002). Inhibitory effect of terbinafine on reactive oxygen species (ROS) generation by Candida albicans. Mycoses 45, 152-155. [DOI] [PubMed] [Google Scholar]
- Schroter, C., Hipler, U.C., Wilmer, A., Kunkel, W., and Wollina, U. (2000). Generation of reactive oxygen species by Candida albicans in relation to morphogenesis. Arch. Dermatol. Res. 292, 260-264. [DOI] [PubMed] [Google Scholar]
- Seyler, R.W., Jr., Olson, J.W., and Maier, R.J. (2001). Superoxide dismutase-deficient mutants of Helicobacter pylori are hypersensitive to oxidative stress and defective in host colonization. Infect. Immun. 69, 4034-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M.W., and Doolittle, R.F. (1992). A comparison of evolutionary rates of the two major kinds of superoxide dismutase. J. Mol. Evol. 34, 175-184. [DOI] [PubMed] [Google Scholar]
- Wilson, R.B., Davis, D., and Mitchell, A.P. (1999). Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181, 1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, H.Y., Kim, S.S., and Rho, H.M. (1999). Overexpression and simple purification of human superoxide dismutase (SOD1) in yeast and its resistance to oxidative stress. J. Biotechnol. 68, 29-35. [DOI] [PubMed] [Google Scholar]