Abstract
Reconstitution of integral membrane proteins into membrane mimetic environments suitable for biophysical and structural studies has long been a challenge. Isotropic bicelles promise the best of both worlds—keeping a membrane protein surrounded by a small patch of bilayer-forming lipids while remaining small enough to tumble isotropically and yield good solution NMR spectra. However, traditional methods for the reconstitution of membrane proteins into isotropic bicelles expose the proteins to potentially destabilizing environments. Reconstituting the protein into liposomes and then adding short-chain lipid to this mixture produces bicelle samples while minimizing protein exposure to unfavorable environments. The result is higher yield of protein reconstituted into bicelles and improved long-term stability, homogeneity, and sample-to-sample reproducibility. This suggests better preservation of protein structure during the reconstitution procedure and leads to decreased cost per sample, production of fewer samples, and reduction of the NMR time needed to collect a high quality spectrum. Furthermore, this approach enabled reconstitution of protein into isotropic bicelles with a wider range of lipid compositions. These results are demonstrated with the small multidrug resistance transporter EmrE, a protein known to be highly sensitive to its environment.
Keywords: isotropic bicelles, membrane mimetic, membrane protein reconstitution, EmrE, NMR
1. Introduction
Structural and biophysical studies of membrane proteins have traditionally lagged behind those of soluble proteins. One of the major challenges of working with membrane proteins is finding a membrane mimetic environment that is conducive to biophysical studies while still maintaining native structure and function [1]. Techniques such as solution NMR have fast-tumbling requirements that are not fulfilled by conventional lipid vesicles. Detergent micelles, isotropic bicelles, and nanodiscs are some of the media available for the solubilization of integral membrane proteins (IMPs). The high curvature and altered lateral pressure of detergent micelles make them a less than ideal membrane mimetic [1-4]. Detergents must be extensively screened to find a suitable match that preserves native structure and function, and gentler detergents such as alkylglycosides, which may better preserve function, are often not conducive to multidimensional NMR studies [5].
Both isotropic bicelles and nanodiscs provide bilayer environments, and each media has its own merits. Nanodiscs are stable particles that can be separated by physical means such as gel filtration chromatography, unlike isotropic bicelles in which the detergent is in constant equilibrium between monomer and bicelle. This same property of isotropic bicelles allows them to reconstitute at any size by varying the long- to short-chain lipid ratio, whereas nanodiscs can only make discretely sized particles. A new membrane scaffold protein must be expressed and purified for each nanodisc size, and they must be carefully tested to determine the proper ratio of scaffold-to-lipid for each preparation [6]. In addition, NMR spectra of IMPs reconstituted into nanodiscs tend to be broadened [1, 2, 7].
Use of isotropic bicelles has been limited by their stability and spectral qualities [1, 2]. Traditional methods for making bicelle samples require harsh conditions that may not preserve proper protein structure and function. Improved bicelle stability would allow for more widespread use. Previous attempts at increasing bicelle stability and sample lifetime have included using ether-linked lipids and/or adding up to 10% of long- and short-chain lipids with charged headgroups such as PS and PE-DTPA [4, 8, 9]. However, recent studies have indicated that non-native ether-linked lipids alter the structure and dynamics of antimicrobial peptides [10], making this a less desirable substitution.
Another challenge with isotropic bicelles is the relatively limited lipid compositions currently in use. More varied lipid compositions have been explored with magnetically aligned large bicelles for solid-state NMR [1]. An expanded lipid composition profile would allow for more physiologically relevant isotropic bicelles, similar to nanodiscs, which can be formed with a range of lipids and lipid extracts [2, 6].
Here, we present an improved reconstitution of integral membrane proteins into small isotropic bicelles that allows reconstitution into bicelles with a much wider range of lipid compositions. This protocol is easily adaptable to any IMP of interest, with no requirement for organic solvent or specific detergents. It also ensures complete detergent removal, eliminating potential instabilities and inhomogeneities caused by residual detergent and providing an optimal sample for biophysical studies. We have recently demonstrated the utility of these bicelles for solution NMR dynamics measurements of EmrE [11]. These cutting edge NMR measurements would not have been possible without the improved sample stability and homogeneity provided by this method.
2. Materials and methods
2.1. EmrE expression and purification
EmrE was expressed in a pET15b plasmid with an N-terminal 6xHis tag (Geoffrey Chang, Scripps Research Institute) and expressed and purified as previously described [11]. Briefly, isotopically labeled EmrE was grown in 15N- or 2H/15N-labeled M9 minimal media. To purify EmrE, cells were lysed by sonication. The membranes were solubilized in 40 mM DM (Affymetrix Anatrace), purified using Ni-NTA resin (Novagen), and cleaved with thrombin (Sigma-Aldrich). Final purification was performed by gel filtration chromatography on a Superdex 200 column (GE Healthcare) equilibrated with 10 mM DM. The purified EmrE was then reconstituted into isotropic bicelles in deoxygenated buffers.
2.2. Sample preparation
2.2.1. Reconstitution into isotropic bicelles by detergent exchange
Purified EmrE was concentrated to 0.5 mL and DM reduced to less than 30 mM, assuming monomeric detergent passed through the filter while micelles were concentrated along with the protein. The resulting sample was run over a Superdex 200 column equilibrated with 25 mM DHPC (Avanti Polar Lipids). EmrE in DHPC was concentrated to 250 μL and long-chain lipid added to a final ratio of 3:1 short- to long-chain lipid (at least 75 mM long-chain lipid and 130-fold excess of long-chain lipid:EmrE). Four freeze-thaw cycles were carried out to ensure homogeneous bicelles.
2.2.2. Reconstitution into isotropic bicelles via liposomes
Long-chain lipid (DLPC, DMPC, DPPC, POPC, DOPC, POPE, POPG, and E. coli polar lipid extract, Avanti Polar Lipids) was hydrated in buffer above the phase transition temperature for 1-2 hours at 20 mg/mL and then sonicated in a high-power bath sonicator (Laboratory Supplies Company, Inc, Hicksville, NY). The lipid vesicles were then incubated with 0.51% octylglucoside (Affymetrix Anatrace) for 15-30 minutes. Purified EmrE in DM was added at a molar ratio of 1:130, EmrE:long-chain lipid. After incubation for an addition 30-60 minutes, Amberlite (Supelco) was added to remove the detergent. Three aliquots of 30 mg Amberlite per mg detergent were added with incubation at room temperature for 1-2 hours, overnight, and another 1-2 hours. The proteoliposomes were collected via ultracentrifugation (50,000 ×g, 1 hr, 20°C) and resuspended in DHPC buffer at a molar ratio of 3:1 short- to long-chain lipid, assuming a 90% recovery of long-chain lipid. The final lipid ratio in the bicelles was confirmed by 1H-NMR using the lipid terminal methyl peaks. The final buffer conditions for all NMR samples were 20 mM potassium phosphate, 30 mM sodium cacodylate, 20 mM NaCl, 0.05% sodium azide, 2 mM TCEP, pH 7, with 2 mM TPP+ and EmrE concentrations in the range of 0.5-1.1 mM.
2.3. Transport assay
H+-driven uptake of the substrate dequalinium2+ was monitored using fluorescence spectroscopy, following the fluorescence signal of the substrate. EmrE was reconstituted into E. coli polar lipid extract as described above with a lipid:EmrE dimer ratio of 640:1. The buffer was 190 mM NH4Cl, 15 mM Tris pH 7 as in [12]. The sample was extruded through a 200 nm filter to create homogeneous vesicles. The proteoliposomes were diluted ten-fold into either the same buffer (no H+ gradient) or a buffer of 140 mM KCl, 10 mM Tricine, 5 mM MgCl2, 10 mM Tris pH 8 (H+ gradient). The final protein concentration was 1 μM. Transport was initiated upon the addition of concentrated dequalinium2+ to the bulk solution. The experiments were carried out in a Varian Cary Eclipse fluorescence spectrometer, at an excitation wavelength of 350 nm and emission wavelength of 460 nm.
2.4. Dynamic light scattering
DLS experiments were carried out on a DynaPro (model 99-E-50, Protein solutions) with the Dynamics V6 software. The instrument was calibrated with an albumin standard (Thermo Scientific) at varying temperatures and concentrations. The NMR samples were diluted four-fold to decrease secondary scattering effects, while still maintaining sufficient lipid concentration to keep the same effective q-value, or ratio of long- to short-chain lipids in the bicelle [13]. Data was collected at 10 and 25°C, for five acquisitions of five minutes each.
2.5. Thin layer chromatography
TLC was carried out using Whatman K6 silica gel 60A plates and a 65:24:4 chloroform:methanol:water solvent system. Isotropic bicelle samples were diluted 10-fold in 2:1 chloroform:methanol for spotting onto the plates. After running the plates, they were dried and developed in an iodine vapor chamber overnight.
2.6. Refractive index measurements
A Reichert-Jung Abbe Mark II digital refractometer with water-bath temperature control was used to measure the refractive indices of isotropic bicelles at the pertinent concentrations and temperatures.
2.7. Viscosity measurements
A calibrated Cannon-Ubbelohde semi-micro viscometer, size 50, was used to determine the viscosity of isotropic bicelles. Both refractive index and viscosity were measured under the same conditions (lipid concentrations and temperatures) as the diffusion measurements and used to calculate the hydrodynamic radius.
2.8. NMR spectroscopy
All experiments were carried out on a 700 MHz Varian Inova spectrometer at 45°C. Lipid ratios were confirmed by integrating the terminal methyl resonances of the short- and long-chain lipids in a 1H spectrum. Standard (1H, 15N)-TROSY HSQC spectra with gradient coherence selection were acquired with 120 and 24 scans per increment, respectively, for non-2H and 2H samples. (1H, 15N)-TRACT [14] was used to measure rotational correlation times. The hydrodynamic radius (rh) is related to the rotational correlation time (τc) by
| (1) |
Data were processed and analyzed using NMRPipe [15] and NMRView [16]. IgorPro (Wavemetrics) was used to fit the diffusion data.
3. Results and discussion
3.1. Reconstitution of an integral membrane protein into isotropic bicelles via liposomes improves bicelle stability and spectral quality
Integral membrane proteins (IMPs) are traditionally reconstituted into isotropic bicelles [3] by two methods: i) direct reconstitution from organic solvent or lyophilized powder; or ii) exchange into DHPC micelles followed by addition of long-chain lipid. Highly stable IMPs can be reconstituted through any method. However, multi-pass α-helical IMPs are generally more sensitive to their environment and handling during purification [17-19]. Organic solvents and lyophilization must be avoided with these proteins, and many are not stable in DHPC micelles. We reconstitute EmrE into vesicles of long-chain lipid and then add DHPC to form bicelles (method iii). This protocol is similar to a strategy used previously to reconstitute the GPCR CXCR1 into large bicelles [20] for solid-state NMR.
EmrE, a 4-transmembrane helix homodimer, is highly sensitive to detergent and lipid environment [18, 21] and serves as a representative system. EmrE is monomeric [22] and unfolded [23] in organic solvent. However, it is a “functional” dimer in the detergent dodecyl maltoside [24, 25] based on ligand-binding, which provides the best proxy for transporter function in a solubilized state. Thus we eliminated organic solvent reconstitution protocols and focused on methods with the potential to preserve EmrE structure and function.
Initially, we reconstituted EmrE into isotropic bicelles via detergent exchange (method ii) (Fig. 1A, B), but there are several disadvantages to this method. Running long gel filtration columns above the critical micellar concentration of DHPC becomes prohibitively expensive, and EmrE has diminished stability upon exchange into DHPC micelles. In addition, residual DM remains in the samples (Fig. 2). The yield, spectral quality, and stability of the samples were variable, with protein precipitating in 2-14 days at 45°C. In addition, minor chemical shift changes were sometimes observed between independently prepared samples. Due to the conformational dynamics of EmrE, NMR spectra are exquisitely sensitive to changes in the bicelle environment, making sample reproducibility essential.
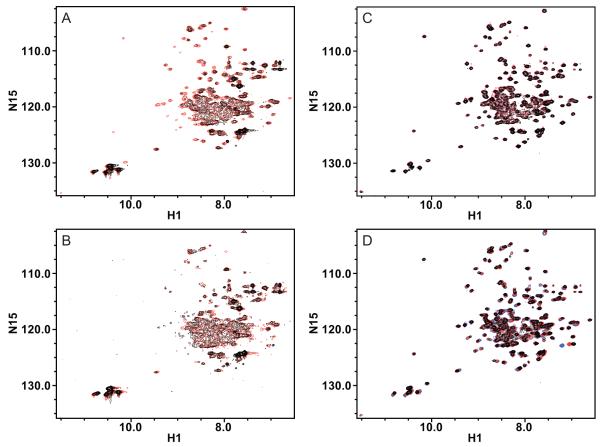
Figure 1.
Overlay of (1H, 15N)-TROSY HSQC spectra of TPP+-bound EmrE in isotropic bicelles reconstituted by different methods. A) 15N-EmrE in DMPC/DHPC bicelles is better resolved and has better signal/noise when reconstituted through liposomes (red) than when reconstituted through detergent exchange (black). B) The spectral quality of 15N-EmrE in DMPC/DHPC bicelles degrades with time for EmrE reconstituted via detergent. After nearly two weeks at 45°C, the spectrum (black) has less signal than the initial spectrum (red). C) The spectrum of 2H/15N/13C-EmrE reconstituted into DLPC/DHPC bicelles via liposomes (red) is virtually unchanged after more than six months (black). D) 2H/15N-EmrE can be stably reconstituted into isotropic bicelles made with different chain lengths (DLPC, blue; DMPC, red; DPPC, black), all capped by DHPC. The minor peak shifts indicate only small changes in EmrE structure with change in bicelle thickness. All spectra were collected at 45°C, in 20 mM KPi, 20 mM NaCl, 30 mM sodium cacodylate, pH 7.
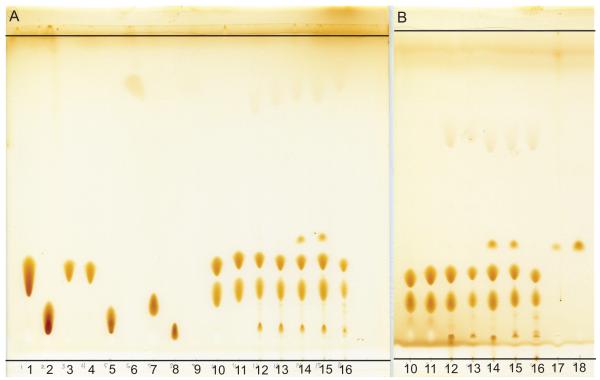
Figure 2.
Thin layer chromatography to qualitatively assess lipid composition and hydrolysis of isotropic bicelle samples used for NMR spectroscopy. The standards include lipids as well as the corresponding breakdown products--lysolipid and fatty acid. A) From left to right: 1) DPPC; 2) 16:0 lyso PC; 3) DMPC; 4) DLPC; 5) 12:0 lyso PC; 6) lauric acid; 7) DHPC; 8) 6:0 lyso PC; 9) hexanoic acid; 10) DLPC/DHPC q=0.33 bicelles; 11) DMPC/DHPC q=0.33 bicelles; 12) initial sample of 15N-EmrE in DMPC/DHPC isotropic bicelles reconstituted through liposomes and 13) after 12 days at 45°C; 14) initial sample of 15N-EmrE in DMPC/DHPC isotropic bicelles reconstituted through detergent exchange and 15) after 12 days at 45°C; and 16) 2H/15N/13C-EmrE after roughly 7 months. B) From left to right, repeat of lanes 10-16; 17) EmrE in 10 mM DM after S200 column during sample purification; and 18) 100 mM DM. Iodine vapor does not appear to be sufficient to visualize hexanoic acid.
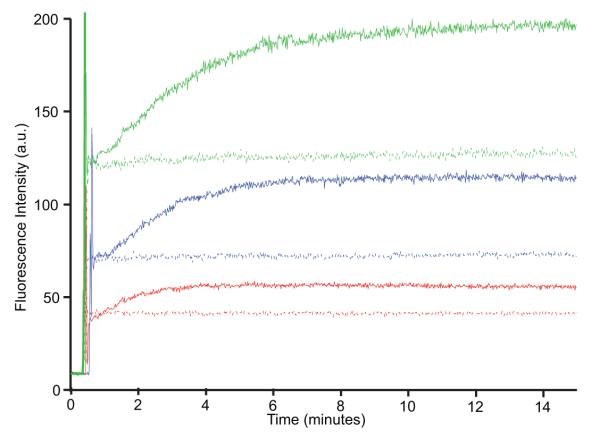
Reconstitution via liposomes (method iii) has two advantages. First, it ensures complete detergent removal before the addition of the short-chain lipid to break up the vesicles (Fig. 1A). This protocol creates isotropic bicelles of well-defined composition and removes the requirement for exchange into DHPC, thus making it suitable for proteins not stable in this particular detergent-like lipid. Second, reconstitution into a liposome allows transport assays to be performed, ensuring that EmrE is fully functional immediately before the final step of bicelle formation. We measured proton-driven uptake of substrate by EmrE reconstituted into E. coli polar lipid liposomes using Amberlite for detergent removal (Fig. 3). These assays use the naturally fluorescent substrate dequalinium, which shows a concentration-dependent increase in fluorescence at 460 nm. No uptake is observed in the absence of a pH gradient. When a pH gradient is introduced, the fluorescence change indicates dequalinium uptake and concentration inside the liposomes, confirming that purification and reconstitution into liposomes yields functional EmrE. Formation of bicelles from the liposomes requires only the addition of DHPC and several freeze-thaw cycles. A spectrum of TPP+-bound EmrE in E. coli polar lipid bicelles (Fig. 4D), the lipid composition used for the transport assays, indicates that the overall structure of EmrE is the same in a variety of lipid environments. It is not possible to measure transport in the final bicelle, but substrate binding affinity and stoichiometry[11] indicate that EmrE is still properly folded and “functional” in bicelles.
Figure 3.
EmrE is functional when reconstituted into liposomes: H+-driven uptake of the substrate dequalinium2+ by EmrE reconstituted into E. coli polar liposomes, as observed by monitoring dequalinium2+ fluorescence. Kinetics timecourses were acquired with excitation at 350 nm and emission at 460 nm. At time zero, proteoliposomes at pH 7 are diluted into a buffer of pH 7 (no proton gradient) or pH 8 (proton gradient). After roughly 30 sec, substrate is added. Assay was carried out with 1 μM EmrE and 5 μM (red), 10 μM (blue), and 20 μM (green) dequalinium2+ and in the presence (solid line) or absence (dashed line) of a H+ gradient.
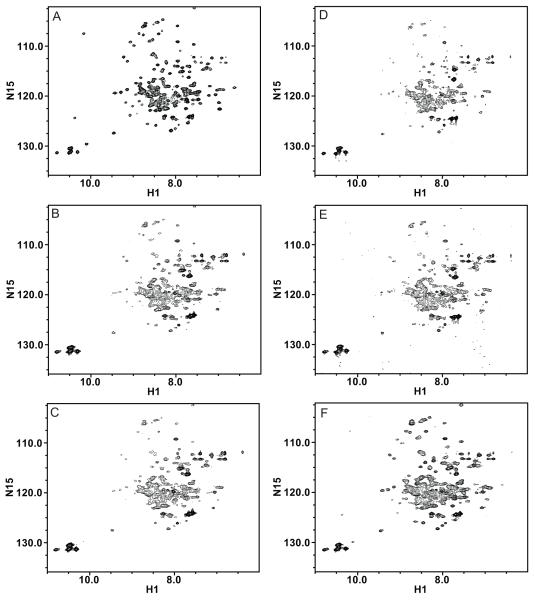
Figure 4.
EmrE was successfully reconstituted with a wide range of long-chain lipid compositions varying in saturation and head-group. (1H, 15N)-TROSY HSQC spectra of TPP+-bound EmrE reconstituted into isotropic bicelles with the following long-chain lipid compositions: A) DPPC, B) POPC, C) DOPC, D) E. coli polar lipid extract, E) 70% POPE, 20% POPG, 10% POPC, and F) 70% POPC, 20% POPE, 10% POPG. The DPPC spectrum was collected on 2H/15N-EmrE, while all others were collected on 1H/15N-EmrE. Spectra were collected under the same conditions as above.
2D TROSY spectra of 15N-EmrE reconstituted into DMPC/DHPC isotropic bicelles by either method show that the liposome method produces significantly better spectra (Fig. 1A). These spectra have improved signal to noise for the same protein concentration and sharper lines, consistent with a more homogeneous sample. In addition, spectra from independently prepared samples overlay exactly, a requirement for specific-labeling-based assignment protocols.
Elevated temperatures are commonly used in NMR to improve linewidth. To test stability under these conditions, the samples were incubated in a water bath at 45°C for nearly two weeks and then a second spectrum was acquired to assess spectral changes. No significant changes were observed for EmrE reconstituted via liposomes and there was no precipitate. For EmrE reconstituted via detergent exchange, precipitate was observed and the spectral quality declined (Fig. 1B).
The long-term stability of the isotropic bicelles reconstituted via liposomes is demonstrated in Figure 1C. After six months of storage at room temperature and weeks of NMR experiments at elevated temperatures, a sample of 2H/15N/13C-EmrE in DLPC/DHPC bicelles is unchanged. Contrary to previous reports [4, 8], these samples are stable for months without requiring expensive ether-linked lipids or special lipid compositions.
We also experimented with the long-chain lipid composition of the bicelles. A series of samples made using different acyl chain lengths reveal only small local peak shifts (Fig. 1D). EmrE is an E. coli protein, so we reconstituted EmrE into liposomes made with E. coli polar extract. These varied in yield and spectral quality, likely due to variability between batches of lipid extract. For a more defined environment, we used two compositions designed to mimic E. coli lipids. EmrE reconstituted well into isotropic bicelle preparations using long-chain mixtures of i) 70% POPC, 20% POPE, 10% POPG, or ii) 70% POPE, 20% POPG, 10% POPC. Overall, there are minimal peak shifts of EmrE with changes in long-chain lipid composition, but the peak-widths vary significantly (Fig. 4). This suggests that we are observing changes in dynamic rate rather than structure. These samples demonstrate the potential for reconstitution of IMPs into isotropic bicelles with a variety of lipid compositions that better mimic native membranes for use in biophysical studies [26, 27].
3.2. Analysis of lipid hydrolysis in isotropic bicelles
We used TLC to assess lipid hydrolysis and residual detergent (Fig. 2). The samples consist primarily of the bicelle lipids. Additional minor lysolipid and fatty acid components indicate some lipid hydrolysis; however, they are still only minor components after 6-months. If hydrolysis is a serious problem, a bicelle with ether-linked lipids, such as DMPC/DIOHPC, can be utilized [9]. However, it is preferable to use ester-linked lipids, as using the non-native ether-lipids may alter the structure and dynamics of peptides in the membrane [10]. An additional component, DM, is visible in the NMR sample reconstituted via detergent exchange (Fig. 2), indicating that the detergent was not fully removed. Precipitation of the NMR samples appears to correlate with small amounts of residual detergent.
3.3. Diffusion of EmrE reconstituted into isotropic bicelles
Disagreement exists in the literature regarding the morphology of isotropic bicelles at small q-values—are they true discs (primarily separated long- and short-chain domains) or mixed micelles? Based on the small apparent hydrodynamic radii (RH) determined via DLS, small q-value (≤0.5) mixtures of long- and short-chain lipids have been differentially diagnosed as mixed micelles [28] or discs[13]. It is difficult to determine shape from DLS data [13] alone and difficult to interpret RH quantitatively. Electron microscopy [13] and AFM [9], have been used to visually assess the morphology of these mixtures. Discoidal structures were observed with both methods, with larger discs and a few wormlike micelles also being observed via AFM under more physiological conditions. Small angle neutron scattering data also supports a disk-like model [29].
NMR spectroscopy has additionally been used to assess bicelle morphology. 31P chemical shifts are different for DMPC and DHPC headgroups, indicating different environments on average and thus supporting significant separation of the lipids into distinct regions [9, 13]. Long- and short-chain lipids are differentially affected by paramagnetic shift reagents, supporting separate regions for the two classes of lipids [29]. Translational diffusion measurements also indicate a distinct separation [30]. Thus, the preponderance of data supports significant domain separation of the long- and short-chain lipids, creating a bilayer-like environment around the protein. Even if lipid separation in the bicelles is incomplete, as is likely the case, this provides a much more native-like environment than a detergent micelle. This is further supported by proper ligand-binding affinity for small multidrug resistance transporters [3, 4] and other IMPs [1, 2] in bicelles.
The hydrodynamic radii of our isotropic bicelles (q = 0.33) were determined using DLS and NMR rotational diffusion data. Table 1 compares the hydrodynamic radii for EmrE samples reconstituted through detergent exchange and via liposomes. There is no significant change in hydrodynamic radius with reconstitution method, and the radii are significantly smaller than those of nanodiscs [6]. Thus, spectral differences between the two bicelle reconstitution methods are not due to differences in overall particle size. Hydrodynamic radii in Table 1 were calculated from the DLS data using both water and isotropic bicelle solution viscosity and refractive index. These parameters have a significant effect on the hydrodynamic radius calculated from DLS. Previous reports in the literature use values for water in these calculations. However, as the bicelles are not at infinite dilution, we also included hydrodynamic radii calculated using the solution parameters measured directly on our bicelle solutions. Using the solution parameters for water, our data matches that of bicelles shown to have a discoidal morphology [13]. Additionally, (i) EmrE is unstable in pure DHPC but very stable in bicelles and (ii) the NMR spectra change significantly with long chain lipid composition (Fig. 4), despite the fact that DHPC is the major component in our bicelles (3:1 DHPC:long-chain lipid). This argues against the mixed micelle model.
Table 1.
Table of hydrodynamic radii of isotropic bicelles determined via DLS and rotational diffusion NMR.
| Sample | Hydrodynamic radius (nm) | ||||
|---|---|---|---|---|---|
| Water Parametersa | Bicelle Parametersb | ||||
| DLS, 10°C | DLS, 25°C | DLS, 10°C | DLS, 25°C | NMR, 45°C c |
|
| Reconstitution via liposomes, 15N EmrE in DMPC/DHPC bicelles |
3.5 ± 0.8 | 3.0 ± 0.4 | 2.0 ± 0.3 | 2.4 ± 0.3 | 3.2 ± 0.3 |
| Reconsitution via detergent exchange, 15N EmrE in DMPC/DHPC bicelles |
3.2 ± 0.8 | 3.0 ± 0.8 | 1.9 ± 0.4 | 2.4 ± 0.6 | 2.9 ± 0.3 |
| Reconstitution via liposomes, 2H/15N/13C EmrE in DLPC/DHPC bicelles |
3.6 ± 0.8 | 3.3 ± 0.5 | 2.2 ± 0.5 | 2.6 ± 0.4 | 2.7 ± 0.3 |
RH values determined from DLS, using the viscosity and refractive index of water.
RH values determined from DLS, using the viscosity and refractive index of the isotropic bicelle solution at the given temperature (see methods).
Calculated from rotational correlation time using equation 1.
4. Conclusions
Conventional methods for reconstitution of integral membrane proteins into isotropic bicelles require exposure of the protein to organic solvent or DHPC micelles. These intermediate environments can be destabilizing to sensitive IMPs. We have found that by first reconstituting EmrE into liposomes and then breaking up the proteoliposomes with DHPC, we can achieve samples of higher spectral quality and greater long-term stability. This method is broadly applicable since the IMP may be reconstituted into liposomes using any method suitable for the protein of interest. Addition of DHPC to form isotropic bicelles from liposomes makes this protocol easy to use and optimize relative to nanodiscs.
The environment-sensitive small multidrug resistance transporter EmrE has been successfully reconstituted into isotropic bicelles of a range of compositions using this protocol. Reconstituting EmrE via liposomes has allowed us to assign the majority of backbone residues and quantitatively measure slow timescale dynamics [11]. The increased sample stability and homogeneity provided by this reconstitution method was critical for the success of these novel NMR experiments on an active transporter. Furthermore, the (1H, 15N)-TROSY HSQC spectra in different lipid environments presented here suggest that EmrE dynamics are significantly affected by lipid composition. Experiments are in progress to further investigate the effect of lipid environment on EmrE structure, dynamics, and function.
Highlights.
Reconstitution without requirement for organic solvent or specific detergent
Increased stability and homogeneity of isotropic bicelle samples
Reconstitution with expanded, more physiologically relevant lipid profile
Significant improvements for applications in biophysical and structural studies
Acknowledgment
We thank Dr. Gregory DeKoster for modifying the (1H, 15N)-TRACT pulse sequence and Dr. Geoffrey Chang for providing us with the EmrE expression plasmid. We thank the Lohman and Schlesinger labs for use of their fluorimeter, DLS and refractometer. This work was supported by an NSF Graduate Research Fellowship (DGE-1143954) to EAM and the Searle Scholars Program (KAHW).
Abbreviations
- NMR
nuclear magnetic resonance
- DM
decylmaltoside
- DHPC
1,2-dihexanoyl-sn-glycero-3-phosphocholine
- DLPC
1,2-dilauroyl-sn-glycero-3-phosphocholine
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphocholine
- DPPC
1,2-dipalmitoyl-sn-glycero-3-phosphocholine
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine
- POPE
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- POPG
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)
- TPP+
tetraphenylphosphonium+
- PS
phosphatidylserine
- PE-DTPA
phosphatidylethanolamine-N-diethylenetriaminepentaacetic acid
- IMP
integral membrane protein
- DLS
dynamic light scattering
- TLC
thin layer chromatography
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Warschawski DE, Arnold AA, Beaugrand M, Gravel A, Chartrand É, Marcotte I. Choosing membrane mimetics for NMR structural studies of transmembrane proteins. BBA - Biomembranes. 2011;1808:1957–1974. doi: 10.1016/j.bbamem.2011.03.016. [DOI] [PubMed] [Google Scholar]
- [2].Raschle T, Hiller S, Etzkorn M, Wagner G. Nonmicellar systems for solution NMR spectroscopy of membrane proteins. Curr Opin Struct Biol. 2010;20:471–479. doi: 10.1016/j.sbi.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Poget SF, Girvin ME. Solution NMR of membrane proteins in bilayer mimics: small is beautiful, but sometimes bigger is better. Biochim Biophys Acta. 2007;1768:3098–3106. doi: 10.1016/j.bbamem.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Poget SF, Cahill SM, Girvin ME. Isotropic bicelles stabilize the functional form of a small multidrug-resistance pump for NMR structural studies. J Am Chem Soc. 2007;129:2432–2433. doi: 10.1021/ja0679836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sanders CR, Sönnichsen F. Solution NMR of membrane proteins: practice and challenges. Magn Reson Chem. 2006;44:S24–40. doi: 10.1002/mrc.1816. Spec No. [DOI] [PubMed] [Google Scholar]
- [6].Bayburt TH, Sligar SG. Membrane protein assembly into Nanodiscs. FEBS Lett. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Raschle T, Hiller S, Yu T-Y, Rice AJ, Walz T, Wagner G. Structural and functional characterization of the integral membrane protein VDAC-1 in lipid bilayer nanodiscs. J Am Chem Soc. 2009;131:17777–17779. doi: 10.1021/ja907918r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Poget SF, Harris R, Cahill SM, Girvin ME. 1H, 13C, 15N backbone NMR assignments of the Staphylococcus aureus small multidrug-resistance pump (Smr) in a functionally active conformation. Biomol NMR Assign. 2010;4:139–142. doi: 10.1007/s12104-010-9228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wu H, Su K, Guan X, Sublette ME, Stark RE. Assessing the size, stability, and utility of isotropically tumbling bicelle systems for structural biology. Biochim Biophys Acta. 2010;1798:482–488. doi: 10.1016/j.bbamem.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bertelsen K, Vad B, Nielsen EH, Hansen SK, Skrydstrup T, Otzen DE, Vosegaard T, Nielsen NC. Long-term-stable ether-lipid vs conventional ester-lipid bicelles in oriented solid-state NMR: altered structural information in studies of antimicrobial peptides. J Phys Chem B. 2011;115:1767–1774. doi: 10.1021/jp110866g. [DOI] [PubMed] [Google Scholar]
- [11].Morrison EA, DeKoster GT, Dutta S, Clarkson M, Vafabakhsh R, Bahl A, Kern D, Ha T, Henzler-Wildman KA. Antiparallel EmrE exports drugs by exchanging between asymmetric structures. Nature. doi: 10.1038/nature10703. In Press. doi 10.1038/nature10703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rotem D, Schuldiner S. EmrE, a multidrug transporter from Escherichia coli, transports monovalent and divalent substrates with the same stoichiometry. J Biol Chem. 2004;279:48787–48793. doi: 10.1074/jbc.M408187200. [DOI] [PubMed] [Google Scholar]
- [13].Glover KJ, Whiles JA, Wu G, Yu N, Deems R, Struppe JO, Stark RE, Komives EA, Vold RR. Structural evaluation of phospholipid bicelles for solution-state studies of membrane-associated biomolecules. Biophysical Journal. 2001;81:2163–2171. doi: 10.1016/s0006-3495(01)75864-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lee D, Hilty C, Wider G, Wüthrich K. Effective rotational correlation times of proteins from NMR relaxation interference. J Magn Reson. 2006;178:72–76. doi: 10.1016/j.jmr.2005.08.014. [DOI] [PubMed] [Google Scholar]
- [15].Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- [16].JOHNSON B, BLEVINS R. NMR VIEW - A COMPUTER-PROGRAM FOR THE VISUALIZATION AND ANALYSIS OF NMR DATA. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- [17].Rosenbusch JP. Stability of membrane proteins: relevance for the selection of appropriate methods for high-resolution structure determinations. J Struct Biol. 2001;136:144–157. doi: 10.1006/jsbi.2001.4431. [DOI] [PubMed] [Google Scholar]
- [18].Tate CG. Practical considerations of membrane protein instability during purification and crystallisation. Methods Mol Biol. 2010;601:187–203. doi: 10.1007/978-1-60761-344-2_12. [DOI] [PubMed] [Google Scholar]
- [19].Popot J-L. Amphipols, nanodiscs, and fluorinated surfactants: three nonconventional approaches to studying membrane proteins in aqueous solutions. Annu Rev Biochem. 2010;79:737–775. doi: 10.1146/annurev.biochem.052208.114057. [DOI] [PubMed] [Google Scholar]
- [20].Park SH, Prytulla S, De Angelis AA, Brown JM, Kiefer H, Opella SJ. High-resolution NMR spectroscopy of a GPCR in aligned bicelles. J Am Chem Soc. 2006;128:7402–7403. doi: 10.1021/ja0606632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Charalambous K, Miller D, Curnow P, Booth PJ. Lipid bilayer composition influences small multidrug transporters. BMC Biochem. 2008;9:31. doi: 10.1186/1471-2091-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Winstone TL, Jidenko M, le Maire M, Ebel C, Duncalf KA, Turner RJ. Organic solvent extracted EmrE solubilized in dodecyl maltoside is monomeric and binds drug ligand. Biochem Biophys Res Commun. 2005;327:437–445. doi: 10.1016/j.bbrc.2004.11.164. [DOI] [PubMed] [Google Scholar]
- [23].Schwaiger M, Lebendiker M, Yerushalmi H, Coles M, Gröger A, Schwarz C, Schuldiner S, Kessler H. NMR investigation of the multidrug transporter EmrE, an integral membrane protein. Eur J Biochem. 1998;254:610–619. doi: 10.1046/j.1432-1327.1998.2540610.x. [DOI] [PubMed] [Google Scholar]
- [24].Butler PJG, Ubarretxena-Belandia I, Warne T, Tate CG. The Escherichia coli multidrug transporter EmrE is a dimer in the detergent-solubilised state. J Mol Biol. 2004;340:797–808. doi: 10.1016/j.jmb.2004.05.014. [DOI] [PubMed] [Google Scholar]
- [25].Schuldiner S. EmrE, a model for studying evolution and mechanism of ion-coupled transporters. Biochim Biophys Acta. 2009;1794:748–762. doi: 10.1016/j.bbapap.2008.12.018. [DOI] [PubMed] [Google Scholar]
- [26].Cho HS, Dominick JL, Spence MM. Lipid domains in bicelles containing unsaturated lipids and cholesterol. J Phys Chem B. 2010;114:9238–9245. doi: 10.1021/jp100276u. [DOI] [PubMed] [Google Scholar]
- [27].Minto RE, Adhikari PR, Lorigan GA. A 2H solid-state NMR spectroscopic investigation of biomimetic bicelles containing cholesterol and polyunsaturated phosphatidylcholine. Chem Phys Lipids. 2004;132:55–64. doi: 10.1016/j.chemphyslip.2004.09.005. [DOI] [PubMed] [Google Scholar]
- [28].van Dam L, Karlsson G, Edwards K. Direct observation and characterization of DMPC/DHPC aggregates under conditions relevant for biological solution NMR. Biochim Biophys Acta. 2004;1664:241–256. doi: 10.1016/j.bbamem.2004.06.005. [DOI] [PubMed] [Google Scholar]
- [29].Luchette PA, Vetman TN, Prosser RS, Hancock RE, Nieh MP, Glinka CJ, Krueger S, Katsaras J. Morphology of fast-tumbling bicelles: a small angle neutron scattering and NMR study. Biochim Biophys Acta. 2001;1513:83–94. doi: 10.1016/s0005-2736(01)00358-3. [DOI] [PubMed] [Google Scholar]
- [30].Andersson A, Mäler L. Size and shape of fast-tumbling bicelles as determined by translational diffusion. Langmuir. 2006;22:2447–2449. doi: 10.1021/la053177l. [DOI] [PubMed] [Google Scholar]