Abstract
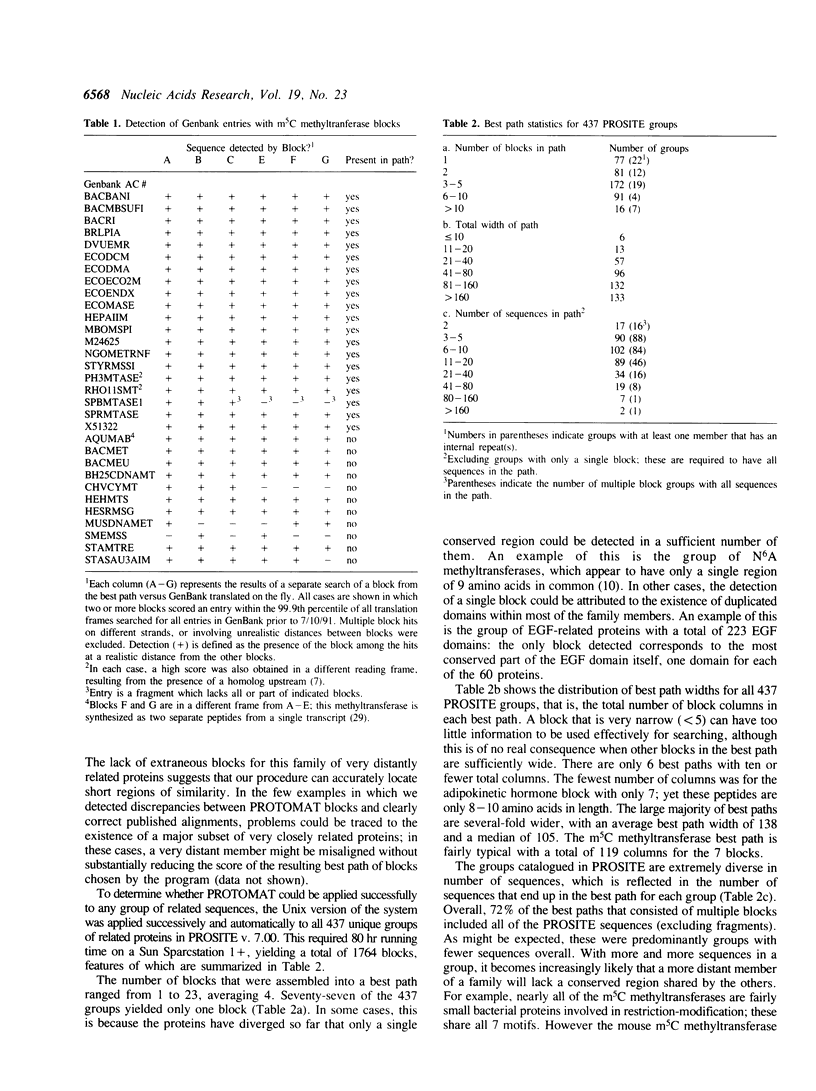
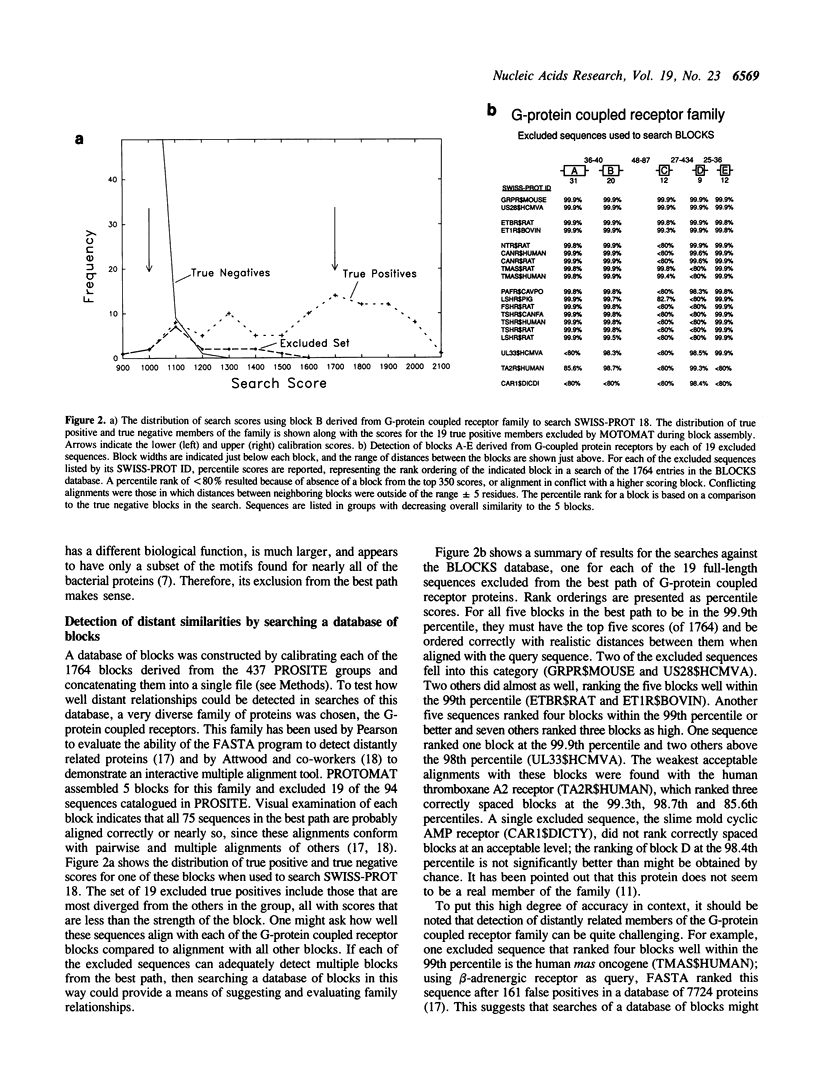
A system is described for finding and assembling the most highly conserved regions of related proteins for database searching. First, an automated version of Smith's algorithm for finding motifs is used for sensitive detection of multiple local alignments. Next, the local alignments are converted to blocks and the best set of non-overlapping blocks is determined. When the automated system was applied successively to all 437 groups of related proteins in the PROSITE catalog, 1764 blocks resulted; these could be used for very sensitive searches of sequence databases. Each block was calibrated by searching the SWISS-PROT database to obtain a measure of the chance distribution of matches, and the calibrated blocks were concatenated into a database that could itself be searched. Examples are provided in which distant relationships are detected either using a set of blocks to search a sequence database or using sequences to search the database of blocks. The practical use of the blocks database is demonstrated by detecting previously unknown relationships between oxidoreductases and by evaluating a proposed relationship between HIV Vif protein and thiol proteases.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Lipman D. J. Protein database searches for multiple alignments. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5509–5513. doi: 10.1073/pnas.87.14.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwood T. K., Eliopoulos E. E., Findlay J. B. Multiple sequence alignment of protein families showing low sequence homology: a methodological approach using database pattern-matching discriminators for G-protein-linked receptors. Gene. 1991 Feb 15;98(2):153–159. doi: 10.1016/0378-1119(91)90168-b. [DOI] [PubMed] [Google Scholar]
- Bairoch A., Boeckmann B. The SWISS-PROT protein sequence data bank. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2247–2249. doi: 10.1093/nar/19.suppl.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A. PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2241–2245. doi: 10.1093/nar/19.suppl.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. Phosphotransferase sequence homology. Nature. 1987 Sep 3;329(6134):21–21. doi: 10.1038/329021a0. [DOI] [PubMed] [Google Scholar]
- Burbaum J. J., Starzyk R. M., Schimmel P. Understanding structural relationships in proteins of unsolved three-dimensional structure. Proteins. 1990;7(2):99–111. doi: 10.1002/prot.340070202. [DOI] [PubMed] [Google Scholar]
- Citron B. A., Milstien S., Gutierrez J. C., Levine R. A., Yanak B. L., Kaufman S. Isolation and expression of rat liver sepiapterin reductase cDNA. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6436–6440. doi: 10.1073/pnas.87.16.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrah P. M., Kay S. A., Teakle G. R., Griffiths W. T. Cloning and sequencing of protochlorophyllide reductase. Biochem J. 1990 Feb 1;265(3):789–798. doi: 10.1042/bj2650789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs R. MacPattern: protein pattern searching on the Apple Macintosh. Comput Appl Biosci. 1991 Jan;7(1):105–106. doi: 10.1093/bioinformatics/7.1.105. [DOI] [PubMed] [Google Scholar]
- Gribskov M., McLachlan A. D., Eisenberg D. Profile analysis: detection of distantly related proteins. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Wallace J. C., Brown J. P. Finding protein similarities with nucleotide sequence databases. Methods Enzymol. 1990;183:111–132. doi: 10.1016/0076-6879(90)83009-x. [DOI] [PubMed] [Google Scholar]
- Karreman C., de Waard A. Agmenellum quadruplicatum M.AquI, a novel modification methylase. J Bacteriol. 1990 Jan;172(1):266–272. doi: 10.1128/jb.172.1.266-272.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller J. W., Baurick K. B., Rutt G. C., O'Malley M. V., Sonafrank N. L., Reynolds R. A., Ebbesson L. O., Vajdos F. F. Pseudomonas cepacia 2,2-dialkylglycine decarboxylase. Sequence and expression in Escherichia coli of structural and repressor genes. J Biol Chem. 1990 Apr 5;265(10):5531–5539. [PubMed] [Google Scholar]
- Patthy L. Detecting homology of distantly related proteins with consensus sequences. J Mol Biol. 1987 Dec 20;198(4):567–577. doi: 10.1016/0022-2836(87)90200-2. [DOI] [PubMed] [Google Scholar]
- Pearson W. R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- Pósfai J., Bhagwat A. S., Pósfai G., Roberts R. J. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989 Apr 11;17(7):2421–2435. doi: 10.1093/nar/17.7.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler G. D., Altschul S. F., Lipman D. J. A workbench for multiple alignment construction and analysis. Proteins. 1991;9(3):180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- Schulz R., Steinmüller K., Klaas M., Forreiter C., Rasmussen S., Hiller C., Apel K. Nucleotide sequence of a cDNA coding for the NADPH-protochlorophyllide oxidoreductase (PCR) of barley (Hordeum vulgare L.) and its expression in Escherichia coli. Mol Gen Genet. 1989 Jun;217(2-3):355–361. doi: 10.1007/BF02464904. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Annau T. M., Chandrasegaran S. Finding sequence motifs in groups of functionally related proteins. Proc Natl Acad Sci U S A. 1990 Jan;87(2):826–830. doi: 10.1073/pnas.87.2.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. F., Smith T. F. Automatic generation of primary sequence patterns from sets of related protein sequences. Proc Natl Acad Sci U S A. 1990 Jan;87(1):118–122. doi: 10.1073/pnas.87.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Searching for patterns in protein and nucleic acid sequences. Methods Enzymol. 1990;183:193–211. doi: 10.1016/0076-6879(90)83014-z. [DOI] [PubMed] [Google Scholar]
- Sternberg M. J. PROMOT: a FORTRAN program to scan protein sequences against a library of known motifs. Comput Appl Biosci. 1991 Apr;7(2):257–260. doi: 10.1093/bioinformatics/7.2.257. [DOI] [PubMed] [Google Scholar]
- Taylor W. R. A flexible method to align large numbers of biological sequences. J Mol Evol. 1988 Dec;28(1-2):161–169. doi: 10.1007/BF02143508. [DOI] [PubMed] [Google Scholar]
- Taylor W. R. Identification of protein sequence homology by consensus template alignment. J Mol Biol. 1986 Mar 20;188(2):233–258. doi: 10.1016/0022-2836(86)90308-6. [DOI] [PubMed] [Google Scholar]
- Vingron M., Argos P. A fast and sensitive multiple sequence alignment algorithm. Comput Appl Biosci. 1989 Apr;5(2):115–121. doi: 10.1093/bioinformatics/5.2.115. [DOI] [PubMed] [Google Scholar]



