We identified SLIT and NTRK-like 1 (SLITRK1) as a candidate gene for Tourette syndrome (TS) based on the mapping of a de novo chr13 inversion, the finding of a rare frameshift mutation in a family with TS, and the association of a functional rare 3′UTR variant (var321) corresponding to highly conserved miRNA binding domain.1 This rare allele was identified in 2 of 174 individuals with TS and was not present in 2148 controls (P = 0.0056). Based on the absence of haplotype sharing between the var321 probands, we concluded that the alleles were likely to represent independent mutations, further supporting the association. In this study, we present additional data based on genome-wide genotyping, multidimensional scaling analysis (MDS), and dense haplotype mapping, providing additional evidence in favor of these initial conclusions.
Two subsequent reports and a recent review have asserted that this reported association might have been the result of population stratification.2–4 These conclusions were based primarily on the observation of a single var321 allele seen among 512 Ashkenazi Jewish control chromosomes,2 which suggested, but did not confirm, a higher frequency among this population than observed in our self-identified White controls,1 which accounted for the majority of the initial control chromosomes screened (1/512 versus 0/3562, Fisher’s exact test P = 0.126). Interestingly, both reports found relatively high rates of var321 among affected Ashkenazi families.2,3 However, in some of these families, the allele was identified in an affected parent but not transmitted to the ascertained proband. Despite other possible explanations such as the high rate of bi-lineal inheritance observed in TS and the low power in these subsequent studies to detect the association using a transmission disequilibrium approach, these findings were interpreted to mean that our initial result was spurious.
The potential confound of population stratification is clearly an important one for rare as well as common variant association studies. In our initial analysis, we relied on patient self-reported ethnicity and haplotype analysis to attempt to rule out cryptic relatedness or shared ethnic origin. However, since publication, the methods available to evaluate this question have improved markedly. Consequently, we have now used an MDS analysis to determine the ethnic genomic profiles of the two initially reported cases and to assess whether either or both cluster with Ashkenazi individuals. We have also identified five additional subjects possessing var321 alleles and conducted additional extensive fine mapping of the SLITRK1 region in all seven carriers in search of haplotype sharing. Both analyses provide further confirmation of our initial conclusions and argue strongly against population stratification confounding our results.
DNAs from 1117 self-reported European descent (Non-Jewish) individuals, 758 self-reported Jewish individuals, and seven unrelated individuals carrying var321 and their available family members were genotyped using either 1M or 317K BeadChips (Illumina, San Diego, CA, USA). The individuals carrying var321 include the two initially reported TS cases (F3 and F4),1 an affected proband from a Spanish family (SP), two affected parents reporting eastern European Jewish descent ascertained by the Tourette Syndrome Association International Genetics Consortium (TSA1 aka TDT08-21-01 and TSA2 aka TDT10-28-02),3 a phenotype-unknown sample from the ‘Caucasian’ human variation panel (HVP),5 and an unaffected Ashkenazi Jewish control (AN) identified by screening 257 female controls from the Hadassah Biological Psychiatry Laboratory. As in our original report, we assigned affected status to individuals with chronic motor tics (CT), obsessive-compulsive disorder (OCD) or TS, given the very strong evidence for a TS-OCD spectrum of disease.6 Control subjects were drawn either from the HapMap study or an investigation of Inflammatory Bowel Disease7 in which patient ascertainment was conducted throughout the United States and Canada. It has been estimated that ~90% of Jews in North America are of Ashkenazi origin,8 indicating that the 758 self-reported Jewish subjects would well represent the Ashkenazi population.
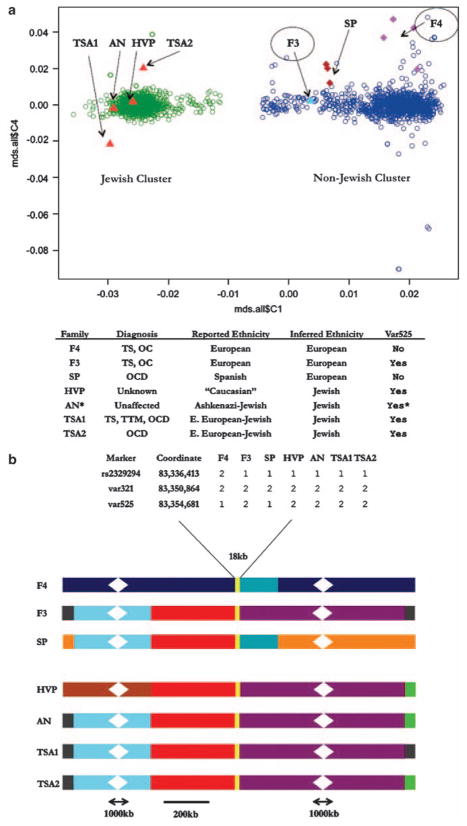
We used the program PLINK to conduct an MDS analysis on a subset of 37 808 high quality SNPs.9 This approach has been shown to be able to distinguish genomic profiles in closely related ethnic groups9 and consequently we reasoned it would reliably differentiate Ashkenazi from non-Ashkenazi samples. Indeed, after one round of outlier removal, we found that the first axis clearly distinguished the two large groups of subjects (Figure 1a). As a confirmation of the utility of the analysis, we found that all three individuals who identified themselves as Jewish or Ashkenazi clustered as anticipated (Figure 1a). Of primary interest, both subjects F3 and F4, who reported no Ashkenazi or Jewish ethnicity, clustered with the European descent (Non-Jewish) group, providing no support for the contention that our initial analysis was flawed. Finally, one newly ascertained proband (SP) mapped with F3 and F4 in the European descent (Non-Jewish) group, and one individual for whom there was no definitive ethnicity data mapped in the Ashkenazi cluster.
Figure 1.
Multidimensional scaling (MDS) and haplotype analysis of var321 carriers. (a) Scatter plot showing axes 1 and 4 of the MDS analysis on 1117 European descent (Non-Jewish), 758 Jewish, and var321 samples. Axis 1 is the horizontal axis and clearly separates the two groups. On the right, the European descent (Non-Jewish) samples (blue) cluster along axis 1 with values >−0.01. On the left, the Jewish samples (green) cluster along axis 1 with values <−0.01. Var321 carriers are indicated by arrows and all fall within the two clusters. The two TS probands from1 are F3 and F4. Var321 carriers without var525 and their family members, SP family (burgundy) and F4 family (purple), are represented by diamonds. Var321–var525 carriers are shown as triangles, predicted non-Jewish (light-blue) and Jewish (red). (b) Diagram of the maximum haplotype sharing between individual haplotypes carrying the var321 allele. Shared colors are haplotype regions that could not be differentiated between samples. Unshared colors are unique haplotype regions. Scale bar is 200 kb, broken regions represent 1000 kb that were omitted to reduce image size. Expanded inset is 18 kb surrounding SLITRK1 and shows phased haplotypes. *Individual is homozygous for var321–var525. TS, Tourette Syndrome; OCD, obsessive-compulsive disorder; OC, obsessive-compulsive symptoms; TTM, trichotillomania; F3 and F4, TS affected from original study; SP, Spanish family with OCD proband and unaffected carrier father; HVP, human variation panel sample; AN, Ashkenazi control; TSA1, TS, OCD, TTM affected and TSA2, OCD affected, from TSAICG study.
Next, we undertook an in-depth study of var321 in all carriers evaluating the extent of maximal haplotype sharing. We used SNP genotyping data, Sanger sequencing of the SLITRK1 genomic region and 48 custom-designed highly polymorphic short tandem repeat (STR) markers. In our previous work, we showed that F3 and F4 shared at most a 87 kb region surrounding SLITRK1. This was based in part on our identification of a second rare cis allele (var525) found 3.8 kb centromeric to var321 only in subject F3 and not present in subject F4. Additional sequencing shows that subjects F3, HVP, AN, TSA1 and TSA2 all carry this rare cis allele var525, whereas F4 and SP do not. In addition, the maximum-shared haplotype between F4 and F3 was refined from our initial report to no more than 18 kb (Figure 1b). In contrast, sharing among individuals carrying the var321-var525 haplotype ranged from 2.1Mb to 12 Mb. AN, the Ashkenazi control, was found to be homozygous for a 4.2Mb haplotype in this region. Genome-wide genotyping data obtained from Illumina 1M arrays did not provide an alternative explanation for this finding as the data did not suggest either that the individual was the product of a consanguineous union or did CNV analysis identify a heterozygous deletion in the SLITRK1 region.
These findings do not provide support for the hypothesis that var321 represents a founder allele in the Ashkenazi population. The presence of at least two clearly distinguishable var321 haplotypes and the small degree of sharing observed between F3 and F4 is most consistent with these being independent mutations, as the alternative would require an extremely ancient common ancestor.
Interestingly, while there is evidence for a larger region of sharing among the individuals carrying a var321–var525 haplotype, its presence in the F3 sample, in light of our MDS analysis, shows that it is not restricted to the Ashkenazi population. Given our current results and the self-reported ethnicity of F3, the most parsimonious explanation is that var321 occurred on this haplotype before the Ashkenazi divergence, but relatively recently in terms of human history. Interestingly, results similar to those reported here have been observed previously in several other movement disorders.10,11 In these cases, identical independent mutation alleles have been identified within and outside the Ashkenazi population and showed similar patterns of haplotype distribution.
Owing to the varying methodologies employed in the studies of var321, it is not possible to construct a precise meta-analysis from the published data. However, the vast majority of alleles reported so far have been found in affected individuals and families, and the denominator in aggregate, both for cases and controls, has reached thousands of individuals. Given the very low allele frequency for var321 in all ethnic groups, which essentially precludes transmission tests of association, the high rate of bi-lineal inheritance in TS, and the observation of incomplete penetrance and complex inheritance for the spectrum of disorders, a large analysis studying sample sizes that are not yet available to the TS genetics community will be required for a more definitive test of replication using an ethnically matched case–control design.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Abelson JF, Kwan KY, O’Roak BJ, Baek DY, Stillman AA, Morgan TM, et al. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 2.Keen-Kim D, Mathews CA, Reus VI, Lowe TL, Herrera LD, Budman CL, et al. Hum Mol Genet. 2006;15:3324–3328. doi: 10.1093/hmg/ddl408. [DOI] [PubMed] [Google Scholar]
- 3.Scharf JM, Moorjani P, Fagerness J, Platko JV, Illmann C, Galloway B, et al. Neurology. 2008;70(Part 2):1495–1496. doi: 10.1212/01.wnl.0000296833.25484.bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sethupathy P, Collins FS. Trends Genet. 2008;24:489–497. doi: 10.1016/j.tig.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Wendland JR, Kruse MR, Murphy DL. Mol Psychiatry. 2006;11:802–804. doi: 10.1038/sj.mp.4001848. [DOI] [PubMed] [Google Scholar]
- 6.Cavanna AE, Servo S, Monaco F, Robertson MM. J Neuropsychiatry Clin Neurosci. 2009;21:13–23. doi: 10.1176/jnp.2009.21.1.13. [DOI] [PubMed] [Google Scholar]
- 7.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, et al. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ostrer H. Nat Rev Genet. 2001;2:891–898. doi: 10.1038/35098506. [DOI] [PubMed] [Google Scholar]
- 9.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valente EM, Povey S, Warner TT, Wood NW, Davis MB. Ann Hum Genet. 1999;63(Part 1):1–8. doi: 10.1046/j.1469-1809.1999.6310001.x. [DOI] [PubMed] [Google Scholar]
- 11.Zabetian CP, Hutter CM, Yearout D, Lopez AN, Factor SA, Griffith A, et al. Am J Hum Genet. 2006;79:752–758. doi: 10.1086/508025. [DOI] [PMC free article] [PubMed] [Google Scholar]