Abstract
Investigations into the genetics of child psychiatric disorders have begun to shed light on molecular and cellular mechanisms of psychopathology. The first strains of success are the result of an increasingly sophisticated appreciation of the allelic architecture of common neuropsychiatric disorders; the consolidation of large patient cohorts; the emergence of genomic tools enabling comprehensive studies of rare as well as common genetic variation; and advances in developmental neuroscience that are fueling the rapid translation of genetic findings. These issues, as well as the opportunities and challenges that now confront gene discovery efforts in genetically complex conditions, are addressed with regard to two paradigmatic childhood disorders: autism and Tourette syndrome.
In many ways it is truly the best of times.
However as excitement justifiably mounts that an inroad into child psychiatric disorders is finally gaining a scientific foothold, a survey of recent genetic findings points to both continuing and emerging challenges. The early application of mature genomic technologies that are providing unequivocal, if not comprehensive, insights into the contribution of common variants to common medical conditions (Altshuler et al., 2008) have so far not revealed definitive findings in child psychiatry, reinforcing the conclusion that the genetics of these disorders are particularly difficult to decipher; current gene discovery efforts in a number of childhood and adult disorders are challenging the biological relevance of the psychiatric diagnostic nosology; and the ability to leverage advances in neuroscience promises to remain both a blessing and a curse: on the one hand, the contribution of neurobiological data to clarify genetic findings and to drive translational efforts is critical. At the same time, the lure of substituting biological plausibility for accepted standards of evidence in genetic or genomic analyses, coupled with the rapid accumulation of massive amounts of sequence and structural variation in affected individuals risks a proliferation of extremely intriguing, but ultimately misguided, neurobiological narratives.
This review will aim to address both sides of the coin: the tremendous promise as well as the challenges facing child psychiatric genetics. The first three sections will address the changing conceptions of the allelic architecture of common disorders; the attendant emerging focus on rare variation in child psychiatry; and the causes and consequences of an increasing reliance on case control association methodology. The review will then turn to consider gene discovery efforts in two paradigmatic childhood disorders: autism and Tourette syndrome, focusing on recent finding, the role of emerging human genetic data in illuminating pathophysiological mechanisms and a view toward the future of these endeavors.
The Allelic Architecture of Child Psychiatric Disorders
Relatively early in the history of genetics of common child psychiatric disorders it became clear that writ large these did not obey simple Mendelian expectations. This consensus led to the ascendance of a “common variant common disease hypothesis” (CVCD) as the predominant paradigm in the field (Chakravarti, 1999; Reich and Lander, 2001; Risch and Merikangas, 1996): The expectation was that any given common allele (defined here as 5% or greater population frequency) would carry moderate effects and was likely to be neither necessary nor sufficient to lead to the clinical phenotype. Indeed the notion was that a combination of risk alleles would contribute to the emergence of pathological traits falling at the extremes of a population distribution. Inherent in this view is the notion that that family members will tend to demonstrate sub-clinical manifestations of a phenotype of interest due to the presence of some, but not all, risk alleles and that one of the primary objectives of human genetic research would be to account for all or most of the common genetic risks shared by the population. The combination of the CVCD paradigm with steadily increasing estimates of the number of genes anticipated to play a role, led to the related conclusion that association methodologies, demonstrating that a particular common allele was overrepresented in the population of affected individuals, would be the most profitable avenue to gene discovery.
A convergence of the common variant paradigm with the technical feasibility of genotyping one or a small number of common variants led to a plethora of candidate gene association analyses throughout the 1990s and into the early part of this century. Indeed for many years, the study of child psychiatric genetics was essentially synonymous with this approach. Unfortunately, the reliability of these studies has proven to be poor (Hirschhorn et al., 2002; Lohmueller et al., 2003). To those outside the field, a steady stream of plausible but routinely contradictory findings began to cast doubt on the entire enterprise.
However, over the past few years, the emergence of highly reproducible associations from genome-wide association (GWAS) studies in diverse clinical areas has clarified many of the reasons for these difficulties (Altshuler et al., 2008; Manolio, 2010): First, the effect size of common alleles contributing to common disorders is, by-in-large, much smaller than anticipated; Second, the prior probability that any chosen candidate gene or allele will be associated with the phenotype of interest is extremely low, even given the most plausible biological hypotheses, a reality that has been reinforced by the very small number of previously suspected loci that have been confirmed by unbiased genome wide analyses; and, finally, the confounds to association studies, particularly attending case control approaches, are more problematic than initially anticipated.
With regard to this last point, several key methodological issues are particularly relevant: for example, the degree to which subtle differences in ancestry among cases and controls, known as population stratification, could derail analyses was appreciated but the methods available initially to control for this confound were rudimentary and later found to be inadequate. The presumption that grouping by observable characteristics or self-reported ethnicity would be sufficient was incorrect. Fortunately the large amount of variation now detected by genome wide genotyping allows for precise matching of cases and controls. And while there remains some debate over how thoroughly even these methods can protect against this confound (McClellan and King, 2010), there is little question that the failure to address ancestry in a rigorous fashion contributed to the large number of non-replicated candidate gene analyses. Similarly, candidate gene association analyses often tended to underestimate the impact of pedestrian confounds such as genotyping error or batch effects.
These difficulties are of more than historical interest: A surprising number of candidate gene studies continue to disregard these issues; while the problem of population stratification is acknowledged (Tost et al., 2010), few so called “imaging genomic” studies, those aimed at investigating the relationship between genetic variation and functional or structural neuroimaging phenotypes, use methods that would be considered state-of-the-art within the genetics community to control for this confound; and even contemporary GWAS studies may still fall prey to overlooked technical artifacts (Sebastiani et al., 2010). Importantly, as will be discussed below, a key consideration for the future is that as many fields, including child psychiatry, increasingly employ case-control methods to study the contribution of rare alleles to disease, similar liabilities must be anticipated, in the analyses of both copy number variation and next generation sequencing data.
The Ascendance of Rare Variant Approaches in Child Psychiatry
Despite the clear numerical predominance of common variant studies in child psychiatry, rare variant approaches have a long history in the field and recent interest in these strategies has risen dramatically. This has been a consequence of an increasing recognition of the relevance of early rare variant findings, the development of CNV and next generation sequencing platforms, and a consensus that GWAS studies have identified a small proportion of the anticipate genetic risk for common disorders and that the “missing inheritance” may be accounted for in part by rare alleles (Goldstein, 2009; Manolio et al., 2009).
Before addressing specific findings in autism and Tourette syndrome, it is worthwhile to differentiate between two complementary but distinct approaches to conceptualizing rare variant analyses: i.e. “outlier” strategies versus methods based on a rare variant common disease (RVCD) hypothesis.
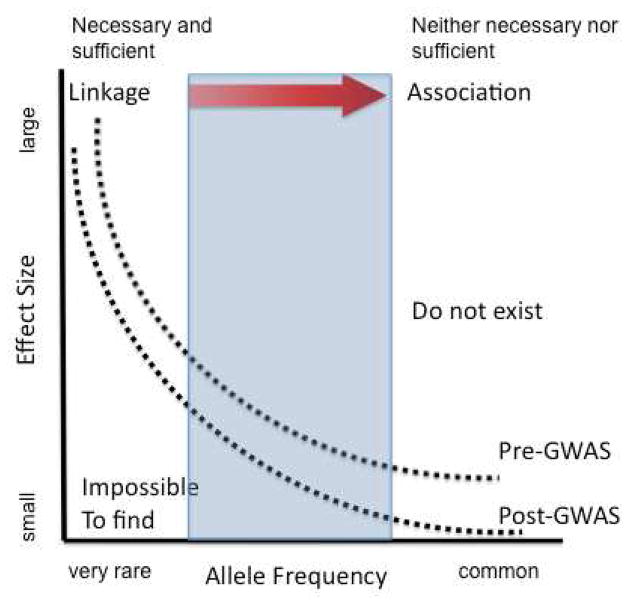
The rationale for outlier studies is that some, potentially very small, proportion of the allelic architecture for a complex disorder will be accounted for by low frequency variants of large effect, including potentially Mendelian forms of a condition. This view is based on the notion that, particularly for early onset disorders, natural selection will tend to ensure a predictable, inverse relationship between allele frequency and effect size (Figure 1). This view disaggregates common phenotypes: disorders are not attacked writ large but with a focus on finding often extremely rare examples that promise to shed light on biological mechanisms relevant to more common examples of the phenotype. This emphasis poses an important contrast to at least the initial rationale for common variant studies by prioritizing any traction with regard to pathophysiology over the effort to clarify population genetic risks.
Figure 1.
The graph represents the general relationship between allele frequency (X-axis) and effect size (Y- axis). The top dotted curve shows the anticipated relationship prior to the advent of genome wide approaches to association and the bottom curve the shift attending empirical data regarding the odds ratios found in the majority of common variant studies. The blue box highlights the region into which recent rare variant findings in ASD fall, representing low frequency alleles but carrying risks that are not of the scale identifed in rare Mendelian disorders. The red arrow at the top of the diagram represents the shift toward association that is required to identify alleles that are neither necessary nor sufficient (either within a given pedigree or the general population) for the emergence of a phenotype
This outlier approach is entirely consistent with, but differs somewhat from studies based on a rare variant common disease (RVCD) hypothesis. Specifically, in contrast to the minimalist expectations regarding the role of rare variation in common disorders reflected in outlier studies, the RVCD paradigm posits specifically that rare variants will carry a significant portion, if not the majority, of population risk for a common disorder. This RVCD hypothesis has been recently pursued through a growing number of CNV analyses and via deep sequencing studies of, for example, blood pressure regulation (Ji et al., 2008), cholesterol and lipid metabolism (Cohen et al., 2004; Johansen et al., 2010) and autism (Bakkaloglu et al., 2008). With the advent of next generation sequencing, such studies promise to proliferate dramatically.
Already, these types of studies have engendered an important reevaluation of the manner in which rare variation may contribute to common disease: historically, rare variation has been synonymous with alleles of very large effect (Figure 1). Indeed, given the methods previously employed to detect rare disease alleles, most discoveries involved Mendelian inheritance, with the attendant demonstration that within the pedigrees being studied, the offending rare allele was necessary and largely sufficient to lead to the phenotype of interest. However, just as has been the case with common variants, recent studies suggest that rare alleles may be carrying smaller than anticipated effects without evidence of being either necessary or sufficient to confer a phenotype (Bucan et al., 2009). A key related point here it that there is an important relationship between the effect size of a given allele, its frequency in the population, and the available methods to detect and confirm the relationship to disease (Figure 1). As common variant findings refocus expectations on alleles with extremely small risks, the spectrum of genetic effects remaining for rare alleles necessarily expands, resulting in a shift in the approaches that would be required to identify this contribution.
The Utility and Pitfalls of Genetic Association
As a consequence, association studies in general, and case-control designs in particular, promise to play a major role in addressing the genetic risks in child psychiatry despite the emerging emphasis on rare variants. While this observation may seem self-evident, the tendency to interpret rare variant findings based on expectations derived from Mendelian disorders can be quite powerful. There is a strong innate skepticism that tends to accompany the observation of rare mutations that are neither necessary nor sufficient to confer a phenotype, as for instance when a putative disease-related rare mutation is not always shared by affected siblings (Bucan et al., 2009; Kumar et al., 2008; Weiss et al., 2008).
An increasing reliance on case-control analyses will also result as a consequence of the emerging ability to readily detect de novo variation on a genome wide scale. While such mutations may carry very large risks, linkage analyses are not plausible given the nature of the mutation. Moreover, the prior conventional wisdom that de novo losses of coding segments provided prima face evidence for disease causality has been definitively laid to rest (Iafrate et al., 2004; Sebat et al., 2004). Consequently, carefully controlled case-control analyses will be required to confirm the relevance of de novo variants in affected cohorts, particularly in those instances in which the penetrance of a given mutation is not 100 percent.
Of course, there is little reason to expect that confounds attending common variant studies will be any less pressing for rare sequence and structural variation. For example, power will certainly remain a critically important issue: The ability to detect association is a function both of effect size (larger for rare variants) and allele frequency (smaller for rare variants) in the study population. This trade off suggests that some rare variants carrying modest effects will be impossible to detect given inherent limitations in the sample sizes that can be consolidated.
Moreover in the case of CNV analyses, typing error may be considerably more problematic for current array based methods compared to SNP genotyping or sequencing due to limitations in prediction accuracy and confirmation methods; there is considerable sample-to-sample variability and the potential for batch-effects is marked even when using identical CNV platforms at different institutions. A recent elegant study identified several other less obvious method-associated technical challenges (Craddock et al., 2010).
Similarly, population stratification, though initially downplayed in studies of rare structural variants, clearly has the potential to confound results (Merikangas et al., 2009). Finally, given the expectation of a very high degree of allelic heterogeneity, rare variant association will often require cumulative counts of variations at a given gene, i.e. an analysis of mutation burden or mutation skew (Bakkaloglu et al., 2008; Cohen et al., 2004; Cohen et al., 2006; Ji et al., 2008; Johansen et al., 2010). In these instances, given the large amount of neutral rare variation in the genome, the differentiation of functional from incidental mutations may be critical to confirm true association (Ji et al., 2008). And this can be a challenging enterprise: the human genome tolerates a tremendous amount of what would have previously been presumed to be clearly deleterious variation (Iafrate et al., 2004; Sebat et al., 2004), and recent evidence suggests accurate interpretation of the impact of particular alleles will be dependent on specific knowledge of the biology of the gene(s) and proteins in question (Ji et al., 2008).
The impact of next generation sequencing
A comprehensive discussion of new genomic technologies is beyond the scope of this review. However, it is important to note that next generation sequencing is already offering unprecedented opportunities to investigate genetic variation, gene expression and epigenetic phenomenon. With regard to gene discovery, the ability to comprehensively identify both sequence and structural variants in large numbers of individuals promises to revolutionize the understanding of the allelic architecture and specific genetic contributors to childhood neuropsychiatric disorders.
While the manner in which these technologies will be most effectively harnessed remains to be seen, several observations are warranted here: first, while there is little question that next generation sequencing will empower large-scale case control studies of rare variation, the field is also likely to see a re-emergence of pedigree based genetic studies. Already sequencing the human exome has been shown to be a powerful approach to mapping rare Mendelian mutations in the setting of tremendous phenotypic heterogeneity and within pedigrees that would have previously been too small to support traditional linkage analyses (Bilguvar et al., 2010); these developments bode well for outlier approaches to common complex disorders. More generally, either multiply-affected families or apparently sporadic pedigrees may well offer the most expedient means to sort through and prioritize massive amounts of rare variation identified via next generation sequencing.
Finally, while the excitement about these new approaches is well justified, it is also a certainty than even the most exhaustive catalogue of genetic variation will leave many questions unanswered and likely serve only as a first important step in illuminating the complex interplay of experience, environment and biology in influencing normal and pathological brain development and function.
Autism and autism spectrum disorders
Autism spectrum disorders are a group of developmental syndromes characterized by fundamental deficits in social communication and language development and accompanied by highly restricted interests, stereotyped repetitive behaviors, or both. The canonical presentation is defined by deficits in all three domains, whereas disorders along the spectrum involve one or more core impairments (Volkmar et al., 2009).
The evidence for a genetic contribution to ASD is very strong, based on family and twin studies (Bailey et al., 1995; Folstein and Rutter, 1977), the overlap of ASD with known genetic disorders (Fombonne et al., 1997), and recent molecular data (addressed below). There is a strong male predominance (Volkmar et al., 2004). Despite ambitious efforts aimed at identifying common and rare alleles, the number of genes or loci that are accepted as carrying definitive risk remains small, and not immune from debate. In contrast, the list of intriguing and plausible candidates (see http://www.mindspec.org/auTSb.html) is proliferating exponentially; a phenomenon that can make deciphering the current literature quite difficult.
The absence of specific and sensitive biological markers for ASD, and the consequent reliance on syndromic categorization and subjective assessment presents predictable challenges, as it does in all areas of psychiatric diagnosis (Volkmar et al., 2009). These issues extend well beyond the scope of the current discussion; however a key phenomenological question that bears directly on the interpretation of the autism genetics literature involves the overlap of ASD and intellectual disability (ID), which is present in approximately 70% of individuals meeting full diagnostic criteria for autism (Chakrabarti and Fombonne, 2001), and approximately 45%–50% for the entire range of ASD diagnoses.
It is important to note here that there is little debate that social impairment and cognitive delay are readily distinguishable in individuals with mild to moderate ID. While this differentiation becomes less reliable in cases of severe to profound delay, debate over this co-occurrence runs deeper and reflects a historical interest in identifying specific risks for autism as opposed to “general disruptions of brain development.” This issue is similarly present with regard to the study of individuals with ASD and seizure, present in 10–25% of individuals with autism (Volkmar and Pauls, 2003) and is particularly relevant to the study of syndromic autism i.e. ASD in the context of a known genetic syndrome or observed in individuals presenting with marked dysmorphology, structural brain abnormalities or other evidence of a genetic syndrome, but absent a known cause.
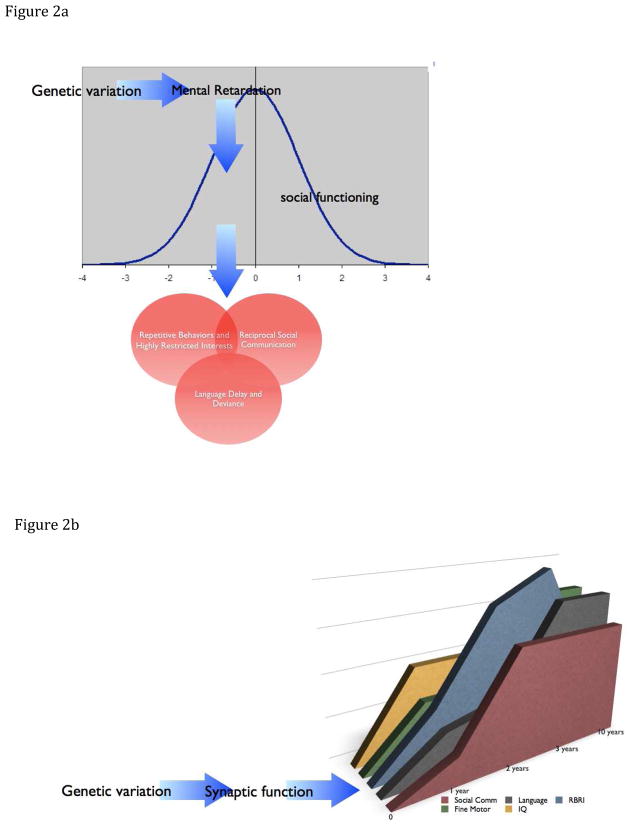
In part these questions reflect an ongoing debate regarding whether social disability, characterized in the context of known genetic disorders, is identical to that observed in idiopathic ASD (Moss and Howlin, 2009), an issue that may be difficult to resolve given the challenges inherent in designing blinded studies of syndromes characterized by distinctive physical features. However, there is a second line of reasoning, based on twin and epidemiological data, that contends that the high rate of ASD in individuals with MR among clinically ascertained populations is an epiphenomenon resulting from a normal distribution of autism traits in the population which is made manifest by the relatively poor “compensatory mechanisms” in individuals with ID (Figure 2a). This view proposes that discovery efforts in cases of overlapping ID an ASD will identify genes for the former; that a different set of genes underlie social ability and disability; and that the optimal strategy for gene discovery with regard to autism would focus on the population of ASD without intellectual disability (Skuse, 2007).
Figure 2.
Two models of the overlap of ASD and ID: 2a represents a paradigm well articulated by Skuse (2007) which holds that the study of syndromic ASD is likely to identify genes resulting in ID and that co-occuring ASD is a consequence of a reduced compensatory capacity in these individuals. In this view, the ASD phenotype is otherwise normally distributed in the population and is influenced by a distinct set of genes. Given this model, the pursuit of true ASD loci would focus on individuals who do not have ID. In the graph, the units on the X axis are standard deviations of a hypothetical measure of social functioning. An alternative model (2b) proposes that rare mutations in genes may alter fundamental neurobiological mechanisms noted here in short hand as “synaptic function” but reflecting a wide range of molecular mechanisms; that these in turn set the stage for a developmental course that is reflected in the development of mutiple funcitonal domains; and that these trajectories will be influenced by other genetic, epigenetic and environmental process leading ultimately to clinical diagnoses that cross categorical boundaries. This model would suggest that the identification of “inciting” rare alleles would point to biology that is relevant to a wide range of observed clinical outcomes including both ID and ASD. In the graph, the X-axis reflects time and the colored bars represent hypothetical trajectories in multiple areas of cognitive, linguistic and behavioral functioning.
While these debates are ongoing, several considerations deserve mention: First in addition to the increasing application of standardized instruments and blinding methods to enhance the diagnostic reliability of ASD in the context of known syndromes, the not infrequent detection of syndromic mutations in individuals with idiopathic ASD is evidence that these variations may lead to behavioral phenomenon that are, for all practical purposes, indistinguishable from idiopathic ASD. Moreover, given that autism and related conditions are syndromic diagnoses and manifestly not unitary biological entities, there would seem to be little question that individuals that are rigorously characterized with ASD have ASD. The more difficult question of whether it is a worthwhile strategy to search for variants contributing to ASD in the context of ID and to pursue the biology of rare syndromes in an effort to understand autism remains contested. As noted above, a central rationale for outlier strategies is that a genetic variation that is definitively identified as causing or contributing to ASD with ID or syndromic ASD offers potential traction with regard to cellular and molecular mechanisms of pathogenesis in those cases. The question of whether this biology is generalizeable remains to be demonstrated. However, there is already solid evidence supporting the use of identical strategies in other common complex and heterogeneous conditions (Ji et al., 2008; Romeo et al., 2009). Moreover, as discussed in more depth below, current evidence already points to some convergence in the molecular mechanisms implicated through the study of syndromic forms of ASD and those suggested by genes that have the shown the strongest evidence so far for contribution to idiopathic ASD (Bourgeron, 2009; Toro et al., 2010). Finally, the observation that identical mutations may lead not only to ID and ASD, but to Schizophrenia and possibly other neuropsychiatric disorders (addressed below) would seem to argue for a model based on the plieotropy of genes underlying fundamental neuronal processes (Figure 2B), a phenomena that is highlighted by recent findings from our group with regard to the genetics of structural brain disorders (Bulgivar et al, 2010).
Early and Rare Variant Findings
These debates bear substantially on the interpretation of the results of gene discovery efforts. An observer inclined to compartmentalize syndromic ASD and ASD in the context of ID would likely contend that the field awaits definitive identification of a risk locus for idiopathic ASD. Someone holding the alternative perspective would likely mark the cloning of the Fragile X Mental Retardation Protein (FMRP) as the first autism gene, given a long standing and well-documented excess of the ASD phenotype observed in boys carrying canonical mutations and the repeated observation of FMRP mutations identified in cohorts of individuals with idiopathic ASD (Brown et al., 1986; Fombonne et al., 1997; Harris et al., 2008; Hernandez et al., 2009; Levitas et al., 1983).
A number of other well-defined syndromes have either been found to have a greater than expected frequency of individuals with ASD or to have core features that overlap with ASD or both (Hoffman and State, 2010). Among the substantial list, in addition to fragile X, the data for an appreciably increased prevalence of mutation carriers among cases of idiopathic ASD is so far best established with regard to tuberous sclerosis (Smalley, 1998; Smalley et al., 1992). Similarly, the overlap among defining features of syndromes such as Angelman or Rett and ASD provides a complementary approach to studying the biology of autism. The contribution of these and other rare syndromes to the development of neurobiological models of ASD pathogenesis is discussed in ensuing sections.
The first rare coding mutation in individuals with putative idiopathic ASD (eg the absence of a known genetic syndrome or clear evidence of a monogenic disorder) was identified in the neuronal adhesion molecule, Neuroligin 4X (Jamain et al., 2003). Based on the prior identification of recurrent deletions on the X chromosome in affected individuals, Thomas Bourgeron’s lab sequenced genes within the interval in 36 sibling pairs and 122 trios and found a truncating frame-shift mutation in NGLN4X arising de novo in an unaffected mother and transmitted to two brothers, one with Asperger syndrome and the second with “typical” autism. Shortly thereafter, mapping of a multi-generational pedigree affected with both ID and ASD by an independent group led to the discovery of a segregating truncating mutation, nearly identical to the initially reported NLGN4X mutation (Laumonnier et al., 2004).
Over the ensuing half-decade, this finding has been further supported by convincing evidence for recurrent de novo mutations in individuals with ASD in SHANK3 a post synaptic scaffolding molecule that falls within the 22q13.3 microdeletion syndrome region (Durand et al., 2007; Gauthier et al., 2009; Lim et al., 1999; Moessner et al., 2007). Moreover recurrent de novo mutations in other interacting molecules, including NRXN1 (Kim et al., 2008; Szatmari et al., 2007) and SHANK2 (Berkel et al., 2010; Pinto et al., 2010b), functional data showing a role for neuroligins in the establishment of both excitatory and inhibitory synapses (Chubykin et al., 2005; Graf et al., 2004; Scheiffele et al., 2000), and interesting, but less conclusive genetic (Jamain et al., 2003) and model systems data (Chadman et al., 2008; Tabuchi et al., 2007) with regard to the closely-related molecule NGNL3, have placed NLGN4X among the most widely accepted genetic findings in idiopathic ASD.
Over the last several years a rapidly expanding list of rare mutations have been described in individuals with ASD, representing so many genes in fact that an exhaustive assessment is not feasible here. However only a small number yet have the property of being observed in rigorous studies by multiple independent investigators. Among these, in addition to NGLN4X, SHANK3 and NRXN1, the neuronal adhesion molecules CNTN4 has been identified by our lab and others via molecular cytogenetic mapping of de novo rearrangements and CNV analyses (Fernandez et al., 2004, 2008; Glessner et al., 2009; Roohi et al., 2009); and, quite recently, SHANK2, was identified both through the identification of de novo disruptive mutations in one patient with MR and another with ASD (Berkel et al., 2010) and a large-scale CNV analysis (Pinto et al., 2010a).
A review of the data regarding another neuronal adhesion protein, encoded by the gene Contactin Associated Protein 2, is illustrative of the general state of the field, with intriguing findings falling just short of definitive evidence: The molecule was first identified as having a role in developmental delay through homozygosity mapping of a rare recessive frame-shift mutation in the Old Order Amish population (Strauss et al., 2006). The report involved a syndrome of intractable epilepsy accompanied by clinically diagnosed autism. Shortly thereafter, independent reports provided evidence for variations in CNTNAP2 in idiopathic ASD. The mapping of a rare de novo chromosomal rearrangement by our lab disrupting this locus in a simplex ASD pedigree and the subsequent identification of multiple rare transmitted, missense substitutions at highly conserved positions in affected multiplex families (Bakkaloglu et al., 2008), the simultaneous findings by two independent groups of common variant association (Alarcon et al., 2008; Arking et al., 2008), and a subsequent report of interaction of CNTNAP2 and FOXP2 along with a association of a common allele in language impairment phenotype (Vernes et al., 2008) generated strong interest in this gene and led to its characterization by some authors as a confirmed idiopathic ASD locus.
However, while the initial mapping of a rare recessive mutation was quite compelling, the finding left open the question of whether and how either common or rare variation in this gene contributes to non-syndromic ASD. For example, the mutation burden analysis conducted by our lab was suggestive, showing an approximately 2-fold increase in very rare missense variants in cases versus controls. However, as noted in the initial publication the results did not reach statistical significance apart from a single recurrent transmitted allele; population stratification could not be ruled out as a confound and we lacked the ability to reliably differentiate functional from incidental mutations in cases and controls. Moreover, the two simultaneous common variant association studies identified different alleles that carried risk for differing phenotypes: one involving the diagnosis of ASD and the other mapping a language quantitative trait locus identified in individuals with ASD. Subsequently, only one of three published GWAS studies in ASD provides any support for association of CNTNAP2 with the diagnosis of ASD and this was modest at best (Anney et al., 2010). Finally, the link to specific language impairment though statistically significant and based on a strong a priori hypothesis was nonetheless identified through a candidate gene association study and replication has yet to be attempted in a genome-wide analysis.
This characterization is not meant to call into question the rigor of the aforementioned studies or to detract from interest in CNTNAP2. The presence of multiple lines of evidence emerging nearly simultaneously from independent groups conducting state-of-the-art studies is unusual in the field and many of the outstanding questions have simply not yet been tested adequately to address the question of independent replication. Nonetheless, this summary highlights issues that are reflective of the current state of the science with regard to many of the most intriguing findings in ASD genetics both with regard to common and rare variants: underscoring the question of the relationship of syndromic findings to common forms of the disorder and highlighting the challenges posed by sample size, power, ancestral matching and distinguishing functional from neutral alleles.
Copy Number Variation
Prior to the advent of high-density microarrays and the ability to detect sub-microscopic variations in chromosomal structure, multiple recurrent chromosomal abnormalities were identified in individuals with ASD. The most common and highly penetrant are maternally inherited duplications of chromosome 15q11–13 (Baker et al., 1994; Hogart et al., 2010). Rare recurrent microscopic deletions at 2q37, 1q21,22q11, 22q13 among others, have been identified in affected individuals (Bucan et al., 2009).
The importance of submicroscopic copy number variation for ASD was first reported by Michael Wigler’s lab through the identification of a marked excess of de novo variations in affected singleton probands (10%) compared to probands from multiplex families (3%) or unaffected controls (1%) (Sebat et al., 2007). A subsequent genome-wide CNV study (Marshall et al., 2008) confirmed a several fold increase in these events, and suggested a cumulative frequency of approximately 5–10 percent in the simplex ASD population. However, another recent large-scale study, while identifying de novo variations in 5.6% of simplex ASD probands, did not find a difference compared to probands from multiplex families (5.5%) (Pinto et al., 2010a). The reasons for the variability observed in this third study have not yet been clarified, although even prior to these results, a hypothesis emerged that the increased frequency identified by Sebat et al (2007) might apply particularly to large events, and consequently, that the increasing resolution of array platforms might tend to obscure some differences between groups.
The study by the Wigler lab also included a description of a 16p11.2 de novo deletion and, subsequently, two groups nearly simultaneously found a significant association of recurrent de novo CNVs (Kumar et al., 2008; Weiss et al., 2008) at this locus in idiopathic ASD, reporting on partially overlapping samples. The finding was confirmed in a subsequent genome wide investigation of structural variation (Marshall et al., 2008). Among the three other recent large-scale CNV studies: the 16p11.2 finding was not replicated in one due, in part, to the finding of a higher than reported rate of 16p11.2 CNVs in the control group (Glessner et al., 2009) and in two others, an independent assessment was precluded by extensive sample overlap with the previously reported cases (Bucan et al., 2009; Pinto et al., 2010b). Interestingly, a recent study of 4284 individuals with MR and multiple congenital anomalies found 16p11.2 CNVs in 0.3%, consistent with that seen previously among ASD cohorts (Bijlsma et al., 2009), and providing further evidence for wide ranging phenotypic manifestations resulting 16p11.2 variations (discussed below).
The 16p11.2 data serve as an important example of the manner in which current findings are challenging notions regarding rare variant contributions to ASD. All four deletions families reported by Kumar et al (and described again in Weiss et al) included two affected children, and in three of these pedigrees, only one of the affected siblings carried the de novo CNV. As noted, based on Mendelian expectations, the observation that within multiple small pedigrees the large de novo deletion was not necessary for the phenotype would raises eyebrows. Indeed, commenting on the lack of 16p11.2 replication in their CNV analysis, Glessner and colleagues write that their results “…indicate that CNVs at the 16p11.2.2 locus may not be sufficient to be causal variants in ASD.” (Glessner et al., 2009) In fact, none of the reports of 16p11.2 have provided data suggesting that the variant is uniformly necessary or sufficient to lead to ASD. The issue addressed by these studies is whether this CNV confers risk. The aforementioned studies and our own preliminary data from a CNV analysis of simplex families, provides mounting evidence that it does.
Recent CNV studies have also provided interesting data regarding other previously identified and novel loci. Glessner et al. (2009) replicated findings at 15q11- 13 and 22q11.21, as well as NRXN1 and CNTN4. As noted, they did not identify a significant difference in cases versus controls for 16q11.2 nor did they observe an association of CNVs at the SHANK3 locus. Their analyses further highlighted multiple novel loci that were found to cluster via gene ontology analyses in the ubiquitin pathway and among molecules categorized as being involved in neuronal development. Pinto et al, in addition to reporting evidence for SHANK2 almost simultaneously with a report by Berkel et al, identified several additional novel loci and provided strong evidence for X linked inherited deletions of DDX53-PTCHD1, with 7 reported males among the cases and none among 4964 controls. Their pathway analyses highlighted genes involved in cellular proliferation, projection and motility, and GTPase/Ras signaling (Pinto et al., 2010b).
Common Variants
Similar to other child psychiatric disorders, the ASD genetics effort was initially characterized by a focus on idiopathic forms of the syndrome, an early inability to identify evidence for single gene inheritance and, subsequently, a widespread preoccupation with the CVCD hypothesis, investigated via non-parametric linkage and candidate gene association studies (Veenstra-Vanderweele et al., 2004).
With regard to non-parametric linkage, the largest study to date included 1,181 multiplex families (Szatmari et al., 2007) and, along with a dozen others using similar approaches, did not identify highly significant evidence for linkage or result in the mapping of a common variation that accounted for the linkage signals identified. It should be noted that these studies are theoretically capable of identifying either common or rare disease alleles. They typically evaluate sib pairs for regions of the genome shared among affected family members more often than would be expected by chance and have the advantages of not requiring a priori specification of a mode of inheritance and of being robust to allelic heterogeneity.
A likely explanation for the lack of definitive results emerging from these investigations to date is the greater than anticipated degree of locus heterogeneity so far observed in ASD. Particularly in light of the modest sample sizes employed, a great diversity of genes all contributing to ASD would tend to obscure evidence for excess sharing at any given locus. However, as noted previously, the reemergence of linkage analyses of all types as important tools in the context of next generation sequencing technologies is a distinct possibility.
With regard to common variants, despite considerable challenges, several studies conducted just prior to the GWAS era resulted in notable findings. These included the identification of EN2 (Benayed et al., 2009; Benayed et al., 2005), the MET oncogene (Campbell et al., 2006; Jackson et al., 2009), and CNTNAP2 (Alarcon et al., 2008; Arking et al., 2008; Vernes et al., 2008). All three have shown some evidence for replication but continue to be the subject of debate due, in part, to their absence from the most promising results emerging from the recent genome wide association studies.
As with other areas of medicine, GWAS have emerged as the gold standard for the identification of common alleles carrying small effects and three relatively large studies in autism have recently been completed. The first studied 780 families and an additional 1204 probands and identified significant association of ASD to an intergenic region of chromosome 5p14.1 mapping between the neuronal adhesion molecules Cadherin 9 and Cadherin 10 (Wang et al., 2009). The second involved on a cohort of 1031 families and found association to an intragenic SNP near the gene Semaphorin 5A (Weiss et al., 2009) and a third used a discovery cohort of 1558 individual and found genome wide evidence for association of MACROD2 (Anney et al., 2010). As a likely reflection of both the heterogeneity of ASD and the challenge of identifying alleles of very modest effect, none of these studies confirmed the others’ findings.
While the field certainly hoped for replication of identical SNPs across these samples, the findings are nonetheless consistent with similar studies of complex conditions, including very modest odds ratios. Despite cohort sizes that would be considered large for child psychiatry, none of these studies were well powered to replicate findings from the other studies (Anney et al., 2010). In addition, a comparison of these results with the candidate gene results mentioned above underscores an ongoing debate regarding both the impact of phenotypic heterogeneity and the best approach to addressing this problem. Both the MET and CNTNAP2 data provide evidence that association is robust either within a specific subgroup of affected individuals or with an endophenotype as opposed to the categorical diagnosis.
There has been a long-standing interest in using phenotypes that fall along the path from genetic variation to the syndromic clinical outcome across all of psychiatry. This is understandable given the clear limitations to our current categorical diagnostic approaches; the identification of more homogenous and biologically relevant entities could very well transform the search for common variants. The challenges in ASD, and for other neuropsychiatric phenotypes, include the difficulty in identifying relevant phenomenon a priori and the related confound of multiple comparisons if a variety of possibilities are examined through the course of a study. Moreover, it is worth noting in the instances where loci contributing to ASD have been most convincingly demonstrated, similar or identical rare mutations confer a very broad range of phenotypes. These results suggest that the effective “distance” between variations in the sequencing or structure of the DNA and resulting brain phenotypes may be quite large. It is difficult to imagine in retrospect what endophenotype would have been useful to detect these mutations, though admittedly the situation may be different for common versus rare variants. However, in other areas of medicine, common variant discovery has been very successful even in highly heterogeneous disorders in leveraging a combination of clinical diagnoses and large cohorts. Given the magnitude of effects identified in those studies and those so far suggested by initial GWAS in ASD, it is clear that the field is just now approaching the sample sizes necessary to begin to answer the question of common variant contributions. Finally, it is worth mentioning that as definitive replicated common variants are identified, the ability to then clarify relevant endophenotypes and a range of genotype-phenotype correlations will be dramatically enhanced.
Molecular findings and diagnostic boundaries
As suggested above, one of the most interesting and thought provoking recent observation in the field has been the wide range of neuropsychiatric manifestations that now appear to emerge from identical rare variants. Indeed the conceptual challenges of integrating the co-occurrence of MR and ASD pales in comparison to the possibility that functionally identical mutations may lead to ASD, MR, seizure disorder, Schizophrenia, ADHD, Tourette syndrome, OCD or some combination of the above. Nonetheless data both from structural and sequencing studies suggests this may be the case: For example 22q11.2 deletions, have long been implicated in the risk for psychosis in addition to the evidence with regard to ASD (Guilmatre et al., 2009; Vassos et al., 2010); CNVs at 16p11.2 were observed in individuals with Schizophrenia (Weiss et al., 2008), a finding that has recently been supported by a large case control association analysis suggesting a particular role for duplications in this interval (McCarthy et al., 2009) and de novo mutations in SHANK3 have been identified in individuals with Schizophrenia as well as those with ASD (Gauthier et al., 2010). Structural variations at CNTNAP2 have been reported not only in ASD and Schizophrenia but Tourette syndrome as well (Friedman et al., 2008; Verkerk et al., 2003), and this overlap has also been observed with regard to NLGN4X (Lawson-Yuen et al., 2008).
At present the data pointing to overlapping risks for Schizophrenia and ASD is particularly compelling (McCarthy et al., 2009), with the prevailing hypothesis that duplications predispose to schizophrenia and a variety of other developmental outcomes including ASD, while deletions do not seem to play a role in the risk for psychosis. The prospect of any shared molecular mechanisms is somewhat ironic. Autism was initially conceptualized as a form of childhood psychosis, but with advances in standardized diagnostic approaches, this idea was rejected (Volkmar and Pauls, 2003). While there is some overlap between ASD and the social withdrawal seen in individuals with schizophrenia, the natural history of the latter is quite distinctive versus the early onset that defines ASD, as are the positive symptoms of schizophrenia: auditory hallucinations and delusions have not been described as more common among individuals with idiopathic ASD.
Not surprisingly, the suggestion of overlap among so many developmental neuropsychiatric disorders opens the door on a variety of interesting debates. As suggested above, the notion of diagnostic substitution has been raised, but this would seems unlikely to explain all of the recent data given the range of signs and symptoms involved and the strikingly distinct developmental profiles. Given the clear demonstration of incomplete penetrance for implicated variants, it is also likely that some of the many observations are incidental findings. Again, the type of large- scale association analysis of 16p11.2 variations reported by McCarthy et al is precisely the methodology that will be required to provide clear answers to this question. The apparently broad range of phenotypic outcomes also poses some interesting challenges to the model that suggests the overlap of MR and ASD is a reflection of genes for the former uncovering the normal distribution of ASD traits in the population (Figure 2a). in contrast, the emergence of a wide range of phenotypes from, for instance, 16p11.2 duplications would seem most consistent with a model of highly pleotrophic effects of mutations that influence fundamental neurobiological processes (Figure 2b).
Emerging Neurobiological Models
At the molecular level, evidence in favor of this latter model has begun to converge at the synapse. The identification of NLGN4X mutations and the independent findings of rare variants in NRXN1, SHANK3 and SHANK2, have further focused attention on the function of neuroligin-neurexin complex and related molecules in the post synaptic density (PSD), a specialized region of the excitatory synapse (Figure 3).
Figure 3.
A highly selected view of the excitatory synapse highlighting genes strongly implicted via rare variant studies of idiopathic ASD (blue boxes) and those identified through the study of syndromic ASD (red boxes)
Perhaps most interestingly, this data converges with the evidence pointing to a key role for the Fragile X Mental Retardation Protein at glutamatergic synapses (Bear et al., 2004; Huber et al., 2002; Nakamoto et al., 2007). The elaboration of this biology has led to an extremely intriguing hypothesis: that the cognitive and social phenotypes may be mediated through deficits in plasticity relating to long-term depression and that this process is potentially reversible via targeting of metabotropic glutamate receptors or related signaling cascades (Bear et al., 2004; Dolen et al., 2010). The notion, supported by recent model systems data (Chang et al., 2008; Dolen et al., 2007), that fragile X syndrome may be amenable to intervention throughout the lifespan is now being translated into clinical trials in individuals with Fragile X as well as with idiopathic ASD.
The intersection of studies of syndromic and idiopathic autism has also focused attention on the PI3K-AKT-mTOR pathway. As noted, there is strong evidence that rare mutations in TSC-1/TSC-2 increase the risk for ASD; the data in this regard for NF-1 and PTEN are also convincing (Butler et al., 2005; Buxbaum et al., 2007). These molecules point to the rapamycin-sensitive mTOR–raptor complex, a key regulator of protein synthesis and cell growth. Further, binding of hepatocyte growth factor to the MET oncogene results in activation of a variety of signaling cascades, a process that is regulated in part by PTEN. These findings point to two intriguing possibilities: first, that targeting of this pathway, as is feasible with rapamycin and other compounds, may provide a novel avenue for treatment (Ehninger et al., 2009; Ehninger et al., 2008); and second that the data may converge with the biology implicated by fragile X to further refine the understanding of the molecular mechanisms leading to human developmental disorders (Narayanan et al., 2007; Sharma et al., 2010).
Of course, the question of whether the pathways implicated by syndromic ASD, which anchor both the mGluR and mTOR hypotheses, may have broader relevance for idiopathic ASD is highlighted by these findings. Ongoing studies, both to further elaborate basic neurobiology and to address the question in the clinic will help provide the answers. What is indisputable is that the conceptual transition reflected in these efforts is remarkable: the notion that mental retardation and ASD associated with FMRP or TSC-1 mutations may not set in stone early in development represents a seismic shift in thinking regarding the opportunities to treat these conditions, and underscores the more general transformative potential of the interplay of human genetic findings and basic neurobiology.
A third area of possible traction in the neurobiology of ASD relates to CNTNAP2. As noted, the initial evidence for the phenotypic consequences of homozygous truncating mutations was quite strong. Moreover, the investigators who mapped the locus in Old Order Amish had the unusual opportunity to examine CNS pathology, due to surgical intervention for the severe epilepsy phenotype. The gross morphological abnormalities included temporal lobe dysplasia, evidence of abnormal cortical migration, and dysmorphic pyramidal neurons. Given many remaining uncertainties regarding the function of CNTNAP2 in the CNS and some evidence for the presence of the protein at the synapse (Bakkaloglu et al., 2008), a convergence with the previously described pathways has not been ruled out. Equally interesting however is the suggestion, based on the expression of CNTNAP2 in post mitotic neurons in the developing human cortex and the pathological data suggests that subtle abnormalities in cortical migration or organization might be an independent avenue to ASD.
When viewed in a more global sense, the recent large scale studies of ASD have begun to suggest other intriguing possibilities; the use of pathway analysis to integrate the very large amount of rare variation data emerging from CNV studies has pointed to the ubiquitin pathway, neuronal adhesion molecules and those involved in cellular proliferation, projection and motility, and GTPase/Ras signalling. A review of the data on 16p11.2 and other specific recurrent CNVs is also focusing attention on dosage sensitivity in conferring risk and shaping developmental outcomes (Toro et al., 2010). The further pursuit of these general leads will be helped tremendously by additional definitive genetic findings that allow for prioritization among the various possibilities, lend greater specificity to testable hypotheses, and provide a clear link to the human phenotype.
Tourette Syndrome
The history of the genetics of Tourette syndrome (TS) has been similar in many respects to that described for autism and related conditions. However, in contrast to the tremendous attention focused on gene discovery in ASD and the voluminous literature, particularly over the last 5 years, the scale of the TS genetics effort has been relatively modest. The types of resources that have facilitated a high level of productivity in other areas of psychiatry and clinical neuroscience, including large patient cohorts and widely accessible biomaterials are just now beginning to be consolidated. Nonetheless, several recent genetic findings have provided promising leads regarding molecular mechanisms; cytogenetic and CNV data point to the important contribution of rare variants; and the first large scale GWAS study will soon be completed.
TS is defined by the persistence of unwanted, brief, repetitive, non-rhythmic motor movements and vocalizations and is a prototypical developmental disorder with an age of onset of between 3 and 8 years (mean ~7 years) and a tendency toward considerable improvement as individuals reach adulthood (Bloch et al., 2006). While the diagnosis requires only the presence of both vocal and motor tics, the vast majority of individuals who present clinically suffer from other psychiatric syndromes: up to 50% of TS probands also suffer from obsessive compulsive disorder (do Rosario, 1997; Ghanizadeh and Mosallaei, 2009); 50–90% of TS probands suffer from attention deficit hyperactivity disorder (Burd et al., 2005; Comings, 2005; Freeman, 2007; Ghanizadeh and Mosallaei, 2009; Leckman, 2003; Roessner et al., 2007; Stewart et al., 2006); and there is an apparently increased risk for both depression and anxiety apart from OCD (Coffey et al., 2000; Kurlan et al., 2002). These high rates of co-morbidity are likely a consequence of a combination of selection bias (as epidemiological studies suggest that only a small percentage of individuals with tics or TS present to clinic); the possibility of a shared genetic liability as has been best documented for OCD (Pauls, 2001); and the possibility of a shared neurobiological substrate among these conditions. Typically probands with TS, chronic persistent tics (either motor or vocal) and tics with OCD as well as family members with these conditions or OCD alone, are typically considered to be affected with a TS spectrum condition for the sake of genetic studies.
Genetic Findings
Twin data, though cumulatively representing a small number of studies and subjects, suggests a significant genetic contribution to TS. Monozygotic twin concordance rates are estimated at 50%–77% compared to 10%–23% for dizygotic twins (Price et al., 1985), with the range dependent on whether TS spectrum conditions are included. However, despite a long-standing consensus that genetics plays a key role in disease etiology, there are, as of yet, no confirmed, replicated genetic risk factors for TS and related conditions.
Early studies focused on large multigenerational pedigrees that seemed to point to single gene autosomal dominant inheritance (Baron et al., 1981; Curtis et al., 1992; Kidd and Pauls, 1982; Pauls and Leckman, 1986). However, over time, as the techniques for mapping Mendelian disorders reached maturity, no TS locus was identified, and a high rate of bilineal inheritance (McMahon et al., 1996) was noted, this hypothesis was abandoned. Subsequent segregation analyses (Hasstedt et al., 1995; Kurlan et al., 1994; Walkup et al., 1996) led to the current characterization of TS as a complex, heterogeneous genetic disorder.
By the late 1990s, the lack of results from parametric analyses resulted in a shift toward nonparametric linkage. In 1999, The Tourette Syndrome Association International Consortium for Genetics reported a study of 92 affected sib pairs which resulted in multipoint maximum-likelihood scores of >2 on chromosomes 4q and chromosome 8 (1999). However, when the study was extended to 238 sib pairs and 18 large families, evidence for linkage in these regions diminished, and suggestive evidence emerged on chromosome 2p (2007). Several other linkage studies, both using parametric and non parametric approaches have resulted in LOD scores either reaching or approaching statistical significance (Breedveld et al., 2010; Curtis et al., 2004; Merette et al., 2000; Paschou et al., 2004); however, as of yet, none has led to the identification of mutations altering the structure or function of transcripts mapping within or near these intervals.
Common variant candidate association studies have been a widely employed and a variety of biologically plausible candidate genes have been assessed, including various dopamine receptors, the dopamine transporter, noradrenergic transcripts, tyrosine hydroxylase, SLC6A3, and a several serotonergic genes (Barr et al., 1996; Brett et al., 1995; Chou et al., 2004). These efforts consisted of samples sizes that would now be considered inadequately powered to detect common variant risks of plausible magnitude. Not surprisingly, they have not yet resulted in consistent and reproducible findings.
While the search for common alleles has predominated, there has also been a steady effort to evaluate the contribution of rare alleles, almost entirely reflected to date by outlier studies. Many of these were reported prior to the CNV era and involved mapping of de novo chromosomal abnormalities: For example, Petek et al reported a de novo duplication disrupting IMMP2L (Inner Mitochondrial Membrane Protein 2L) and four independent studies described rearrangements at the chromosome 18q22 region with breakpoints mapping approximately within 1Mb of each other. However no missense or nonsense mutations been identified in IMMP2L or in chromosome 18 disrupted or flanking transcripts, though the number of screened individuals has been very small.
Three similar findings are notable for their overlap with the ASD literature: An insertion of chromosome 2p21-p23 at 7q35-q36 was found to disrupt CNTNAP2 in three affected individuals from one family (Verkerk et al., 2003) and (Lawson-Yuen et al., 2008) reported a pedigree with a transmitted NLGN4X deletion involving exons 4, 5, and 6. The proband had autism as well as motor tics, while his sibling who also carried the deletion was diagnosed with TS and ADHD. The carrier mother was reported to have a learning disorder, anxiety, and depression. Again, large scale sequencing of TS probands has not been reported for either transcript. A very recent CNV
In a study from our laboratory that has become the subject of some contention, Abelson and colleagues (Abelson et al., 2005) reported a de novo chromosome 13 inversion in a sporadic TS pedigree. Subsequent sequence analysis of 174 unrelated probands of the flanking transcript Slit and Trk-like, Family Member 1 SLITRK1, mapping 350kb from this de novo breakpoint, revealed a single nucleotide deletion predicted to result in a prematurely truncated protein that segregated with TS and trichotillomania (compulsive hair pulling) (TTM) in a small family. Additionally, our group found two independent occurrences of a rare variant (var321) in a highly conserved region of the 3′UTR corresponding to the binding site for the microRNA hsa-miR-189. in vitro methods were used to demonstrate the functional consequences both of the coding and 3′ UTR variations and haplotype analysis was performed to test whether the var321 mutations were independent. Screening of several thousand controls resulted in a nominally significant association of the miRNA variant with TS (p=.006).
Subsequent genetic studies of SLITRK1 have yielded inconsistent results. Re-sequencing of individuals with TS has not revealed additional obviously pathogenic coding mutations (Chou et al., 2007; Deng et al., 2006; Zimprich et al., 2008). A common variant candidate gene study in a small cohort found significant association (Miranda et al., 2009) and several rare missense mutations restricted to individuals with TTM were identified in (Zuchner et al., 2006); however the small sample size of the former and the absence of a bona fide mutation burden analysis is the latter limited the conclusions that could be drawn from either study. Two publications specifically evaluated the rare variant var321(Keen-Kim et al., 2006; Scharf et al., 2008). Given the extreme low allele frequency, neither had the sample size necessary to conduct meaningful statistical analyses, however, both noted that the variant, while present in affected family members, appeared to be neither necessary nor sufficient for TS in some of these pedigrees and found a relatively high proportion of affected families of Ashkenazi descent that carried var321, leading them to contend that the initial report was an incidental finding confounded by population stratification.
As discussed at length in the previous sections, the criteria implicitly employed in these studies is appropriately applied to alleles purported to carry Mendelian risks but provides little evidence for or against association of var321. Moreover, our laboratory subsequently tested the hypothesis that population stratification due to occult Ashkenazi ancestry led to the initial finding(O’Roak et al., 2010). Using a combination of genome-wide genotyping data, a multi-dimensional scaling analysis and dense haplotype mapping we found no evidence to support the conclusion that occult ethnicity confounded our results. Instead the additional data provided further evidence that the two instances of var321 were on distinct haplotypes suggesting either recurrent independent mutations or the sharing of an ancient, very rare allele by affected individuals, with either alternative providing additional support for the relationship with TS.
The transition from outlier cytogenetic studies to CNV analyses in TS has so far been slow. Only a single study genome wide study has been published of structural variation and this included 111 individuals (Sundaram et al., 2010). Despite careful attention to potential confounds including population stratification and batch effects, the small sample size limited the ability to arrive at definitive conclusions. However, recurrent CNVs were identified at loci previously implicated in ASD and Schizophrenia, including NRNX1 and the chromosome 1q21 region.
Finally, a recent from our laboratory using parametric linkage and an outlier strategy has suggested a potential role for histaminergic neurotransmission in TS (Ercan- Sencicek et al., 2010). A very rare family with a father, 8 offspring meeting DSM-IV-TR criteria for TS and no evidence for bilineal inheritance was identified. Genome-wide analysis identified a single region reaching the maximum theoretical LOD score for the pedigree (Lod =2.1). Sequencing of all genes within the chromosome 15 interval led to the discovery of a single rare coding mutation, a premature termination codon (W317X) in the gene L-histidine decarboxylase (HDC), the rate-limiting enzyme in histamine biosynthesis. Given experimental evidence for incomplete or absent nonsense-mediated decay, and the knowledge that wild-type (wt) HDC forms an active ~110-kDa homodimer, the mutant protein was evaluated for possible dominant negative effects, and this was confirmed through in vitro studies.
Genetic Findings and Emerging Models
Given the paucity of specific genetic findings, the ability of gene discovery efforts to define molecular models remains limited, and a comprehensive consideration of the data regarding neuroanatomical substrates of TS is beyond the scope of this review. Briefly, attention has been focused on the cortical striatal-thalamo-cortical circuitry that mediates the integration of movement, sensation, emotion and intention. This long-standing interest is supported by analogy to other movement disorders, model systems studies, neuroimaging data and recent neuropathological findings pointing to abnormalities in cholingeric and GABAergic interneurons (Kalanithi et al., 2005; Kataoka et al., 2010).
In addition, there has been a long-standing interest in the role of dopaminergic (DA) neurotransmission. In addition to data from other movement disorders, this interest is driven in part by the clinical observation that DA blockade is the most effective and reliable means in the current pharmacological armamentarium to transiently reduce tics (Scahill et al., 2006) and that DA agonists may promote tics and other stereotypies. However, neither genetic or neuroimaging studies have been definitive and the precise nature of cellular or molecular pathogenic mechanisms remains illusive (Harris and Singer, 2006; Singer, 2005).
As noted, strong genetic findings in TS are sparse and consequently the relevance of the biology of outlier cases remains uncertain: SLITRK1 has been pursued on several fronts. Our laboratory in collaboration with Angeliki Louvi reported on the conserved and developmentally regulated pattern of expression in CTSC circuits, highlighting both mRNA and protein expression in cholinergic interneurons and the striosomal compartment (Stillman et al., 2009); Kajiwara and colleagues found evidence that the demonstrated regulation of neurite outgrowth (Aruga and Mikoshiba, 2003) is mediated by binding to 14-3-3 molecules (Kajiwara et al., 2009); and Katayama et al (Katayama et al., 2010) described an anxiety phenotype and evidence for increased noradrenergic neurotransmission in the mouse knock-out. A particularly intriguing recent result has been the finding of an obsessive-compulsive phenotype resulting from the mouse knockout of the closely related molecule SLITRK5 (Shmelkov et al., 2010).
The identification of a highly penetrant mutation in the gene L-histitide Decarboxylase (HDC) in a dense pedigree with TS provides a direct link to prior hypotheses regarding the involvement of DA pathways in TS: histaminergic (HA) neurotransmission is mediated by four known G-protein coupled receptors; HA in the CNS is known to modulate sleep wake, cognition, movement and behavior; and, while the biological implications are not yet clear, both histamine 2 (H2R) and histamine 3 (H3R) receptors are enriched in the human and rodent striatum (Haas et al., 2008). H3R is of particular interest as it acts as a pre-synaptic auto-receptor on HA containing projection neurons, with activation leading to decreased synthesis and HA release. It also functions as a pre-synaptic regulator of a variety of neurotransmitters, including DA, from non-HA containing neurons; and as a post-synaptic receptor that has been shown to co-localize with and modulate signaling through both D1 and D2 receptors in the striatum.
While HDC null mice are viable and exhibit no structural brain abnormalities they have been known for some time to have decreased brain HA and to show increased sensitivity to stereotypic behaviors upon administration of DA agonists (Kubota et al., 2002). Such stimulant-induced stereotypic movements, including rearing, sniffing, and biting have previously been proposed as a model of human tics (Saka and Graybiel, 2003). Taken together these data point to the possibility that decreased HA synthesis and/or release in the CNS, whether mediated by HDC mutation or other processes, could predispose to or augment stereotypic behaviors. Consistent with this hypothesis, studies of selective H3R antagonists and inverse agonists which increase histaminergic neurotransmission have been shown to moderate the locomotor effects of stimulants in rodent models (Fox et al., 2005), although a very recent study has called into questioned the extent of the effect (Burban et al., 2010).
Finally, H3R compounds have already entered early clinical trials, have a reportedly favorable side effect profile, and are being evaluated for a variety of neuropsychiatric indications. The convergence of the human genetic and model systems data and the potential availability of clinically useful compounds being investigated in related conditions suggests a surprisingly direct avenue to test empirically the generalizabilty of the biology implicated by gene discovery in a single highly unusual pedigree.
Summary, conclusions and future prospects
The foregoing has highlighted both the tremendous promise as well as the considerable challenges that continue to face the study of the genetics of child psychiatric disorders, and ASD and TS in particular.
With regard to ASD, this review points to several overarching conclusions: 1) that despite tremendous progress, many current findings are not yet as definitive as in other areas of medicine; 2) that some of this uncertainty reflects the inherent difficulty in establishing association of rare alleles – either those that are de novo or conferring moderate risks; 3) that it would be entirely premature to minimize the importance of common variant studies, as ASD samples are just now reaching a point where well-powered analyses can realistically investigate the contributing common alleles carrying plausible risks; and 4) that even a relatively small number of unambiguous genetic findings, whether with regard to common or rare variants, holds the potential to help elaborate relevant molecular mechanisms.
With regard to TS, the conclusions are quite similar, but, as noted, the field has been comparatively slow to leverage emerging technologies due to limited patient resources. Nonetheless, the findings to date point to the convergence of risks with other neurodevelopmental disorders, a theme that is extending across all of psychiatry. Moreover, the recent HDC findings from our group highlight a more general theme regarding the opportunities that may be afforded by simultaneous rapid progress across scientific disciplines. Given parallel advances in neurobiology and pharmacology, the speed with which genetic findings may be found to translate into clinically relevant investigations also promises to increase.
Finally, the review has focused on the tremendous promise that gene discovery has to drive neurobiological studies yielding novel and important insights into the molecular mechanisms of child psychopathology. As noted throughout, the challenges of arriving at a necessary level of certainty regarding genetic findings remains considerable in both ASD and TS. At the same time, the tools necessary to do so are clearly present. The consolidation of very large samples, the ability to feasibly query every base of the human exome and the imminent ability to do so cross the entire human genome, the maturation of proven methods to confirm association in multigenic complex disorders, and the manner in which new technologies are empowering both pedigree-based and case-control studies, all suggest that the renewed optimism that characterizes the field is well founded.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.A complete genome screen in sib pairs affected by Gilles de la Tourette syndrome. The Tourette Syndrome Association International Consortium for Genetics. Am J Hum Genet. 65:1428–1436. doi: 10.1086/302613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Genome scan for Tourette disorder in affected-sibling-pair and multigenerational families. Am J Hum Genet. 80:265–272. doi: 10.1086/511052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abelson JF, Kwan KY, O’Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, et al. Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 4.Alarcon M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, Sebat J, Wigler M, Martin CL, Ledbetter DH, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anney R, Klei L, Pinto D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Sykes N, Pagnamenta AT, et al. A genomewide scan for common alleles affecting risk for autism. Hum Mol Genet. 2010 doi: 10.1093/hmg/ddq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, Rea A, Guy M, Lin S, Cook EH, Chakravarti A. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aruga J, Mikoshiba K. Identification and characterization of Slitrk, a novel neuronal transmembrane protein family controlling neurite outgrowth. Mol Cell Neurosci. 2003;24:117–129. doi: 10.1016/s1044-7431(03)00129-5. [DOI] [PubMed] [Google Scholar]
- 9.Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- 10.Baker P, Piven J, Schwartz S, Patil S. Brief report: duplication of chromosome 15q11-13 in two individuals with autistic disorder. J Autism Dev Disord. 1994;24:529–535. doi: 10.1007/BF02172133. [DOI] [PubMed] [Google Scholar]
- 11.Bakkaloglu B, O’Roak BJ, Louvi A, Gupta AR, Abelson JF, Morgan TM, Chawarska K, Klin A, Ercan-Sencicek AG, Stillman AA, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82:165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baron M, Shapiro E, Shapiro A, Rainer JD. Genetic analysis of Tourette syndrome suggesting major gene effect. Am J Hum Genet. 1981;33:767–775. [PMC free article] [PubMed] [Google Scholar]
- 13.Barr CL, Wigg KG, Zovko E, Sandor P, Tsui LC. No evidence for a major gene effect of the dopamine D4 receptor gene in the susceptibility to Gilles de la Tourette syndrome in five Canadian families. Am J Med Genet. 1996;67:301–305. doi: 10.1002/(SICI)1096-8628(19960531)67:3<301::AID-AJMG6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 14.Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 15.Benayed R, Choi J, Matteson PG, Gharani N, Kamdar S, Brzustowicz LM, Millonig JH. Autism-associated haplotype affects the regulation of the homeobox gene, ENGRAILED 2. Biol Psychiatry. 2009;66:911–917. doi: 10.1016/j.biopsych.2009.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benayed R, Gharani N, Rossman I, Mancuso V, Lazar G, Kamdar S, Bruse SE, Tischfield S, Smith BJ, Zimmerman RA, et al. Support for the homeobox transcription factor gene ENGRAILED 2 as an autism spectrum disorder susceptibility locus. Am J Hum Genet. 2005;77:851–868. doi: 10.1086/497705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berkel S, Marshall CR, Weiss B, Howe J, Roeth R, Moog U, Endris V, Roberts W, Szatmari P, Pinto D, et al. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat Genet. 2010;42:489–491. doi: 10.1038/ng.589. [DOI] [PubMed] [Google Scholar]
- 18.Bijlsma EK, Gijsbers AC, Schuurs-Hoeijmakers JH, van Haeringen A, Fransen van de Putte DE, Anderlid BM, Lundin J, Lapunzina P, Perez Jurado LA, Delle Chiaie B, et al. Extending the phenotype of recurrent rearrangements of 16p11.2: deletions in mentally retarded patients without autism and in normal individuals. Eur J Med Genet. 2009;52:77–87. doi: 10.1016/j.ejmg.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 19.Bilguvar K, Ozturk AK, Louvi A, Kwan KY, Choi M, Tatli B, Yalnizoglu D, Tuysuz B, Caglayan AO, Gokben S, et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467:207–210. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bloch MH, Peterson BS, Scahill L, Otka J, Katsovich L, Zhang H, Leckman JF. Adulthood outcome of tic and obsessive-compulsive symptom severity in children with Tourette syndrome. Arch Pediatr Adolesc Med. 2006;160:65–69. doi: 10.1001/archpedi.160.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bourgeron T. A synaptic trek to autism. Curr Opin Neurobiol. 2009;19:231–234. doi: 10.1016/j.conb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Breedveld GJ, Fabbrini G, Oostra BA, Berardelli A, Bonifati V. Tourette disorder spectrum maps to chromosome 14q31.1 in an Italian kindred. Neurogenetics. 2010 doi: 10.1007/s10048-010-0244-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brett PM, Curtis D, Robertson MM, Gurling HM. Exclusion of the 5- HT1A serotonin neuroreceptor and tryptophan oxygenase genes in a large British kindred multiply affected with Tourette’s syndrome, chronic motor tics, and obsessive-compulsive behavior. Am J Psychiatry. 1995;152:437–440. doi: 10.1176/ajp.152.3.437. [DOI] [PubMed] [Google Scholar]
- 24.Brown WT, Jenkins EC, Cohen IL, Fisch GS, Wolf-Schein EG, Gross A, Waterhouse L, Fein D, Mason-Brothers A, Ritvo E, et al. Fragile X and autism: a multicenter survey. Am J Med Genet. 1986;23:341–352. doi: 10.1002/ajmg.1320230126. [DOI] [PubMed] [Google Scholar]
- 25.Bucan M, Abrahams BS, Wang K, Glessner JT, Herman EI, Sonnenblick LI, Alvarez Retuerto AI, Imielinski M, Hadley D, Bradfield JP, et al. Genomewide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burban A, Sadakhom C, Dumoulin D, Rose C, Le Pen G, Frances H, Arrang JM. Modulation of prepulse inhibition and stereotypies in rodents: no evidence for antipsychotic-like properties of histamine H3-receptor inverse agonists. Psychopharmacology (Berl) 2010;210:591–604. doi: 10.1007/s00213-010-1863-2. [DOI] [PubMed] [Google Scholar]
- 27.Burd L, Freeman RD, Klug MG, Kerbeshian J. Tourette Syndrome and learning disabilities. BMC Pediatr. 2005;5:34. doi: 10.1186/1471-2431-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butler MG, Dasouki MJ, Zhou XP, Talebizadeh Z, Brown M, Takahashi TN, Miles JH, Wang CH, Stratton R, Pilarski R, Eng C. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J Med Genet. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buxbaum JD, Cai G, Chaste P, Nygren G, Goldsmith J, Reichert J, Anckarsater H, Rastam M, Smith CJ, Silverman JM, et al. Mutation screening of the PTEN gene in patients with autism spectrum disorders and macrocephaly. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:484–491. doi: 10.1002/ajmg.b.30493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell DB, Sutcliffe JS, Ebert PJ, Militerni R, Bravaccio C, Trillo S, Elia M, Schneider C, Melmed R, Sacco R, et al. A genetic variant that disrupts MET transcription is associated with autism. Proc Natl Acad Sci U S A. 2006;103:16834–16839. doi: 10.1073/pnas.0605296103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chadman KK, Gong S, Scattoni ML, Boltuck SE, Gandhy SU, Heintz N, Crawley JN. Minimal aberrant behavioral phenotypes of neuroligin-3 R451C knockin mice. Autism Res. 2008;1:147–158. doi: 10.1002/aur.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakrabarti S, Fombonne E. Pervasive developmental disorders in preschool children. JAMA. 2001;285:3093–3099. doi: 10.1001/jama.285.24.3093. [DOI] [PubMed] [Google Scholar]
- 33.Chakravarti A. Population genetics--making sense out of sequence. Nat Genet. 1999;21:56–60. doi: 10.1038/4482. [DOI] [PubMed] [Google Scholar]
- 34.Chang S, Bray SM, Li Z, Zarnescu DC, He C, Jin P, Warren ST. Identification of small molecules rescuing fragile X syndrome phenotypes in Drosophila. Nat Chem Biol. 2008;4:256–263. doi: 10.1038/nchembio.78. [DOI] [PubMed] [Google Scholar]
- 35.Chou IC, Tsai CH, Lee CC, Kuo HT, Hsu YA, Li CI, Tsai FJ. Association analysis between Tourette’s syndrome and dopamine D1 receptor gene in Taiwanese children. Psychiatr Genet. 2004;14:219–221. doi: 10.1097/00041444-200412000-00010. [DOI] [PubMed] [Google Scholar]
- 36.Chou IC, Wan L, Liu SC, Tsai CH, Tsai FJ. Association of the Slit and Trk-like 1 gene in Taiwanese patients with Tourette syndrome. Pediatr Neurol. 2007;37:404–406. doi: 10.1016/j.pediatrneurol.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 37.Chubykin AA, Liu X, Comoletti D, Tsigelny I, Taylor P, Sudhof TC. Dissection of synapse induction by neuroligins: effect of a neuroligin mutation associated with autism. J Biol Chem. 2005;280:22365–22374. doi: 10.1074/jbc.M410723200. [DOI] [PubMed] [Google Scholar]
- 38.Coffey BJ, Biederman J, Smoller JW, Geller DA, Sarin P, Schwartz S, Kim GS. Anxiety disorders and tic severity in juveniles with Tourette’s disorder. J Am Acad Child Adolesc Psychiatry. 2000;39:562–568. doi: 10.1097/00004583-200005000-00009. [DOI] [PubMed] [Google Scholar]
- 39.Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 40.Cohen JC, Pertsemlidis A, Fahmi S, Esmail S, Vega GL, Grundy SM, Hobbs HH. Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc Natl Acad Sci U S A. 2006;103:1810–1815. doi: 10.1073/pnas.0508483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Comings D, Chen TJ, Blum K, Mengucci JF, Blum SH, Meshkin B. Neurogenetic interactions and aberrant behavioral co-morbidity of attention deficit hyperactivity disorder (ADHD): dispelling myths. Theor Biol Med Model. 2005;2 doi: 10.1186/1742-4682-2-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Craddock N, Hurles ME, Cardin N, Pearson RD, Plagnol V, Robson S, Vukcevic D, Barnes C, Conrad DF, Giannoulatou E, et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Curtis D, Brett P, Dearlove AM, McQuillin A, Kalsi G, Robertson MM, Gurling HM. Genome scan of Tourette syndrome in a single large pedigree shows some support for linkage to regions of chromosomes 5, 10 and 13. Psychiatr Genet. 2004;14:83–87. doi: 10.1097/01.ypg.0000107927.32051.f5. [DOI] [PubMed] [Google Scholar]
- 44.Curtis D, Robertson MM, Gurling HM. Autosomal dominant gene transmission in a large kindred with Gilles de la Tourette syndrome. Br J Psychiatry. 1992;160:845–849. doi: 10.1192/bjp.160.6.845. [DOI] [PubMed] [Google Scholar]
- 45.Deng H, Le WD, Xie WJ, Jankovic J. Examination of the SLITRK1 gene in Caucasian patients with Tourette syndrome. Acta Neurol Scand. 2006;114:400–402. doi: 10.1111/j.1600-0404.2006.00706.x. [DOI] [PubMed] [Google Scholar]
- 46.do Rosario M, Miguel Filho EC. Obsessive-compulsive disorder and Tourette syndrome: is there a relationship? Sao Paulo Med J. 1997;115:1410–1411. doi: 10.1590/s1516-31801997000200008. [DOI] [PubMed] [Google Scholar]
- 47.Dolen G, Carpenter RL, Ocain TD, Bear MF. Mechanism-based approaches to treating fragile X. Pharmacol Ther. 2010 doi: 10.1016/j.pharmthera.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 48.Dolen G, Osterweil E, Rao BS, Smith GB, Auerbach BD, Chattarji S, Bear MF. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsater H, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ehninger D, de Vries PJ, Silva AJ. From mTOR to cognition: molecular and cellular mechanisms of cognitive impairments in tuberous sclerosis. J Intellect Disabil Res. 2009;53:838–851. doi: 10.1111/j.1365-2788.2009.01208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ehninger D, Han S, Shilyansky C, Zhou Y, Li W, Kwiatkowski DJ, Ramesh V, Silva AJ. Reversal of learning deficits in a Tsc2+/− mouse model of tuberous sclerosis. Nat Med. 2008;14:843–848. doi: 10.1038/nm1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ercan-Sencicek AG, Stillman AA, Ghosh AK, Bilguvar K, O’Roak BJ, Mason CE, Abbott T, Gupta A, King RA, Pauls DL, et al. L-histidine decarboxylase and Tourette’s syndrome. N Engl J Med. 2010;362:1901–1908. doi: 10.1056/NEJMoa0907006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fernandez T, Morgan T, Davis N, Klin A, Morris A, Farhi A, Lifton RP, State MW. Disruption of contactin 4 (CNTN4) results in developmental delay and other features of 3p deletion syndrome. Am J Hum Genet. 2004;74:1286–1293. doi: 10.1086/421474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fernandez T, Morgan T, Davis N, Klin A, Morris A, Farhi A, Lifton RP, State MW. Disruption of Contactin 4 (CNTN4) results in developmental delay and other features of 3p deletion syndrome. Am J Hum Genet. 2008;82:1385. doi: 10.1016/j.ajhg.2008.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Folstein S, Rutter M. Genetic influences and infantile autism. Nature. 1977;265:726–728. doi: 10.1038/265726a0. [DOI] [PubMed] [Google Scholar]
- 56.Fombonne E, Du Mazaubrun C, Cans C, Grandjean H. Autism and associated medical disorders in a French epidemiological survey. J Am Acad Child Adolesc Psychiatry. 1997;36:1561–1569. doi: 10.1016/S0890-8567(09)66566-7. [DOI] [PubMed] [Google Scholar]
- 57.Fox GB, Esbenshade TA, Pan JB, Radek RJ, Krueger KM, Yao BB, Browman KE, Buckley MJ, Ballard ME, Komater VA, et al. Pharmacological properties of ABT-239 [4-(2-{2-[(2R)-2-Methylpyrrolidinyl]ethyl}-benzofuran-5- yl)benzonitrile]: II. Neurophysiological characterization and broad preclinical efficacy in cognition and schizophrenia of a potent and selective histamine H3 receptor antagonist. J Pharmacol Exp Ther. 2005;313:176–190. doi: 10.1124/jpet.104.078402. [DOI] [PubMed] [Google Scholar]
- 58.Freeman RD. Tic disorders and ADHD: answers from a world-wide clinical dataset on Tourette syndrome. Eur Child Adolesc Psychiatry. 2007;16(Suppl 1):15–23. doi: 10.1007/s00787-007-1003-7. [DOI] [PubMed] [Google Scholar]
- 59.Friedman JI, Vrijenhoek T, Markx S, Janssen IM, van der Vliet WA, Faas BH, Knoers NV, Cahn W, Kahn RS, Edelmann L, et al. CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Mol Psychiatry. 2008;13:261–266. doi: 10.1038/sj.mp.4002049. [DOI] [PubMed] [Google Scholar]
- 60.Gauthier J, Champagne N, Lafreniere RG, Xiong L, Spiegelman D, Brustein E, Lapointe M, Peng H, Cote M, Noreau A, et al. De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0906232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gauthier J, Spiegelman D, Piton A, Lafreniere RG, Laurent S, St-Onge J, Lapointe L, Hamdan FF, Cossette P, Mottron L, et al. Novel de novo SHANK3 mutation in autistic patients. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:421–424. doi: 10.1002/ajmg.b.30822. [DOI] [PubMed] [Google Scholar]
- 62.Ghanizadeh A, Mosallaei S. Psychiatric disorders and behavioral problems in children and adolescents with Tourette syndrome. Brain Dev. 2009;31:15–19. doi: 10.1016/j.braindev.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 63.Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 65.Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 2004;119:1013–1026. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guilmatre A, Dubourg C, Mosca AL, Legallic S, Goldenberg A, Drouin-Garraud V, Layet V, Rosier A, Briault S, Bonnet-Brilhault F, et al. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009;66:947–956. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haas HL, Sergeeva OA, Selbach O. Histamine in the nervous system. Physiol Rev. 2008;88:1183–1241. doi: 10.1152/physrev.00043.2007. [DOI] [PubMed] [Google Scholar]
- 68.Harris K, Singer HS. Tic disorders: neural circuits, neurochemistry, and neuroimmunology. J Child Neurol. 2006;21:678–689. doi: 10.1177/08830738060210080901. [DOI] [PubMed] [Google Scholar]
- 69.Harris SW, Hessl D, Goodlin-Jones B, Ferranti J, Bacalman S, Barbato I, Tassone F, Hagerman PJ, Herman H, Hagerman RJ. Autism profiles of males with fragile X syndrome. Am J Ment Retard. 2008;113:427–438. doi: 10.1352/2008.113:427-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hasstedt SJ, Leppert M, Filloux F, van de Wetering BJ, McMahon WM. Intermediate inheritance of Tourette syndrome, assuming assortative mating. Am J Hum Genet. 1995;57:682–689. [PMC free article] [PubMed] [Google Scholar]
- 71.Hernandez RN, Feinberg RL, Vaurio R, Passanante NM, Thompson RE, Kaufmann WE. Autism spectrum disorder in fragile X syndrome: a longitudinal evaluation. Am J Med Genet A. 2009;149A:1125–1137. doi: 10.1002/ajmg.a.32848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 73.Hoffman EJ, State MW. Progress in cytogenetics: implications for child psychopathology. J Am Acad Child Adolesc Psychiatry. 2010;49:736–751. doi: 10.1016/j.jaac.2010.03.016. quiz 856–737. [DOI] [PubMed] [Google Scholar]
- 74.Hogart A, Wu D, LaSalle JM, Schanen NC. The comorbidity of autism with the genomic disorders of chromosome 15q11.2-q13. Neurobiol Dis. 2010;38:181–191. doi: 10.1016/j.nbd.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci U S A. 2002;99:7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 77.Jackson PB, Boccuto L, Skinner C, Collins JS, Neri G, Gurrieri F, Schwartz CE. Further evidence that the rs1858830 C variant in the promoter region of the MET gene is associated with autistic disorder. Autism Res. 2009;2:232–236. doi: 10.1002/aur.87. [DOI] [PubMed] [Google Scholar]
- 78.Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, Bourgeron T. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ji W, Foo JN, O’Roak BJ, Zhao H, Larson MG, Simon DB, Newton-Cheh C, State MW, Levy D, Lifton RP. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40:592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johansen CT, Wang J, Lanktree MB, Cao H, McIntyre AD, Ban MR, Martins RA, Kennedy BA, Hassell RG, Visser ME, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42:684–687. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kajiwara Y, Buxbaum JD, Grice DE. SLITRK1 binds 14-3-3 and regulates neurite outgrowth in a phosphorylation-dependent manner. Biol Psychiatry. 2009;66:918–925. doi: 10.1016/j.biopsych.2009.05.033. [DOI] [PubMed] [Google Scholar]
- 82.Kalanithi PS, Zheng W, Kataoka Y, DiFiglia M, Grantz H, Saper CB, Schwartz ML, Leckman JF, Vaccarino FM. Altered parvalbumin-positive neuron distribution in basal ganglia of individuals with Tourette syndrome. Proc Natl Acad Sci U S A. 2005;102:13307–13312. doi: 10.1073/pnas.0502624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kataoka Y, Kalanithi PS, Grantz H, Schwartz ML, Saper C, Leckman JF, Vaccarino FM. Decreased number of parvalbumin and cholinergic interneurons in the striatum of individuals with Tourette syndrome. J Comp Neurol. 2010;518:277–291. doi: 10.1002/cne.22206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Katayama K, Yamada K, Ornthanalai VG, Inoue T, Ota M, Murphy NP, Aruga J. Slitrk1-deficient mice display elevated anxiety-like behavior and noradrenergic abnormalities. Mol Psychiatry. 2010;15:177–184. doi: 10.1038/mp.2008.97. [DOI] [PubMed] [Google Scholar]
- 85.Keen-Kim D, Mathews CA, Reus VI, Lowe TL, Herrera LD, Budman CL, Gross- Tsur V, Pulver AE, Bruun RD, Erenberg G, et al. Overrepresentation of rare variants in a specific ethnic group may confuse interpretation of association analyses. Hum Mol Genet. 2006;15:3324–3328. doi: 10.1093/hmg/ddl408. [DOI] [PubMed] [Google Scholar]
- 86.Kidd KK, Pauls DL. Genetic hypotheses for Tourette syndrome. Adv Neurol. 1982;35:243–249. [PubMed] [Google Scholar]
- 87.Kim HG, Kishikawa S, Higgins AW, Seong IS, Donovan DJ, Shen Y, Lally E, Weiss LA, Najm J, Kutsche K, et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kubota Y, Ito C, Sakurai E, Watanabe T, Ohtsu H. Increased methamphetamine-induced locomotor activity and behavioral sensitization in histamine-deficient mice. J Neurochem. 2002;83:837–845. doi: 10.1046/j.1471-4159.2002.01189.x. [DOI] [PubMed] [Google Scholar]
- 89.Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, Badner JA, Gilliam TC, Nowak NJ, Cook EH, Jr, Dobyns WB, Christian SL. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 90.Kurlan R, Como PG, Miller B, Palumbo D, Deeley C, Andresen EM, Eapen S, McDermott MP. The behavioral spectrum of tic disorders: a community-based study. Neurology. 2002;59:414–420. doi: 10.1212/wnl.59.3.414. [DOI] [PubMed] [Google Scholar]
- 91.Kurlan R, Eapen V, Stern J, McDermott MP, Robertson MM. Bilineal transmission in Tourette’s syndrome families. Neurology. 1994;44:2336–2342. doi: 10.1212/wnl.44.12.2336. [DOI] [PubMed] [Google Scholar]
- 92.Laumonnier F, Bonnet-Brilhault F, Gomot M, Blanc R, David A, Moizard MP, Raynaud M, Ronce N, Lemonnier E, Calvas P, et al. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet. 2004;74:552–557. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lawson-Yuen A, Saldivar JS, Sommer S, Picker J. Familial deletion within NLGN4 associated with autism and Tourette syndrome. Eur J Hum Genet. 2008;16:614–618. doi: 10.1038/sj.ejhg.5202006. [DOI] [PubMed] [Google Scholar]
- 94.Leckman JF. Phenomenology of tics and natural history of tic disorders. Brain Dev. 2003;25(Suppl 1):S24–28. doi: 10.1016/s0387-7604(03)90004-0. [DOI] [PubMed] [Google Scholar]
- 95.Levitas A, Hagerman RJ, Braden M, Rimland B, McBogg P, Matus I. Autism and the fragile X syndrome. J Dev Behav Pediatr. 1983;4:151–158. doi: 10.1097/00004703-198309000-00002. [DOI] [PubMed] [Google Scholar]
- 96.Lim S, Naisbitt S, Yoon J, Hwang JI, Suh PG, Sheng M, Kim E. Characterization of the Shank family of synaptic proteins. Multiple genes, alternative splicing, and differential expression in brain and development. J Biol Chem. 1999;274:29510–29518. doi: 10.1074/jbc.274.41.29510. [DOI] [PubMed] [Google Scholar]
- 97.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 2003;33:177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 98.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363:166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 99.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, Perkins DO, Dickel DE, Kusenda M, Krastoshevsky O, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 103.McMahon WM, van de Wetering BJ, Filloux F, Betit K, Coon H, Leppert M. Bilineal transmission and phenotypic variation of Tourette’s disorder in a large pedigree. J Am Acad Child Adolesc Psychiatry. 1996;35:672–680. [PubMed] [Google Scholar]
- 104.Merette C, Brassard A, Potvin A, Bouvier H, Rousseau F, Emond C, Bissonnette L, Roy MA, Maziade M, Ott J, Caron C. Significant linkage for Tourette syndrome in a large French Canadian family. Am J Hum Genet. 2000;67:1008–1013. doi: 10.1086/303093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Merikangas AK, Corvin AP, Gallagher L. Copy-number variants in neurodevelopmental disorders: promises and challenges. Trends Genet. 2009;25:536–544. doi: 10.1016/j.tig.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 106.Miranda DM, Wigg K, Kabia EM, Feng Y, Sandor P, Barr CL. Association of SLITRK1 to Gilles de la Tourette Syndrome. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:483–486. doi: 10.1002/ajmg.b.30840. [DOI] [PubMed] [Google Scholar]
- 107.Moessner R, Marshall CR, Sutcliffe JS, Skaug J, Pinto D, Vincent J, Zwaigenbaum L, Fernandez B, Roberts W, Szatmari P, Scherer SW. Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Moss J, Howlin P. Autism spectrum disorders in genetic syndromes: implications for diagnosis, intervention and understanding the wider autism spectrum disorder population. J Intellect Disabil Res. 2009;53:852–873. doi: 10.1111/j.1365-2788.2009.01197.x. [DOI] [PubMed] [Google Scholar]
- 109.Nakamoto M, Nalavadi V, Epstein MP, Narayanan U, Bassell GJ, Warren ST. Fragile X mental retardation protein deficiency leads to excessive mGluR5-dependent internalization of AMPA receptors. Proc Natl Acad Sci U S A. 2007;104:15537–15542. doi: 10.1073/pnas.0707484104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Narayanan U, Nalavadi V, Nakamoto M, Pallas DC, Ceman S, Bassell GJ, Warren ST. FMRP phosphorylation reveals an immediate-early signaling pathway triggered by group I mGluR and mediated by PP2A. J Neurosci. 2007;27:14349–14357. doi: 10.1523/JNEUROSCI.2969-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.O’Roak BJ, Morgan TM, Fishman DO, Saus E, Alonso P, Gratacos M, Estivill X, Teltsh O, Kohn Y, Kidd KK, et al. Additional support for the association of SLITRK1 var321 and Tourette syndrome. Mol Psychiatry. 2010;15:447–450. doi: 10.1038/mp.2009.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Paschou P, Feng Y, Pakstis AJ, Speed WC, DeMille MM, Kidd JR, Jaghori B, Kurlan R, Pauls DL, Sandor P, et al. Indications of linkage and association of Gilles de la Tourette syndrome in two independent family samples: 17q25 is a putative susceptibility region. Am J Hum Genet. 2004;75:545–560. doi: 10.1086/424389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pauls DL. Update on the genetics of Tourette syndrome. Adv Neurol. 2001;85:281–293. [PubMed] [Google Scholar]
- 114.Pauls DL, Leckman JF. The inheritance of Gilles de la Tourette’s syndrome and associated behaviors. Evidence for autosomal dominant transmission. N Engl J Med. 1986;315:993–997. doi: 10.1056/NEJM198610163151604. [DOI] [PubMed] [Google Scholar]
- 115.Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010a;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010b doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Price RA, Kidd KK, Cohen DJ, Pauls DL, Leckman JF. A twin study of Tourette syndrome. Arch Gen Psychiatry. 1985;42:815–820. doi: 10.1001/archpsyc.1985.01790310077011. [DOI] [PubMed] [Google Scholar]
- 118.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 119.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 120.Roessner V, Becker A, Banaschewski T, Freeman RD, Rothenberger A. Developmental psychopathology of children and adolescents with Tourette syndrome--impact of ADHD. Eur Child Adolesc Psychiatry. 2007;16(Suppl 1):24–35. doi: 10.1007/s00787-007-1004-6. [DOI] [PubMed] [Google Scholar]
- 121.Romeo S, Yin W, Kozlitina J, Pennacchio LA, Boerwinkle E, Hobbs HH, Cohen JC. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. J Clin Invest. 2009;119:70–79. doi: 10.1172/JCI37118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Roohi J, Montagna C, Tegay DH, Palmer LE, DeVincent C, Pomeroy JC, Christian SL, Nowak N, Hatchwell E. Disruption of contactin 4 in three subjects with autism spectrum disorder. J Med Genet. 2009;46:176–182. doi: 10.1136/jmg.2008.057505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Saka E, Graybiel AM. Pathophysiology of Tourette’s syndrome: striatal pathways revisited. Brain Dev. 2003;25(Suppl 1):S15–19. doi: 10.1016/s0387-7604(03)90002-7. [DOI] [PubMed] [Google Scholar]
- 124.Scahill L, Erenberg G, Berlin CM, Jr, Budman C, Coffey BJ, Jankovic J, Kiessling L, King RA, Kurlan R, Lang A, et al. Contemporary assessment and pharmacotherapy of Tourette syndrome. NeuroRx. 2006;3:192–206. doi: 10.1016/j.nurx.2006.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Scharf JM, Moorjani P, Fagerness J, Platko JV, Illmann C, Galloway B, Jenike E, Stewart SE, Pauls DL. Lack of association between SLITRK1var321 and Tourette syndrome in a large family-based sample. Neurology. 2008;70:1495–1496. doi: 10.1212/01.wnl.0000296833.25484.bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 127.Sebastiani P, Solovieff N, Puca A, Hartley SW, Melista E, Andersen S, Dworkis DA, Wilk JB, Myers RH, Steinberg MH, et al. Genetic Signatures of Exceptional Longevity in Humans. Science. 2010 doi: 10.1126/science.1190532. [DOI] [PubMed] [Google Scholar]
- 128.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 130.Sharma A, Hoeffer CA, Takayasu Y, Miyawaki T, McBride SM, Klann E, Zukin RS. Dysregulation of mTOR signaling in fragile X syndrome. J Neurosci. 2010;30:694–702. doi: 10.1523/JNEUROSCI.3696-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shmelkov SV, Hormigo A, Jing D, Proenca CC, Bath KG, Milde T, Shmelkov E, Kushner JS, Baljevic M, Dincheva I, et al. Slitrk5 deficiency impairs corticostriatal circuitry and leads to obsessive-compulsive-like behaviors in mice. Nat Med. 2010;16:598–602. 591–602. doi: 10.1038/nm.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Singer HS. Tourette’s syndrome: from behaviour to biology. Lancet Neurol. 2005;4:149–159. doi: 10.1016/S1474-4422(05)01012-4. [DOI] [PubMed] [Google Scholar]
- 133.Skuse DH. Rethinking the nature of genetic vulnerability to autistic spectrum disorders. Trends Genet. 2007;23:387–395. doi: 10.1016/j.tig.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 134.Smalley SL. Autism and tuberous sclerosis. J Autism Dev Disord. 1998;28:407–414. doi: 10.1023/a:1026052421693. [DOI] [PubMed] [Google Scholar]
- 135.Smalley SL, Tanguay PE, Smith M, Gutierrez G. Autism and tuberous sclerosis. J Autism Dev Disord. 1992;22:339–355. doi: 10.1007/BF01048239. [DOI] [PubMed] [Google Scholar]
- 136.Stewart SE, Illmann C, Geller DA, Leckman JF, King R, Pauls DL. A controlled family study of attention-deficit/hyperactivity disorder and Tourette’s disorder. J Am Acad Child Adolesc Psychiatry. 2006;45:1354–1362. doi: 10.1097/01.chi.0000251211.36868.fe. [DOI] [PubMed] [Google Scholar]
- 137.Stillman AA, Krsnik Z, Sun J, Rasin MR, State MW, Sestan N, Louvi A. Developmentally regulated and evolutionarily conserved expression of SLITRK1 in brain circuits implicated in Tourette syndrome. J Comp Neurol. 2009;513:21–37. doi: 10.1002/cne.21919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Strauss KA, Puffenberger EG, Huentelman MJ, Gottlieb S, Dobrin SE, Parod JM, Stephan DA, Morton DH. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354:1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 139.Sundaram SK, Huq AM, Wilson BJ, Chugani HT. Tourette syndrome is associated with recurrent exonic copy number variants. Neurology. 2010;74:1583–1590. doi: 10.1212/WNL.0b013e3181e0f147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, Powell CM, Sudhof TC. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Toro R, Konyukh M, Delorme R, Leblond C, Chaste P, Fauchereau F, Coleman M, Leboyer M, Gillberg C, Bourgeron T. Key role for gene dosage and synaptic homeostasis in autism spectrum disorders. Trends Genet. 2010;26:363–372. doi: 10.1016/j.tig.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 143.Tost H, Kolachana B, Hakimi S, Lemaitre H, Verchinski BA, Mattay VS, Weinberger DR, Meyer-Lindenberg A. A common allele in the oxytocin receptor gene (OXTR) impacts prosocial temperament and human hypothalamiclimbic structure and function. Proc Natl Acad Sci U S A. 2010;107:13936–13941. doi: 10.1073/pnas.1003296107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Vassos E, Collier DA, Holden S, Patch C, Rujescu D, St Clair D, Lewis CM. Penetrance for copy number variants associated with schizophrenia. Hum Mol Genet. 2010;19:3477–3481. doi: 10.1093/hmg/ddq259. [DOI] [PubMed] [Google Scholar]
- 145.Veenstra-Vanderweele J, Christian SL, Cook EH., Jr Autism as a paradigmatic complex genetic disorder. Annu Rev Genomics Hum Genet. 2004;5:379–405. doi: 10.1146/annurev.genom.5.061903.180050. [DOI] [PubMed] [Google Scholar]
- 146.Verkerk AJ, Mathews CA, Joosse M, Eussen BH, Heutink P, Oostra BA. CNTNAP2 is disrupted in a family with Gilles de la Tourette syndrome and obsessive compulsive disorder. Genomics. 2003;82:1–9. doi: 10.1016/s0888-7543(03)00097-1. [DOI] [PubMed] [Google Scholar]
- 147.Vernes SC, Newbury DF, Abrahams BS, Winchester L, Nicod J, Groszer M, Alarcon M, Oliver PL, Davies KE, Geschwind DH, et al. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008;359:2337–2345. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Volkmar FR, Lord C, Bailey A, Schultz RT, Klin A. Autism and pervasive developmental disorders. J Child Psychol Psychiatry. 2004;45:135–170. doi: 10.1046/j.0021-9630.2003.00317.x. [DOI] [PubMed] [Google Scholar]
- 149.Volkmar FR, Pauls D. Autism. Lancet. 2003;362:1133–1141. doi: 10.1016/S0140-6736(03)14471-6. [DOI] [PubMed] [Google Scholar]
- 150.Volkmar FR, State M, Klin A. Autism and autism spectrum disorders: diagnostic issues for the coming decade. J Child Psychol Psychiatry. 2009;50:108–115. doi: 10.1111/j.1469-7610.2008.02010.x. [DOI] [PubMed] [Google Scholar]
- 151.Walkup JT, LaBuda MC, Singer HS, Brown J, Riddle MA, Hurko O. Family study and segregation analysis of Tourette syndrome: evidence for a mixed model of inheritance. Am J Hum Genet. 1996;59:684–693. [PMC free article] [PubMed] [Google Scholar]
- 152.Wang K, Zhang H, Ma D, Bucan M, Glessner JT, Abrahams BS, Salyakina D, Imielinski M, Bradfield JP, Sleiman PM, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Weiss LA, Arking DE, Daly MJ, Chakravarti A. A genome-wide linkage and association scan reveals novel loci for autism. Nature. 2009;461:802–808. doi: 10.1038/nature08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 155.Zimprich A, Hatala K, Riederer F, Stogmann E, Aschauer HN, Stamenkovic M. Sequence analysis of the complete SLITRK1 gene in Austrian patients with Tourette’s disorder. Psychiatr Genet. 2008;18:308–309. doi: 10.1097/YPG.0b013e3283060f6f. [DOI] [PubMed] [Google Scholar]
- 156.Zuchner S, Cuccaro ML, Tran-Viet KN, Cope H, Krishnan RR, Pericak-Vance MA, Wright HH, Ashley-Koch A. SLITRK1 mutations in trichotillomania. Mol Psychiatry. 2006;11:887–889. doi: 10.1038/sj.mp.4001898. [DOI] [PubMed] [Google Scholar]