Abstract
Methylseleninic acid (MSA) has been shown to have potent anticancer activity and is an excellent compound for studying the anticancer effects of selenium in vitro. To gain insights into the effects of MSA in prostate cancer, we characterized the global transcriptional response of LNCaP, an androgen-sensitive human prostate cancer cell line, to MSA by using high-density cDNA microarrays. We identified 951 genes whose expression shows striking dose- and time-dependent changes in response to 3-30 μM MSA over the time course of 48 h. Transcript levels of many cell cycle-regulated genes change in response to MSA, suggesting that MSA inhibits proliferation. Consistent with these gene expression changes, cell proliferation, monitored by carboxyfluoroscein succinimidyl ester staining, was decreased after MSA treatment, and an accumulation of cells at G0/G1 phase was detected by flow cytometry. Surprisingly, MSA also modulated expression of many androgen-regulated genes, suppressed androgen receptor (AR) expression at both mRNA and protein level, and decreased levels of prostate specific antigen secreted into the medium. Low concentrations of MSA also induced significant increases in transcript levels of phase 2 detoxification enzymes and induced NADPH dehydrogenase, quinone 1 enzymatic activity, a surrogate marker of global phase 2 enzyme activity. Our results suggest that MSA may protect against prostate cancer by inhibiting cell proliferation, by modulating the expression of AR and AR-regulated genes and by inducing carcinogen defenses.
INTRODUCTION
Increasing evidence suggests that selenium compounds have promise as prostate cancer preventive agents. Several epidemiological studies have shown an inverse association between selenium levels in the serum or toenails and the subsequent risk of developing prostate cancer (Willett et al., 1983; Yoshizawa et al., 1998; Helzlsouer et al., 2000; Nomura et al., 2000; Brooks et al., 2001a). Animal and human intervention trials have shown that a daily supplementation with selenium-containing compounds reduces the risk of several malignancies, particularly human prostate cancer (Ip and White, 1987; el-Bayoumy, 1994; Reddy et al., 1994; Clark et al., 1996, 1998; Medina et al., 2001; Rao et al., 2001; Davis et al., 2002; Duffield-Lillico et al., 2002). The Nutritional Prevention of Cancer Trial, for instance, showed significantly lower incidence of prostate cancer diagnosis in subjects randomized to receive 200 μg of selenized yeast after 6.4 and 7.4 yr of follow-up, as well as reduced total cancer incidence (Clark et al., 1996; Duffield-Lillico et al., 2002). Although this study has been criticized for its use of secondary endpoints, it has provided compelling rationale for the recently initiated Selenium and Vitamin E Cancer Prevention Trial (SELECT), a 12-year prospective, randomized trial involving 32,000 men (Hoque et al., 2001; Klein et al., 2001).
The inverse relationship between selenium intake and prostate cancer risk has prompted a great deal of interest in understanding the mechanisms of selenium chemoprevention. Diverse forms of selenium have been shown to affect a variety of biological processes important in carcinogenesis (Ip, 1998; Combs, 2001; El-Bayoumy, 2001; Fleming et al., 2001; Ganther, 2001; Kim and Milner, 2001; Lu and Jiang, 2001; Youn et al., 2001). Selenium compounds have been shown to inhibit cell proliferation and induce apoptosis, and these are thought to be major mechanisms by which selenium prevents tumor initiation or progression (Ip et al., 2000a; Combs, 2001; Ganther, 2001; Lu, 2001). Selenium compounds also protect cells against oxidative stress and genetic damage, and block tumor angiogenesis (El-Bayoumy, 2001; Lu and Jiang, 2001). However, a comprehensive understanding of the mechanisms underlying selenium's anticancer effects is currently lacking.
Monomethylated forms of selenium are highly potent and efficacious chemopreventive agents. Methylselenocysteine (MSC) and methylseleninic acid (MSA) have been shown to be more active in cancer prevention than inorganic selenite, or selenomethionine, the form of selenium being used in SELECT (Ip et al., 1991; Ip, 1998; Combs, 2001; Hoque et al., 2001; Klein et al., 2001). It is believed that they are the direct precursors of methylselenol, possibly the key metabolite responsible for selenium's anticancer activity. Whereas MSC requires the action of cysteine conjugate β-lyase or related lyases to be converted to methylselenol, MSA does not (Andreadou et al., 1996; Ganther and Lawrence, 1997; Ip, 1998; Ip et al., 2000b). It is 10 times more potent than MSC in affecting biological processes in vitro, probably because of limited β-lyase activity in cultured eukaryotic cells (Ip et al., 2000b). Therefore, MSA is an ideal compound for studying the anticancer effects of selenium in vitro.
DNA microarrays provide a genome-wide view of the biological processes affected by cellular perturbations and offer an opportunity to gain new insights into the mechanisms by which preventive agents exert their effects (Williams and Brooks, 2001). Herein, we have undertaken a systematic evaluation of the changes in gene expression that result from treatment of the androgen-sensitive prostate cancer cell line LNCaP with MSA. We identified 1128 clones representing 951 genes whose expression levels are affected by MSA in a time- and dose-dependent manner. The transcriptional profiles and confirmatory experiments suggest that MSA causes cell accumulation at G0/G1 modulates the expression of androgen receptor (AR) and its regulated genes, and induces enzymes that detoxify carcinogens.
MATERIALS AND METHODS
Cell Culture and Treatment
LNCaP cells were cultured in RPMI 1640 medium with 2 mM l-glutamine, 100 U/ml penicillin/100 μg/ml streptomycin (Invitrogen, Carlsbad, CA), and 5% defined fetal bovine serum that contributed 13 nM selenium to the medium (Hyclone Laboratories, Logan, UT). When cells reached ∼40-60% confluence, the medium was changed, and 12-24 h later the cells were treated with 3, 10, or 30 μM MSA (pH adjusted to 7.0) (Selenium Technologies, Lubbock, TX). The doses used in this study were chosen based on previous studies using MSA in vitro and reported selenium levels in human serum (Ip et al., 2000b; Nomura et al., 2000; Brooks et al., 2001a; Jiang et al., 2001; Sinha et al., 2001; Dong et al., 2002; Zhu et al., 2002). At several time points after exposure, total RNA was harvested as described below. Untreated cells cultured in parallel were used as controls for each time point.
Total RNA Isolation
Medium was aspirated from each 150-mm cell culture plate, and 5 ml of TRIzol solution (Invitrogen) was added. After 5 min of gentle agitation, lysates were extracted with chloroform, and the organic and aqueous layers were separated using Phase Lock Gel (Eppendorf-5 Prime, Inc., Boulder, CO). Total RNA was precipitated with isopropanol and further purified with RNeasy mini kit (QIAGEN, Valenica, CA). The concentration of total RNA was determined using an MBA 2000 spectrometer (PerkinElmer Life Sciences, Boston, MA), and the integrity of total RNA was assessed using a 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA).
cDNA Microarray Hybridizations
Fluorescently labeled cDNA probes were prepared from 70 μg of total RNA isolated from MSA-treated cells (Cy5 labeled) and control cells (Cy3 labeled) by reverse transcription with an Oligo dT primer 5′-TTTTTTTTTTTTTTTT-3′ (QIAGEN) as described previously (Zhao et al., 2002). Labeled probes from MSA-treated and control cells for each time point were mixed and hybridized overnight to spotted cDNA microarrays with 42,941 elements (Stanford Functional Genomics Facility). Microarray slides were then washed to remove unbound probe and analyzed as described previously (Zhao et al., 2002).
Data Processing and Analysis
Fluorescence intensities for each fluoroprobe were acquired using an Axon scanner 4000B and analyzed with GenePix Pro3.0 software (Axon Instruments, Union City, CA). Spots of poor quality were removed from further analysis by visual inspection. Data files containing fluorescence ratios were entered into the Stanford Microarray Database where biological data were associated with fluorescence ratios, and genes were selected for further analysis (Sherlock et al., 2001). Only spots with a signal intensity >150% above background in both Cy5 and Cy3 channels in at least 80% of the microarray experiments were used in the subsequent analysis. We arbitrarily selected transcripts whose expression level varied at least twofold after treatment compared with controls in at least three of the experiments examined. Prior work has shown that twofold variations in expression reliably reflect changes in expression levels measured by other methods (Blader et al., 2001; Jones and Arvin, 2003). The genes in the resulting data table were ordered by their patterns of gene expression by using hierarchical clustering analysis (Eisen et al., 1998) and visualized using Treeview software (http://rana.lbl.gov/Eisen-Software.htm). The data for all 1128 clones as well as the primary data are available at http://www.stanford.edu/~hongjuan/MSA.
Cell Proliferation and Cell Cycle Assay
Cell proliferation was determined using 5- or 6-(N-succinimidyloxycarbonyl)-3′,6′-O,O′-diacetylfluorescein (CFSE) (Dojindo Molecular Technologies, Gaithersburg, MD) staining (Lyons, 2000; Groszer et al., 2001) Untreated cells were stained with 1 μM CFSE in RPMI 1640 medium at 37°C for 10 min before being seeded in 60-mm plates with fresh media. After cells were cultured overnight, the media were again changed to eliminate residual CFSE that may have leaked from the cells. Half of the plates were treated with MSA for different lengths of time and harvested by trypsinization, and the remaining untreated plates cultured in parallel were used as controls. The absolute intensity of CFSE within each cell was measured by flow cytometry, and the average intensity of CFSE within the population calculated using Flow Jo software (http://www.flowjo.com/v4/html/overview.html).
Cell cycle distribution was determined by propidium iodide (PI) (Sigma-Aldrich, St. Louis, MO) staining. After aspirating the media, treated and control cells were collected by trypsinization and washed with 1× phosphate-buffered saline. Duplicate samples were collected for each growth condition. Cells were fixed with 70% ice-cold ethanol overnight and stained with PI (20 μg/ml) in presence of RNase A (300 μg/ml) at 37°C for 30 min. The DNA content of the cells was determined by flow cytometry, and cell cycle distribution was analyzed with Flow Jo software.
Western Blotting
Treated and control cells were lysed with 1 ml of radioimmunoprecipitation assay buffer (pH 7.4, 50 mM Tris-HCl, 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin). The cell lysate was passed through a 21-gauge needle to shear the cellular DNA. Protein concentration was determined using a BCA protein assay kit (Pierce Chemical, Rockford, IL). Ten to 15 μg of protein was separated using a 4-20% Tris-HCl precast gel (Bio-Rad, Hercules, CA), and transferred to a Hybond-P membrane (Amersham Life Sciences, Arlington Heights, IL). AR was detected with a rabbit polyclonal antibody against the amino terminus of human AR, sc-816 (Santa Cruz Biotechnology, Santa Cruz, CA) and visualized with an ECL Plus kit (Amersham Biosciences, Piscataway, NJ). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was detected with a monoclonal mouse anti-rabbit antibody, MoAb 6C5, which reacts with human GAPDH (Research Diagnostics, Flanders, NJ). AR and GAPDH signal intensities were quantified with a GS-700 densitometer (Bio-Rad).
Determination of Secreted Prostate-specific Antigen (PSA) Levels
Media from MSA-treated and control cells cultured on a 24-well plate was aspirated and stored at -80°C. PSA concentration in the thawed medium was measured using a human prostate specific antigen ELISA kit (Alpha Diagnostic International, San Antonio, TX) and was normalized to total protein of cells cultured in the same well where the medium was taken.
NADPH Dehydrogenase, Quinone 1 (NQO1) Enzymatic Activity Assay
After aspirating the media, treated and control cells cultured in a 96-well plate were lysed with 200 μl of 0.08% digitonin (Sigma-Aldrich)/2 mM EDTA (pH 8.0) at 37°C for 30 min. NQO1 enzymatic activity was assessed in triplicate by the menadione-coupled reduction of tetrazolium dye as described previously (Brooks et al., 2001b). Enzymatic activity for each sample was averaged across the triplicate and normalized to total cell protein in each sample.
RESULTS
MSA Affects Gene Expression in LNCaP Cells in a Dose- and Time-dependent Manner
To study systematically the effects of MSA in human prostate cancer cells in vitro, we characterized the temporal program of gene expression induced by treating LNCaP cells with three different concentrations of MSA. Thirty-one samples (10 samples/concentration over the course of 48 h plus one sample from untreated cells) were analyzed on microarrays containing ∼42,941 features representing ∼29,587 different human genes as inferred from UNIGENE clusters. The 1128 clones representing 951 genes displayed changes in expression levels of at least twofold after MSA treatment compared with controls in at least three samples. Many of the transcripts represent poorly characterized genes or expressed sequence tags. The data for the 1128 transcripts were ordered by their patterns of gene expression by hierarchical clustering (Eisen et al., 1998) (Figure 1). The complete data set, including raw data, is available at http://www.stanford.edu/~hongjuan/MSA.
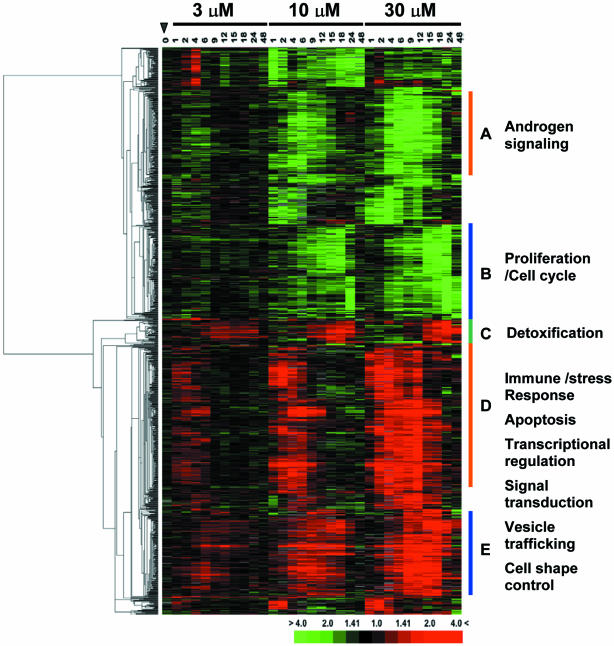
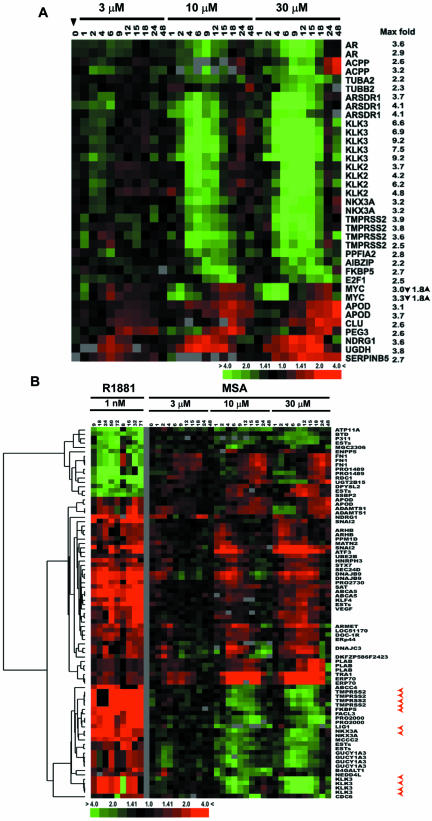
Figure 1.
Hierarchical clustering analysis of MSA-responsive genes in LNCaP cells. Each column represents data from a single time point after treatment with MSA, and each row represents expression levels for a single gene across the time course. The 1128 transcripts were up-regulated (red) or down-regulated (green) after exposure to 3, 10, or 30 μM MSA as indicated at the top of the image. The degree of color saturation corresponds with the ratio of gene expression shown at the bottom of the image. For comparison, the gene expression pattern of untreated cells at time 0 is shown at the closed arrowhead. The data from each treatment condition were arranged in a time ascending order (1, 2, 4, 6, 9, 12, 15, 18, 24, and 48 h) as indicated on top of the image. The gene tree shown at the left of the image corresponds to the degree of similarity (Pearson correlation) of the pattern of expression for genes across the experiments. Genes in cluster A-E show different temporal response to MSA in a dose-dependent manner. Full transcript identities and raw data are available at http://www.Stanford.edu/~hongjuan/MSA.
MSA produced discrete, reproducible, time- and dose-dependent changes in gene expression in LNCaP cells. Expression changes were largely similar among cells treated with 3, 10, and 30 μM MSA; however, with higher concentrations of MSA, changes in gene expression were larger in both the magnitude and duration. The number of transcripts whose expression increased or decreased was similar (541 and 587, respectively). Approximately one-half of the transcripts showed changes within 1-2 h after treatment with peak variation occurring within 8 h and returned to baseline expression levels by 24 h (Figure 1, clusters A and D). Many of the functionally characterized genes in cluster A are known to be involved in androgen signaling pathways. The remaining transcripts were delayed in their response, with expression changes that peaked between 12 and 24 h and that remained apparent at 48 h (Figure 1, clusters B, C, and E). These included genes involved in cell cycle regulation (cluster B) and phase 2 detoxification enzymes (cluster C). Known genes in clusters D and E are involved in diverse biological processes, including immune and stress responses (IGSF3, IGSF4, and NFIL3), apoptosis regulation (BIRC2, BIRC3, and TNFAIP3), transcriptional regulation (ATF3, ELF3, and MAD), signal transduction (JAK1, ARHB, and SH3BP5), tumor suppression (MEN1, ING1, and IRF1), vesicle trafficking (SEC24D, STX1A, and RAB31), and cell shape control (KLHL2, WASF1, and MAP1B).
MSA Changes Expression of Cell Cycle-regulated Genes
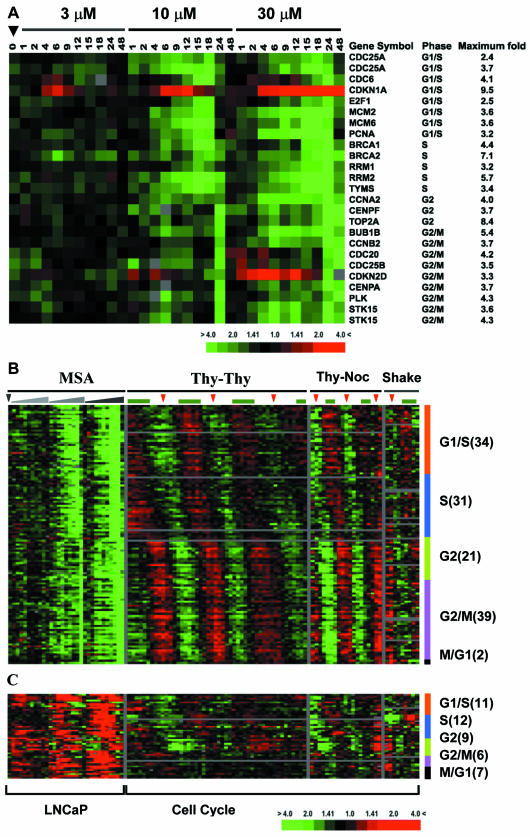
MSA has been shown to inhibit cell growth through its effects on the cell cycle in several model systems, although not in the LNCaP cell line. A subset of the 1128 transcripts (Figure 1, cluster B) modulated by MSA in LNCaP cells represent known cell cycle-regulated genes (Figure 2A). To gain insight into the effect of MSA on cell cycle-regulated genes, we compared these 1128 transcripts to a set of 1134 transcripts (representing >850 genes) that vary periodically as synchronized HeLa cells pass through the cell cycle (Whitfield et al., 2002). In the latter data set, all 1134 transcripts were grouped according to the phase in the cell cycle where their expression peaked. Between the MSA and cell cycle data sets, 172 transcripts were found in common. The 127 transcripts that showed decreased expression were distributed throughout all phases of the cell cycle and included genes involved in DNA replication initiation (CDC6, MCM2, and MCM6), DNA repair (PCNA), and cell cycle control (CDC25A and E2F1) expressed in G1/S phase, DNA replication (RRM1, RRM2, and TYMS) expressed in S phase, chromosome condensation and organization (TOP2A and CENPA), mitotic spindle checkpoint (CDC20 and BUB1B), and centrosome duplication (PLK and STK15) expressed in G2 and M phase (Figure 2B). There were 45 clones in common between the data sets that were up-regulated by MSA that, again, were distributed throughout all phases of the cell cycle. These transcripts show periodic expression in HeLa cells with an expression pattern that was the inverse of the genes that are down-regulated by MSA. In this set of transcripts are known inhibitors of proliferation, most notably CDKN1A (p21), CDKN2D (p19), and CDKN1C (p57), all of which are potent negative regulators of G1 cyclin/cdk complexes (Sherr and Roberts, 1999; Gitig and Koff, 2000). This suggests that induction of this set of genes by MSA may modulate decreased proliferation in LNCaP cells.
Figure 2.
Cell cycle-regulated genes modulated by MSA. Genes that occur more than once are represented by multiple clones on arrays. (A) Transcripts representing previously characterized cell cycle-regulated genes. (B) Cell cycle-regulated transcripts identified by Whitfield et al. (2002) that are down-regulated by MSA. The number of transcripts belonging to different cell cycle phases is shown at the right of the image. The effect of MSA on expression of these genes is shown to the left organized in the same order as in A. The pattern of these genes across multiple cell cycles in HeLa cells is shown to the right. Thy-Thy indicates a double thymidine block to synchronize cells at S phase before release. Thy-Noc indicates a thymidinenocodazole block to synchronize cells at mitosis before release. Shake indicates cells collected with an automated cell shaker that were used as synchronized in mitosis. The green bar above each column represents S phase, and the red arrowheads indicate mitosis as estimated by flow cytometry or bromodeoxyuridine labeling. (C) Cell cycle-regulated transcripts identified by Whitfield et al. (2002) that are up-regulated by MSA.
The distribution of transcripts affected by MSA across all phases of the cell cycle suggested that MSA might cause LNCaP cells to exit the cell cycle, rather than induce an arrest at a specific cell cycle phase or slow cell cycle progression. In the HeLa cell cycle experiments, cell cycle arrest was associated with high expression of transcripts typically expressed during the phase of the cell cycle at which arrest occurs (see Thy-Thy, Thy-Noc, and Shake off in Figure 2B). In LNCaP treated with MSA, on the other hand, expression variations of cell cycle-regulated transcripts were not selectively associated with any particular phase of the cell cycle; cell cycle-regulated transcripts typically expressed in a particular phase of the cell cycle (i.e., G1, S, or G2/M) all showed decreased expression and the transcripts that displayed increased expression are known to inhibit cell proliferation. These expression changes, therefore, suggest that cells are exiting the cell cycle in response to MSA, rather than arresting at a particular phase in the cell cycle.
MSA Inhibits Cell Proliferation by Induction of Cell Accumulation at G0/G1
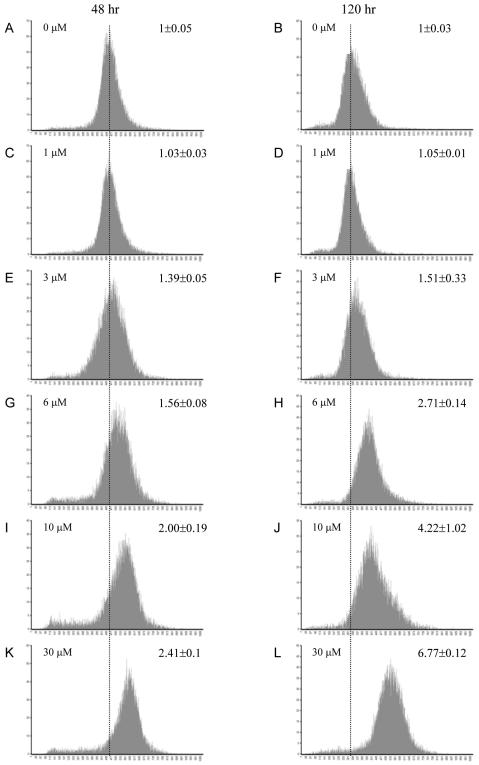
Based on the expression changes in the cell cycle-regulated genes, we assessed the effect of MSA on the proliferation of the LNCaP cells after pulse exposure to CFSE. CFSE diffuses freely into cells where it is converted to a fluorescently tagged membrane impermeable dye that is retained in the cytoplasm. With each round of cell division, the retained CFSE is partitioned equally to daughter cells and the relative intensity of the dye becomes decreased by half. At concentrations between 3 and 30 μM, MSA produced a dose-dependent inhibition of LNCaP cell growth, evident by the significantly higher mean intensity of CFSE in treated cells compared with controls (Figure 3). CFSE levels in MSA-treated cells remained high relative to control cells up to 48 h and then the inhibitory effect began to diminish (our unpublished data). Exchange of the medium at 72 h and retreatment with MSA produced growth inhibition out to 120 h similar in magnitude to that produced by the first treatment. Therefore, as predicted from gene expression profiling, MSA inhibits LNCaP cell growth and cells retain sensitivity to this inhibition with repeated treatments.
Figure 3.
Cell proliferation monitored by CFSE staining and flow cytometry with and without MSA exposure. The y-axis represents the number of cells, and the x-axis represents the intensity of CFSE in the cells. Cells harvested 48 h after CESF staining (left) and 120 h (right). Media with fresh MSA were exchanged at 72 h after CFSE staining. The concentration of MSA used to treat the cells is shown at the top left corner of each graph. The mean average intensity of CFSE in treated cells was normalized against that of the control cells and is shown at the top right corner of each graph. Each graph represents data from triplicate samples.
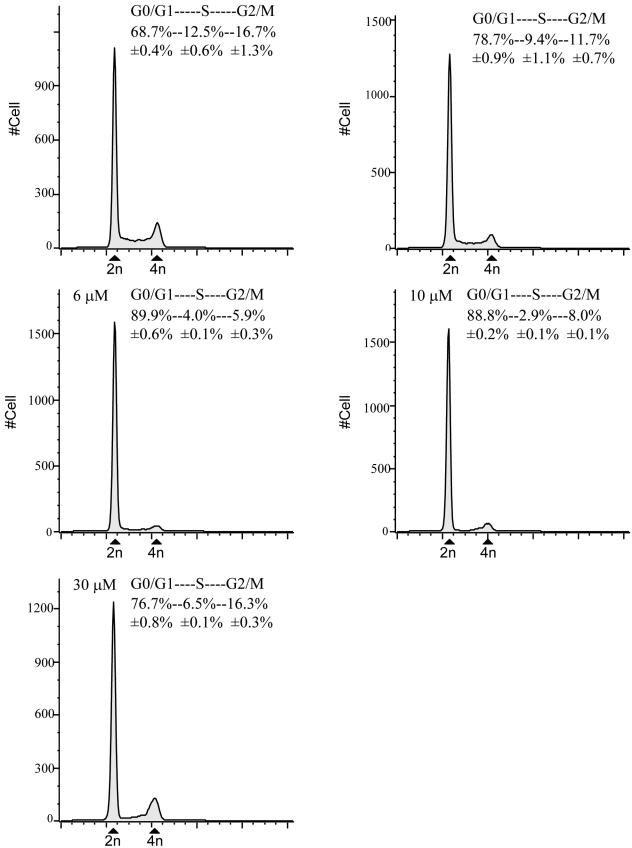
To evaluate whether the decreased proliferation we observed was most consistent with cell cycle arrest or exit from the cell cycle, we performed flow cytometry on MSA-treated and untreated LNCaP cells. The proportion of cells at G0/G1, S, and G2/M phase was determined after 24-h exposure to different concentrations of MSA. Cells treated with 3, 6, 10, and 30 μM MSA all showed an increase in the percentage of cells at G0/G1 phase with a corresponding depletion of cells in S and G2/M phase (Figure 4). The most pronounced effects were seen with 6 and 10 μM MSA, where the fraction of cells in S and G2/M phase decreased by 66 and 63%, respectively. We did not see evidence of apoptosis at any of the doses tested. These results are most consistent with MSA inducing either G1 arrest or causing cells to exit the cell cycle (G0).
Figure 4.
Cell cycle distribution of asynchronous LNCaP cells 24 h after treatment with MSA determined by flow cytometry. The percentage of cells in each phase of the cell cycle represents data from duplicate experiments. The concentration of MSA for each treatment group was shown in the top left corner of each graph.
MSA Modulates Transcript Levels of AR and Androgen-responsive Genes
To our surprise, we found that MSA modulated the expression of AR and a group of well-characterized androgen-regulated genes in a time- and dose-dependent manner. Two clones representing AR showed decreased transcript levels in response to MSA, and 19 known androgen-regulated genes showed altered transcript levels. MSA suppressed expression of 12 androgen-induced genes (KLK3, KLK2, ACPP, NKX3A, TMPRSS2, E2F1, ARSDR1, FKBP5, TUBA2, TUBB2, PPFIA1, and AIBZIP) and increased expression of six of seven genes normally suppressed by androgen (APOD, CLU, PEG3, UGD, NDRG1, and SERPINB5) (Figure 5A). Myc transcript levels, previously shown to be suppressed by androgen, showed a biphasic response to MSA.
Figure 5.
Androgen-responsive genes modulated by MSA. Genes that occur more than once are represented by multiple clones on arrays. (A) MSA-induced expression changes of known androgen-regulated genes. (B) MSA-affected transcripts that are present in a list of androgen-responsive transcripts identified by DePrimo et al. (2002). On the left are gene expression patterns from two separate time courses induced by treatment of LNCaP cells with the synthetic androgen R1881. On the right are expression patterns of this same set of genes after MSA treatment. The red arrowheads point to well-characterized androgen-regulated genes.
We compared our MSA-regulated data set to a recently reported set of 103 androgen-regulated genes (Nelson et al., 2002) and found that 18 of 26 genes found in both data sets showed a reciprocal response to MSA (Table 1). Intriguingly, when compared with a set of 567 androgen-regulated transcripts we had identified previously (DePrimo et al., 2002), 85 of the MSA-regulated transcripts representing 61 genes were found in common, and only one-half of the transcripts were reciprocally regulated (Figure 5B). Therefore, comparison of the MSA expression data set to this larger androgen-regulated data set suggested that MSA has mixed effects on androgen-responsive genes.
Table 1.
Comparison of gene expression changes induced by MSA and androgen reported by Nelson et al. (2002)
| Expression change
|
|||||
|---|---|---|---|---|---|
| Gene symbol
|
Androgen
|
MSA Max fold
|
|||
| Description | 24 hr | 48 hr | Biological process | ||
| CDC14B | Cell division cycle 14 homolog B | 3.0 ↑ | 3.1 ↑ | 2.9 ↓ | Proliferation/differentiation/apoptosis |
| ID2a | Hes6 neuronal differentiation gene ortholog | 1.6 ↑ | 3.8 ↑ | 2.7 ↑ | Proliferation/differentiation/apoptosis |
| NDRG1b | N-myc downstream regulated | 13.7 ↑ | 14.8 ↑ | 3.6 ↓ | Proliferation/differentiation/apoptosis |
| KLK2 | Kallikrein 2, prostatic | 8.8 ↑ | 9.0 ↑ | 6.2 ↓ | Protease/protease Inhibitor |
| KLK3b | Kallikrein 3, prostate specific antigen | 7.9 ↑ | 10.2 ↑ | 9.2 ↓ | Protease/protease Inhibitor |
| TMPRSS2b | Transmembrane protease, serine 2 | 15.5 ↑ | 18.3 ↑ | 3.9 ↓ | Protease/protease Inhibitor |
| GUCY1A3b | Guanylate cyclase 1, soluble, alpha 3 | 2.9 ↑ | 3.3 ↑ | 4.3 ↓ | Signal transduction |
| INPP4B | Inositol polyphosphate-4-phosphatase, type II | 2.3 ↑ | 4.6 ↑ | 2.3 ↓ | Signal transduction |
| PEG3 | Paternally expressed 3 | 3.2 ↓ | 4 ↓ | 2.6 ↑ | Signal transduction |
| FN1b | Fibronectin 1 | 2.5 ↓ | 4.4 ↓ | 4.1 ↑ | Structure/motility/adhesion |
| H1F0 | Histone family, member 0 | 2.9 ↑ | 3.2 ↑ | 7.4 ↓ | Structure/motility/adhesion |
| B4GALT1b | BetaGlcNAc beta 1,4-galactosyltransferase | 3.3 ↑ | 3.3 ↑ | 3.5 ↓ | Metabolism |
| FACL3b | Fatty-acid-Coenzyme A ligase, long-chain 3 | 2.7 ↑ | 3.7 ↑ | 2.4 ↓ | Metabolism |
| SATa | Spermidine/spermine N1-acetyltransferase | 3.7 ↑ | 7.3 ↑ | 2.3 ↑ | Metabolism |
| SCD | Stearoyl-CoA desaturase | 5.9 ↑ | 4.5 ↑ | 5.8 ↓ | Metabolism |
| UGDH | UDP-glucose dehydrogenase | 2.9 ↑ | 4.0 ↑ | 3.8 ↓ | Metabolism |
| KLF4a | Kruppel-like factor 4 | 2.3 ↑ | 3.0 ↑ | 2.4 ↑ | Transcription regulation |
| MYCa | V-myc myelocytomatosis viral oncogene homolog | 2.7 ↓ | 2.8 ↓ | 3.3 ↓ 1.8 ↑ | Transcription regulation |
| NKX3Ab | NK3 transcription factor homolog A (Drosophila) | 14.9 ↑ | 14.1 ↑ | 3.2 ↓ | Transcription regulation |
| ABCC4b | ATP-binding cassette, sub-family C | 5.5 ↑ | 7.8 ↑ | 2.5 ↓ | Transport/trafficking |
| FKBP5b | FK506 binding protein 5 | 24.4 ↑ | 25.4 ↑ | 2.7 ↓ | Transport/trafficking |
| SEC24Da,b | SEC24 related gene family, member D | 3.0 ↑ | 2.6 ↑ | 2.9 ↑ | Transport/trafficking |
| RDC1b | G protein-coupled receptor | 7.8 ↓ | 4.5 ↓ | 2.4 ↑ | Stress response |
| DNAJB9a,b | DnaJ (Hsp40)homolog, subfamily B | 4.0 ↑ | 3.6 ↑ | 4.2 ↑ | Stress response |
| SGKa | Serum/glucocorticoid regulated kinase | 4.4 ↑ | 2.4 ↑ | 2.5 ↑ | Stress response |
| ST7a | Suppression of tumorigenicity 7 | 2.7 ↓ | 4.2 ↓ | 2.5 ↓ | Other functions |
Genes show similar expression changes under the influence of androgen and MSA.
Genes that are also represented in dataset from DePrimo et al. (2002).
MSA Represses AR Protein Expression and the Level of Secreted PSA
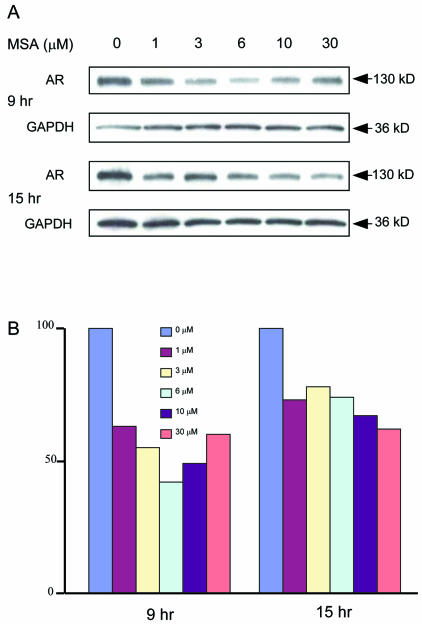
To characterize further the effects of MSA on the androgen axis, we performed Western blotting to compare AR protein levels from treated and untreated LNCaP cells (Figure 6A). The decreased AR transcript levels we observed on the microarrays were associated with decreased AR protein levels at 9 and 15 h after MSA exposure, even at relatively low doses (1 μM). AR protein levels decreased 40-60% after 9 h of MSA exposure, and 30-40% after 15-h exposure. There did not seem to be a significant difference in the degree of AR down-regulation for different MSA concentrations at 15 h; however, 6 μM MSA produced more striking suppression of AR protein levels at 9 h (Figure 6B).
Figure 6.
MSA decreases AR protein expression. (A) AR protein level after 9 and 15 h of exposure to different concentrations of MSA by western blotting analysis. GAPDH from each sample is shown as an internal control. (B) Quantitation of AR protein levels by using a densitometer. The signal intensity of AR was normalized to GAPDH in each same sample. AR intensity of treated cells was normalized against that of the untreated control cells.
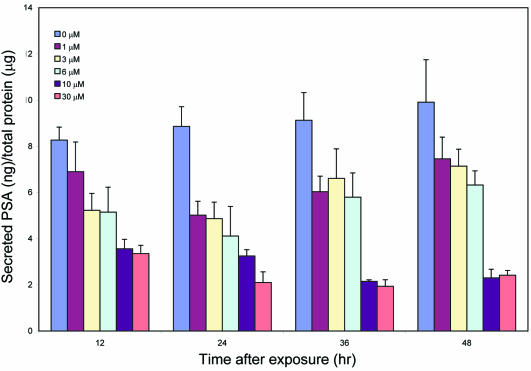
To evaluate further the effects of MSA on androgen-regulated genes, we determined the level of secreted PSA in the cell culture media after exposure of cells to MSA (Figure 7). A dose-dependent decrease in secreted PSA level was detected within 12 h after MSA exposure and continued out to 48 h. Therefore, protein levels of PSA, a well-known androgen target, show modulation similar to that observed for transcript levels using microarray analysis.
Figure 7.
MSA decreases levels of PSA secreted into the media in LNCaP cells. PSA levels in the cell culture medium measured by ELISA and normalized against the total protein of the cultured cells. Each column represents data from experiments performed in triplicate.
MSA Up-Regulates Detoxification Enzymes
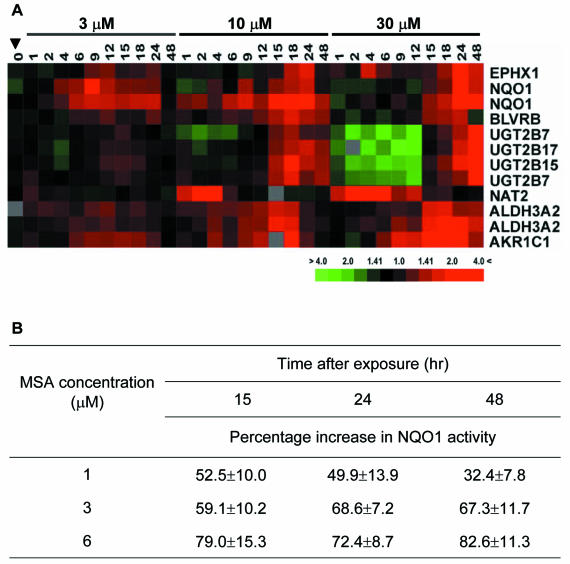
Phase 2 detoxification enzymes function in metabolizing and inactivating xenobiotics and toxins and thereby protect cells against carcinogens. We noted 12 transcripts representing seven genes encoding phase 2 enzymes were up-regulated by MSA (Figure 8A). The mRNA levels of NQO1, a surrogate marker of global phase 2 enzyme activity, were induced by as little as 3 μM MSA. At higher concentrations, several other phase 2 enzymes were induced coordinately with NQO1. We tested whether MSA also increases the enzymatic activity of NQO1 in LNCaP cells by a colorimetric assay involving the mendione-coupled reduction of tetrazolium dye (Brooks et al., 2001b). Treated and control LNCaP cells were harvested at 15, 24, or 48 h after exposed to 1, 3, or 6 μM MSA. The NQO1 activity in each sample was normalized to the total protein of that sample, and the percentage of increase of NQO1 activity compared with control is shown in Figure 8B. NQO1 activity was induced similarly by all three concentrations of MSA and increased over time. Therefore, the increases in NQO1 transcript levels observed in the microarray experiments correlated well with induction of NQO1 enzymatic activity.
Figure 8.
MSA induces expression of several phase 2 enzymes. Genes that occur more than once are represented by multiple clones on arrays. (A) Transcript levels of phase 2 enzymes after treatment with 3, 10, and 30 μM MSA. (B) Percentage increase of NQO1 enzymatic activity after treatment with 1, 3, and 6 μM MSA compared with untreated cells. Results shown represent the average of triplicate experiments.
DISCUSSION
MSA induces striking dose- and time-dependent changes in gene expression in LNCaP cells, suggesting that selenium acts by diverse mechanisms as a putative prostate cancer preventive agent. MSA decreases proliferation of LNCaP cells, possibly by causing cells to exit the cell cycle, alters the expression of many genes in the androgen axis, including AR and many androgen-responsive genes, and induces expression of phase 2 detoxification enzymes, an effect that could be particularly relevant to human prostate cancer chemoprevention. Our findings support the hypothesis that monomethylated selenium may be responsible, at least in part, for the potential anticancer activity of selenium supplements.
Several reports using a variety of model systems have shown that selenium inhibits cell proliferation, and this inhibition is thought to underlie selenium chemoprevention (Ip et al., 2000a; Combs, 2001; Ganther, 2001; Lu, 2001). Decreased proliferation has been attributed to cell cycle arrest, although in prostate cancer cell lines no consistent pattern of arrest has been observed. After treatment with sodium selenite or selenomethionine, growth arrest has been reported in the G1 and G2/M phases of the cell cycle, depending on the prostate cancer cell line in which these compounds were tested (Redman et al., 1998; Menter et al., 2000; Venkateswaran et al., 2002; Bhamre et al., 2003). This lack of consistency may be due to innate differences between the cell lines or to differences in metabolism of the forms of selenium used in these studies. Based on compelling evidence that methylselenol is largely responsible for the chemopreventive activities of selenium compounds, we used MSA in our studies because it can be converted directly into methylselenol in vitro (Ip et al., 2000b). MSA produced a dose-dependent inhibition of cell growth of LNCaP with an accumulation of cells in G0/G1 phase. Similar inhibition of proliferation and accumulation of cells in G0/G1 has been observed in breast cancer and endothelial cells treated with MSA (Sinha et al., 2001; Wang et al., 2001; Dong et al., 2002).
We noted that a striking decrease in expression of many cell cycle-regulated genes from all phases of the cell cycle accompanied growth inhibition in LNCaP cells. Microarray analysis has been used in mammary cancer cells and PC-3 prostate cancer cells, and down-regulation of cell cycle-regulated genes has been observed along with increased expression of CDK inhibitors (Dong et al., 2002, 2003). In these reports, decreased proliferation had been attributed to cell cycle arrest due to modulation of key regulators of the cell cycle, many of which are seen in our data set. Comparison of our data set to genes whose expression varies periodically as HeLa cells pass through the cell cycle provides a broader view of the effects of MSA on the cell cycle. The coordinate, decreased expression of genes involved in all phases of the cell cycle coupled with the increased expression of CDK-inhibitors (CDKN1A, CDKN2D, and CDKN1C) suggest MSA causes LNCaP cells to exit the cell cycle, rather than inducing an arrest at a specific phase in the cell cycle. Whether this is the primary mechanism by which selenium compounds inhibit cell growth awaits further study. Certainly, assessment of the effects of other forms of selenium on the expression of cell cycle genes in prostate cells could provide additional information on the means by which selenium compounds inhibit prostate cancer growth. Ultimately, it will be necessary to evaluate the effects of selenium on prostate cancer growth in vivo, and the cell cycle-regulated genes identified in this and other studies could serve as biomarkers of response.
Perhaps the most striking observation from our microarray experiments is that MSA produced changes in transcript levels of AR and AR-regulated genes. Androgens are critical to prostate carcinogenesis, and androgen deprivation therapy is a mainstay of prostate cancer treatment. MSA suppresses the expression of AR at both mRNA and protein levels, decreases transcript levels of PSA, and decreases PSA protein excretion into the media. A small set of well-characterized androgen-regulated genes, including those with androgen response regulatory elements, show expression changes that are reciprocal to those induced by androgen. Comparison of the MSA data set with a large data set of genes modulated in response to androgens shows that many, but not all, androgen-regulated genes show expression changes opposite to what is seen after treatment with androgens. Some genes were regulated similarly in the two data sets, suggesting that MSA has mixed effects on the transcription of AR-regulated genes. It is possible that genes that are regulated similarly by MSA and androgens are not direct targets of androgen signaling pathways. For instance, androgen treatment of LNCaP cells is known to produce cellular stress by inducing an oxidative burst, and induction of stress response genes has been observed with expression profiling after androgen treatment (Xu et al., 2001; DePrimo et al., 2002). Therefore, the transcripts regulated similarly by androgens and MSA (DNAJB9, ATF3, and VEGF) might reflect cellular stress or other pathways that have been activated secondarily.
Effects of selenium on AR and AR-regulated genes in prostate cancer cell lines have not been observed with other selenium compounds; in fact, two reports have shown that selenomethionine does not have an effect on AR function or PSA secretion in LNCaP cells (Zhang et al., 2002; Bhamre et al., 2003). One possible explanation for the lack of effect of selenomethionine on androgen-regulated genes is its poor conversion to methylselenol in vitro. Intriguingly, men supplemented with selenized yeast do show small but significant decreases in their serum PSA levels compared with control subjects, suggesting the possibility that selenium compounds can affect AR-regulated genes in vivo where they can be metabolized to methylselenol (El-Bayoumy et al., 2002). In addition, effects of MSA on AR-regulated genes in PC-3 cells were not observed by Dong et al. (2002, 2003), suggesting that MSA may affect transcription of AR-regulated genes through AR.
It is tempting to speculate that MSA blocks proliferation in prostate cells through its effects on AR and AR-regulated genes. Consistent with our findings, Venkateswaran et al. (200) observed that selenomethionine did not affect the growth of wild-type (AR-null) PC-3 prostate cancer cell lines, but did inhibit growth of PC-3 cells stably expressing AR. However, three other groups have observed growth inhibition by selenium compounds in prostate cancer cell lines that do not express AR (Redman et al., 1998; Menter et al., 2000; Dong et al., 2003). Additional work will be necessary to understand the role of MSA on androgen signaling pathways and cell growth.
Our studies suggest that enhancement of detoxification is another mechanism that underlies the chemopreventive effects of MSA. MSA up-regulates mRNA levels of several phase 2 enzymes, including EPHX1, NQO1, NAT2, and members of the UGTB family, as well as the enzymatic activity of NQO1. We have observed similar induction of NQO1 enzymatic activity in LNCaP cells treated with sodium selenite and selenium dioxide (Brooks et al., 2002), demonstrating that several forms of selenium are capable of inducing phase 2 enzymatic activity in prostate cells. Induction of phase 2 enzymatic activity has been proposed as a promising avenue of prostate cancer prevention after the discovery that virtually all human prostate cancers and precursor lesions (PIN) lose expression of the phase 2 enzyme glutathione S-transferase π (GSTP1) (DePrimo et al., 2001; Nelson et al., 2001). Global induction of phase 2 enzymes by selenium compounds might compensate for the loss of GSTP1 expression that occurs early in prostate carcinogenesis thereby and protect vulnerable prostatic epithelial cells against genome damage.
In summary, we have characterized the global transcriptional response program of LNCaP to MSA. The expression changes we observed imply that MSA exerts its anticancer activity through diverse mechanisms, including inhibition of cell proliferation, modulation of the expression of AR and its regulated genes, and induction of enzymes involved in carcinogen detoxification. Therefore, this data set provides a potential resource for understanding the modes of action of MSA and serves as a source for candidate biomarkers of selenium's effects that could be measured in vivo. Discovery of such markers could help in the design and interpretation of selenium intervention trials currently in progress.
Acknowledgments
We thank Dr. Zijie Sun for generously providing LNCaP cells, Dr. John Higgins for critical review of this manuscript, the Stanford Functional Genomics Facility for production of the high-quality microarrays used in this study, and the Stanford Microarray Database group for support on data storage and analysis. This work was supported by the Department of Defense (DAMD17-98-1-8555), the Doris Duke Foundation (T98064) and the Oxnard Foundation to J.D.B. M.L.W. is supported by a National Research Service Award Postdoctoral Fellowship from the National Human Genome Research Institute (HG00220) and by a grant from the Scleroderma Research Foundation.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-07-0501. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-07-0501.
Abbreviations used: AR, androgen receptor; CFSE, carboxyfluoroscein succinimidyl ester; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MSA, methylseleninic acid; MSC, methylselenocysteine; NQO1, NADPH dehydrogenase, quinone 1; PSA, prostate-specific antigen.
References
- Andreadou, I., Menge, W.M., Commandeur, J.N., Worthington, E.A., and Vermeulen, N.P. (1996). Synthesis of novel Se-substituted selenocysteine derivatives as potential kidney selective prodrugs of biologically active selenol compounds: evaluation of kinetics of beta-elimination reactions in rat renal cytosol. J. Med. Chem. 39, 2040-2046. [DOI] [PubMed] [Google Scholar]
- Bhamre, S., Whitin, J.C., and Cohen, H.J. (2003). Selenomethionine does not affect PSA secretion independent of its effect on LNCaP cell growth. Prostate 54, 315-321. [DOI] [PubMed] [Google Scholar]
- Blader, I.J., Manger, I.D., and Boothroyd, J.C. (2001). Microarray analysis reveals previously unknown changes in Toxoplasma gondii-infected human cells. J. Biol. Chem. 276, 24223-24231. [DOI] [PubMed] [Google Scholar]
- Brooks, J.D., Goldberg, M.F., Nelson, L.A., Wu, D., and Nelson, W.G. (2002). Identification of potential prostate cancer preventive agents through induction of quinone reductase in vitro. Cancer Epidemiol. Biomarkers Prev. 11, 868-875. [PubMed] [Google Scholar]
- Brooks, J.D., Metter, E.J., Chan, D.W., Sokoll, L.J., Landis, P., Nelson, W.G., Muller, D., Andres, R., and Carter, H.B. (2001a). Plasma selenium level before diagnosis and the risk of prostate cancer development. J. Urol. 166, 2034-2038. [PubMed] [Google Scholar]
- Brooks, J.D., Paton, V.G., and Vidanes, G. (2001b). Potent induction of phase 2 enzymes in human prostate cells by sulforaphane. Cancer Epidemiol. Biomarkers Prev. 10, 949-954. [PubMed] [Google Scholar]
- Clark, L.C., et al. (1996). Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin. A randomized controlled trial. Nutritional Prevention of Cancer Study Group. J. Am. Med. Assoc. 276, 1957-1963. [PubMed] [Google Scholar]
- Clark, L.C., et al. (1998). Decreased incidence of prostate cancer with selenium supplementation: results of a double-blind cancer prevention trial. Br. J. Urol. 81, 730-734. [DOI] [PubMed] [Google Scholar]
- Combs, G.F., Jr. (2001). Considering the mechanisms of cancer prevention by selenium. Adv. Exp. Med. Biol. 492, 107-117. [DOI] [PubMed] [Google Scholar]
- Davis, C.D., Zeng, H., and Finley, J.W. (2002). Selenium-enriched broccoli decreases intestinal tumorigenesis in multiple intestinal neoplasia mice. J. Nutr. 132, 307-309. [DOI] [PubMed] [Google Scholar]
- DePrimo, S.E., Diehn, M., Nelson, J.B., Reiter, R.E., Matese, J., Fero, M., Tibshirani, R., Brown, P.O., and Brooks, J.D. (2002). Transcriptional programs activated by exposure of human prostate cancer cells to androgen. Genome Biol. 3, RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePrimo, S.E., Shinghal, R., Vidanes, G., and Brooks, J.D. (2001). Prevention of prostate cancer. Hematol. Oncol. Clin. North Am. 15, 445-457. [DOI] [PubMed] [Google Scholar]
- Dong, Y., Ganther, H.E., Stewart, C., and Ip, C. (2002). Identification of molecular targets associated with selenium-induced growth inhibition in human breast cells using cDNA microarrays. Cancer Res. 62, 708-714. [PubMed] [Google Scholar]
- Dong, Y., Zhang, H., Hawthorn, L., Ganther, H.E., and Ip, C. (2003). Delineation of the molecular basis for selenium-induced growth arrest in human prostate cancer cells by oligonucleotide array. Cancer Res. 63, 52-59. [PubMed] [Google Scholar]
- Duffield-Lillico, A.J., Reid, M.E., Turnbull, B.W., Combs, G.F., Jr., Slate, E.H., Fischbach, L.A., Marshall, J.R., and Clark, L.C. (2002). Baseline characteristics and the effect of selenium supplementation on cancer incidence in a randomized clinical trial: a summary report of the Nutritional Prevention of Cancer Trial. Cancer Epidemiol. Biomarkers Prev. 11, 630-639. [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Bayoumy, K. (1994). Evaluation of chemopreventive agents against breast cancer and proposed strategies for future clinical intervention trials. Carcinogenesis 15, 2395-2420. [DOI] [PubMed] [Google Scholar]
- El-Bayoumy, K. (2001). The protective role of selenium on genetic damage and on cancer. Mutat. Res. 475, 123-139. [DOI] [PubMed] [Google Scholar]
- El-Bayoumy, K., Richie, J.P., Jr., Boyiri, T., Komninou, D., Prokopczyk, B., Trushin, N., Kleinman, W., Cox, J., Pittman, B., and Colosimo, S. (2002). Influence of selenium-enriched yeast supplementation on biomarkers of oxidative damage and hormone status in healthy adult males: a clinical pilot study. Cancer Epidemiol. Biomarkers Prev. 11, 1459-1465. [PubMed] [Google Scholar]
- Fleming, J., Ghose, A., and Harrison, P.R. (2001). Molecular mechanisms of cancer prevention by selenium compounds. Nutr. Cancer 40, 42-49. [DOI] [PubMed] [Google Scholar]
- Ganther, H.E. (2001). Selenium metabolism and mechanisms of cancer prevention. Adv. Exp. Med. Biol. 492, 119-130. [DOI] [PubMed] [Google Scholar]
- Ganther, H.E., and Lawrence, J.R. (1997). Chemical transformations of selenium in living organisms. Improved forms of selenium for cancer prevention. Tetrahedron 53, 12299-112310. [Google Scholar]
- Gitig, D.M., and Koff, A. (2000). Cdk pathway: cyclin-dependent kinases and cyclin-dependent kinase inhibitors. Methods Mol. Biol. 142, 109-123. [DOI] [PubMed] [Google Scholar]
- Groszer, M., Erickson, R., Scripture-Adams, D.D., Lesche, R., Trumpp, A., Zack, J.A., Kornblum, H.I., Liu, X., and Wu, H. (2001). Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science 294, 2116-2118. [DOI] [PubMed] [Google Scholar]
- Helzlsouer, K.J., Huang, H.Y., Alberg, A.J., Hoffman, S., Burke, A., Norkus, E.P., Morris, J.S., and Comstock, G.W. (2000). Association between alpha-tocopherol, gamma-tocopherol, selenium, and subsequent prostate cancer. J. Natl. Cancer Inst. 92, 2018-2023. [DOI] [PubMed] [Google Scholar]
- Hoque, A., et al. (2001). Molecular epidemiologic studies within the Selenium and Vitamin E Cancer Prevention Trial (SELECT). Cancer Causes Control 12, 627-633. [DOI] [PubMed] [Google Scholar]
- Ip, C. (1998). Lessons from basic research in selenium and cancer prevention. J. Nutr. 128, 1845-1854. [DOI] [PubMed] [Google Scholar]
- Ip, C., Hayes, C., Budnick, R.M., and Ganther, H.E. (1991). Chemical form of selenium, critical metabolites, and cancer prevention. Cancer Res. 51, 595-600. [PubMed] [Google Scholar]
- Ip, C., Thompson, H.J., and Ganther, H.E. (2000a). Selenium modulation of cell proliferation and cell cycle biomarkers in normal and premalignant cells of the rat mammary gland. Cancer Epidemiol. Biomarkers Prev. 9, 49-54. [PubMed] [Google Scholar]
- Ip, C., Thompson, H.J., Zhu, Z., and Ganther, H.E. (2000b). In vitro and in vivo studies of methylseleninic acid: evidence that a monomethylated selenium metabolite is critical for cancer chemoprevention. Cancer Res. 60, 2882-2886. [PubMed] [Google Scholar]
- Ip, C., and White, G. (1987). Mammary cancer chemoprevention by inorganic and organic selenium: single agent treatment or in combination with vitamin E and their effects on in vitro immune functions. Carcinogenesis 8, 1763-1766. [DOI] [PubMed] [Google Scholar]
- Jiang, C., Wang, Z., Ganther, H., and Lu, J. (2001). Caspases as key executors of methyl selenium-induced apoptosis (anoikis) of DU-145 prostate cancer cells. Cancer Res. 61, 3062-3070. [PubMed] [Google Scholar]
- Jones, J.O., and Arvin, A.M. (2003). Microarray analysis of host cell gene transcription in response to Varicella-Zoster virus infection of human t cells and fibroblasts in vitro and scidhu skin xenografts in vivo. J. Virol. 77, 1268-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.S., and Milner, J. (2001). Molecular targets for selenium in cancer prevention. Nutr. Cancer 40, 50-54. [DOI] [PubMed] [Google Scholar]
- Klein, E.A., Thompson, I.M., Lippman, S.M., Goodman, P.J., Albanes, D., Taylor, P.R., and Coltman, C. (2001). SELECT: the next prostate cancer prevention trial. Selenum and Vitamin E Cancer Prevention Trial. J. Urol. 166, 1311-1315. [DOI] [PubMed] [Google Scholar]
- Lu, J. (2001). Apoptosis and angiogenesis in cancer prevention by selenium. Adv. Exp. Med. Biol. 492, 131-145. [DOI] [PubMed] [Google Scholar]
- Lu, J., and Jiang, C. (2001). Antiangiogenic activity of selenium in cancer chemoprevention: metabolite-specific effects. Nutr. Cancer 40, 64-73. [DOI] [PubMed] [Google Scholar]
- Lyons, A.B. (2000). Analysing cell division in vivo and in vitro using flow cytometric measurement of CFSE dye dilution. J. Immunol. Methods 243, 147-154. [DOI] [PubMed] [Google Scholar]
- Medina, D., Thompson, H., Ganther, H., and Ip, C. (2001). Se-methylselenocysteine: a new compound for chemoprevention of breast cancer. Nutr. Cancer 40, 12-17. [DOI] [PubMed] [Google Scholar]
- Menter, D.G., Sabichi, A.L., and Lippman, S.M. (2000). Selenium effects on prostate cell growth. Cancer Epidemiol. Biomarkers Prev. 9, 1171-1182. [PubMed] [Google Scholar]
- Nelson, P.S., Clegg, N., Arnold, H., Ferguson, C., Bonham, M., White, J., Hood, L., and Lin, B. (2002). The program of androgen-responsive genes in neoplastic prostate epithelium. Proc. Natl. Acad. Sci. USA 99, 11890-11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, W.G., De Marzo, A.M., Deweese, T.L., Lin, X., Brooks, J.D., Putzi, M.J., Nelson, C.P., Groopman, J.D., and Kensler, T.W. (2001). Preneoplastic prostate lesions: an opportunity for prostate cancer prevention. Ann. N.Y. Acad. Sci. 952, 135-144. [DOI] [PubMed] [Google Scholar]
- Nomura, A.M., Lee, J., Stemmermann, G.N., and Combs, G.F., Jr. (2000). Serum selenium and subsequent risk of prostate cancer. Cancer Epidemiol. Biomarkers Prev. 9, 883-887. [PubMed] [Google Scholar]
- Rao, C.V., Wang, C.Q., Simi, B., Rodriguez, J.G., Cooma, I., El-Bayoumy, K., and Reddy, B.S. (2001). Chemoprevention of colon cancer by a glutathione conjugate of 1,4-phenylenebis(methylene)selenocyanate, a novel organoselenium compound with low toxicity. Cancer Res. 61, 3647-3652. [PubMed] [Google Scholar]
- Reddy, B.S., Upadhyaya, P., Simi, B., and Rao, C.V. (1994). Evaluation of organoselenium compounds for potential chemopreventive properties in colon carcinogenesis. Anticancer Res. 14, 2509-2514. [PubMed] [Google Scholar]
- Redman, C., Scott, J.A., Baines, A.T., Basye, J.L., Clark, L.C., Calley, C., Roe, D., Payne, C.M., and Nelson, M.A. (1998). Inhibitory effect of selenomethionine on the growth of three selected human tumor cell lines. Cancer Lett. 125, 103-110. [DOI] [PubMed] [Google Scholar]
- Sherlock, G., et al. (2001). The Stanford microarray database. Nucleic Acids Res. 29, 152-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr, C.J., and Roberts, J.M. (1999). CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13, 1501-1512. [DOI] [PubMed] [Google Scholar]
- Sinha, R., Unni, E., Ganther, H.E., and Medina, D. (2001). Methylseleninic acid, a potent growth inhibitor of synchronized mouse mammary epithelial tumor cells in vitro. Biochem. Pharmacol. 61, 311-317. [DOI] [PubMed] [Google Scholar]
- Venkateswaran, V., Klotz, L.H., and Fleshner, N.E. (2002). Selenium modulation of cell proliferation and cell cycle biomarkers in human prostate carcinoma cell lines. Cancer Res. 62, 2540-2545. [PubMed] [Google Scholar]
- Wang, Z., Jiang, C., Ganther, H., and Lu, J. (2001). Antimitogenic and proapoptotic activities of methylseleninic acid in vascular endothelial cells and associated effects on PI3K-AKT, ERK, JNK and p38 MAPK signaling. Cancer Res. 61, 7171-7178. [PubMed] [Google Scholar]
- Whitfield, M.L., et al. (2002). Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 13, 1977-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett, W.C., Polk, B.F., Morris, J.S., Stampfer, M.J., Pressel, S., Rosner, B., Taylor, J.O., Schneider, K., and Hames, C.G. (1983). Prediagnostic serum selenium and risk of cancer. Lancet 2, 130-134. [DOI] [PubMed] [Google Scholar]
- Williams, E.D., and Brooks, J.D. (2001). New molecular approaches for identifying novel targets, mechanisms, and biomarkers for prostate cancer chemopreventive agents. Urology 57, 100-102. [DOI] [PubMed] [Google Scholar]
- Xu, L.L., Su, Y.P., Labiche, R., Segawa, T., Shanmugam, N., McLeod, D.G., Moul, J.W., and Srivastava, S. (2001). Quantitative expression profile of androgen-regulated genes in prostate cancer cells and identification of prostate-specific genes. Int. J. Cancer 92, 322-328. [DOI] [PubMed] [Google Scholar]
- Yoshizawa, K., Willett, W.C., Morris, S.J., Stampfer, M.J., Spiegelman, D., Rimm, E.B., and Giovannucci, E. (1998). Study of prediagnostic selenium level in toenails and the risk of advanced prostate cancer. J. Natl. Cancer Inst. 90, 1219-1224. [DOI] [PubMed] [Google Scholar]
- Youn, B.W., Fiala, E.S., and Sohn, O.S. (2001). Mechanisms of organoselenium compounds in chemoprevention: effects on transcription factor-DNA binding. Nutr. Cancer 40, 28-33. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Ni, J., Messing, E.M., Chang, E., Yang, C.R., and Yeh, S. (2002). Vitamin E succinate inhibits the function of androgen receptor and the expression of prostate-specific antigen in prostate cancer cells. Proc. Natl. Acad. Sci. USA 99, 7408-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, H., Hastie, T., Whitfield, M.L., Borresen-Dale, A.L., and Jeffrey, S.S. (2002). Optimization and evaluation of T7 based RNA linear amplification protocols for cDNA microarray analysis. BMC Genomics 3, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Z., Jiang, W., Ganther, H.E., and Thompson, H.J. (2002). Mechanisms of cell cycle arrest by methylseleninic acid. Cancer Res. 62, 156-164. [PubMed] [Google Scholar]