Abstract
Aim
Genetic polymorphisms have the potential to influence drug metabolism and vary among ethnic groups. This study evaluated the correlation of genetic polymorphisms with nevirapine pharmacokinetics exposure in Malawians.
Materials & methods
CYP450 2B6, 2D6, 3A4 and 3A5, ABCB1 and constitutive androstane receptor and pregnane X receptor, were analyzed for polymorphisms in 26 subjects.
Results
Allele frequencies (variant) were: CYP2B6 514G>T (0.31) CYP2D6*4 (0.02); CYP2D6*17 (0.35); CYP3A4*1B (0.77); CYP3A5*3 (0.25); ABCB1 2677G>T (0.0), ABCB1 3435C>T (0.21), NR1I3 13711152T>C (0.02), NR1I2 44477T>C (0.10), NR1I2 63396C>T (0.33), NR1I2 6-bp indel (del: 0.17). CYP2B6 516G>T (non-wild-type/wild-type) correlated with nevirapine pharmacokinetic parameters; geometric mean ratios (95% CI): 1.75 (1.27–2.40) for area under the concentration time curve (AUC)0–12 h, 1.58 (1.03–2.42) for C0, and 0.53 (0.31–0.91) for clearance. In a multivariable model, nevirapine AUC increased by 1.5% per year of age (p < 0.0001), CYP2B6 516 T allele increased AUC by 92% (p < 0.0001), and CYP3A5*3 decreased AUC by 31% (p = 0.0027).
Conclusion
Allele frequencies were similar to other sub-Saharan African populations. The T allele for CYP2B6 516 was significantly associated with nevirapine exposure.
Keywords: CYP2B6, CYP450, Malawi, nevirapine, nuclear receptor, P-glycoprotein, pharmacokinetics
Genetic polymorphisms have been identified in genes that encode drug-metabolizing enzymes and transporters such as the CYP450s (CYPs; CYPs) and P-glycoproteins (MDR1; ABCB1) [1]. These polymorphisms may explain interindividual differences in drug pharmacokinetics [1]. Patients with genotypes that produce less active enzymes in major drug metabolic pathways have reduced metabolism and therefore higher drug exposures and potentially more adverse effects. Patients with activity-enhancing genotypes clear drugs faster, which can lead to subtherapeutic concentrations and increased risk of therapy failure [1]. Specifically, CYP polymorphisms have been associated with interpatient variation in antiretroviral plasma concentrations, and the allele frequencies vary among ethnic groups [2,3].
HIV-infected patients on antiretroviral therapy have demonstrated altered plasma concentrations due to differing CYP450 enzyme genotypes [2,4,5]. These genotypes have also shown differing frequencies based on ethnicity [6]. A Ugandan population has been shown to have significantly higher drug exposure compared to western populations; this can partially be explained by different frequencies of genetic polymorphisms in drug-metabolizing enzymes and transporters [7].
Antiviral therapy is expanding in developing countries using doses optimized for western populations. We recently found that steady-state nevirapine concentrations were significantly increased in a group of Malawian adults and children compared to concentrations seen in a mostly western Caucasian cohort of adults and children [8–11]. The 1.6- to 2.2-fold increase in drug exposure could not be explained by weight, age or other observable patient-specific factors. Nevirapine is non-nucleoside reverse transcriptase inhibitor used for the treatment of HIV infections in adults and children. It is metabolized by the CYP450 system, specifically CYP3A4/5, CYP2D6 and CYP2B6 [12], and data suggest that polymorphisms in the genes encoding these enzymes can alter nevirapine concentrations [2,7,13–18]. The clinical impact of the CYP2B6 polymorphism, 516G>T, has been described in various settings. The use of single-dose nevirapine to prevent perinatal vertical transmission has been shown to have prolonged exposures in women with the CYP2B6 516 T allele, which greatly increases the risk of developing non-nucleoside reverse transcriptase inhibitor resistance mutations [18].
Nevirapine autoinduces its metabolism by activating CYP3A4/5 and CYP2B6 expression, and data show that nevirapine may do this through the activation of the nuclear receptor, constitutive androstane receptor (CAR; NR1I3) [19]. The nuclear receptor pregnane X receptor (PXR; NR1I2) regulates basal expression of CYP3A4/5 [20]. Polymorphisms in the genes coding for these nuclear receptors have been shown to influence the expression of the CYPs and therefore influence nevirapine exposure [19–21].
Frequencies of polymorphisms in CYP3A4/5, CYP2B6, ABCB1, PXR and CAR can vary between ethnic groups [22], which might contribute to the difference in drug exposure we previously observed between Malawians and western subjects [2,8,9]. However, the frequencies of the polymorphisms in the abovementioned genes have not been reported in a Malawian population.
Materials & methods
Study design
This was a nonrandomized pharmacogenetic observational study in HIV-infected Malawian children and adults who participated in pharmacokinetic studies evaluating nevirapine [8,9]. The University of North Carolina Institutional Review Board and the Malawi National Health Sciences Research Committee approved the study protocol. All research subjects provided consent or assent with guardian consent. The objective was to determine the frequencies of CYP, ABCB1 and nuclear receptor polymorphisms in 30 Malawian adults and children and to explore correlations between their genotypes and nevirapine (Virammune®) exposure.
Samples & DNA extraction
Nevirapine concentrations were measured in blood plasma by a validated HPLC-UV method as previously described [8,9,23].
DNA was isolated from whole blood using the QIAamp® DNA Mini kit (Qiagen Sciences, MD, USA). Genotyping was performed using PCR and Pyrosequencing® as previously described [24]. Primers and PCR conditions for ABCB1 2677 (rs2032582) and 3435 (rs1045642), CYP2B6 516G>T (Q172H; rs3745274), CYP2D6*4 (rs3892097), CYP3A4*1B (rs2740574), and CYP3A5*3 (rs776746) were previously described [24–27]. For CYP2D6*17 (rs28371706), DNA was amplified using the forward primer: 5′-GAG GCG CTG GTG ACC CAC-3′ and biotinylated reverse primers: 5′-biotin-CTG TCC CCA CCG CTG CTT-3′ at an annealing temperature of 65°C. The internal primer was 5′-CGC CTG TGC CCA TCA-3′ and the sequence to analyze was C/TCCAGATCC.
NR1I2 44477T>C (rs1523130), 63396C>T (rs2472677) and the 6-bp deletion (6-bp indel; rs3842689) were genotyped by previously described methods [20]. The trans NR1I3 polymorphism, rs10494390 (NT_004487.18:g.13711152T>C), was amplified from 50 ng of genomic DNA by the forward 5′-ACCTTCGGGGTTGAATTTT-3′ and reverse 5′-TGTAGCATTTGGGTTTTGGA-3′ primers.
Data analysis/statistics
Genotype data from the Malawi sample was compared to genotype data of Yoruban (sub-Saharan African), African–American and Caucasians of European descent obtained from the HapMap database. Fisher’s Exact Test was used to determine whether the Malawian sample’s genotype frequencies differed significantly from the archived data. The Cochran–Armitage Trend Test was employed to determine if a genotype trend could be detected between groups.
The pharmacokinetic parameters were generated by noncompartmental analysis using WinNonlin (v5.2, Pharsight, Inc., NC, USA). Area under the concentration time curve (AUC) 0–12 h was extrapolated from partial dosing interval data (adults: 0–8 h, children: 0–6 h). Weight-adjusted (kg) apparent oral clearance (CL/F) was determined to confirm AUC results of the pediatric subjects. Trough concentrations (C0) are predose concentrations. Statistical analysis was performed using SAS (v9.1.3, SAS Institute, NC, USA). Geometric mean ratios (GMRs) of non-wild-type (WT) versus WT genotypes were calculated using linear regression of natural log transformed pharmacokinetic parameters with α significance levels of 5% and 0.625%; the latter to account for multiple statistical tests for the eight genotypes tested. Multiple linear regression was performed to develop a multiple variable model (α = 0.05) with AUC as outcome and genotypes and demographics as covariates. WT genotypes are defined as genotypes that are homozygous for the WT allele, and non-WT genotypes are defined as genotypes that are either heterozygous or homozygous for the variant (VAR) allele. WT alleles are defined by having functional enzyme gene products, often designated as *1 for the CYPs, or having the highest allele frequency in this study sample.
Results
DNA was isolated from 24 Malawian subjects, of whom 11 were adults with a median (range) age of 40 (26–46) years, weight of 72 (60–80) kg, BMI of 25.4 (19.8–32.5) kg/m2 and five were male. Thirteen were children with median (range) age of 6.7 (1.3–13.6) years, weight of 18 (9–30.5) kg, BMI z-score of 0.31 (−5.39–1.28) and six were male [8,9]. Subjects were also taking trade formulations of lamivudine (Epivir®) and stavudine (Zerit®) at the time of pharmacokinetic sampling. Nevirapine was dosed using standard dosing in adults at 200 mg twice daily. The pediatric group was dosed using standard weight-based dosing, in which children up to 8 years of age are dosed 7 mg/kg twice daily and children 8 years of age and older are dose 4 mg/kg twice daily.
Genotype frequencies of 26 Malawians are presented in Table 1; genetic sampling included two subjects without pharmacokinetic data. The genotypes could not be reliably tested for Hardy–Weinberg equilibrium due to a small sample size. The Malawian genotype frequencies did not differ from Yorubans. The Malawian’s CYP2D6*17 and ABCB1 2677G>T genotype data differed from that of the African–American individuals; other genotypes tested did not differ. The Malawian genotype frequencies differed from Caucasians in regards to all genotypes tested except for CYP2B6 516G>T (p = 0.31). The VAR allele frequencies of CYP3A4*1B and CYP2D6*17 were higher and CYP3A5*3, CYP2D6*4, ABCB1 2677G>T, ABCB1 3435C>T, NR1I3 13711152T>C, NR1I2 44477T>C, 63396C>T and 6-bp indel were lower in the Malawians than Caucasians.
Table 1.
Genotype frequencies for polymorphisms in CYPs, ABCB1 and nuclear receptors in Malawian subjects in comparison to sub-Saharan African, African–American and Caucasian populations using Fisher’s Exact Test and Cochran–Armitage Trend Test.
| Allele | Allele SNP
|
Genotype frequency
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| WT | VAR | Malawian† | YRI: sub-Saharan | ASW: African–American | CEU: Caucasian | ||||
| CYP2B6 516 | G | T | GG: 0.54 | GG: 0.34 | (n = 146) | GG: 0.47 | (n = 57) | GG: 0.53 | (n = 113) |
| rs3745274 | GT: 0.31 | GT: 0.49 | F: p = 0.14 | GT: 0.46 | F: p = 0.30 | GT: 0.4 | F: p = 0.31 | ||
| TT: 0.15 | TT: 0.18 | TT: 0.07 | TT: 0.07 | ||||||
|
| |||||||||
| CYP2D6*4 | G | A | GG: 0.96 | GG: 0.93 | (n = 118)‡ | GG: 0.82 | (n = 144)§ | GG: 0.58 | (n = 118)‡ |
| rs3892097 | GA: 0.04 | GA: 0.7 | F: p = 1 | GA: 0.17 | F: p = 0.17 | GA: 0.37 | F: p < 0.001 | ||
| AA: 0.0 | AA: 0.0 | AA: 0.01 | AA: 0.05 | T: p < 0.001 | |||||
|
| |||||||||
| CYP2D6*17 | C | T | CC: 0.38 | CC: 0.61 | (n = 118)‡ | CC: 0.75 | (n = 118)§ | CC: 1.0 | (n = 120)‡ |
| rs28371706 | CT: 0.54 | CT: 0.31 | F: p = 0.07 | CT: 0.2 | F: p < 0.001 | CT: 0.0 | F: p < 0.001 | ||
| TT: 0.08 | TT: 0.08 | TT: 0.04 | T: p = 0.002 | TT: 0.0 | T: p < 0.001 | ||||
|
| |||||||||
| CYP3A4*1B | A | G | AA: 0.04 | AA: 0.03 | (n = 118)‡ | NR | AA: 0.95 | (n = 120)‡ | |
| rs2740574 | AG: 0.38 | AG: 0.44 | F: p = 0.86 | AG: 0.05 | F: p < 0.001 | ||||
| GG: 0.58 | GG: 0.53 | GG: 0.0 | T: p < 0.001 | ||||||
|
| |||||||||
| CYP3A5*3 | A | G | AA: 0.5 | AA: 0.68 | (n = 147) | AA: 0.37 | (n = 57) | AA: 0.0 | (n = 111) |
| rs776746 | AG: 0.5 | AG: 0.31 | F: p = 0.22 | AG: 0.54 | F: p = 0.24 | AG: 0.07 | F: p < 0.001 | ||
| GG: 0.0 | GG: 0.01 | GG: 0.09 | GG: 0.93 | T: p < 0.001 | |||||
|
| |||||||||
| ABCB1 2677 | G | T | GG: 1.0 | GG: 1.0 | (n = 120)‡ | GG: 0.81 | (n = 57) | GG: 0.26 | (n = 113) |
| rs2032582 | GT: 0.0 | GT: 0.0 | F: NS | GT: 0.18 | F: p = 0.04 | GT: 0.55 | F: p < 0.001 | ||
| TT: 0.0 | TT: 0.0 | TT: 0.02 | T: p = 0.02 | TT: 0.19 | T: p < 0.001 | ||||
|
| |||||||||
| ABCB1 3435 | C | T | CC: 0.62 | CC: 0.8 | (n = 145) | CC: 0.63 | (n = 56) | CC: 0.15 | (n = 113) |
| rs1045642 | CT: 0.35 | CT: 0.19 | F: p = 0.09 | CT: 0.34 | F: p = 1 | CT: 0.56 | F: p < 0.001 | ||
| TT: 0.04 | TT: 0.01 | TT: 0.04 | TT: 0.29 | T: p < 0.001 | |||||
|
| |||||||||
| NR1I3 13711152 | T | C | TT 0.96 | TT: 0.92 | (n = 147) | TT: 0.81 | (n = 57) | TT: 0.65 | (n = 113) |
| rs10494390 | TC 0.04 | TC: 0.07 | F: p = 0.74 | TC: 0.19 | F: p = 0.09 | TC: 0.33 | F: p = 0.002 | ||
| CC 0.0 | CC: 0.01 | CC: 0 | CC: 0.03 | T: p = 0.002 | |||||
|
| |||||||||
| NR1I2 44477 | T | C | TT: 0.8¶ | TT: 0.85 | (n = 147) | TT: 0.75 | (n = 56) | TT: 0.13 | (n = 112) |
| rs1523130 | TC: 0.2¶ | TC: 0.14 | F: p = 0.61 | TC: 0.25 | F: p = 0.77 | TC: 0.46 | F: p < 0.001 | ||
| CC: 0.0¶ | CC: 0.01 | CC: 0.0 | CC: 0.42 | T: p < 0.001 | |||||
|
| |||||||||
| NR1I2 63396 | C | T | CC: 0.5 | CC: 0.34 | (n = 62) | NR | CC: 0.14 | (n = 64) | |
| rs2472677 | CT: 0.35 | CT: 0.6 | F: p = 0.07 | CT: 0.44 | F: p = 0.001 | ||||
| TT: 0.15 | TT: 0.06 | TT: 0.42 | T: p < 0.001 | ||||||
|
| |||||||||
| NR1I2 6-bp indel | GAGAAG | – | +/+ 0.69 | NR | NR | NR | |||
| rs3842689 | +/− 0.27 | ||||||||
| −/− 0.04 | |||||||||
WT and VAR nucleotides listed for reference. Comparison population data from International HapMap Project [12], unless otherwise indicated.
Genotype data from n = 26 available.
Genotype data from n = 20 available.
ASW: African ancestry in southwest USA; CEU: Utah residents with northern and western European ancestry from the CEPH collection; F: Fisher’s Exact Test; NR: Not reported; NS: Nonsignificant; T: Cochran–Armitage Trend Test; VAR: Variant allele; WT: Wild-type allele; YRI: Yoruba in Ibadan, Nigeria.
The results of the correlation analysis are listed in Table 2. The pharmacokinetic data were log normally distributed. With α = 0.05, the CYP2B6 516 T allele was significantly associated with increased AUC, C0 and CL/F. With α = 0.00625 for multiple comparison correction, this significance was lost . The NR1I2 6-bp indel deletion allele was significantly associated with decreased AUC0–12 h and increased CL/F with α = 0.05; however, this lost significance with α = 0.00625.
Table 2.
Pharmacokinetic parameter geometric mean ratios (non-wild-type or variant versus wild-type).
| Polymorphism | NVP AUC0–12 h | NVP AUC0–6 h† | NVP C0 | CL/F‡ |
|---|---|---|---|---|
| CYP2B6 516G>T | 1.75* | 1.71* | 1.58** | 0.53** |
| rs3745274 | (1.27–2.40) | (1.25–2.34) | (1.03–2.42) | (0.31–0.91) |
|
| ||||
| CYP3A4*1B§ | 0.86 | 0.87 | 0.87 | 0.96 |
| rs2740574 | (0.59–1.27) | (0.60–1.27) | (0.55–1.39) | (0.53–1.74) |
|
| ||||
| CYP3A5*3 | 0.91 | 0.92 | 0.93 | 1.03 |
| rs776746 | (0.62–1.33) | (0.63–1.35) | (0.59–1.48) | (0.57–1.86) |
|
| ||||
| CYP2D6*17 | 1.13 | 1.11 | 1.06 | 0.94 |
| rs28371706 | (0.76–1.68) | (0.75–1.64) | (0.65–1.70) | (0.51–1.72) |
|
| ||||
| ABCB1 3435C>T | 1.10 | 1.07 | 0.98 | 0.83 |
| rs1045642 | (0.75–1.63) | (0.73–1.58) | (0.61–1.57) | (0.46–1.50) |
|
| ||||
| NR1I2 44477T>C | 0.80 | 0.81 | 0.90 | 1.21 |
| rs1523130 | (0.50–1.28) | (0.51–1.27) | (0.51–1.59) | (0.58–2.49) |
|
| ||||
| NR1I2 63396C>T | 0.80 | 0.79 | 0.82 | 1.35 |
| rs2472677 | (0.55–1.17) | (0.55–1.14) | (0.52–1.30) | (0.76–2.42) |
|
| ||||
| NR1I2 6-bp indel | 0.67** | 0.71 | 0.66 | 1.86** |
| rs3842689 | (0.45–0.999) | (0.48–1.05) | (0.41–1.07) | (1.02–3.39) |
The geometric mean ratios (95% CI) for nevirapine AUC0–12 h, AUC0–6 h, C0 and CL/F comparing data from subjects (n = 24) with at least one variant allele to subjects with wild-type alleles for each SNP. ABCB1 2766G>T was not evaluated because all subjects were homozygous wild-type. CYP2D6*4 and CAR 13711152C>T were not evaluated because only one subject had the variant allele.
p < 0.01;
p < 0.05.
Partial AUC from 0–6 h for verifying extrapolated AUC0–12 h.
Weight-adjusted apparent oral clearance.
Correlation analysis was performed using *1B/*1B versus *1/*1B and *1/*1 as only one subject was *1/*1.
AUC: Area under the concentration time curve; CL/F: Apparent oral clearance; NVP: Nevirapine.
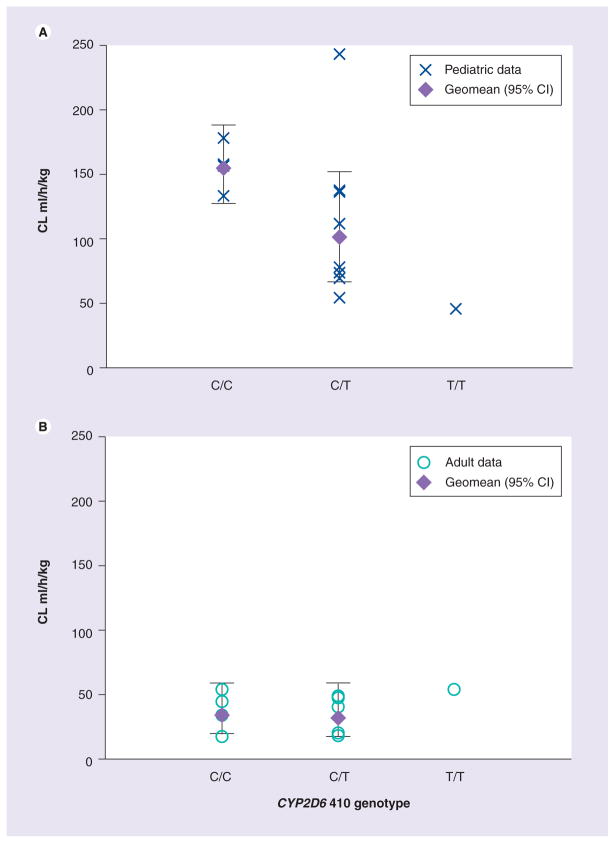
Figure 1 shows a decrease in weight-based CL/F with the number of CYP2D6*17 alleles within the pediatric group. This was not seen in the adult subjects (Figure 1b). In order to confirm the effect of CYP2D6*17 on CL/F in the pediatric group, the GMR for CL/F was determined separately in the pediatric and adult groups. The GMR (VAR/WT; 95% CI) for CYP2D6*17 was 0.59 (0.36–0.98; p = 0.04) and 1.02 (0.61–1.70; p = 0.9) for children and adults, respectively, demonstrating that CYP2D6*17 is associated with decreased clearance only in the pediatric group of our sample.
Figure 1. Weight-adjusted clearance versus CYP2D6*17 genotype (*17 = T allele) in the pediatric group and adult group.
(A) Pediatric group, (B) Adult group.
CL: Clearance.
The following multiple variable model was determined for nevirapine AUC: Ln(AUC0–12 h) = 10.67 + 0.65 (2B6Var) – 0.37 (3A5Var) + 0.015 (age). The CYP2B6 516 T allele increases AUC0–12 h by 92% (95% CI: 51–145%), CYP3A5*3 allele decreases AUC0–12 h by 31% (95% CI: 12–46%) and age increases AUC0–12 h by 1.5% (95% CI: 0.7–2%) per year. Table 3 shows the p-values for each of the variables tested. The effect of the PXR 6-bp indel on nevirapine AUC was not significant in the final multiple variable model despite significance (α = 0.05) seen in the univariate analysis. A similar model was determined for weight-adjusted CL/F (not shown).
Table 3.
Multiple variable model determination.
| Covariates | p-value | Model |
|---|---|---|
| CYP2B6 516G>T | 0.0001 | Y |
| CYP3A4*1B | 0.5101 | N |
| CYP3A5*3 | 0.0128 | Y |
| CYP2D6*17 | 0.237 | N |
| ABCB1 3435C>T | 0.6735 | N |
| NR1I2 44477T>C | 0.8535 | N |
| NR1I2 63396C>T | 0.3883 | N |
| NR1I2 6-bp indel | 0.3344 | N |
| Age | 0.0017 | Y |
| Weight | 0.7563 | N |
| Sex | 0.2216 | N |
| Dose | 0.2215 | N |
N: Excluded from model; Y: Included in model.
Discussion
CYP2B6 genotype is associated with intra-population differences in Malawians. Consistent with previous studies [7], the T allele at CYP2B6 516 correlated with increased exposure and decreased clearance of nevirapine. Since our study and others have demonstrated association of the T allele with increased nevirapine exposures and varying frequencies of CYP polymorphisms among ethnic groups, antiretrovirals must be evaluated in all potential treatment populations [2,7].
Drug-metabolizing enzyme and transporter genotype data from our sample agree with other African populations as reported in the HapMap database (Luhya in Webuye, Kenya; Maasai in Kinyawa, Kenya and the Yoruban in Ibadan, Nigeria) [22]. For example, CYP2B6 516 T allele frequencies ranged from 0.315 to 0.42 for the African groups in the HapMap database; the Malawian frequency was 0.31. For CYP3A5*3, the HapMap database reports a wide range of allele frequencies: 0.128–0.486 (Malawians frequency: 0.25), which demonstrates sub-Saharan African ethnic groups could have dissimilar CYP allele frequencies. This may be clinically relevant to drugs metabolized by these CYP enzymes.
Following the multiple comparisons correction, CYP2B6 516 T allele correlated with increased AUC. CYP2B6 and age were independently associated with nevirapine AUC, C0 and CL/F. CYP3A5*3 is an allele that causes decreased function of CYP3A5 and is expected to decrease clearance of nevirapine, thereby increasing its exposure. Although there was no correlation in the univariate analysis, CYP3A5*3 was associated with decreased AUC when controlling for CYP2B6 and age. CYP3A5*3 correlating with lower nevirapine exposure may be a result of heterozygous individuals in our sample expressing more enzyme than homozygous WT individuals and/or CYP3A5*3 being in linkage with another functional allele [28]. In addition, our sample did not include individuals who were CYP3A5*3/*3, which is a genotype found only in Africans based on the HapMap data, which may introduce bias in our model.
The CYP2D6*17 allele appears to be specific to African populations and is associated with decreased CYP2D6 enzyme activity [3]. Only within the pediatric group did CYP2D6*17 correlate with a decrease in weight-adjusted nevirapine CL/F, suggesting that CYP2D6 may play a larger role in nevirapine metabolism in children. However, this would need to be confirmed in a larger study population accounting for other CYP2D6 SNPs and other polymorphisms in linkage disequilibrium.
CAR and PXR are functionally linked with the expression of various CYPs [19]. Nevirapine preferentially induces CYP2B6 through activation of CAR [19], and polymorphisms in PXR are associated with higher basal expression and reduced induction of CYP3A4 activity [20]. Polymorphisms in NR1I3 and NR1I2 may attenuate the induction of CYP2B6 and CYP3A5 leading to higher nevirapine exposures; however, our study did not demonstrate this. The NR1I2 6-bp indel, which is in linkage disequilibrium with NR1I2 44477T>C, was associated with lower nevirapine exposures, which may indicate increased basal expression of CYP3A4.
The multivariable model showed that nevi-rapine concentrations were influenced by CYP2B6 and CYP3A5 genotypes in addition to age. Currently there is limited clinical utility for this model, due to a lack of widely available genotypic testing for these enzymes in all populations. In addition, there is limited data linking genotype, concentration and safety and/or efficacy. Despite this, the clinical implications of this model include that it is possible to predict nevirapine exposures based on genotype. With more advanced modeling methods, nevi-rapine concentrations could be predicted based on genotype and other known patient factors in order to predict adverse effects such as rash due to supratherapeutic concentrations or virologic failure due to subtherapeutic concentrations. However, these models will also be limited to the groups in which they are studied. In addition, this is a potentially useful tool for research purposes in which outliers of drug concentrations exist with no obvious decreased adherence or extra dosing.
The US Department of Health and Human Services (US DHHS) currently recommends a target nevirapine trough plasma concentration of 3 μg/ml based on data demonstrating a five-fold increase in virologic failure risk with lower trough concentrations [29,30,101]. These data were developed in a Caucasian population with the same doses currently being used in African populations. The Malawian and other sub-Saharan African HIV-infected patients would benefit greatly from efficacy and safety studies focusing on their populations since there is an overwhelming amount of data demonstrating that these groups have genetic differences that affect how the drugs are metabolized and eliminated.
Factors such as nutritional status and diet can also affect drug metabolism. Low protein and high carbohydrate consumption are associated with decreased hepatic CYP metabolism, while high protein diets are associated with increased metabolism [31]. Our subjects had normal nutritional status based on BMI, except for one underweight child. Western diets are generally higher in animal protein than those in sub-Saharan Africa, while diets in Malawi are considerably lower in animal protein and include carbohydrate-rich protein sources [102]. This dietary factor may contribute to the differences observed in nevirapine exposure between Malawians and westerners.
Despite our limited sample size, our novel pilot data provide evidence that ethnic diversity of drug-metabolizing enzyme genetics may play a role in antiretroviral drug exposures. Larger studies are needed to define population allele frequencies and to evaluate the clinical impact of drug-metabolizing enzymes, transporter and nuclear receptor polymorphisms.
Executive summary.
Nevirapine pharmacokinetics in Malawians
Previously studied HIV-infected adults and children had 1.6- to 2-fold higher exposures compared to published data in western cohorts.
Drug exposures can be influenced by polymorphisms in genes encoding enzymes, transporters and receptors that are involved in drug metabolism and elimination.
Aim
We sought to identify the allele frequencies of known genetic polymorphisms in drug-metabolizing enzymes, transporters and nuclear receptor genes in Malawians and evaluate the influence of these genetic polymorphisms on nevirapine pharmacokinetics.
Allele frequency & genotype correlation with nevirapine pharmacokinetics
Malawian allele frequencies were similar to other sub-Saharan Africans, but differed from Caucasians.
The CYP2B6 516 T allele was significantly correlated with increased nevirapine exposures.
CYP2D6*17 was associated with increased clearance only in children.
In a multivariable model, nevirapine AUC increased by 1.5% with each year increase in age, increased by 92% with the presence of the CYP2B6 516 T allele and decreased by 31% with the presence of CYP3A5*3.
Conclusion
Genetic polymorphisms influence nevirapine metabolism in Malawians.
Larger studies are needed to evaluate the clinical impact of genetic variability among ethnic groups.
Footnotes
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
This study was supported in part by the Pharmacogenetics for Every Nation Initiative, the Pharmacogenetics Research Network (NIH U01 GM63340), the UNC Translational and Clinical Sciences Institute (UL1RR025747), NIH Grant GM60346, Cancer Center Support Grant (NIH P30 CA21765), and by the American Lebanese Syrian Associated Charities (ALSAC). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Contributor Information
Kevin C Brown, University of North Carolina at Chapel Hill, Eshelman School of Pharmacy, 3202 Kerr Hall, Chapel Hill, NC 27599-7569, USA.
Mina C Hosseinipour, University of North Carolina at Chapel Hill, School of Medicine, Chapel Hill, NC, USA and UNC Project, Lilongwe, Malawi.
Janelle M Hoskins, University of North Carolina at Chapel Hill, Eshelman School of Pharmacy, 3202 Kerr Hall, Chapel Hill, NC 27599-7569, USA and UNC Institute of Pharmacogenomics & Individualized Therapy, Chapel Hill, NC, USA.
Ranjit K Thirumaran, St Jude Children’s Research Hospital, Department of Pharmaceutical Sciences, Mempis, TN, USA.
Hsiao-Chuan Tien, UNC FPG Child Development Institute, Chapel Hill, NC, USA.
Ralf Weigel, Lighthouse Clinic, Lilongwe, Malawi.
Jean Tauzie, Lighthouse Clinic, Lilongwe, Malawi.
Ida Shumba, Lighthouse Clinic, Lilongwe, Malawi.
Jatinder K Lamba, University of Minnesota, College of Pharmacy, Minneapolis, MN, USA and University of Minnesota, Institute of Human Genetics, Minneapolis, MN, USA.
Erin G Schuetz, St Jude Children’s Research Hospital, Department of Pharmaceutical Sciences, Mempis, TN, USA.
Howard L McLeod, University of North Carolina at Chapel Hill, Eshelman School of Pharmacy, 3202 Kerr Hall, Chapel Hill, NC 27599-7569, USA and UNC Institute of Pharmacogenomics & Individualized Therapy, Chapel Hill, NC, USA.
Angela DM Kashuba, University of North Carolina at Chapel Hill, Eshelman School of Pharmacy, 3202 Kerr Hall, Chapel Hill, NC 27599-7569, USA and University of North Carolina at Chapel Hill, Center for AIDS Research, Chapel Hill, NC, USA.
Amanda H Corbett, University of North Carolina at Chapel Hill, Eshelman School of Pharmacy, 3202 Kerr Hall, Chapel Hill, NC 27599-7569, USA.
References
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Evans WE, McLeod HL. Pharmacogenomics – drug disposition, drug targets, and side effects. N Engl J Med. 2003;348(6):538–549. doi: 10.1056/NEJMra020526. [DOI] [PubMed] [Google Scholar]
- 2▪▪.Rotger M, Colombo S, Furrer H, et al. Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacogenet Genomics. 2005;15(1):1–5. doi: 10.1097/01213011-200501000-00001. First paper to describe CYP2B6 polymorphisms and effects on nevirapine therapy. [DOI] [PubMed] [Google Scholar]
- 3.Bradford LD. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics. 2002;3(2):229–243. doi: 10.1517/14622416.3.2.229. [DOI] [PubMed] [Google Scholar]
- 4.Haas DW, Wu H, Li H, et al. MDR1 gene polymorphisms and phase 1 viral decay during HIV-1 infection: an adult aids clinical trials group study. J Acquir Immune Defic Syndr. 2003;34(3):295–298. doi: 10.1097/00126334-200311010-00006. [DOI] [PubMed] [Google Scholar]
- 5.Haas DW, Ribaudo HJ, Kim RB, et al. Pharmacogenetics of efavirenz and central nervous system side effects: an adult aids clinical trials group study. AIDS. 2004;18(18):2391–2400. [PubMed] [Google Scholar]
- 6.Klein K, Lang T, Saussele T, et al. Genetic variability of CYP2D6 in populations of African and Asian origin: allele frequencies, novel functional variants, and possible implications for anti-HIV therapy with efavirenz. Pharmacogenet Genomics. 2005;15(12):861–873. doi: 10.1097/01213011-200512000-00004. [DOI] [PubMed] [Google Scholar]
- 7▪▪.Penzak SR, Kabuye G, Mugyenyi P, et al. Cytochrome p450 2B6 (CYP2B6) G516T influences nevirapine plasma concentrations in HIV-infected patients in Uganda. HIV Med. 2007;8(2):86–91. doi: 10.1111/j.1468-1293.2007.00432.x. First paper to present nevirapine concentration differences associated with CYP2B6 genotype in a group of Africans. [DOI] [PubMed] [Google Scholar]
- 8▪.Hosseinipour MC, Corbett AH, Kanyama C, et al. Pharmacokinetic comparison of generic and trade formulations of lamivudine, stavudine and nevirapine in HIV-infected Malawian adults. AIDS. 2007;21(1):59–64. doi: 10.1097/QAD.0b013e3280117ca0. Describes the adult pharmacokinetic study on which this present pharmacogenetic study was based. [DOI] [PubMed] [Google Scholar]
- 9▪.Corbett AH, Hosseinipour MC, Nyirenda J, et al. Pharmacokinetics of generic and trade formulations of lamivudine, stavudine and nevirapine in HIV-infected Malawian children. Antiviral Ther. 2010;15(1):83–90. doi: 10.3851/IMP1488. Describes the pediatric pharmacokinetic study on which this present pharmacogenetic study was based. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Heeswijk RP, Veldkamp AI, Mulder JW, et al. The steady-state pharmacokinetics of nevirapine during once daily and twice daily dosing in HIV-1-infected individuals. AIDS. 2000;14(8):F77–F82. doi: 10.1097/00002030-200005260-00001. [DOI] [PubMed] [Google Scholar]
- 11.Luzuriaga K, Bryson Y, McSherry G, et al. Pharmacokinetics, safety, and activity of nevirapine in human immunodeficiency virus type 1-infected children. J Infect Dis. 1996;174(4):713–721. doi: 10.1093/infdis/174.4.713. [DOI] [PubMed] [Google Scholar]
- 12.Erickson DA, Mather G, Trager WF, Levy RH, Keirns JJ. Characterization of the in vitro biotransformation of the HIV-1 reverse transcriptase inhibitor nevirapine by human hepatic cytochromes P-450. Drug Metab Disp. 1999;27(12):1488–1495. [PubMed] [Google Scholar]
- 13.Wyen C, Hendra H, Vogel M, et al. Impact of CYP2B6 983T>C polymorphism on non-nucleoside reverse transcriptase inhibitor plasma concentrations in HIV-infected patients. J Antimicrob Chemother. 2008;61(4):914–918. doi: 10.1093/jac/dkn029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14▪▪.Saitoh A, Sarles E, Capparelli E, et al. CYP2B6 genetic variants are associated with nevirapine pharmacokinetics and clinical response in HIV-1-infected children. AIDS. 2007;21(16):2191–2199. doi: 10.1097/QAD.0b013e3282ef9695. First paper to present nevirapine concentration differences associated with CYP2B6 genotype in children. [DOI] [PubMed] [Google Scholar]
- 15.Ramachandran G, Ramesh K, Hemanth Kumar AK, et al. Association of high T allele frequency of CYP2B6 G516T polymorphism among ethnic south Indian HIV-infected patients with elevated plasma efavirenz and nevirapine. J Antimicrob Chemother. 2009;63(4):841–843. doi: 10.1093/jac/dkp033. [DOI] [PubMed] [Google Scholar]
- 16.Mahungu T, Smith C, Turner F, et al. Cytochrome p450 2B6 516G-->T is associated with plasma concentrations of nevirapine at both 200 mg twice daily and 400 mg once daily in an ethnically diverse population. HIV Med. 2009;10(5):310–317. doi: 10.1111/j.1468-1293.2008.00689.x. [DOI] [PubMed] [Google Scholar]
- 17.Haas DW, Gebretsadik T, Mayo G, et al. Associations between CYP2B6 polymorphisms and pharmacokinetics after a single dose of nevirapine or efavirenz in African–Americans. J Infect Dis. 2009;199(6):872–880. doi: 10.1086/597125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18▪.Chantarangsu S, Cressey TR, Mahasirimongkol S, et al. Influence of CYP2B6 polymorphisms on the persistence of plasma nevirapine concentrations following a single intra-partum dose for the prevention of mother to child transmission in HIV-infected Thai women. J Antimicrob Chemother. 2009;64(6):1265–1273. doi: 10.1093/jac/dkp351. Presents a major public health concern regarding CYP2B6 polymorphisms and the use of single-dose nevirapine for preventing vertical transmission, which is currently a standard practice in many African nations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faucette SR, Zhang TC, Moore R, et al. Relative activation of human pregnane x receptor versus constitutive androstane receptor defines distinct classes of CYP2B6 and CYP3A4 inducers. J Pharmacol Exp Ther. 2007;320(1):72–80. doi: 10.1124/jpet.106.112136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamba J, Lamba V, Strom S, Venkataramanan R, Schuetz E. Novel single nucleotide polymorphisms in the promoter and intron 1 of human pregnane X receptor/NR1I2 and their association with CYP3A4 expression. Drug Metab Disp. 2008;36(1):169–181. doi: 10.1124/dmd.107.016600. [DOI] [PubMed] [Google Scholar]
- 21.Lamba J, Lamba V, Schuetz E. Genetic variants of PXR (NR1I2) and CAR (NR1I3) and their implications in drug metabolism and pharmacogenetics. Curr Drug Metab. 2005;6(4):369–383. doi: 10.2174/1389200054633880. [DOI] [PubMed] [Google Scholar]
- 22▪▪.International HapMap Consortium. The international HapMap project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. This group hosts an online database, which is the most extensive source for genotyping data among different ethnic groups. [DOI] [PubMed] [Google Scholar]
- 23.Rezk NL, Tidwell RR, Kashuba AD. Simultaneous determination of six HIV nucleoside analogue reverse transcriptase inhibitors and nevirapine by liquid chromatography with ultraviolet absorbance detection. J Chromatog. 2003;791(1–2):137–147. doi: 10.1016/s1570-0232(03)00224-1. [DOI] [PubMed] [Google Scholar]
- 24.Marsh S, King CR, Garsa AA, McLeod HL. Pyrosequencing of clinically relevant polymorphisms. Methods Mol Biol (Clifton, NJ) 2005;311:97–114. doi: 10.1385/1-59259-957-5:097. [DOI] [PubMed] [Google Scholar]
- 25.Garsa AA, McLeod HL, Marsh S. CYP3A4 and CYP3A5 genotyping by pyrosequencing. BMC Med Genet. 2005;6:19. doi: 10.1186/1471-2350-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohrbacher M, Kirchhof A, Geisslinger G, Lötsch J. Pyrosequencing-based screening for genetic polymorphisms in cytochrome P450 2B6 of potential clinical relevance. Pharmacogenomics. 2006;7(7):995–1002. doi: 10.2217/14622416.7.7.995. [DOI] [PubMed] [Google Scholar]
- 27.Zackrisson AL, Lindblom B. Identification of CYP2D6 alleles by single nucleotide polymorphism analysis using pyrosequencing. Eur J Clin Pharmacol. 2003;59(7):521–526. doi: 10.1007/s00228-003-0654-7. [DOI] [PubMed] [Google Scholar]
- 28.Kuehl P, Zhang J, Lin Y, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27(4):383–391. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 29.De Vries-Sluijs TE, Dieleman JP, Arts D, et al. Low nevirapine plasma concentrations predict virological failure in an unselected HIV-1-infected population. Clin Pharmacokinetics. 2003;42(6):599–605. doi: 10.2165/00003088-200342060-00009. [DOI] [PubMed] [Google Scholar]
- 30.Veldkamp AI, Weverling GJ, Lange JM, et al. High exposure to nevirapine in plasma is associated with an improved virological response in HIV-1-infected individuals. AIDS. 2001;15(9):1089–1095. doi: 10.1097/00002030-200106150-00003. [DOI] [PubMed] [Google Scholar]
- 31.Ensom MHH, Blouin RA. Dietary influences on drug disposition. In: Burton ME, Shaw LM, Schentag JJ, Evans WE, editors. Applied Pharmacokinetics and Pharmacodynamics: Principles of therapeutic drug monitoring. Lippincott Williams and Wilkins; Baltimore, MA, USA: 2006. pp. 242–256. [Google Scholar]
Websites
- 101.Panel on antiretroviral guidelines for adults and adolescents. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. Department of health and human services; Jan 10, 2011. [Accessed 15 May 2011]. pp. 1–166. www.Aidsinfo.Nih.Gov/contentfiles/adultandadolescentgl.Pdf. [Google Scholar]
- 102.FAO Malawi nutrition profile. Nutrition and Consumer Protection Division. Food and Agriculture Organization of the United Nations; 2008. [Accessed 26 May 2010]. www.fao.org/ag/agn/nutrition/mwi_en.stm. [Google Scholar]
- 103.NCBI dbSNP. www.ncbi.nlm.nih.gov/snp.