Abstract
The endoplasmic reticulum chaperone gp96 is required for the cell surface expression of a narrow range of proteins, including toll-like receptors (TLRs) and integrins. To identify a more comprehensive repertoire of proteins whose cell surface expression is dependent on gp96, we developed plasma membrane profiling (PMP), a technique that combines SILAC labeling with selective cell surface aminooxy-biotinylation. This approach allowed us to compare the relative abundance of plasma membrane (PM) proteins on gp96-deficient versus gp96-reconstituted murine pre-B cells. Analysis of unfractionated tryptic peptides initially identified 113 PM proteins, which extended to 706 PM proteins using peptide prefractionation. We confirmed a requirement for gp96 in the cell surface expression of certain TLRs and integrins and found a marked decrease in cell surface expression of four members of the extended LDL receptor family (LDLR, LRP6, Sorl1 and LRP8) in the absence of gp96. Other novel gp96 client proteins included CD180/Ly86, important in the B-cell response to lipopolysaccharide. We highlight common structural motifs in these client proteins that may be recognized by gp96, including the beta-propeller and leucine-rich repeat. This study therefore identifies the extended LDL receptor family as an important new family of proteins whose cell surface expression is regulated by gp96.
Keywords: gp96, plasma membrane, biotin, SILAC, LDLR, Sorl1, LRP6, LRP8, CD180
Introduction
Gp96 (also known as gpr94 or HSP90b1) is a ubiquitously expressed chaperone and the only ER resident member of the HSP90 family.1 Gp96 has several biological functions, of which the best recognized is its role in the folding and assembly of membrane proteins of the secretory pathway. In the absence of gp96 these proteins are targeted for ER-associated degradation (ERAD).1 However, the chaperone activity of gp96 is selective, with targets including integrins, toll-like receptors (TLRs), insulin-like growth factors (IGFs), thyroglobulin and the platelet glycoprotein Ib-IX-V complex.2−8 The selectivity of gp96 as a chaperone is illustrated by the case of the integrin family.3 While many integrins require chaperoning by gp96, some α- and β-integrins are gp96 independent.2,9 The first reported gp96 clients came from the immunoglobulin family.10 However, serum immunoglobulins are normal in mice with a B-cell selective gp96-depletion, suggesting that folding of immunoglobulins is gp96-independent.11 Other than the presence of disulfide bonds, no common structural feature of gp96 clients has emerged.12
To gain a more complete understanding of cell surface receptors whose expression is dependent on gp96, we developed an unbiased proteomics approach for the selective enrichment of plasma membrane proteins. Selective oxidation and aminooxy-biotinylation of sialylated PM glycoproteins, followed by off-line high pH reversed-phase HPLC (HpRP-HPLC) fractionation allowed the mass spectrometry identification of up to 700 PM proteins, as defined by gene ontology (GO) terms. Using this technique, we performed a SILAC-based differential analysis of plasma membrane proteins whose expression is dependent upon gp96. A comparison of gp96-knockout and gp96-reconstituted murine pre-B cells3,13 found that the cell surface expression of 29 proteins was decreased >3.0 fold in the absence of gp96. We confirmed the reported gp96 dependence of integrins α4, αV, αL, β2 and toll-like receptors TLR6 and TLR4, and the lack of dependence of the α6 and β1 integrins.2−4,9,11 We now report that gp96 is required for cell surface expression of four members of the LDLR family: (i) the founding member LDLR, which plays a central role in regulating the concentration of plasma LDL and when absent leads to familial hypercholesterolaemia,14 (ii) the Wnt signaling protein lipoprotein receptor-related protein-6 (LRP6) which participates with Frizzled proteins as coreceptors in Wnt signaling,15 (iii) the sortilin related receptor Sorl1 implicated in the pathogenesis of Alzheimer’s disease,16 and (iv) LRP8 (apolipoprotein E receptor 2) required for neural development through Reelin signaling.17 None of the remaining six members of the LDLR family were detected by mass spectrometry. Our data suggest that LDL receptor family members are dependent on gp96 for optimal cell surface expression.
Materials and Methods
Cells and Cell Culture
All cell lines were derived from the murine pre-B-cell line 70Z/3, as described previously. The NF-κB reporter cell line GTPT3 was engineered to express three NF-κB-dependent markers for previous studies of IKKγ mutants.18 GTPT3 cells were mutagenised with ICR191 (Sigma), and individual clones derived. The resulting M36 clone has two frameshift mutations in the ORF of gp96, resulting in production of a truncated nonfunctional protein.13 M36/gp96+ cells were derived by stable transduction of M36 cells with gp96.13 M36 and M36/gp96+ cells were used for the majority of the analysis in our study. The E4.126 clone was independently derived from 70Z/3 cells by ICR191 mutagenesis, and also carries two frameshifting mutations in gp96.3,13 All cells were grown in SILAC RPMI 1640 medium (Thermo Pierce, Cramlington, U.K.) supplemented with 10% dialyzed fetal calf serum (FCS) (JRH Biosciences, Lenexa, KS), Penicillin at 100 units/mL, Streptomycin at 0.1 mg/mL (Sigma, Dorset, U.K.) and 50 μM Mercaptoethanol (Sigma). Media was supplemented with either light (Arg 0, Lys 0, Sigma) or heavy (Arg 10, Lys 8, Cambridge Isotope Laboratories, Andover, MA) amino acids at 50 mg/L and l-proline at 280 mg/L.19
Preparation of PM Proteins Using Aminooxy-biotin
We have optimized our procedure20 for enrichment of PM proteins for proteomic analysis, as described below (Figure 1). M36 cells were grown in light media, and M36/gp96+ cells in heavy media for seven divisions. 1.2 × 108 of each cell type were pooled in a 1:1 ratio and washed twice in ice-cold PBS pH 7.4 (Sigma). All of the following steps were performed on ice. An oxidation/biotinylation mix21 comprising 1 mM sodium meta-periodate, 100 mM aminooxy-biotin (Biotium Inc., Hayward, CA) and 10 mM aniline (Sigma) in ice-cold PBS pH 6.7 was vortexed with aniline added immediately prior to vortexing. Surface sialic acid residues were oxidized then biotinylated by resuspending cells in 6 mL mix (1 mL per 4 × 107 cells) with rocking in the dark. The oxidation reaction was quenched by glycerol to 1 mM final concentration. Cells were washed once in PBS pH 7.4/5% (v/v) FCS, then once in PBS pH 7.4. PBS contained 1 mM CaCl2 and 0.5 mM MgCl2. Biotinylated cells were centrifuged at 400× g at 4 °C for 5 min, and the resulting cell pellet was resuspended and incubated at 4 °C for 30 min in lysis buffer (1% Triton X-100 (high purity, Thermo), 150 mM NaCl, 1× protease inhibitor (complete, without EDTA (Roche)), 5 mM iodoacetamide (Sigma), 0.1 mg/mL PMSF and 10 mM Tris-HCl pH 7.6). Nuclei were removed by centrifugation at 4 °C, initially at 2800× g then twice at 16000× g. Biotinylated proteins were enriched by incubating for 2 h at 4 °C with 144 μL high affinity streptavidin agarose beads (6 μL beads per 107 input cells, Pierce). Extensive washing was performed using a vacuum manifold and Snap Cap spin columns (Pierce). All washes or incubations in the following description were with 600 μL buffer; all centrifugation steps were performed at 1000× g for 1 min. Beads were initially washed 20 × with lysis buffer, 20× with PBS/0.5% (w/v) SDS and incubated for 20 min at RT with PBS/0.5% (w/v) SDS/100 mM DTT, then centrifuged. Further washing was performed 20× with UC buffer (6 M urea, 100 mM Tris-HCl pH 8.5), followed by alkylation for 20 min at RT with UC buffer containing 50 mM iodoacetamide. Beads were washed (20× per step with centrifugation after each step), using UC buffer, 5 M NaCl, 100 mM Na2CO3, PBS then water, resuspended in 400 μL 50 mM NH4HCO3 containing 5 μg modified sequencing grade trypsin (Promega), then transferered to a protein LoBind tube (Eppendorf), where biotinylated glycoproteins were digested on-beads overnight. Beads were transferred to a Snap Cap spin column and tryptic peptides collected by centrifugation at 1000× g for 1 min. Beads were rinsed once with 50 mM NH4HCO3, and tryptic fractions pooled. Ten percent of the resultant digest was desalted and concentrated by StageTip22 for immediate analysis. The remaining tryptic peptide sample was fractionated by HpRP-HPLC (see below). To elute glycopeptides, beads were washed with PBS, then water, then G7 buffer (New England Biolabs, Hitchin, U.K.). Beads were incubated for 5 h in 400 μL G7 buffer containing 30000 units of glycerol free PNGase (New England Biolabs). Glycopeptides were collected by centrifugation at 1000× g for 1 min, beads were washed once with G7 buffer, and eluates pooled and concentrated on a StageTip.22
Figure 1.

Plasma membrane profiling workflow. Heavy and light labeled cells are mixed early in the procedure and sialylated glycoproteins oxidized and biotinylated. The enriched glycoproteins are digested and bound N-linked glycopeptides are released using PNGase F. Eluates are prepared for LC–MS/MS.
Biotinylation was confirmed by staining aliquots of cells prior to and after biotinylation with streptavidin-allophycocyanin (eBioscience, San Diego, CA). The incorporation of heavy label was checked by analysis of a lysate of 3 × 106 heavy labeled cells, generated using SDS/DTT/Tris (SDT) buffer and Filter Aided Sample Processing (FASP).23 Incorporation was >98% for both arginine and lysine-containing peptides.
High pH reverse-phase high pressure liquid chromatography (HpRP-HPCL) fractionation and mass spectrometric analysis
A total of 100 μg of tryptic peptide was subjected to HpRP-HPLC fractionation using a Dionex Ultimate 3000 powered by an ICS-3000 SP pump with an Agilent ZORBAX Extend-C18 column (4.6 mm × 250 mm, 5 μm particle size). Mobile phases (H20, 0.1% NH4OH or MeCN, 0.1% NH4OH) were adjusted to pH 10.5 with the addition of formic acid and peptides were resolved using a linear 40 min 0.1–40% MeCN gradient over 40 min at a 400 μL/min flow rate and a column temperature of 15 °C. Eluting peptides were collected in 15 s fractions.
Fractions were dried down using an Eppendorf Concentrator and resuspended in 8 μL MS solvent (3% MeCN, 0.1% TFA). Fractions 25 to 152 inclusive were analyzed and in each case 3 μL was injected and subjected to LC–MS/MS using a NanoAcquity uPLC (Waters, MA) coupled to an LTQ-OrbiTrap XL (Thermo, FL, UA). Peptides were eluted using a gradient rising from 7 to 25% MeCN by 30 min, 40% MeCN by 39 min and 85% MeCN by 42 min. MS data was acquired between 400 and 2000 m/z at 60000 fwhm with lockmass enabled (445.120025 m/z). CID spectra were acquired in the LTQ with MSMS switching operating in a top 6 DDA fashion triggered at 500 counts. Unfractionated peptides were eluted using a gradient rising from 3 to 25% MeCN by 90 min, 40% MeCN by 110 min and 85% MeCN by 115 min. Spectra were acquired using the same MS parameters.
Database Searching and Data Processing
Raw MS files were processed in parallel using MaxQuant versions 1.0.13.13 and 1.1.1.14.24,25 Data were searched against the International Protein Index mouse database release 3.68 using MASCOT Daemon 2.3.2 and Andromeda version 1.1.1.14.25 Fragment ion tolerance was set to 0.5 Da with a maximum of 2 missed tryptic cleavage sites. Carbamidomethyl cysteine was defined as a fixed modification, oxidized methionine, N-terminal acetylation and deamidation (NQ) were selected as variable modifications. Reversed decoy databases were used and the false discovery rate for both peptides and proteins were set at 0.01. Protein quantitation utilized razor and unique peptides and required a minimum of 2 ratio counts and normalized protein ratios reported. Peptide requantify was enabled in all analyses apart from where indicated. Significance A values were calculated and Gene Ontology Cellular Compartment (GOCC) terms added using Perseus version 1.1.1.13 (downloaded from http://maxquant.org).
To assess the number of PM proteins identified, we summed the proteins with GOCC terms plasma membrane (PM), cell surface but not PM (CS) and extracellular but not PM/CS (XC). We previously identified a subset of proteins annotated by GO as integral to the membrane, but with no subcellular assignment and a short GOCC (ShG) term (a 4-part term containing the terms “integral to membrane”, “intrinsic to membrane”, “membrane part”, “cell part” or a 5-part term containing “membrane” in addition to these terms).20 Where a majority of proteins identified from a given sample are annotated PM/CS/XC, it is likely that a substantial proportion of the proteins with a short GOCC term are also integral PM proteins—including these proteins provides a useful upper estimate of the number of identified PM proteins.
Structural Annotation and Protein–Protein Interactions
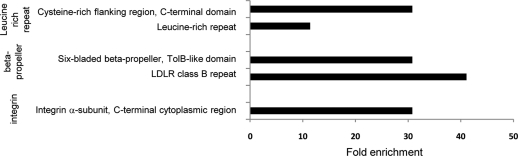
We used the method of Adachi et al.26 to determine statistically enriched protein domains in our data set. Briefly, Interpro (release 34.0) was used to annotate protein structural domains, and Cytoscape27 plugin BiNGO28 was used for domain enrichment analysis. The test data set was the InterPro annotations of all proteins we identified by ≥2 peptides and found downregulated >3 fold in the absence of gp96. The reference set was the InterPro annotations of all proteins we identified by ≥2 peptides. The InterPro ontology was derived from the interpro.xml file, InterPro release 34.0 (ftp://ftp.ebi.ac.uk/pub/databases/interpro/). The enrichment analysis was performed using a Hypergeometric test, and we selected all InterPro domains that were significant with p < 0.001 after correcting for multiple term testing by Benjamini and Hochberg false discovery rate. Fold enrichment was calculated by comparing the test set against the reference set.
Cytoscape version 2.8.2 (www.cytoscape.org) was used with the Mimi plugin29 to determine the known interactions of proteins down- or up-regulated >3 fold in the absence of gp96.
Flow Cytometry
All of the following steps were performed on ice to prevent receptor internalization. Cells were washed in ice-cold PBS pH 7.4 containing 1 mM CaCl2 and 0.5 mM MgCl2 (Sigma), incubated for 15 min with 10 mg/mL Fc block (Vivaglobulin, CSL Behring UK Ltd.) and stained (30 min, 4 °C) using the following antimouse monoclonal antibodies: anti-CD44 FITC, anti-CD18 PE, anti-H2Kb PE (Caltag Laboratories, Burlingame, CA) and unconjugated anti-LDLR (R and D systems, Abingdon, U.K.). Cells were washed in PBS. Cells that had been stained with the unconjugated anti-LDLR primary antibody were stained (30 min, 4 °C) with antirat Cy5 (Invitrogen, Carlsbad, CA) then washed with PBS. Cells were fixed in 1% paraformaldehyde and analyzed on a FACS Calibur using Cellquest (BD Pharmingen) and FCSPress v1.3 (www.fcspress.com).
Results
Enrichment of Plasma Membrane Proteins by PMP
To identify proteins whose cell surface expression was dependent on gp96 we further developed and improved our plasma membrane enrichment technique, termed “plasma membrane profiling” (Figure 1).20 Aminooxy-biotinylated PM proteins were prepared from M36 cells (light media) and M36/gp96+ cells (heavy media), as described above. After capture on streptavidin beads, proteins were digested with trypsin. Ten percent of this tryptic peptide sample was desalted and concentrated to confirm enrichment of PM proteins. We identified 135 proteins, of which 89 were annotated “plasma membrane” (PM). An additional 2 proteins were annotated “cell surface” (CS), and 3 proteins “extracellular” (XC), giving a total of 94 PM/CS/XC proteins, representing 70% of all identified proteins with a GOCC annotation (Table 1). Including the 19 proteins with a “short GO” (ShG) annotation (described above) gave a useful upper estimate of the number of identified PM proteins, 113 PM/CS/XC/ShG proteins (84%). A total of 7 proteins were annotated “nucleus”, 4 of which were additionally annotated PM. The 3 nuclear proteins not annotated PM are likely to represent contaminants (Table 1).
Table 1. Proteins Enriched using PMP were Identified by LC–MS/MS and their Subcellular Location Inferred from GOCC Termsa.
| unfractionated | all fractions Mascot | all fractions Andromeda | all fractions combined | |
|---|---|---|---|---|
| Total proteins identified | 135 | 1709 | 1551 | 1826 |
| Total proteins quantified | 93 | 1181 | 1137 | 1271 |
| Annotations | ||||
| Membrane | 127 | 1352 | 1241 | 1433 |
| Plasma membrane or Cell surface (PM/CS) | 91 | 481 | 452 | 509 |
| Extracellular but not PM/CS (XC) | 3 | 48 | 39 | 54 |
| Short GO description (ShG) | 19 | 141 | 129 | 143 |
| Nuclear but not PM/CS/XC/ShG | 3 | 352 | 318 | 377 |
| Total PM/CS/XC | 94 | 529 | 491 | 563 |
| Total PM/CS/XC/ShG | 113 | 670 | 620 | 706 |
| % PM/CS/XC of all identified + annotated proteins | 70% | 34% | 35% | 35% |
| % PM/CS/XC/ShG of all identified+annotated proteins | 84% | 44% | 44% | 43% |
Numbers of proteins are shown with the indicated annotations. The percent PM/CS/XC is calculated as (PM/CS/XC annotations/(total protein identifications – proteins without GOCC annotation)).
HPLC Fractionation of Tryptic Peptides Results in a Substantial Increase in Identified Plasma Membrane Proteins
As a significant number of PM proteins were identified at high purity from the unfractionated tryptic peptide sample, we performed HpRP-HPLC fractionation on the remaining 90% of sample. One-hundred twenty-eight fractions were analyzed (Figure S1, Supporting Information), and this data processed in parallel with the PNGase fraction using MaxQuant version 1.0.13.13 (referred to as “Mascot”) and MaxQuant version 1.1.1.14 (referred to as “Andromeda”). Fractionation substantially increased the total and PM protein identifications—combining the Mascot and Andromeda data, a total of 563 proteins annotated PM/CS/XC and 706 proteins annotated PM/CS/XC/ShG were identified (Table 1, Table S1, Supporting Information). Four-hundred fifty proteins annotated PM/CS/XC and 565 proteins annotated PM/CS/XC/ShG were quantified, out of a total of 1271 quantified proteins.
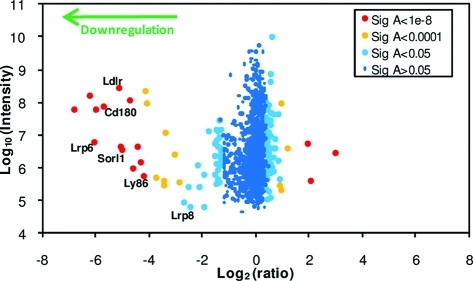
Forty-six of the 1271 proteins were downregulated >3 fold (Table S2, Supporting Information), 29 of which were identified by ≥2 peptides (Figure 2 and Table 2). Of these 29 proteins, 24 were annotated PM/CS/XC and 26 PM/CS/XC/ShG (93%).
Figure 2.
Scatterplot of all proteins identified by ≥2 peptides. Fold change (x-axis) is shown as log2, and summed ion intensity (y-axis) is shown as log10. Significance A values were calculated using Perseus version 1.1.1.13.
Table 2. Proteins Identified by ≥2 Peptides and Down- or Up-regulated >3 Fold in gp96 Deficient Cellsa.
| ratio |
peptides |
% coverage |
significance
A |
GOCC |
structure |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IPI number | protein name | Mct | Adr | Mct | Adr | Mct | Adr | Mct | Adr | M | P | C | X | N | S | βP | Lrr | Egf | Ig |
| down-regulated | |||||||||||||||||||
| IPI00121334 | Integrin alpha-4 | 113.2 | 49.2 | 12 | 10 | 13.7 | 11.5 | 9.2 × 10–21 | 6.1 × 10–12 | M | P | C | βP | ||||||
| IPI00828582 | Integrin alpha L | 73.3 | 43.3 | 27 | 25 | 28.4 | 26 | 2.0 × 10–17 | 2.9 × 10–11 | M | P | βP | |||||||
| IPI00271594 | Low-density lipoprotein receptor-related protein 6 | 65.6 | 21.9 | 12 | 10 | 9.2 | 7.6 | 1.3 × 10–16 | 5.2 × 10–8 | M | P | βP | Egf | ||||||
| IPI00130117 | Integrin alpha 9 | 63.5 | 37.8 | 10 | 10 | 11.8 | 12 | 2.1 × 10–16 | 1.4 × 10–10 | M | P | βP | |||||||
| IPI00343568 | CD180 antigen | 52.5 | 22.4 | 9 | 8 | 19.2 | 16.8 | 4.3 × 10–15 | 4.2 × 10–8 | M | P | Lrr | |||||||
| IPI00785217 | Low-density lipoprotein receptor | 35.0 | 25.2 | 22 | 16 | 27.8 | 22.5 | 1.7 × 10–12 | 1.3 × 10–8 | M | P | X | βP | Egf | |||||
| IPI00553419 | Desmoplakin | 32.7 | 9.9 | 9 | 5 | 3.2 | 2 | 4.4 × 10–12 | 5.4 × 10–5 | M | P | ||||||||
| IPI00776230 | Sortilin-related receptor | 32.3 | 29.5 | 9 | 8 | 5.5 | 4.9 | 5.2 × 10–12 | 2.4 × 10–9 | M | X | βP | Egf | ||||||
| IPI00320605 | Integrin beta-2 | 26.3 | 25.1 | 23 | 21 | 37.5 | 35.5 | 7.9 × 10–11 | 1.3 × 10–8 | M | P | C | Egf | ||||||
| IPI00918724 | Latrophilin-1 | 24.6 | 24.0 | 2 | 3 | 1.6 | 2.4 | 1.9 × 10–10 | 2.0 × 10–8 | M | P | ||||||||
| IPI00229475 | Junction plakoglobin | 21.8 | 17.2 | 9 | 8 | 14.1 | 12.6 | 8.2 × 10–10 | 5.4 × 10–7 | M | P | N | |||||||
| IPI00113539 | Fibronectin | 19.6 | 39.8 | 5 | 2 | 2.4 | 1.1 | 3.0 × 10–9 | 8.0 × 10–11 | M | P | X | |||||||
| IPI00128026 | Lymphocyte antigen 86 | 18.2 | 2 | 1 | 16.7 | 11.1 | 7.1 × 10–9 | X | Ig | ||||||||||
| IPI00857195 | Integrin alpha V | 17.3 | 17.1 | 28 | 27 | 33.6 | 32.3 | 1.3 × 10–8 | 5.6 × 10–7 | M | P | βP | |||||||
| IPI00266264 | Integrin beta-3 | 16.8 | 15.8 | 20 | 18 | 32.7 | 28.2 | 1.7 × 10–8 | 1.1 × 10–6 | M | P | Egf | |||||||
| IPI00468203 | Annexin A2 | 13.1 | 9.7 | 3 | 3 | 11.5 | 11.5 | 2.4 × 10–7 | 6.4 × 10–5 | M | P | X | |||||||
| IPI00309419 | Leucine-rich repeat-containing protein 4C | 11.0 | 15.1 | 2 | 3 | 3.8 | 5.3 | 1.4 × 10–6 | 1.7 × 10–6 | M | P | Lrr | Ig | ||||||
| IPI00405227 | Vinculin | 11.0 | 13.0 | 4 | 2 | 4.5 | 2.4 | 1.5 × 10–6 | 6.4 × 10–6 | M | P | ||||||||
| IPI00227288 | Extracellular LRR and Fn type-III domain protein 1 | 10.3 | 10.1 | 5 | 4 | 8.2 | 7 | 2.6 × 10–6 | 4.5 × 10–5 | M | S | Lrr | |||||||
| IPI00828535 | Attractin | 8.3 | 8.2 | 4 | 5 | 4 | 4.9 | 1.9 × 10–5 | 0.0002 | M | P | βP | Egf | ||||||
| IPI00224294 | N-glycan processing alpha-mannosidase IIx | 7.1 | 6.6 | 2 | 3 | 2.3 | 3.1 | 6.6 × 10–5 | 0.0009 | Egf | |||||||||
| IPI00223457 | EGF-containing fibulin-like extracellular matrix protein 1 | 6.5 | 2 | 4.7 | 0.0001 | X | Egf | ||||||||||||
| IPI00648457 | Adhesion molecule with Ig like domain 1 | 5.8 | 12.5 | 3 | 2 | 5 | 3.2 | 0.0003 | 8.5 × 10–6 | M | S | Lrr | Ig | ||||||
| IPI00410868 | Low-density lipoprotein receptor-related protein 8 | 5.4 | 2 | 2 | 2.2 | 2.2 | 0.0031 | M | P | X | βP | Egf | |||||||
| IPI00380390 | Toll-like receptor 6 | 4.5 | 33.1 | 4 | 3 | 5.2 | 4.8 | 0.0019 | 6.6 × 10–10 | M | P | Lrr | |||||||
| IPI00480329 | E1A-binding protein p400 | 4.4 | 1 | 2 | 0.7 | 1.1 | 0.0096 | M | N | ||||||||||
| IPI00129526 | Endoplasmin | 3.8 | 5.3 | 6 | 5 | 8.9 | 7.7 | 0.0047 | 0.0035 | M | P | ||||||||
| IPI00605095 | Leucine-rich repeats and Ig-like domains protein 2 | 3.8 | 2 | 1 | 2.5 | 1.4 | 0.0052 | M | P | Lrr | Ig | ||||||||
| IPI00342158 | Nuclear pore membrane glycoprotein 210 | 3.4 | 3.3 | 13 | 10 | 8 | 6.5 | 0.0092 | 0.0364 | M | N | ||||||||
| up-regulated | |||||||||||||||||||
| IPI00387201 | Tumor necrosis factor receptor superfamily member 19 | 0.3 | 0.2 | 4 | 3 | 13.2 | 10.8 | 1.8 × 10–16 | 1.9 × 10–15 | M | P | X | |||||||
| IPI00130804 | Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase | 0.2 | 2 | 1 | 8 | 3.4 | 4.8 × 10–18 | M | |||||||||||
| IPI00399440 | Rapamycin-insensitive companion of mTOR | 0.1 | 1 | 2 | 1 | 2 | 4.1 × 10–30 | ||||||||||||
Ratio is shown as the fold downregulation, calculated by Mascot (Mct) or Andromeda (Adr). Peptides refers to total unique or razor peptides. Significance A values were calculated by MaxQuant or Perseus. For a given protein, GOCC annotations are shown: M (membrane), P (plasma membrane), C (cell surface), X (extracellular), N (nucleus) and S (short GO term). Structural features were determined by literature search and Interpro release 34.0: beta-propeller (βP), leucine rich repeat (Lrr), EGF or EGF-like domain (Egf), immunoglobulin or immunoglobulin-like domain (Ig). Other abbreviations: Fibronectin (Fn). We performed this experiment as a single replicate. Confirmation by flow cytometry used at least three biological replicates (Figure 3).
Plasma Membrane Profiling Confirms the Critical Role of gp96 in Cell Surface Expression of Integrins and TLRs
Our data was validated by confirming the dependence on gp96 for cell surface expression of integrins α4, αV, αL, and β2,2,3,9,11 and gp96 independence of integrins β1, α6 and α52 (Table 2 and Table S1, Supporting Information). Furthermore, flow cytometric analysis of integrin β2 (CD18) demonstrated decreased cell surface expression to the level of the unstained control in two independently derived gp96-deficient cell lines (Figure 3). We also showed that integrin α9 is gp96 dependent (Table 2). The normal cell surface expression of integrin α1 and decreased expression of integrin β3 in gp96-deficient cells contrasts with a previous report.2
Figure 3.
Flow cytometry confirms that LDLR is gp96 dependent. (a) M36 and M36/gp96+ cells or (b) the independent gp96 knockout E4.126 and control GTPT3 cells were stained with monoclonal antibodies against the indicated cell surface targets. At least three replicate stains were performed for each cell surface target, confirming the above observations (data not shown).
Two TLRs were identified. Surface localization of TLR6 was dependent on gp96 (Table 2), as reported.3,4,30 TLR4 was identified by two peptides (Table S1, Supporting Information) however no protein ratio was reported by either Mascot or Andromeda. One of the two peptides was quantified by both algorithms, with fold down-regulation ratios of 34.2 (Mascot) and 14.1 (Andromeda), suggesting that surface expression of TLR4 is dependent on gp96, as reported.3,4
Several proteins were not dependent on gp96 for their surface expression, including CD44, ICAM1, CXCR4, IL7R as well as MHC class I allotypes3,11 and this could be confirmed by flow cytometry for CD44 and H2-Kb (Figure 3).
Finally, gp96 (Endoplasmin) was detected at the PM in M36/gp96+ cells and showed decreased plasma membrane expression (3.8–5.3 fold) in M36 cells (Table 2). This corresponds to Western blotting analysis of gp96 from M36 lysates which showed a truncated protein at low abundance.13 The two frameshifting mutations in the ORF of gp96 in M36 cells explain the loss of wild-type protein.13
Downregulation of LDLR Family Members in gp96 Deficient Cells
Most notable of the novel proteins whose cell surface expression is decreased in the absence of gp96 were 4 proteins of the extended LDLR family: the LDLR itself (25.2–35.0 fold), LRP6 (21.9–65.6 fold), Sorl1 (29.5–32.3 fold) and LRP8 (apolipoprotein E receptor 2) (5.4 fold). The other 6 members of the LDLR family (VLDLR, MEGF7, LRP1, LRP1B, LRP2, LRP5) were not detected. This likely reflects either low expression levels in pre-B cells, or solubility problems since these proteins are large, ranging from 211 to 522 kDa, and may therefore not be efficiently enriched by our PMP approach. The absolute dependence of cell surface LDLR expression on gp96 was readily confirmed by flow cytometry in both gp96 deficient cells (M36 and E4.126) (Figure 3). No other antibodies suitable for flow cytometry were available for other mouse protein family members. Given the structural similarities of other members of the LDLR family to LDLR and ApoER2, it seems likely that these six proteins are also gp96-dependent, though confirmation is required given the selectivity of gp96 for certain α-integrins.
All members of the LDLR family and more distantly related receptors are characterized by the presence of at least one YWTD-motif containing beta-propeller domain, with flanking EGF-like cysteine-rich repeats.12 Both beta-propellers and EGF repeats have been identified as structural motifs present in other known gp96 clients. All α-integrins contain a single seven-bladed beta-propeller, and β-integrins contain four cysteine-rich EGF repeats31 Among the other proteins dependent on gp96 for cell surface expression, attractin contains a beta-propeller (of the Kelch-repeat type rather than WD-repeat type); Laminin alpha 5, attractin, N-glycan processing alpha-mannosidase IIx and EGF-containing fibulin-like extracellular matrix protein 1 contain EGF-like domains (Table 2).
To determine which protein structures were over represented within the downregulated proteins as compared to all proteins identified by ≥2 peptides, we used the method of Adachi et al.26 Significantly enriched (p < 0.001) protein domains or repeats were part of one of three structures: the beta-propeller, the leucine-rich repeat, or α integrin (Figure 4). All α-integrins and all members of the extended LDLR family contain β-propeller motifs, suggesting that the beta-propeller and leucine-rich repeat may require gp96 for proper folding. However, there must be further complexity as some α-integrins containing a β-propeller, and some TLR proteins containing the leucine-rich repeat are not dependent on gp96 for cell surface expression.
Figure 4.
Fold enrichment of structures over-represented (p < 0.001) in proteins downregulated in the absence of gp96 (Table 2), in comparison to all proteins identified by ≥2 peptides.
CD180/Ly86 is a Novel gp96 Client
In addition to LDLR family members, CD180 (RP105, Ly78) and Ly86 (MD1) both show decreased cell surface expression in the absence of gp96 (Table 2). CD180 is related to TLRs, contains leucine-rich repeats32,33 and binds Ly86 (MD1).34 Gp96 may be required for CD180/MD1 complex assembly, by analogy to TLR4/MD2, as in the absence of gp96, TLR4 and MD2 cannot associate.3 Together CD180/Ly86 affect the immune response to lipopolysaccharide (LPS) by interfereing with an LPS-derived signal delivered through the CD14-MD2-TLR4 complex.35 Additional LRR-containing proteins downregulated in gp96-deficient cells include: Leucine-rich repeat-containing protein 4C, Extracellular leucine-rich repeat and fibronectin type-III domain-containing protein 1, Adhesion molecule with Ig like domain 1, Leucine-rich repeats and immunoglobulin-like domains protein 2 (Table 2).
Other Novel gp96 Clients or Interacting Partners
Ly86 may show decreased cell surface expression in gp96 deficient cells due to interaction with its LRR containing partner, CD180. To investigate whether the surface localization of some proteins was aberrant because their binding partners are dependent on gp96 we searched all proteins identified by ≥2 peptides, down- or up-regulated >3 fold (Table 2) using Cytoscape 2.8.2 to identify interacting partners (Figure S2, Supporting Information). Fibronectin is a ligand for integrin αV/β3,36 which might explain its downregulation (19.6–39.8 fold) in gp96 deficient cells.
Correspondence of Mascot and Andromeda Ratios
Protein ratios generally corresponded well when data were processed using either Mascot or Andromeda (Table S1, Supporting Information) except when proteins were downregulated >3 fold, in which case the ratios generated using either software corresponded less well (Table 2 and Table S2, Supporting Information). Gp96 is critical for proper folding and functional expression of a number of integrins and TLRs; in gp96-deficient cells, these proteins are minimally expressed at the cell surface as assessed by flow cytometry.3,5 An optional step in the MaxQuant workflow is the “requantify” strategy where ratios are calculated for precursors not assembled into SILAC pairs. This strategy is useful for quantifying low abundance peptides with high ratios, where one of the SILAC partners is at or below noise level,24 as is likely here for light labeled peptide derived from gp96 cell surface clients (M36 cells). When we reprocessed data with requantify turned off, there was much better correspondence between Mascot and Andromeda ratios (Figure S3, Supporting Information) albeit with a reduced number of quantified proteins.
Discussion
We developed plasma membrane profiling to compare the relative abundance of plasma membrane proteins in gp96-deficient versus gp96-reconstituted murine pre-B cells. We identified 706 PM proteins and found a requirement for gp96 in the cell surface expression of 29 of these proteins, including integrins, TLRs, and four members of the extended LDL receptor family (LDLR, LRP6, Sorl1 and LRP8). Our data indicates that gp96 is required for assembly of this functionally important class of proteins and suggests that it may be required to chaperone folding of some beta-propeller domains and leucine-rich repeats.
Our enrichment for PM proteins using PMP compares well with other reports for plasma membrane protein preparations. For example, sucrose density gradient with 2-phase partitioning identified 428 proteins from rat liver with 67% showing plasma membrane annotation.37 Cationic silica microbeads identified a total of 450 proteins from lung tissue with 81% plasma membrane annotation.38 We previously used cationic silica microbeads to enrich PM proteins, to determine novel substrates of the MARCH9 transmembrane E3 ligase39 and identified 157 PM proteins from an immortalised B-lymphocyte cell line (16% of identified and annotated proteins). In our experience, this technique is less reliable than the aminooxy-biotin, giving lower yields and purity of plasma membrane proteins. In comparison to the other techniques tried, including crude membrane preparation, biotinylation of surface proteins with N-hydroxysulfosuccinimydyl-S,S-biotin and cationic silica microbeads, PMP gives us the best PM protein enrichment, the greatest number of PM protein identifications and best reproducibility between biological repeats.20 Cell surface capture (CSC) with biocytin hydrazide ligation of oxidized cell surface glycoproteins identified 96–341 proteins, of which 95% were present at the cell surface, as defined by the presence of at least one transmembrane or GPI anchor domain, a consensus glycosylation motif and an asparagine to aspartic acid deamidation site.40 A major advantage of PMP over CSC is the potential for increased protein sequence coverage and thus improved quantitative accuracy. Whereas CSC identified 1–3 glycopeptides per protein identified, in this study and previous, we identify 1–95 peptides per protein. Other potential advantages of PMP using aminooxy-biotin over CSC with biocytin hydrazide include the more stable product formation with the oxime ligation of aminooxy-biotin to cell surface aldehyde compared to the hydrazone ligation of biocytin hydrazide.41 The use of aniline as a catalyst enables use of aminooxy-biotin at relatively low concentration (100 μM).21
Following fractionation of the tryptic peptide sample, the percentage of proteins annotated PM/CS/XC fell from 70 to 35%. The increased identification of non-PM contaminants seen following high resolution fractionation suggests that contaminants form a minor part of the total protein content and highlights the efficiency of PM protein enrichment. Some of these contaminants might include proteins endogenously biotinylated at low level such as histones.42 The decreased percentage of proteins annotated PM/CS/XC in the fractionated sample also suggests that a proportion of the 143 proteins annotated “short GO” may not be of PM origin. Curation of the murine GO database is however likely to be incomplete—for example, protein S100-A14 (Table S2, Supporting Information) was not annotated with GOCC terms by Perseus, but resides at the plasma membrane.43
An essential role for gp96 in the cell surface expression of four of the ten members of the extended LDLR family adds these functionally and clinically important proteins to the integrins and TLRs in their requirement for gp96 for cell surface expression. Cellular cholesterol uptake is likely to be significantly diminished when cell surface expression of LDLR and other family members is reduced, and may explain the observed increase in proteins implicated in cholesterol metabolism in gp96-deficient cells (Table S1, Supporting Information, Table 2). These include scavenger receptor class B type I (upregulated 1.9 fold) which mediates the bidirectional transfer of cholesterol between cells and HDL cholesterol,44 hydroxymethylglutaryl-CoA synthase (Hmgcs1, upregulated 1.8 fold) which condenses acetyl-CoA with acetoacetyl-CoA to form HMG-CoA, the substrate for HMG-CoA reductase,45 acetoacetyl-CoA synthetase and fatty acid synthase. Together these proteins may compensate for the reduction in cellular cholesterol uptake through the induction of alternative uptake or synthesis pathways. In addition, the mitochondrial/peroxisomal protein delta(3,5)-delta(2,4)-dienoyl-CoA isomerase (Ech1) was upregulated 4.1 fold in gp96 deficient cells (Table 2)—Ech1 acts in the alternative pathway for the beta-oxidation of unsaturated fatty acids, a source of acetyl-CoA.46 Increased Ech1 expression might also provide acetyl-CoA as a substrate for Hmgcs1 and cholesterol synthesis, although other metabolic pathways could also generate this important intermediate.
Since gp96 appears likely to play a key role in chaperoning the assembly of members of the extended LDLR, integrin and TLR protein families, are there shared structural features which shed light on the protein domains recognized by this chaperone and explain its selectivity? A key candidate here is the beta-propeller domain, which is formed from twisted β-sheet structural repeats arranged toroidally around a central axis. The extracellular domain of α-subunit integrins contains a seven-bladed beta-propeller which complexes with the I/A domain to form the ligand-binding head of the integrin.47 All members of the extended LDLR family share the same six-bladed beta-propeller domain, formed by six contiguous YWTD repeats. The core members of this family share further structural features, with ligand-binding cysteine-rich repeats followed by the beta-propeller, flanked by EGF-like cysteine-rich repeats.48 While these domains can be found individually in other proteins, it is only in the LDLR family that they coexist in the same protein.49 Clearly, not all propeller domain-containing proteins require gp96 for correct folding, but the unique interface between the propeller and adjacent modules may require gp96-assisted assembly. LDLR family members also have a specific requirement for the molecular ER chaperone boca (Drosophila melanogaster), and its mammlian orthologue Mesd, for folding and trafficking to the cell surface.50−52 In the absence of this chaperone they are retained in the ER, where gp96 is also most abundant. Boca/mesd is specifically required for maturation of the LDLR family beta-propeller/EGF modules,53−55 and it will therefore be interesting to determine whether Mesd interacts with gp96, and the relative roles of these different chaperones in LDLR assembly. It will also be necessary to confirm biochemically that the changes we observe in surface expression of LDLR family members are due to the absence of specific gp96 chaperone activity, as opposed to changes in transcription or translation of individual client genes. As the known functions of gp96 include chaperoning the folding of proteins, targeting of misfolded proteins to ERAD and storing calcium,1 transcriptional effect would seem unlikely. The known gp96 clients, integrins and TLRs, fail to reach the cell surface in the absence of gp96 due to misfolding1−8—it is therefore likely that LDLR family members are similarly affected in the absence of gp96.
The TLRs play a critical role in innate immunity and are type I transmembrane proteins with common structural features, including an N-terminal ectodomain containing multiple leucine-rich repeats for ligand binding, a short helix embedded in the membrane and a C-terminal cytosolic tail containing the Toll-interleukin-1 receptor (TIR) domain for recruiting signaling adaptors. Though not containing any beta-propeller domains, the widespread, structural leucine-rich repeat of 20–30 amino acids has a characteristic repetitive sequence pattern, built from tandem repeats to form curved solenoid structures, particularly well suited for protein–protein interactions.56 A possible role for gp96 may therefore be chaperoning the assembly of proteins with repeating units, such as repeated leucine-rich repeats, or repeated subunits of a beta-propeller. Gp96-deficient (M36 and E4.126) cells are resistant to infection by vesicular stomatitis virus (VSV) and viruses pseudotyped with VSV-G13 suggesting that one or more of the extended LDLR family, or indeed other gp96-dependent proteins identified in this analysis may be the receptor for VSV-G.
In summary, Gp96 is required for cell surface expression of at least four members of the extended LDL receptor family, as well CD180/Ly86 and most integrins and TLRs. Among the proteins whose cell surface expression was gp96 dependent, we found a significant enrichment for beta-propeller and leucine rich repeats, suggesting an important role of gp96 in the folding of these modules.
Acknowledgments
This work was supported by the Wellcome Trust. M.P.W. is a Wellcome Trust postdoctoral fellow, and P.J.L. is a Wellcome Trust senior clinial fellow.
Supporting Information Available
Tables S1–S2 and Figures S1–S3. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Supplementary Material
References
- Eletto D.; Dersh D.; Argon Y. GRP94 in ER quality control and stress responses. Semin. Cell Dev. Biol. 2010, 21 (5), 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staron M.; Yang Y.; Liu B.; Li J.; Shen Y.; Zuniga-Pflucker J. C.; Aguila H. L.; Goldschneider I.; Li Z. gp96, an endoplasmic reticulum master chaperone for integrins and Toll-like receptors, selectively regulates early T and B lymphopoiesis. Blood 2009, 115 (12), 2380–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randow F.; Seed B. Endoplasmic reticulum chaperone gp96 is required for innate immunity but not cell viability. Nat. Cell Biol. 2001, 3 (10), 891–896. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Liu B.; Dai J.; Srivastava P. K.; Zammit D. J.; Lefrancois L.; Li Z. Heat shock protein gp96 is a master chaperone for toll-like receptors and is important in the innate function of macrophages. Immunity 2007, 26 (2), 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding C. V. gp96 leads the way for toll-like receptors. Immunity 2007, 26 (2), 141–143. [DOI] [PubMed] [Google Scholar]
- Ostrovsky O.; Ahmed N. T.; Argon Y. The chaperone activity of GRP94 toward insulin-like growth factor II is necessary for the stress response to serum deprivation. Mol. Biol. Cell 2009, 20 (6), 1855–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrovsky O.; Eletto D.; Makarewich C.; Barton E. R.; Argon Y. Glucose regulated protein 94 is required for muscle differentiation through its control of the autocrine production of insulin-like growth factors. Biochim. Biophys. Acta 2009, 1803 (2), 333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staron M.; Wu S.; Hong F.; Stojanovic A.; Du X.; Bona R.; Liu B.; Li Z. Heat-shock protein gp96/grp94 is an essential chaperone for the platelet glycoprotein Ib-IX-V complex. Blood 2011, 117 (26), 7136–7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales C.; Wu S.; Yang Y.; Hao B.; Li Z. Drosophila glycoprotein 93 Is an ortholog of mammalian heat shock protein gp96 (grp94, HSP90b1, HSPC4) and retains disulfide bond-independent chaperone function for TLRs and integrins. J. Immunol. 2009, 183 (8), 5121–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick J.; Dul J. L.; Argon Y. Sequential interaction of the chaperones BiP and GRP94 with immunoglobulin chains in the endoplasmic reticulum. Nature 1994, 370 (6488), 373–375. [DOI] [PubMed] [Google Scholar]
- Liu B.; Li Z. Endoplasmic reticulum HSP90b1 (gp96, grp94) optimizes B-cell function via chaperoning integrin and TLR but not immunoglobulin. Blood 2008, 112 (4), 1223–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May P.; Herz J. LDL receptor-related proteins in neurodevelopment. Traffic 2003, 4 (5), 291–301. [DOI] [PubMed] [Google Scholar]
- Bloor S.; Maelfait J.; Krumbach R.; Beyaert R.; Randow F. Endoplasmic reticulum chaperone gp96 is essential for infection with vesicular stomatitis virus. Proc. Natl. Acad. Sci. U.S.A. 2010, 107 (15), 6970–6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutar A. K.; Naoumova R. P. Mechanisms of disease: genetic causes of familial hypercholesterolemia. Nat. Clin. Pract. Cardiovasc. Med. 2007, 4 (4), 214–225. [DOI] [PubMed] [Google Scholar]
- Logan C. Y.; Nusse R. The Wnt signaling pathway in development and disease. Ann. Rev. Cell Dev. Biol. 2004, 20, 781–810. [DOI] [PubMed] [Google Scholar]
- Rogaeva E.; Meng Y.; Lee J. H.; Gu Y.; Kawarai T.; Zou F.; Katayama T.; Baldwin C. T.; Cheng R.; Hasegawa H.; Chen F.; Shibata N.; Lunetta K. L.; Pardossi-Piquard R.; Bohm C.; Wakutani Y.; Cupples L. A.; Cuenco K. T.; Green R. C.; Pinessi L.; Rainero I.; Sorbi S.; Bruni A.; Duara R.; Friedland R. P.; Inzelberg R.; Hampe W.; Bujo H.; Song Y. Q.; Andersen O. M.; Willnow T. E.; Graff-Radford N.; Petersen R. C.; Dickson D.; Der S. D.; Fraser P. E.; Schmitt-Ulms G.; Younkin S.; Mayeux R.; Farrer L. A.; George-Hyslop P. St The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat. Genet. 2007, 39 (2), 168–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu S.; Renfro A.; Quattrocchi C. C.; Sheldon M.; D’Arcangelo G. Reelin promotes hippocampal dendrite development through the VLDLR/ApoER2-Dab1 pathway. Neuron 2004, 41 (1), 71–84. [DOI] [PubMed] [Google Scholar]
- Bloor S.; Ryzhakov G.; Wagner S.; Butler P. J.; Smith D. L.; Krumbach R.; Dikic I.; Randow F. Signal processing by its coil zipper domain activates IKK gamma. Proc. Natl. Acad. Sci. U.S.A. 2008, 105 (4), 1279–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall S. C.; Hughes C.; Stewart M. H.; Doble B.; Bhatia M.; Lajoie G. A. Prevention of amino acid conversion in SILAC experiments with embryonic stem cells. Mol. Cell. Proteomics 2008, 7 (9), 1587–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weekes M. P.; Antrobus R.; Lill J. R.; Duncan L. M.; Hor S.; Lehner P. J. Comparative analysis of techniques to purify plasma membrane proteins. J. Biomol. Tech. 2010, 21 (3), 108–115. [PMC free article] [PubMed] [Google Scholar]
- Zeng Y.; Ramya T. N.; Dirksen A.; Dawson P. E.; Paulson J. C. High-efficiency labeling of sialylated glycoproteins on living cells. Nat. Methods 2009, 6 (3), 207–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J.; Mann M.; Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2 (8), 1896–1906. [DOI] [PubMed] [Google Scholar]
- Wisniewski J. R.; Zougman A.; Nagaraj N.; Mann M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6 (5), 359–362. [DOI] [PubMed] [Google Scholar]
- Cox J.; Matic I.; Hilger M.; Nagaraj N.; Selbach M.; Olsen J. V.; Mann M. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 2009, 4 (5), 698–705. [DOI] [PubMed] [Google Scholar]
- Cox J.; Neuhauser N.; Michalski A.; Scheltema R. A.; Olsen J. V.; Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10 (4), 1794–1805. [DOI] [PubMed] [Google Scholar]
- Adachi J.; Kumar C.; Zhang Y.; Mann M. In-depth analysis of the adipocyte proteome by mass spectrometry and bioinformatics. Mol. Cell. Proteomics 2007, 6 (7), 1257–1273. [DOI] [PubMed] [Google Scholar]
- Shannon P.; Markiel A.; Ozier O.; Baliga N. S.; Wang J. T.; Ramage D.; Amin N.; Schwikowski B.; Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13 (11), 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S.; Heymans K.; Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21 (16), 3448–3449. [DOI] [PubMed] [Google Scholar]
- Gao J.; Ade A. S.; Tarcea V. G.; Weymouth T. E.; Mirel B. R.; Jagadish H. V.; States D. J. Integrating and annotating the interactome using the MiMI plugin for cytoscape. Bioinformatics 2009, 25 (1), 137–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.; Yang Y.; Qiu Z.; Staron M.; Hong F.; Li Y.; Wu S.; Hao B.; Bona R.; Han D.; Li Z. Folding of Toll-like receptors by the HSP90 paralogue gp96 requires a substrate-specific cochaperone. Nat. Commun. 2010, 1, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barczyk M.; Carracedo S.; Gullberg D. Integrins. Cell Tissue Res. 2010, 339 (1), 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake K.; Yamashita Y.; Ogata M.; Sudo T.; Kimoto M. RP105, a novel B cell surface molecule implicated in B cell activation, is a member of the leucine-rich repeat protein family. J. Immunol. 1995, 154 (7), 3333–3340. [PubMed] [Google Scholar]
- Ogata H.; Su I.; Miyake K.; Nagai Y.; Akashi S.; Mecklenbrauker I.; Rajewsky K.; Kimoto M.; Tarakhovsky A. The toll-like receptor protein RP105 regulates lipopolysaccharide signaling in B cells. J. Exp. Med. 2000, 192 (1), 23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake K.; Shimazu R.; Kondo J.; Niki T.; Akashi S.; Ogata H.; Yamashita Y.; Miura Y.; Kimoto M. Mouse MD-1, a molecule that is physically associated with RP105 and positively regulates its expression. J. Immunol. 1998, 161 (3), 1348–1353. [PubMed] [Google Scholar]
- Gorczynski R. M.; Kai Y.; Miyake K. MD1 expression regulates development of regulatory T cells. J. Immunol. 2006, 177 (2), 1078–1084. [DOI] [PubMed] [Google Scholar]
- Danen E. H.; van Rheenen J.; Franken W.; Huveneers S.; Sonneveld P.; Jalink K.; Sonnenberg A. Integrins control motile strategy through a Rho-cofilin pathway. J. Cell. Biol. 2005, 169 (3), 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R.; Li X.; Liu Z.; Peng X.; Hu W.; Wang X.; Chen P.; Xie J.; Liang S. Integration of a two-phase partition method into proteomics research on rat liver plasma membrane proteins. J. Proteome Res. 2006, 5 (3), 634–642. [DOI] [PubMed] [Google Scholar]
- Durr E.; Yu J.; Krasinska K. M.; Carver L. A.; Yates J. R.; Testa J. E.; Oh P.; Schnitzer J. E. Direct proteomic mapping of the lung microvascular endothelial cell surface in vivo and in cell culture. Nat. Biotechnol. 2004, 22 (8), 985–992. [DOI] [PubMed] [Google Scholar]
- Hor S.; Ziv T.; Admon A.; Lehner P. J. Stable isotope labeling by amino acids in cell culture and differential plasma membrane proteome quantitation identify new substrates for the MARCH9 transmembrane E3 ligase. Mol. Cell. Proteomics 2009, 8 (8), 1959–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollscheid B.; Bausch-Fluck D.; Henderson C.; O’Brien R.; Bibel M.; Schiess R.; Aebersold R.; Watts J. D. Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat. Biotechnol. 2009, 27 (4), 378–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia J.; Raines R. T. Hydrolytic stability of hydrazones and oximes. Angew. Chem., Int. Ed. 2008, 47 (39), 7523–7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroishi T.; Rios-Avila L.; Pestinger V.; Wijeratne S. S.; Zempleni J. Biotinylation is a natural, albeit rare, modification of human histones. Mol. Genet. Metab. 2011, 104 (4), 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q.; Chen H.; Luo A.; Ding F.; Liu Z. S100A14 stimulates cell proliferation and induces cell apoptosis at different concentrations via receptor for advanced glycation end products (RAGE). PLoS One 2011, 6 (4), e19375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergeer M.; Korporaal S. J.; Franssen R.; Meurs I.; Out R.; Hovingh G. K.; Hoekstra M.; Sierts J. A.; Dallinga-Thie G. M.; Motazacker M. M.; Holleboom A. G.; Van Berkel T. J.; Kastelein J. J.; Van Eck M.; Kuivenhoven J. A. Genetic variant of the scavenger receptor BI in humans. N. Engl. J. Med. 2011, 364 (2), 136–145. [DOI] [PubMed] [Google Scholar]
- Rokosz L. L.; Boulton D. A.; Butkiewicz E. A.; Sanyal G.; Cueto M. A.; Lachance P. A.; Hermes J. D. Human cytoplasmic 3-hydroxy-3-methylglutaryl coenzyme A synthase: expression, purification, and characterization of recombinant wild-type and Cys129 mutant enzymes. Arch. Biochem. Biophys. 1994, 312 (1), 1–13. [DOI] [PubMed] [Google Scholar]
- Allenbach L.; Poirier Y. Analysis of the alternative pathways for the beta-oxidation of unsaturated fatty acids using transgenic plants synthesizing polyhydroxyalkanoates in peroxisomes. Plant Physiol. 2000, 124 (3), 1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson R. S.; Corbi A. L.; Berman L.; Springer T. Primary structure of the leukocyte function-associated molecule-1 alpha subunit: an integrin with an embedded domain defining a protein superfamily. J. Cell Biol. 1989, 108 (2), 703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon H.; Blacklow S. C. Structure and physiologic function of the low-density lipoprotein receptor. Annu. Rev. Biochem. 2005, 74, 535–562. [DOI] [PubMed] [Google Scholar]
- Bork P.; Downing A. K.; Kieffer B.; Campbell I. D. Structure and distribution of modules in extracellular proteins. Q. Rev. Biophys. 1996, 29 (2), 119–167. [DOI] [PubMed] [Google Scholar]
- Culi J.; Mann R. S. Boca, an endoplasmic reticulum protein required for wingless signaling and trafficking of LDL receptor family members in Drosophila. Cell 2003, 112 (3), 343–354. [DOI] [PubMed] [Google Scholar]
- Hsieh J. C.; Lee L.; Zhang L.; Wefer S.; Brown K.; DeRossi C.; Wines M. E.; Rosenquist T.; Holdener B. C. Mesd encodes an LRP5/6 chaperone essential for specification of mouse embryonic polarity. Cell 2003, 112 (3), 355–367. [DOI] [PubMed] [Google Scholar]
- Li Y.; Chen J.; Lu W.; McCormick L. M.; Wang J.; Bu G. Mesd binds to mature LDL-receptor-related protein-6 and antagonizes ligand binding. J. Cell Sci. 2005, 118 (Pt 22), 5305–5314. [DOI] [PubMed] [Google Scholar]
- Culi J.; Springer T. A.; Mann R. S. Boca-dependent maturation of beta-propeller/EGF modules in low-density lipoprotein receptor proteins. EMBO J. 2004, 23 (6), 1372–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koduri V.; Blacklow S. C. Requirement for natively unstructured regions of mesoderm development candidate 2 in promoting low-density lipoprotein receptor-related protein 6 maturation. Biochemistry 2007, 46 (22), 6570–6577. [DOI] [PubMed] [Google Scholar]
- Liu C. C.; Pearson C.; Bu G. Cooperative folding and ligand-binding properties of LRP6 beta-propeller domains. J. Biol. Chem. 2009, 284 (22), 15299–15307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bella J.; Hindle K. L.; McEwan P. A.; Lovell S. C. The leucine-rich repeat structure. Cell. Mol. Life Sci. 2008, 65 (15), 2307–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.