Abstract
• Background and Aims Weedy rice (Oryza sativa f. spontanea) is one of the most notorious weeds occurring in rice-planting areas worldwide. The objectives of this study are to determine the genetic diversity and differentiation of weedy rice populations from Liaoning Province in North-eastern China and to explore the possible origin of these weedy populations by comparing their genetic relationships with rice varieties (O. sativa) and wild rice (O. rufipogon) from different sources.
• Methods Simple sequence repeat (SSR) markers were used to estimate the genetic diversity of 30 weedy rice populations from Liaoning, each containing about 30 individuals, selected rice varieties and wild O. rufipogon. Genetic differentiation and the relationships of weedy rice populations were analysed using cluster analysis (UPGMA) and principle component analysis (PCA).
• Key Results The overall genetic diversity of weedy rice populations from Liaoning was relatively high (He = 0·313, I = 0·572), with about 35 % of the genetic variation found among regions. The Liaoning weedy rice populations were closely related to rice varieties from Liaoning and japonica varieties from other regions but distantly related to indica rice varieties and wild O. rufipogon.
• Conclusions Weedy rice populations from Liaoning are considerably variable genetically and most probably originated from Liaoning rice varieties by mutation and intervarietal hybrids. Recent changes in farming practices and cultivation methods along with less weed management may have promoted the re-emergence and divergence of weedy rice in North-eastern China.
Keywords: Oryza sativa, O. spontanea, O. rufipogon, weedy rice, molecular markers, simple sequence repeat (SSR), genetic diversity, differentiation, origin
INTRODUCTION
Weeds persist in agricultural ecosystems competing for nutrients, water, sunlight and other resources with crops, and consequently cause a constant challenge for crop production (Dekker, 1997). Appropriate management of weeds occurring in agroecosystems will harmonize the systems and enhance the sustainable crop production. A central characteristic of weeds, which contributes to their success in agroecosystems, is the genetic variability and plasticity found within and among weed populations (Green et al., 2001). This enables weeds to infest a wide range of diverse habitats. Thus, a full understanding of the genetic diversity of weeds is a major prerequisite for their effective management. In addition, elucidating the origin and evolutionary processes of weeds is helpful for designing effective management strategies for weed control (Pyšek and Prach, 2003). As proposed by Baker (1974) and de Wet and Harlan (1975), there are three principal processes for the origin of weeds: (a) wild colonizing plants adapting to continuous habitat disturbances, along with the domestication of crop species; (b) derivatives of hybrids between cultivated plants and their reproductively compatible wild relatives; and (c) abandoned domesticates evolving towards a less intimate association with man. In addition, back-mutation and segregation of distant intervarietal hybrids also play certain roles in the origin of weeds (Burger et al., 2006).
Weedy rice (Oryza sativa f. spontanea) of the Poaceae is a weed accompanying rice and is widely distributed in rice-planting areas all over the world, particularly in South and South-east Asia, South and North America, and southern Europe (Ferrero et al., 1999; Mortimer et al., 2000; Noldin, 2000). Weedy rice is taxonomically classified as the same species as cultivated rice (O. sativa), but is strongly characterized by its seed shattering and dormancy, which apparently increase the distribution of this species. As a notorious weed occurring in rice fields, it commonly causes yield reduction and affects the quality of rice grains (Hoagland and Paul, 1978). Most weedy rice strains possess seeds with red pericarps; thus it is also referred to as red rice (Gealy et al., 2003) although some strains have white pericarps (Arrieta-Espinoza1 et al., 2005). Morphologically, weedy rice is highly variable and appears to be an intermediate between wild and cultivated rice. Long-term sympatric distribution has led to similarities between weedy and cultivated rice through natural hybridization and introgression, making the control of weedy rice very difficult when compared with other weeds. Furthermore, there might be potential environmental consequences of weedy rice because of the commercial release of genetically modified (GM) rice (Chen et al., 2004). Many GM rice varieties possess transgenes coding for traits such as resistance to insect herbivores, diseases, herbicides or tolerance to environmental stresses (Lu and Snow, 2005). These fitness-related transgenes have the potential to be introgressed into weedy rice through hybridization (outcrossing) with GM rice varieties (Chen et al., 2004), compounding the difficulties in managing weedy rice.
Weedy rice was commonly found in rice fields of China (Jiang et al., 1985). During the past several decades, weedy rice was effectively managed and became negligible in Chinese rice agriculture systems because of the manual transplanting technology that was adopted predominantly in rice cultivation, in addition to intensive inputs of human labour for weed control (Zhang, 2000). However, in recent years, weedy rice has re-emerged in many rice-planting areas in China, particularly in the North-east, and Jiangsu and Hainan Provinces (Sun et al., 2005; Wang et al., 2005; Yu et al., 2005). Weedy rice has spread rapidly in these regions where direct seeding or related technologies were adopted and gradually became popular, accompanied by less weed management (Yu et al., 2005). The occurrences of weedy rice in North-eastern China have already caused considerable problems for rice production in the region (Ma et al., 2005). If it continues to spread, larger weed problems for rice production may occur, particularly where exchange of rice varieties (sometimes with weedy rice seeds) becomes more frequent between regions and less manpower is allocated for weed control in general.
Despite the fast emergence of weedy rice in China, knowledge of its genetic diversity along with procedures and mechanisms of its reoccurrence in these regions is still limited, which hinders the design of effective practical tools and methods for weedy rice management. In order to fill the knowledge gaps, it is necessary to study weedy rice populations occurring in the region with a proper sampling strategy and to compare them with sets of rice varieties and wild rice from other regions using more powerful tools for characterization. This will enable us to understand the level and distribution of the genetic diversity of weedy rice populations over the regions and to explore the possible origin of weedy rice occurring in these regions for their effective control.
The rapid development of molecular markers provides an effective tool for studying genetic diversity and population differentiation of plant species (Parker et al., 1998; O'Hanlon et al., 2000). Simple sequence repeats (SSRs), also referred as microsatellites, are frequently used as allele-specific and co-dominant markers in plants to study genetic diversity and evolutionary relationships. SSRs are distributed widely in eukaryotic genomes and are suitable for studying genetic variation in populations because high levels of polymorphism can generally be detected, even for closely related individuals (Chen et al., 1997; Song et al., 2003).
During a field survey, a large number of weedy rice individuals from different rice-planting areas in North-eastern China were collected. Additionally, a wide range of cultivated rice varieties from the same region and other parts of China, as well as from other countries, in addition to some wild rice (O. rufipogon) accessions from different sources were assembled. Extensive molecular analyses were conducted to investigate variation patterns of these weedy rice populations, rice varieties and wild rice accessions. The objectives of this study were to (a) determine the level and distribution of genetic diversity of weedy rice populations in North-eastern China; (b) investigate differentiation of weedy rice populations from different localities; and (c) explore the possible origin of weedy rice occurring in North-eastern China by comparing genetic relationships of weedy rice populations with a wide range of cultivated and wild rice accessions from different sources.
MATERIALS AND METHODS
Plant materials
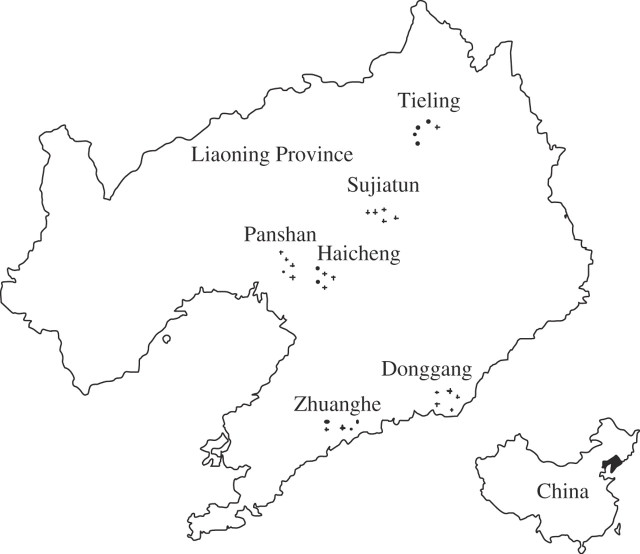
After an extensive survey of weedy rice infestation in all rice-planting areas of North-eastern China, six regions in Liaoning Province were selected as sampling sites (Fig. 1, Table 1), where weedy rice was found by farmers in different years (from 1985 to 2001). Information on the occurrence of weedy rice during the past 16 years was obtained by field investigations and questionnaires to local farmers who owned the rice fields with weedy rice and to regional agricultural extension workers. These regions represented almost all of the rice-planting areas in Liaoning and included some of the areas with the most severe weedy rice occurrence in North-eastern China. Seed samples of weedy rice were collected from rice fields of these regions from late August to early September 2004. Five rice fields (infested by weedy rice) with a distance of at least 1 km from each other were randomly selected for sampling within each region. About 30 weedy rice individuals were randomly sampled from each field. All individuals from the same field were regarded as members of one population. One to three panicles from each individual plant were collected separately, placed in a paper seed-bag and naturally air dried. The area of each sampled field was about 5000 m2. The rice variety grown in the same field was also collected a few days later after the weedy rice was sampled because the cultivated rice usually matured about a week later than weedy rice.
Fig. 1.
Sampling sites of weedy rice (Oryza sativa f. spontanea) populations in Liaoning Province of China.
Table 1.
Population samples of weedy rice (Oryza sativa f. spontanea) collected from Liaoning Province included in the SSR analysis
| Locality in Liaoning Province | Population code for each region | No. of individuals collected in each population | Year when weedy rice was found |
|---|---|---|---|
| Tieling County | tl1, tl2, tl3, tl4, tl5 | 30−31 | 1998 |
| Sujiatun County | sjt1, sjt2, sjt3, sjt4, sjt5 | 30−31 | 1994 |
| Panshan County | ps1, ps2, ps3, ps4, ps5 | 30−31 | 2001 |
| Haicheng County | hc1, hc2, hc3, hc4, hc5 | 30−31 | 2000 |
| Donggang County | dg1, dg2, dg3, dg4, dg5 | 30−32 | 1985 |
| Zhuanghe County | zh1, zh2, zh3, zh4, zh5 | 30−32 | 1997 |
For the analysis of the genetic relationships of weedy rice populations and the comparison of cultivated and wild rice, samples of rice varieties and wild O. rufipogon from different origins were assembled and included in the analysis (see details in Table 2). Cultivated rice was divided into five groups based on their origins: (a) group ‘c-LN’, from Liaoning Province; (b) group ‘jap-Ch’, japonica varieties from other regions of China; (c) group ‘in-Ch’, indica varieties from other regions of China; (d) group ‘c-JK’, varieties from Japan and South Korea; and (e) group ‘c-Ph’, varieties from the Philippines. Wild rice was divided into three groups that were collected from China (coded as ‘w-Ch’), South-east Asia (‘w-SEA’) and South Asia (‘w-SA’).
Table 2.
Samples of the cultivated rice (Oryza sativa) and perennial common wild rice (O. rufipogon) of different origins used in the SSR analysis
| Origin of cultivated and wild rice with their group codes (in parentheses) | Variety name or accession/sample code (for wild rice) |
|---|---|
| O. sativa | |
| Liaoning Province, China (c-LN) | Danhannuo-3, Danjing-4, Danjing-8, Dongshi-8, Fengmin-2000, Gangyuan-3, Han-9710, Huanghai-2, Jingyue-2, Liao-263, Liao-912, Liaojing-137, Liaojing-263, Liaojing-288, Liaojing-294, Liaojing-92-34, Liaokai-79, Liaoxing-1, Liaoyou-7, Liaoyou-14, Liaoyou-0201, Liaoyou-2000, Liaozuo-2000-4, Qingfengnuo, Pingzhi-15, Shennong-606, Tiejing-7, Yanjing-68, Zhongzuo-9052, Zhuangyu-3 (30 varieties) |
| Jiangsu, Jiangxi, Shaanxi, Sichuan, Zhejiang Provinces, China, (japonica rice group, jap-Ch) | Jiangyinzao, Manyedao, Baimiguannian, Hubeizao, Sichengoujiangmidao, Madaozi, Xiangu, Zagu, Zhushituo, Zihong (10 varieties) |
| Anhui, Guizhou, Hubei, Hunan, Jiangsu Provinces, China (indica rice group, in-Ch) | Tianzuozao, Yingqiunuo, Baikeshangu, Funingxiaobairen, Weinanhongmangdao, Danuo, Guidongxian, Huangguanuo, Niangunuo, Tutounuodao (10 varieties) |
| Japan and South Korea (c-JK) | AkitaKomachi, Koshihikari, Nipponbare, Kinuhikari, Yirihu, Taisheyuo-Mochi (six varieties) |
| The Philippines (c-Ph) | IR36, IR64 (two varieties) |
| O. rufipogon | |
| Guangdong, Hunan, Jiangxi Provinces, China (w-Ch) | G-1, G-2, G-3, G-4, H-1, H-2, H-3, H-4, J-1, J-2, J-3, J-4, J-5 (three accessions) |
| Thailand, Cambodia, Malaysia (w-SEA) | 104646, 105735, 106036 (three accessions) |
| India, Sri Lanka, Nepal, Bangladesh (w-SA) | 101974, 105424, 105698, 105887 (four accessions) |
DNA extraction and SSR analysis
One good seed was randomly selected from each of the individual weedy rice samples, rice varieties and wild rice accessions for germination. Seeds of weedy and wild rice were treated in an oven at 50 °C (dry-heating) for 2 d before germination to break seed dormancy. Total genomic DNA was extracted from 7-day-old fresh seedlings following a modified cetyltrimethyl ammonium bromide (CTAB) protocol (Saghai-Maroof et al., 1984). Twenty SSR primer pairs designed from cultivated rice were used to assay genetic variation of all included materials, based on the RiceGenes Database (http://gramene.org). Detailed information of the primer pairs is given in Table 3. The polymerase chain reactions (PCRs) were performed in a PTC 10096v thermocycler (MJ Research Inc., Watertown, MA, USA) programmed as described by Wu and Tanksley (1993). A denaturation period of 4 min at 94 °C was followed by 36 cycles of 40 s at 94 °C, 30 s at 55 °C and 40 s at 72 °C, and then 10 min at 72 °C for the final extension. Reactions were carried out in a volume of 20 μL containing 1× buffer, 1 mm each of dATP, dCTP, dGTP and dTTP, 10 mm of SSR primer, 50 ng of genomic DNA and 0·6 U of Taq polymerase (TaKaRa Inc.). The PCR products were separated on 6 % polyacrylamide denaturing gels and silver stained as described by Song et al. (2003). The molecular size of amplified alleles for each SSR locus was determined based on its migration relative to the pUC19 DNA/MspI (HpaII) marker (Fermentas International Inc.), as well as the three control varieties IR36, IR64 and Nipponbare that were loaded in each gel as references. Banding patterns identified in the rice varieties IR36, IR64 and Nipponbare are available at the RiceGenes Database.
Table 3.
SSR primer pairs used for DNA amplification in this study with their parameters of genetic diversity of overall weedy rice (Oryza sativa f. spontanea) populations from Liaoning Province
| Primer code | LO | SSR motif | Forward (5′−3′) | Reverse (5′−3′) | Na | He | Molecular weight (bp) |
|---|---|---|---|---|---|---|---|
| RM11 | 7 | (GA)17 | tctcctcttcccccgatc | atagcgggcgaggcttag | 7 | 0·414 | 121−147 |
| RM14 | 1 | (GA)17 | ccgaggagaggagttcgac | gtgccaatttcctcgaaaaa | 3 | 0·016 | 174−191 |
| RM17 | 12 | (GA)21 | tgccctgttattttcttctctc | ggtgatcctttcccatttca | 4 | 0·357 | 162−187 |
| RM19 | 12 | (ATC)10 | caaaaacagagcagatgac | ctcaagatggacgccaaga | 1 | 0·000 | 216 |
| RM21 | 11 | (GA)21 | acagtattccgtaggcacgg | gctccatgagggtggtagag | 10 | 0·494 | 132−160 |
| RM44 | 8 | (GA)16 | acgggcaatccgaacaacc | tcgggaaaacctaccctacc | 9 | 0·713 | 100−140 |
| RM55 | 3 | (GA)17 | ccgtcgccgtagtagagaag | tcccggttattttaaggcg | 5 | 0·056 | 219−238 |
| RM84 | 1 | (TCT)10 | taagggtccatccacaagatg | ttgcaaatgcagctagagtac | 4 | 0·452 | 110−128 |
| RM167 | 11 | GGAA(GA)16GGGG | gatccagcgtgaggaacacgt | agtccgaccacaaggtgcgttgtc | 4 | 0·549 | 124−150 |
| RM180 | 7 | (ATT)10 | ctacatcggcttaggtgtagcaacacg | acttgctctacttgtggtgagggactg | 2 | 0·492 | 110−111 |
| RM205 | 9 | (GA)25 | ctggttctgtatgggagcag | ctggcccttcacgtttcagtg | 2 | 0·114 | 154−156 |
| RM211 | 2 | (GA)18 | ccgatctcatcaaccaactg | cttcacgaggatctcaaagg | 2 | 0·003 | 143−156 |
| RM212 | 1 | (GA)24 | ccactttcagctactaccag | cacccatttgtctctcattatg | 3 | 0·010 | 118−136 |
| RM215 | 9 | (GA)16 | caaaatggagcagcaagagc | tgagcacctccttctctgtag | 4 | 0·342 | 146−152 |
| RM219 | 9 | (GA)17 | cgtcggatgatgtaaagcct | catatcggcattcgcctg | 7 | 0·723 | 192−214 |
| RM230 | 8 | (GA)13 | gccagaccgtggatgttc | caccgcagtcacttttcaag | 3 | 0·385 | 253−257 |
| RM253 | 6 | (GA)25 | tccttcaagagtgcaaaacc | gcattgtcatgtcgaagcc | 7 | 0·294 | 133−146 |
| RM276 | 6 | (AG)8A3(GA)33 | ctcaacgttgacacctcgtg | tcctccatcgagcagtatca | 6 | 0·458 | 75−144 |
| RM280 | 4 | (GA)16 | acacgatccactttgcgc | tgtgtcttgagcagccagg | 4 | 0·009 | 151−175 |
| RM289 | 5 | G11(GA)16 | ttccatggcacacaagcc | ctgtgcacgaacttccaaag | 4 | 0·375 | 85−110 |
LO, location on chromosome number; Na, observed number of alleles; He, expected heterozygosity.
SSR banding score and statistical analysis
The amplified SSR DNA bands representing different alleles were scored as different genotypes because of the co-dominant nature of SSR markers. As a result, the bands were recorded as homozygous genotypes (AA, BB, CC…) or heterozygous genotypes (AB, AC, BC…). Genetic parameters such as average observed allele number (Na), the percentage of polymorphic loci (P), observed heterozygosity (Ho), expected heterozygosity (He) and Shannon's diversity index (I) were calculated to estimate the level of genetic diversity. The F-statistics (Fis, Fit and Fst) (Wright, 1978) were computed for polymorphic loci to test for the departure from Hardy–Weinberg equilibrium and to estimate genetic differentiation among weedy rice populations. The outcrossing rate [t = (1− Fit)/(1 + Fit)] was calculated based on the Fit values to estimate indirectly the mating pattern of weedy rice populations (Wright, 1978). All of the calculations were performed using POPGENE version 1·31 (Yeh et al., 1999). To estimate correlation between the level of genetic diversity and the year when weedy rice was found, a linear correlation analysis (y = a + bx) was conducted using STATISTICA version 6·0 (StatSoft Inc., OK, USA).
A hierarchical analysis of molecular variance (AMOVA) of weedy rice populations was calculated to partition genetic diversity within and among regions. For AMOVA, the genotype banding patterns were converted into a ‘1’ (present) and ‘0’ (absence) matrix and subject to analysis, using ARLEQUIN version 3·0 (Excoffier et al., 2005).
Genetic relationships among weedy rice populations and different groups of cultivated rice (c-LN, jap-Ch, in-Ch, c-JK and c-Ph) and wild O. rufipogon (w-Ch, w-SEA and w-SA) based on their origins were estimated by genetic distance using Nei's unbiased genetic distance coefficient [GDij = −lnΣpipj/(Σpi2Σpj2)1/2] (Nei, 1978). The genetic distance was calculated by the GENETIX 4·02 computer package (Belkhir, 1999). The resulting genetic distance matrix was used for a cluster analysis according to the unweighted pair-group method with arithmetic averages (UPGMA), using the software program NTSYSpc version 2·0 (Rohlf, 1998).
To assess further the genetic relationships of weedy rice populations and selected rice varieties (from Liaoning, japonica and indica ecotypes from China) as individual plants, a principle component analysis (PCA) was conducted based on the SSR variation patterns (converted into the 1 and 0 matrix). The correlation matrix was selected to calculate coefficients of the first three principal components, using MINITAB version 14·13 (Minitab Inc., State College, PA, USA).
In addition, to detect the proportion of the SSR alleles shared among weedy rice populations, Liaoning cultivated rice varieties, rice varieties from other regions (excluding Liaoning Province) and wild O. rufipogon, the genetic distance (Dps) as defined by the proportion of shared alleles [ps = ΣkΣamin(fa,i, fa,j)/D] at the 20 SSR loci (Bowcock et al., 1994) was calculated, where fa,i/a,j is the frequency of alleles in population i/j, and D is the number of loci. The Dps was calculated using the Microsatellite Analyser (MSA) version 4·00 (Dieringer et al., 2002).
RESULTS
Genetic diversity of weedy rice populations
Nineteen out of the selected 20 rice SSR primer pairs showed allelic polymorphism in weedy rice populations from Liaoning Province. A total of 91 alleles with an average of 4·55 per locus (ranging from 75 to 257 bp) were generated by the 20 SSR primers (Table 3). The highest number of alleles was scored at the locus RM21 (10 alleles) and the highest level of genetic diversity was detected at the locus RM219 (He = 0·723, Table 3). In addition, a few rare alleles were observed exclusively in some weedy populations, such as the Panshan (RM17, 187 bp), Sujiatun (RM21, 145 bp; RM44, 140 bp), Tieling (RM44, 140 bp) and Haicheng (RM280, 175 bp) populations.
Judging from the overall genetic parameters (Table 4), considerable genetic diversity was found in Liaoning weedy rice populations (the He of all weedy rice populations was 0·313, I was 0·572) but the genetic diversity was not evenly distributed across populations and regions. The highest level of genetic diversity was found in the Panshan region (He = 0·380, I = 0·728), whereas the lowest was scored in the Donggang region (He = 0·011, I = 0·030). At the population level, the greatest and least genetic diversity was also found in Panshan and Donggang populations, respectively (data not shown).
Table 4.
Genetic diversity of 30 weedy rice (Oryza sativa f. spontanea) populations from six regions in Liaoning Province estimated based on polymorphisms of the 20 SSR loci (numbers in parentheses indicate standard deviations)
| Region | Na | P (%) | Ho | He | I | Fis | Fit | Fst | t |
|---|---|---|---|---|---|---|---|---|---|
| Tieling | 2·350 (1·424) | 70 | 0·007 (0·024) | 0·254 (0·243) | 0·434 (0·428) | 0·968 | 0·974 | 0·178 | 0·013 |
| Sujiatun | 2·900 (1·889) | 80 | 0·002 (0·005) | 0·151 (0·165) | 0·296 (0·292) | 0·986 | 0·989 | 0·242 | 0·006 |
| Panshan | 4·100 (2·150) | 95 | 0·030 (0·037) | 0·380 (0·281) | 0·728 (0·575) | 0·894 | 0·921 | 0·252 | 0·041 |
| Haicheng | 2·650 (1·387) | 85 | 0·002 (0·007) | 0·236 (0·230) | 0·397 (0·364) | 0·985 | 0·990 | 0·337 | 0·005 |
| Donggang | 1·550 (0·826) | 40 | 0·001 (0·001) | 0·011 (0·016) | 0·030 (0·043) | 0·968 | 0·969 | 0·043 | 0·016 |
| Zhuanghe | 2·400 (1·501) | 70 | 0·011 (0·034) | 0·197 (0·202) | 0·346 (0·359) | 0·930 | 0·946 | 0·228 | 0·028 |
| Overall | 4·550 (2·417) | 95 | 0·009 (0·014) | 0·313 (0·239) | 0·572 (0·457) | 0·958 | 0·973 | 0·348 | 0·014 |
Na, average number of alleles; P, percentage of polymorphic loci; Ho, observed heterozygosity; He, expected heterozygosity; I, Shannon diversity index; Fis, Fit and Fst, estimates of F-statistics of sub- and regional populations (Hartl and Clark 1989); t, outcrossing rate = (1 − Fit)/(1 + Fit).
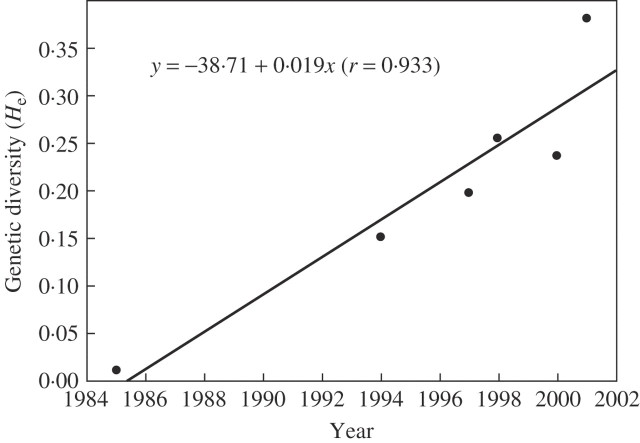
The analytical data further showed a positive correlation between genetic diversity and the year when weedy rice was found in the regions (r = 0·933, P < 0·01, Fig. 2). Among the six regions, the Panshan weedy populations that were found in the most recent years (since 2001) possessed the highest level of genetic diversity. In contrast, the Donggang populations that were found relatively early in 1985 showed the lowest level of genetic diversity.
Fig. 2.
Linear correlation between genetic diversity (He) and the years when weedy rice was found in different localities of Liaoning Province.
Differentiation of weedy rice populations
The values of Fst revealed nearly 35 % of the total genetic variation existing among regions (Table 4). Genetic differentiation among populations of a region was not evenly distributed across the sampled area (Fst = 0·043−0·337). The AMOVA analysis also confirmed this differentiation pattern, with 35·14 % (P < 0·001) of total genetic variation among regions, 18·65 % (P < 0·001) among populations within regions and 46·21 % (P < 0·001) within populations (Table 5). The F-statistics showed high positive values (close to 1) of Fis and Fit in weedy rice populations within and among regions (Table 4), indicating the lack of heterozygosity in weedy rice populations, and the significant departure of allelic frequencies from the Hardy–Weinberg equilibrium. The overall outcrossing rate of weedy rice populations was considerably low (1·4 %), with the highest value (4·1 %) observed in the Panshan populations (Table 4).
Table 5.
Analysis of molecular variance (AMOVA) based on the 20 SSR loci of 909 weedy rice (Oryza sativa f. spontanea) individuals
| Source of variation | d.f. | SSD | CV | % Total | P-value |
|---|---|---|---|---|---|
| Among regions | 5 | 1981·54 | 2·35 | 35·14 | <0·001 |
| Among populations within regions | 24 | 979·60 | 1·25 | 18·65 | <0·001 |
| Within populations | 879 | 2711·98 | 3·09 | 46·21 | <0·001 |
| Total | 908 | 5673·12 | 6·69 |
d.f., degree of freedom; SSD, sum of squared deviations; CV, variance component estimates; % total, percentage of total variation)
Genetic relationships of weedy rice populations
In order to understand the genetic relationships of weedy rice populations to cultivated rice and wild O. rufipogon from different sources and origins, the genetic distance index was calculated among the groups. Table 6 summarizes the genetic distance values obtained by the pairwise method. Weedy rice populations showed a close genetic distance with rice varieties from Liaoning, compared with that with cultivated and wild rice from other origins. To illustrate the genetic relationships among the various groups, a Nei's UPGMA dendrogram based on the genetic distance was constructed (Fig. 3), involving weedy rice populations, rice varieties from Liaoning and other areas (China, Japan, South Korea and the Philippines) and wild O. rufipogon from China and other Asian countries. The UPGMA dendrogram showed that all weedy rice populations and the group of rice varieties from Liaoning Province were clustered closely into a large group, closely associated with the group of japonica rice varieties. The weedy–japonica rice groups were then clustered with the Japanese–South Korean rice varieties and Chinese wild O. rufipogon. In the dendrogram, wild O. rufipogon from South-east and South Asian countries and indica rice varieties from China and the Philippines clustered relatively distant from the Liaoning weedy rice populations (Fig. 3).
Table 6.
Genetic distance (Nei, 1978) among the weedy rice (Oryza sativa f. spontanea) populations, groups of cultivated (O. sativa) and wild rice (O. rufipogon) with different origins
| G. D. | dg1 | dg2 | dg3 | dg4 | dg5 | hc1 | hc2 | hc3 | hc4 | hc5 | ps1 | ps2 | ps3 | ps4 | ps5 | sjt1 | sjt2 | sjt3 | sjt4 | sjt5 | tl1 | tl2 | tl3 | tl4 | tl5 | zh1 | zh2 | zh3 | zh4 | zh5 | c-LN | jap-Ch | in-Ch | c-JK | c-Ph | w-Ch | w-SEA | w-SA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dg1 | – | |||||||||||||||||||||||||||||||||||||
| dg2 | 0 | – | ||||||||||||||||||||||||||||||||||||
| dg3 | 0 | 0·001** | – | |||||||||||||||||||||||||||||||||||
| dg4 | 0 | 0·001** | 0·001* | – | ||||||||||||||||||||||||||||||||||
| dg5 | 0 | 0·000† | 0·000** | 0·000† | – | |||||||||||||||||||||||||||||||||
| hc1 | 0·362 | 0·362 | 0·371 | 0·349 | 0·369 | – | ||||||||||||||||||||||||||||||||
| hc2 | 0·286 | 0·284 | 0·284 | 0·272 | 0·290 | 0·172 | – | |||||||||||||||||||||||||||||||
| hc3 | 0·402 | 0·400 | 0·401 | 0·387 | 0·407 | 0·125 | 0·055 | – | ||||||||||||||||||||||||||||||
| hc4 | 0·388 | 0·386 | 0·385 | 0·368 | 0·392 | 0·223 | 0·049 | 0·132 | – | |||||||||||||||||||||||||||||
| hc5 | 0·276 | 0·273 | 0·283 | 0·260 | 0·281 | 0·110 | 0·110 | 0·095 | 0·132 | – | ||||||||||||||||||||||||||||
| ps1 | 0·208 | 0·204 | 0·210 | 0·210 | 0·213 | 0·268 | 0·151 | 0·169 | 0·283 | 0·209 | – | |||||||||||||||||||||||||||
| ps2 | 0·376 | 0·370 | 0·377 | 0·372 | 0·382 | 0·457 | 0·243 | 0·307 | 0·254 | 0·235 | 0·212 | – | ||||||||||||||||||||||||||
| ps3 | 0·235 | 0·233 | 0·235 | 0·230 | 0·239 | 0·299 | 0·157 | 0·236 | 0·217 | 0·207 | 0·132 | 0·139 | – | |||||||||||||||||||||||||
| ps4 | 0·406 | 0·405 | 0·406 | 0·393 | 0·412 | 0·403 | 0·148 | 0·239 | 0·178 | 0·218 | 0·288 | 0·231 | 0·175 | – | ||||||||||||||||||||||||
| ps5 | 0·188 | 0·185 | 0·192 | 0·181 | 0·193 | 0·195 | 0·078 | 0·120 | 0·174 | 0·079 | 0·084 | 0·170 | 0·135 | 0·127 | – | |||||||||||||||||||||||
| sjt1 | 0·419 | 0·413 | 0·426 | 0·408 | 0·424 | 0·316 | 0·227 | 0·162 | 0·334 | 0·162 | 0·191 | 0·433 | 0·440 | 0·390 | 0·168 | – | ||||||||||||||||||||||
| sjt2 | 0·304 | 0·298 | 0·310 | 0·296 | 0·309 | 0·319 | 0·195 | 0·202 | 0·269 | 0·130 | 0·152 | 0·261 | 0·325 | 0·311 | 0·099 | 0·072 | – | |||||||||||||||||||||
| sjt3 | 0·357 | 0·351 | 0·362 | 0·349 | 0·362 | 0·253 | 0·175 | 0·129 | 0·285 | 0·126 | 0·151 | 0·333 | 0·349 | 0·329 | 0·114 | 0·030 | 0·045 | – | ||||||||||||||||||||
| sjt4 | 0·349 | 0·343 | 0·354 | 0·342 | 0·354 | 0·304 | 0·179 | 0·141 | 0·280 | 0·145 | 0·142 | 0·328 | 0·351 | 0·303 | 0·117 | 0·011* | 0·055 | 0·020* | – | |||||||||||||||||||
| sjt5 | 0·334 | 0·330 | 0·341 | 0·325 | 0·339 | 0·258 | 0·200 | 0·137 | 0·347 | 0·154 | 0·134 | 0·398 | 0·379 | 0·422 | 0·130 | 0·047 | 0·081 | 0·067 | 0·059 | – | ||||||||||||||||||
| tl1 | 0·180 | 0·179 | 0·185 | 0·177 | 0·184 | 0·193 | 0·225 | 0·211 | 0·350 | 0·135 | 0·143 | 0·307 | 0·271 | 0·357 | 0·105 | 0·209 | 0·151 | 0·152 | 0·162 | 0·158 | – | |||||||||||||||||
| tl2 | 0·233 | 0·231 | 0·239 | 0·228 | 0·236 | 0·225 | 0·263 | 0·276 | 0·316 | 0·148 | 0·213 | 0·321 | 0·317 | 0·437 | 0·164 | 0·304 | 0·193 | 0·223 | 0·249 | 0·238 | 0·042* | – | ||||||||||||||||
| tl3 | 0·327 | 0·322 | 0·333 | 0·318 | 0·332 | 0·254 | 0·203 | 0·186 | 0·240 | 0·083 | 0·196 | 0·275 | 0·330 | 0·313 | 0·115 | 0·134 | 0·071 | 0·087 | 0·104 | 0·137 | 0·082 | 0·069 | – | |||||||||||||||
| tl4 | 0·223 | 0·219 | 0·227 | 0·217 | 0·227 | 0·234 | 0·148 | 0·157 | 0·257 | 0·104 | 0·120 | 0·287 | 0·256 | 0·307 | 0·058 | 0·139 | 0·074 | 0·096 | 0·113 | 0·075 | 0·081 | 0·107 | 0·055 | – | ||||||||||||||
| tl5 | 0·186 | 0·184 | 0·190 | 0·186 | 0·192 | 0·201 | 0·122 | 0·131 | 0·232 | 0·119 | 0·065* | 0·203 | 0·194 | 0·274 | 0·044 | 0·161 | 0·094 | 0·098 | 0·107 | 0·103 | 0·055 | 0·082 | 0·071 | 0·036 | – | |||||||||||||
| zh1 | 0·087 | 0·087 | 0·088 | 0·085 | 0·089 | 0·318 | 0·218 | 0·305 | 0·360 | 0·252 | 0·184 | 0·331 | 0·228 | 0·386 | 0·150 | 0·363 | 0·260 | 0·292 | 0·295 | 0·284 | 0·099 | 0·152 | 0·239 | 0·168 | 0·119 | – | ||||||||||||
| zh2 | 0·075 | 0·074 | 0·073 | 0·073 | 0·075 | 0·437 | 0·247 | 0·355 | 0·377 | 0·302 | 0·192 | 0·340 | 0·245 | 0·415 | 0·186 | 0·389 | 0·286 | 0·327 | 0·310 | 0·308 | 0·137 | 0·163 | 0·260 | 0·200 | 0·135 | 0·026 | – | |||||||||||
| zh3 | 0·112 | 0·112 | 0·111 | 0·108 | 0·114 | 0·397 | 0·177 | 0·278 | 0·292 | 0·237 | 0·163 | 0·274 | 0·224 | 0·325 | 0·126 | 0·304 | 0·209 | 0·253 | 0·232 | 0·232 | 0·121 | 0·139 | 0·190 | 0·137 | 0·091 | 0·041 | 0·019 | – | ||||||||||
| zh4 | 0·143 | 0·141 | 0·147 | 0·142 | 0·148 | 0·194 | 0·141 | 0·194 | 0·260 | 0·134 | 0·116 | 0·237 | 0·150 | 0·261 | 0·064 | 0·289 | 0·196 | 0·208 | 0·221 | 0·218 | 0·048 | 0·099 | 0·144 | 0·087 | 0·054 | 0·073 | 0·114 | 0·104 | – | |||||||||
| zh5 | 0·124 | 0·124 | 0·128 | 0·120 | 0·126 | 0·279 | 0·236 | 0·318 | 0·375 | 0·216 | 0·193 | 0·333 | 0·254 | 0·345 | 0·124 | 0·344 | 0·240 | 0·275 | 0·277 | 0·265 | 0·058 | 0·111 | 0·199 | 0·144 | 0·115 | 0·044 | 0·078* | 0·078* | 0·065 | – | ||||||||
| c-LN | 0·240 | 0·237 | 0·240 | 0·245 | 0·245 | 0·453 | 0·193 | 0·282 | 0·321 | 0·290 | 0·095 | 0·147 | 0·107 | 0·212 | 0·110 | 0·370 | 0·252 | 0·277 | 0·264 | 0·328 | 0·232 | 0·317 | 0·280 | 0·191 | 0·124 | 0·213 | 0·217 | 0·177 | 0·109 | 0·240 | – | |||||||
| jap-Ch | 0·477 | 0·471 | 0·477 | 0·476 | 0·483 | 0·560 | 0·409 | 0·432 | 0·531 | 0·362 | 0·271 | 0·394 | 0·318 | 0·393 | 0·268 | 0·366 | 0·292 | 0·300 | 0·309 | 0·399 | 0·356 | 0·398 | 0·336 | 0·316 | 0·315 | 0·406 | 0·449 | 0·397 | 0·346 | 0·394 | 0·312 | – | ||||||
| in-Ch | 1·067 | 1·067 | 1·070 | 1·064 | 1·067 | 1·186 | 1·190 | 1·285 | 1·162 | 1·245 | 1·015 | 1·025 | 0·811 | 1·232 | 1·095 | 1·336 | 1·086 | 1·275 | 1·222 | 1·293 | 1·099 | 1·183 | 1·327 | 1·273 | 1·111 | 1·055 | 1·071 | 1·075 | 1·099 | 1·018 | 1·089 | 0·882 | – | |||||
| c-JK | 0·573 | 0·572 | 0·572 | 0·574 | 0·576* | 0·888 | 0·613 | 0·819 | 0·704 | 0·744 | 0·539 | 0·585 | 0·494 | 0·733 | 0·580 | 0·895 | 0·748 | 0·774 | 0·781 | 0·859 | 0·687 | 0·724 | 0·783 | 0·671 | 0·595 | 0·540 | 0·542 | 0·519 | 0·555 | 0·530 | 0·464* | 0·662* | 1·526 | – | ||||
| c-Ph | 2·432 | 2·432** | 2·474 | 2·469 | 2·478 | 1·441** | 1·632** | 1·642 | 1·465** | 1·650 | 1·820 | 1·550 | 1·298 | 1·715** | 1·899 | 1·936** | 2·157** | 1·883** | 1·794** | 2·396** | 1·956 | 2·204 | 1·964 | 2·292 | 1·889 | 1·959 | 2·448 | 2·348 | 1·729 | 2·153 | 1·722 | 1·442† | 0·860 | 2·403 | – | |||
| w-Ch | 0·785 | 0·789 | 0·792 | 0·779 | 0·790 | 0·867 | 0·806 | 0·974 | 0·923 | 0·857 | 0·793 | 0·929 | 0·716 | 0·786 | 0·760 | 1·015 | 1·023 | 0·964 | 0·930 | 1·077 | 0·778 | 0·896 | 1·008 | 0·937 | 0·864 | 0·722 | 0·791 | 0·784 | 0·712 | 0·648 | 0·792 | 0·738 | 1·050 | 0·595 | 1·455† | – | ||
| w-SEA | 1·425* | 1·442 | 1·430* | 1·416 | 1·434 | 1·338* | 1·323 | 1·467 | 1·412* | 1·456 | 1·408 | 1·341* | 1·201 | 1·421 | 1·381* | 2·093* | 1·806 | 1·988* | 1·959 | 1·658 | 1·301 | 1·459 | 1·721* | 1·472* | 1·452 | 1·229* | 1·413 | 1·396 | 1·204 | 1·206* | 1·420* | 1·696 | 1·134 | 1·883 | 1·477† | 1·014** | – | |
| w-SA | 1·496 | 1·494 | 1·494 | 1·482 | 1·498* | 1·620 | 1·387 | 1·667* | 1·424* | 1·482* | 1·541* | 1·300 | 1·220 | 1·247 | 1·380 | 1·922* | 1·827 | 1·749* | 1·728* | 2·109 | 1·562 | 1·661 | 1·784 | 1·758* | 1·599 | 1·435 | 1·468 | 1·452 | 1·351 | 1·367 | 1·273 | 1·161** | 1·563** | 1·204 | 1·793 | 0·693* | 0·443† | – |
Symbols are the results of significance tests (1000 permutations) of genetic distances between pairs of groups. †P < 0.05); **P < 0.05); *P < 0.01); others without symbols = P = 0.001). Levels of genetic distance among nearly all different clusters were significant.
Fig. 3.
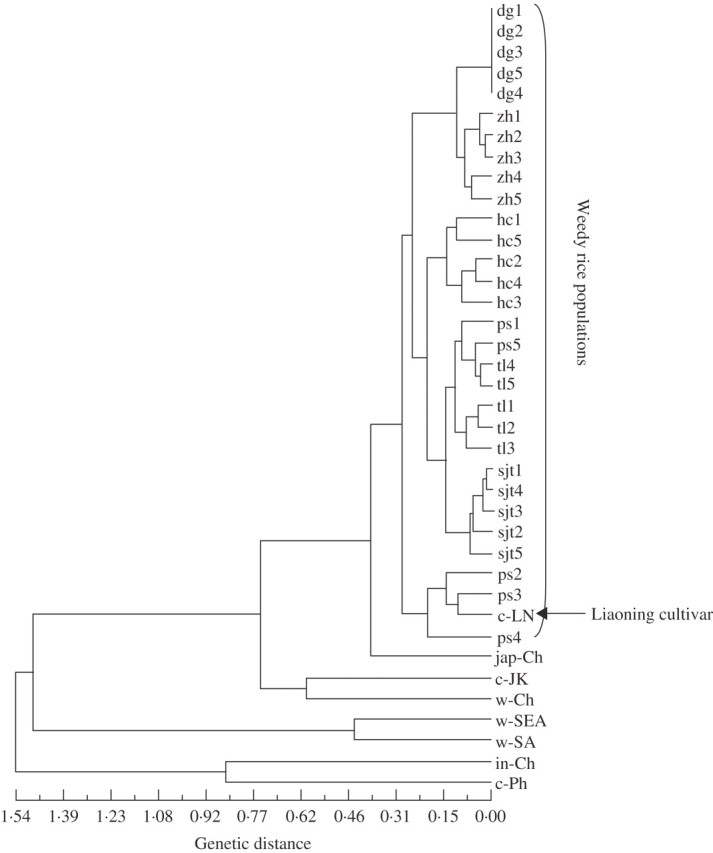
Dendrogram (UPGMA) constructed based on polymorphisms of 20 SSR loci in 30 weedy rice (Oryza sativa f. spontanea) populations, different groups of rice varieties (O. sativa) and wild rice (O. rufipogon), using Nei's unbiased genetic distance coefficients (Nei, 1978, see Table 6). Codes for the weedy rice populations, rice varieties and wild rice refer to those in Tables 1 and 2. The arrow in the dendrogram indicates the group of rice varieties from Liaoning Province.
Within the weedy rice cluster, the dendrogram showed that weedy rice populations from the same regions in Liaoning clustered together, with the exception of the Panshan populations that were scattered into two groups. This pattern suggested a close genetic relationship of weedy rice occurring within the same regions, but considerable genetic differentiation in weedy rice among regions. Three weedy rice populations from Panshan grouped in the same cluster with Liaoning rice varieties. In addition, weedy rice populations from regions with a close spatial distance showed a relatively close genetic relationship (Fig. 3).
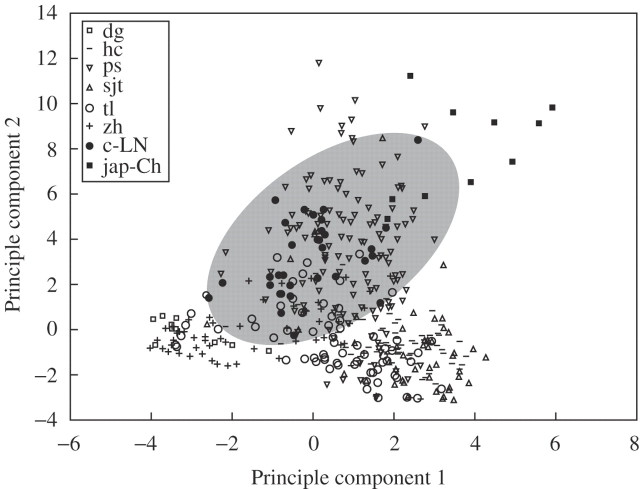
In order to understand the genetic relationships of weedy and cultivated rices as individuals, PCA was conducted based on the SSR data matrix of the 20 loci for 959 individuals, including all weedy rice populations, 30 rice varieties from Liaoning Province and ten each of japonica and indica rice varieties (as references) from other areas in China. The scatter plot of the first and second principle components showed a clear genetic variation and differentiation pattern of Liaoning weedy rice populations (Fig. 4). For example, weedy rice individuals from Panshan scattered over a very large area in the plot, suggesting their high level of genetic diversity, and weedy rice individuals from each region tended to be clustered together, showing their differentiation among regions. The scatter plot clearly illustrated that rice varieties from Liaoning Province were distributed among the individuals of weedy rice, with the japonica rice varieties closely associated with weedy rice individuals (Fig. 4) and the indica rice varieties distantly related to weedy rice individuals (data not shown). Considerable overlap between Liaoning rice varieties and most Panshan, many Tieling and some Zhuanghe-Haicheng weedy individuals was observed.
Fig. 4.
Scatter plot of the first and second principle components based on variation of 20 SSR loci for 949 individuals of weedy rice and cultivars from Liaoning Province and other regions (japonica) in China, showing a considerable area of overlap (shaded) of the Liaoning rice varieties with many weedy rice individuals. The letters dg, hc, ps, sjt, tl and zh in the top left corner denote the individuals of weedy rice from Donggang, Haicheng, Panshan, Sujiatun, Tieling and Zhuanghe regions, respectively; c-LN and jap-Ch denote rice varieties from Liaoning and other regions in China.
To confirm the relatedness of weedy rice populations with other sets of rice materials, particularly the varieties from Liaoning, the proportion of shared alleles among weedy rice populations, Liaoning cultivated rice varieties, rice varieties from other regions and wild O. rufipogon was calculated (Table 7). The results showed that weedy rice populations shared a much higher proportion of alleles with Liaoning rice varieties (64·4–79·8 %) than with rice varieties from other regions (31·4–49·1 %) and wild O. rufipogon samples (19·0–28·3 %).
Table 7.
Proportion of shared alleles [ps = ∑k∑amin(fa,i, fa,j)/D] at 20 SSR loci among the weedy rice populations, cultivated varieties and wild O. rufipogon as estimated by genetic distance (Dps) following Bowcock et al. (1994)
| dg | hc | ps | sjt | tl | Zh | c-LN | |
|---|---|---|---|---|---|---|---|
| dg | – | ||||||
| hc | 0.652 | – | |||||
| ps | 0.637 | 0.754 | – | ||||
| sjt | 0.661 | 0.766 | 0.679 | – | |||
| tl | 0.705 | 0.783 | 0.735 | 0.793 | – | ||
| zh | 0.828 | 0.714 | 0.716 | 0.699 | 0.806 | – | |
| c-LN | 0.670 | 0.645 | 0.798 | 0.644 | 0.682 | 0.735 | – |
| CV* | 0.314 | 0.388 | 0.491 | 0.369 | 0.400 | 0.388 | 0.401 |
| Wild† | 0.197 | 0.241 | 0.283 | 0.190 | 0.235 | 0.246 | 0.241 |
The codes of the weedy populations and cultivated rice varieties from Liaoning Province refer to Table 1 and Table 2; *Cultivated rice varieties from other regions excluding Liaoning Province; †The studied O. rufipogon samples from different regions.
DISCUSSION
Relatively high genetic diversity is found in the Liaoning weedy rice populations
The present results based on the polymorphic pattern of 20 selected SSR loci demonstrated that Liaoning weedy rice populations possessed relatively high genetic diversity (He = 0·313, I = 0·572), although with considerable variation among populations. This result is different from that of a previous study by Yu et al. (2005), where low genetic diversity (He = 0·053) was found in a total of 42 weedy rice individuals from Liaoning Province using SSR marker genotyping. The discrepancy in the genetic diversity between this study and the previous report is probably due to differences in sample size (with 909 weedy individuals in this study). In addition, differences in sampling sites and areas may also considerably influence the result of genetic diversity in the analysed samples. In principle, a high level of genetic diversity of weedy rice populations provides a broad genetic basis potential for their adaptation to a wide range of agroecosystems, which may increase the complexity of their control (Dekker, 1997; Holt and Hochberg, 1997). On the other hand, weedy rice also contains many agronomically useful strains for rice breeding (Chen and Suh, 2004). Broad genetic variation in the weedy rice gene pool may provide more opportunities for selecting beneficial genetic resources for rice breeding.
The results also showed that the genetic diversity of the Liaoning weedy rice populations was not evenly distributed. Considerable variation was found across populations and regions, with the highest diversity in Panshan (He = 0·380) and the lowest (He = 0·011) in Donggang populations. This might be attributed to (a) different farming practices (e.g. the strength of weed control), seed sources (e.g. company-supplied or self-maintained seeds) and the number of rice varieties used in different regions; and (b) the origin of weedy rice from different procedures, for example mutation or hybridization segregation. Weedy rice is an autogamous species with an extremely low outcrossing rate and restricted pollen-mediated gene flow (Gealy et al., 2003; Chen et al., 2004). The findings of the high Fis and Fit values and low outcrossing rate (t) of weedy rice populations in this study strongly supported this viewpoint. A slightly higher outcrossing rate was found in a few weedy rice populations, which allows more opportunities for natural hybridization and introgression among weedy rice individuals and with cultivated rice. Such a low frequency of hybridization and introgression could play an important role in the long-term evolution of weedy rice populations and in the maintenance of a certain amount of genetic diversity. Interestingly, the level of genetic diversity seems to be associated with outcrossing rates of weedy rice populations, supporting the deduction that gene flow from cultivated rice might contribute to the genetic diversity of weedy rice. This viewpoint is supported by similar studies on genetic variation of wild rice (O. rufipogon), in which wild rice populations adjacent to rice fields had a higher genetic diversity than those at some distance from cultivated rice (Song et al., 2003; Cai et al., 2004). This indicates that introgression from cultivated rice can considerably shape genetic diversity of its wild relatives.
The results from this study revealed a positive correlation between the level of genetic diversity and the time when weedy rice was found in rice fields. There was an interesting trend of genetic diversity in the six surveyed regions, namely weedy rice populations discovered by farmers in recent years (1998–2001) possessed a relatively high level of genetic diversity, whereas those found longer ago had a relatively low level. The differences in genetic diversity among weedy populations might be associated with the weed management procedures. Farmers in the rice-planting regions usually remove weeds (including weedy rice) manually. Consequently, farmers pulled out the most obvious off-types of weedy rice they encountered. This procedure might considerably reduce variation of weedy rice if it infested rice fields for a longer period of time. After a certain period, weedy rice individuals morphologically similar to cultivated rice were left in the fields. Selective removal by humans will tend to even up weedy rice within a population, thus reducing its diversity, as indicated by the lower diversity in early years seen in Fig. 2. As a consequence, extensive control of weedy rice even manually minimized the genetic diversity of weedy rice populations. Cost-effective weed management in rice fields is still necessary to keep weedy rice populations at a low level, even though the farming and cultivation practices of rice fields are shifting to a less managed mode.
Considerable genetic differentiation is present among the Liaoning weedy rice populations
The results based on both Fst and AMOVA demonstrated that major genetic variation existed within Liaoning weedy rice populations. However, considerable differentiation was found among populations within and among regions. The observed differentiation of weedy rice populations is probably caused by limited exchange of genetic materials among weedy rice populations because of the inbreeding nature of weedy rice with an extremely low outcrossing rate. In principle, considerable gene flow is an evolutionary force that tends to maintain genetic homogeneity among populations (Slatkin, 1987) and, in contrast, limited gene flow may promote substantial genetic differentiation among populations. The results of the present study showed that the outcrossing rate (t = 0·6–4·1 %) of different populations was very low, which will considerably restrict pollen-mediated gene flow, supporting the viewpoint that limited gene flow had an impact on differentiation of weedy rice populations. In addition, because weedy rice is always surrounded by rice cultivars in fields, genetic introgression from different cultivated rice varieties through time may increase variation among weedy rice populations. Ellstrand et al. (1999) and Song et al. (2006) also addressed the importance of introgression from crop species, which may have a substantial impact on differentiation and evolutionary processes in wild and weedy populations.
The possible origin of Liaoning weedy rice based on its genetic relationships
The results showed that weedy rice populations from Liaoning Province had a very close genetic relationship with Liaoning rice varieties. This is clearly reflected by the cluster analysis of the grouped weedy variety–wild rice materials and the PCA of weedy individuals and cultivated rice (Liaoning, indica and japonica). Both cluster analysis and PCA showed a relatively close genetic relationship of weedy rice populations with japonica rice varieties, but a relatively distant relationship with indica rice varieties. This result suggests that weedy rice populations in Liaoning Province are most likely to be the japonica ecotype. The origin of the japonica ecotype of weedy rice can be easily explained by the strong influence of introgression with Liaoning rice varieties, all of which are japonica ecotypes. In addition, the present results showed that wild rice O. rufipogon either from China or other countries possessed a distant genetic relationship with Liaoning weedy rice, indicating no or extremely limited introgression of Liaoning weedy rice with the wild rice.
All the above results have ruled out the possibility that Liaoning weedy rice directly originated from wild rice O. rufipogon or from its hybrids. This observation gains supports from the analytical data showing that Liaoning weedy rice populations shared a much lower proportion of alleles at the 20 SSR loci with O. rufipogon samples than rice varieties from Liaoning Province. Actually, wild rice species have no distribution in the temperate regions such as North-eastern China (Song et al., 2005) because of the cold winters which affect their survival. As an alternative process, Liaoning weedy rice most probably originated from Liaoning rice varieties, possibly through back-mutation (towards seed shattering and dormancy) or selection of recombined segregants between varietal hybrids with seed shattering and dormancy traits. The occurrences of mutation and novel recombinants in weedy rice is reflected by the presence of rare and unique alleles in certain weedy rice populations. The tight correlation between weedy rice populations and Liaoning rice varieties revealed by the cluster analysis and PCA supports this explanation of the origin of weedy rice from Liaoning Province. The results showing a considerably high proportion of alleles shared by weedy rice populations and rice varieties from Liaoning Province in this study strongly support the above conclusion. Similar results have been found in other studies on the origins of weedy rice, where weedy rice strains were generated by natural hybridization between different rice varieties or between indica and japonica ecotypes (Ishikawa et al., 2005). Judging from the genetic diversity patterns of this study, a further hypothesis could be put forward that the origin of weedy rice from North-eastern China is probably through the selection of mutation of cultivated rice and segregants of intervarietal hybridization, without the direct involvement of wild O. rufipogon. The current changes towards less weed management in rice fields may have promoted the fast accumulation of weedy rice. As a consequence, proper weed management such as rotation of rice and other upland crops in the same field and herbicide treatment before rice cultivation to control weedy rice populations is necessary in rice farming.
Conclusions
In conclusion, the genetic diversity of weedy rice from Liaoning Province was relatively high although considerable variation was observed among different populations across regions. There seems to be a positive correlation between the abundance of genetic diversity and the time when weedy rice was found. Weedy rice populations from various regions showed considerable genetic differentiation. The uneven distribution of genetic diversity and genetic differentiation among Liaoning weedy rice populations is probably associated with several factors, such as strength of weed management in the regions, limited gene flow among weedy populations and introgression with different rice varieties over time. It is evident that Liaoning weedy rice populations are closely related genetically to rice varieties from Liaoning but distantly related to varieties and wild O. rufipogon from other regions. This result rules out the possibility that Liaoning weedy rice has its origin from wild rice or its hybrids, which supports the hypothesis that Liaoning weedy rice most probably originated from Liaoning cultivated rice. The recent changes of farming practices and cultivation methods with application of direct seeding and seedling broadcasting technologies with less weed management may have promoted the re-emergence and genetic diversification of weedy rice in North-eastern China. The abundant genetic diversity of weedy rice populations accompanied by the changes of farming practices will complicate weedy rice control in the future and consequently threaten rice production. Thus, effective methodologies for weed control and management must be developed to prevent weedy rice from extensive spreading and infestation across all rice-planting regions in China.
Acknowledgments
This research was supported by the Ministry of Science and Technology (grant no. 2006CBI00205), Shanghai Science and Technology Commission (grant no. 03DZ19309 and 03DJ14014) and by the Italian Ministry of Environment and Environmental Protection, within the research project ‘Application of biotechnology to the protection of the environment, in collaboration with China’ (years 2005–2007).
LITERATURE CITED
- Arrieta-Espinoza1 G, Sánchez1 E, Vargas S, Lobo J, Quesada T, Espinoza AM. (2005) The weedy rice complex in Costa Rica. I. Morphological study of relationships between commercial rice varieties, wild Oryza relatives and weedy types. Genetic Resources and Crop Evolution 52575–587. [Google Scholar]
- Baker HG. (1974) The evolution of weeds. Annual Review of Ecology and Systematics 51–24. [Google Scholar]
- Belkhir K. (1999) GENETIX, Version 4·02 a Windows program for population genetic analysis(Laboratoire Genome, Populations: interactions UPR 9060 du CNRS, Universite Montpellier 2, Montpellier, France).
- Bowcock AM, Ruíz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL. (1994) High resolution human evolutionary trees with polymorphic microsatellites. Nature 368455–457. [DOI] [PubMed] [Google Scholar]
- Burger JC, Lee S, Ellstrand NC. (2006) Origin and genetic structure of feral rye in the western United States. Molecular Ecology 152527–2539. [DOI] [PubMed] [Google Scholar]
- Cai H-W, Wang X-K, Morishima H. (2004) Comparison of population genetic structures of common wild rice (Oryza rufipogon Griff.), as revealed by analyses of quantitative traits, allozymes, and RFLPs. Heredity 92409–417. [DOI] [PubMed] [Google Scholar]
- Chen LJ and Suh HS. (2004) Study and utilization on weedy rice. In Yang QW (Ed.). Proceedings of the First National Conference on Wild Rice in China, Nanchang, Jiangxi, China pp. 258–264.
- Chen LJ, Lee DS, Song ZP, Suh HS, Lu B-R. (2004) Gene flow from cultivated rice (Oryza sativa) to its weedy and wild relatives. Annals of Botany 9367–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Temnykh S, Xu Y, Cho YG, McCouch SR. (1997) Development of a microsatellite framework map providing genome-wide coverage in rice (Oryza sativa L.). Theoretical and Applied Genetics 95553–567. [Google Scholar]
- Dekker J. (1997) Weed diversity and weed management. Weed Science 37237–46. [Google Scholar]
- Dieringer D and Schlötterer C. (2002) Microsatellite analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Molecular Ecology Notes 3167–169. [Google Scholar]
- Ellstrand NC, Prentice HC, Hancock JF. (1999) Gene flow and introgression from domesticated plants into their wild relatives. Annual Review of Ecology and Systematics 30539–563. [Google Scholar]
- Arlequin version 3·0: an integrated software package for population genetics data analysis Excoffier L, Laval G, Schneider S. (2005) (Computational and Molecular Population Genetics Lab (CMPG), Institute of Zoology, University of Berne, Switzerland) http://cmpg.unibe.ch/software/arlequin3.
- Ferrero A, Vidotto F, Balsari P, Airoldi G. (1999) Mechanical and chemical control of red rice (Oryza sativa L. Var. sylvatica) in rice (Oryza sativa L.) per-planting. Crop Protection 18245–251. [Google Scholar]
- Gealy DR, Mitten DH, Rutger JN. (2003) Gene flow between red rice (Oryza sativa) and herbicide-resistant rice (O. sativa): implications for weed management. Weed Technology 17627–645. [Google Scholar]
- Green JM, Barker JHA, Marshall EJP, Froud-Williams RJ, Peters NCB, Arnold GM, et al. (2001) Microsatellite analysis of the inbreeding grass weed Barren Brome (Anisantha sterilis) reveals genetic diversity at the within- and between-farm scales. Molecular Ecology 101035–1045. [DOI] [PubMed] [Google Scholar]
- Hartl DL and Clark AG. (1989) Principles of population genetics 2nd edn (Sinauer Associates Inc. Publishers, Sunderland, MA).
- Hoagland RE and Paul RV. (1978) A comparative SEM study of red rice and several commercial rice (Oryza sativa) varieties. Weed Science 26619–625. [Google Scholar]
- Holt RD and Hochberg ME. (1997) When is biological control evolutionarily stable (or is it?). Ecology 781673–1683. [Google Scholar]
- Ishikawa R, Toki N, Imai K, Sato YI, Yamagishi H, Shinamoto Y, et al. (2005) Origin of weedy rice grown in Bhutan and the force of genetic diversity. Genetic Resources and Crop Evolution 52395–403. [Google Scholar]
- Jiang H, Wu JL, Wang GL. (1985) Study on weedy rice from Lianyungang, China. Crop Resource of China 24–7 (In Chinese.). [Google Scholar]
- Lu B-R and Snow AA. (2005) Gene flow from genetically modified rice and its environmental consequences. BioScience 55669–678. [Google Scholar]
- Ma DR, Chen WF, Xu XJ, Zhang WZ. (2005) Origin and control management of weedy rice in Liaoning. Chinese Agricultural Science Bulletin 21358–360. [Google Scholar]
- Mortimer M, Pandey S, Piggin C. (2000) Weedy rice: approaches to ecological appraisal and implications for research priorities. In Baki BB, Chin DV, Mortimer M (Eds.). Proceedings of Wild and Weedy Rice in Rice Ecosystems in Asia. A review(International Rice Research Institute, Los Banos, Philippines) pp. 97–105.
- Nei M. (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hanlon PC, Peakall R, Briese DT. (2000) A review of new PCR-based genetic markers and their utility to weed ecology. Weed Research 40239–254. [Google Scholar]
- Parker PG, Snow AA, Schug MD, Booton GC, Fuerst PA. (1998) What molecules can tell us about populations: choosing and using a molecular marker? Ecology 79361–382. [Google Scholar]
- Pyšek P and Prach K. (2003) Research into plant invasions in a crossroads region: history and focus. Biological Invasions 5337–348. [Google Scholar]
- Rohlf FJ. (1998) NTSYSpc: numerical taxonomy and multivariate analysis system, version 2·02(Exeter Software, Setauket, New York).
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. (1984) Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proceedings of the National Academy of Sciences of the USA 818014–8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. (1987) Gene flow and the geographic structure of natural populations. Science 236787–792. [DOI] [PubMed] [Google Scholar]
- Song ZP, Xu X, Wang B, Chen JK, Lu B-R. (2003) Genetic diversity in the northernmost Oryza rufipogon populations estimated by SSR markers. Theoretical and Applied Genetics 1071492–1499. [DOI] [PubMed] [Google Scholar]
- Song ZP, Li B, Chen JK, Lu B-R. (2005) Genetic diversity and conservation of common wild rice (Oryza rufipogon) in China. Plant Species Biology 2083–92. [Google Scholar]
- Song ZP, Rong J, Zhu WY, Xu X, Chen JK, Lu B-R. (2006) Impact of introgression from cultivated rice on differentiation of its wild relative Oryza rufipogon (Poaceae). Evolutionary Ecology (in press).
- Song ZP, Zhu WY, Rong J, Xu X, Chen JK, Lu B-R. (2006) Evidences of introgression from cultivated rice to Oryza rufipogon (Poaceae) populations based on SSR fingerprinting: implications for wild rice differentiation and conservation. Evolutionary Ecology (doi: 10.1007/s10682-006-9113-0).
- Sun JD, Xiao YC, Huang XF, Jiao WC, Chen JC, Zhou YY. (2005) Primary study on occurrence and control of weedy rice in japonica rice fields. Weed Science of China 221–23 (In Chinese.). [Google Scholar]
- Wang WX, Zhu TH, Shao GS, Xuan SN. Advances in the study on taxonomy, origin, utilization of weedy rice. Weed Science of China 21–5 (In Chinese.).
- de Wet JMJ and Harlan JR. (1975) Weeds and domesticates; evolution in man-made habitats. Economic Botany 2999–107. [Google Scholar]
- Wright S. (1978) Variability within and among natural populations(University of Chicago Press, Chicago).
- Wu KS and Tanksley SD. (1993) Abundance, polymorphism and genetic mapping of microsatellites in rice. Molecular and General Genetics 241225–235. [DOI] [PubMed] [Google Scholar]
- Yeh FC, Yang RC, Boyle T. (1999) Microsoft Window-based freeware for population genetic analysis (POPGENE), version 1·31.
- Yu GQ, Bao Y, Shi CH, Dong CQ, Ge S. (2005) Genetic diversity and population differentiation of Liaoning weedy rice detected by RAPD and SSR markers. Biochemical Genetics 43261–270. [DOI] [PubMed] [Google Scholar]
- Zhang CX. (2000) Wild and weedy rice in China. In Baki BB, Chin DV, Mortimer M (Eds.). Proceedings of Wild and Weedy Rice in Rice Ecosystems in Asia. A review(International Rice Research Institute, Los Banos, Philippines) pp. 35.