Abstract
The Saccharomyces cerevisiae high osmolarity glycerol (HOG) mitogen-activated protein kinase pathway is required for osmoadaptation and contains two branches that activate a mitogen-activated protein kinase (Hog1) via a mitogen-activated protein kinase kinase (Pbs2). We have characterized the roles of common pathway components (Hog1 and Pbs2) and components in the two upstream branches (Ste11, Sho1, and Ssk1) in response to elevated osmolarity by using whole-genome expression profiling. Several new features of the HOG pathway were revealed. First, Hog1 functions during gene induction and repression, cross talk inhibition, and in governing the regulatory period. Second, the phenotypes of pbs2 and hog1 mutants are identical, indicating that the sole role of Pbs2 is to activate Hog1. Third, the existence of genes whose induction is dependent on Hog1 and Pbs2 but not on Ste11 and Ssk1 suggests that there are additional inputs into Pbs2 under our inducing conditions. Fourth, the two upstream pathway branches are not redundant: the Sln1-Ssk1 branch has a much more prominent role than the Sho1-Ste11 branch for activation of Pbs2 by modest osmolarity. Finally, the general stress response pathway and both branches of the HOG pathway all function at high osmolarity. These studies demonstrate that cells respond to increased osmolarity by using different signal transduction machinery under different conditions.
INTRODUCTION
Unicellular organisms such as Saccharomyces cerevisiae live in environments that are constantly changing. In their natural environment, yeast cells adapt to a variety of fluctuating stress conditions such as nutrient availability and external osmolarity. Cells respond to increased external osmolarity in a number of ways, including accumulation of a compatible solute inside the cell. S. cerevisiae uses glycerol, whose intracellular accumulation leads to retention of cellular water to prevent dehydration and protein aggregation (Reed et al., 1987). During conditions of increased osmolarity, both the general stress response pathway and the high osmolarity glycerol (HOG) pathway are used to stimulate transcriptional responses that counteract the stress (for reviews, see Gustin et al., 1998; Sprague, 1998; Estruch, 2000; O'Rourke et al., 2002).
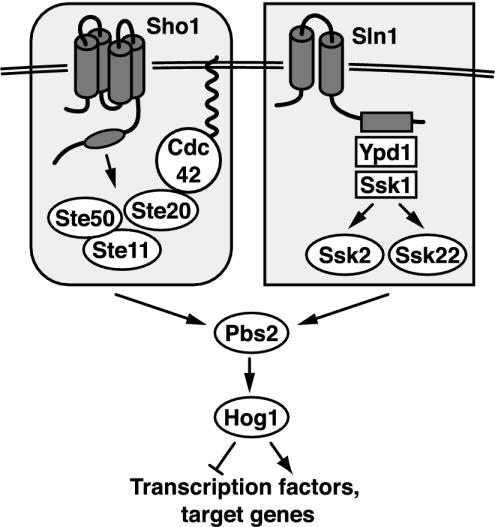
The HOG pathway (Figure 1) uses two membrane-spanning proteins (Sho1 and Sln1) to respond to high external osmolarity. Signals from each are transduced by unique components and converge to activate the mitogen-activated protein kinase kinase (MAPKK) Pbs2. The Sho1 branch requires Cdc42, Ste20 (a PAK homolog), and Ste50 to activate the mitogen-activated protein kinase kinase kinse (MAPKKK) Ste11 (Maeda et al., 1995; Posas and Saito, 1997; O'Rourke and Herskowitz, 1998; van Drogen et al., 2000; Raitt et al., 2000; Reiser et al., 2000). The Sln1 protein uses Ypd1 and Ssk1 to activate two partially redundant MAPKKKs, Ssk2 and Ssk22 (Posas et al., 1996; Posas and Saito, 1998). Any of the three osmoresponsive MAPKKKs (Ste11, Ssk2, or Ssk22) is able to activate Pbs2, which then phosphorylates the MAPK Hog1, resulting in translocation of Hog1 to the nucleus and an increase in its kinase activity (Brewster et al., 1993; Ferrigno et al., 1998; Reiser et al., 1999; Zhan and Guan, 1999; Bilsland-Marchesan et al., 2000; Proft et al., 2001). Hog1 regulates gene expression through several transcription factors, Hot1, Sko1 and Smp1, and perhaps also through Msn1, Msn2, and Msn4 (Schüller et al., 1994; Proft and Serrano, 1999; Rep et al., 1999; de Nadal et al., 2003). Whole-genome analyses of the response to high osmotic stress demonstrate both unique and overlapping roles for some of these transcription factors (Rep et al., 2000). It has recently been shown that Hog1 is localized to promoters of some regulated genes through interaction with the abovementioned transcription factors (Alepuz et al., 2001; Proft and Struhl, 2002).
Figure 1.
Model of the yeast hyperosmotic-response MAPK pathway. See INTRODUCTION for details.
The two upstream branches of the HOG pathway use very different components and exhibit some functional specializations. The Sln1-Ypd1-Ssk1 branch uses a multistep phosphorelay system, which is widely used in bacteria for environmental monitoring (Stock et al., 2000), whereas the Sho1 branch uses a novel protein containing an SH3 domain. The Sho1 branch makes use of several proteins (Ste20, Ste50, and Ste11) that are components of two other mitogen-activated protein kinase (MAPK) cascades, the pheromone-response and pseudohyphal/invasive growth pathways (Posas and Saito, 1997; O'Rourke and Herskowitz, 1998; Posas et al., 1998). In some respects, the Sho1 and Sln1 branches are functionally redundant. Both can stimulate phosphorylation of Pbs2, phosphorylation and nuclear translocation of Hog1, induction of several reporter genes, and growth on high-osmolarity media (Posas and Saito, 1997; Van Wuytswinkel et al., 2000). Some distinctions between these two branches have, however, been noted; they seem to have slightly differing sensitivities to the concentration of external solute, as assayed by Hog1 phosphorylation (Maeda et al., 1995; Van Wuytswinkel et al., 2000). In addition, the entire Sho1 branch upstream of Pbs2 is essential for pseudohyphal development (O'Rourke and Herskowitz, 1998; Ramezani Rad et al., 1998). Thus, stimulation of the Sho1 and Sln1 branches may lead to differential activation of gene targets.
We have sought to determine the roles of different components of the HOG pathway in generating the transcriptional output during response to high extracellular osmolarity. We have therefore characterized the relationship between external solute concentration and transcriptional response in wild-type strains and in strains blocked at various positions in the HOG pathway. Our work provides insights into the mechanisms of the cellular response to increased osmolarity and clarifies the roles of individual HOG pathway components.
MATERIALS AND METHODS
Strains, Media, and Genetic Techniques
Yeast strains were derived from an ADE2 strain in the W303 strain background (trp1 leu2 ura3 his3 can1 GAL+ psi+). Yeast strains were IH4506 (MATa), IH4553 (MATa hog1::hisG), IH4522 (MATa pbs2::LEU2), IH4508 (MATa ssk1::HIS3cg), IH4510 (MATa sho1::TRP1), IH4537 (MATa ste11::TRP1cg), IH4531 (MATa ssk1::HIS3cg ste11::TRP1cg), and IH4514 (MATa ssk1::HIS3cg sho1::TRP1cg). Yeast strains were grown in YEPD medium (1% yeast extract, 2% Bacto-Peptone, 2% glucose) at 30°C. Synthetic complete solid media (Rose et al., 1990) were used for selecting gene replacements during strain construction. d-Sorbitol and KCl (Fisher Scientific, Pittsburgh, PA) were used at final concentrations as indicated. For the α-factor time course, α-factor in 0.01 M HCl was added to a final concentration of 38.5 μM. Yeast transformations were carried out using the lithium acetate procedure (Schiestl and Gietz, 1989). Gene disruptions using Candida glabrata markers (HIS3cg or TRP1cg) were constructed by polymerase chain reaction-mediated gene disruption as described previously (Sakumoto et al., 1999). Gene disruptions were confirmed by phenotypic analysis and polymerase chain reactions with gene-specific primers.
Culture Conditions for Time Courses
Overnight cultures were grown to OD600 5.0 in rich YEPD medium (100 ml in a 500-ml flask). Once the cultures were at the proper density, 31 ml of culture was transferred to a triple baffled 2.8 l Fernbach flask containing 1 l YEPD (for a final OD600 of 0.15). This culture was grown at 30°C until it reached OD600 0.6 (typically 3-4 h), at which time a 250-ml aliquot was harvested for the 0-min time point. 0.5 l YEPD containing KCl or sorbitol was then added, yielding a 1.25-l culture containing the indicated solute concentration. At various time points, 250-ml aliquots were harvested for RNA isolation. For time series involving 10 samples, two cultures were used for growing the required quantity of cells. Cells were recovered by vacuum filtration using a Fisher 1 l filter unit with nitrocellulose filters (0.45-μm pore size). After the media was separated from the cells, the nitrocellulose filter and cells were placed into a 50-ml plastic tube and frozen in liquid nitrogen. The samples were then stored at -80°C until processed. Using this procedure, cells were harvested from culture flasks and transferred to liquid nitrogen within 2 min. The detailed yeast growth protocol can be found on http://www.microarrays.org.
RNA Isolation and Microarray Techniques
Protocols and information on reagents were obtained from http://www.microarrays.org/. DNA microarrays were made in-house by spotting polymerase chain reaction products onto glass microscope slides. Open reading frame (ORF) DNA microarrays, total RNA isolation, and mRNA purification followed published procedures (DeRisi et al., 1997), except that 4 μg of mRNA was used for cDNA synthesis in the presence of amino-allyl dUTP before labeling of the cDNA with Cy3 or Cy5 (Shoemaker et al., 2001). After RNA isolation, cDNA synthesis, and Cy5 dye labeling, samples were hybridized to a DNA microarray along with a control wild-type cDNA sample labeled with Cy3, which was derived from yeast exponentially growing in YEPD medium of standard osmolarity.
Data Acquisition and Analysis
Microarrays were scanned with GenePix 4000A or 4000B microarray scanners (Axon Instruments, Foster City, CA). Genes were assigned to the resulting microarray images with GenePix Prosoftware. After removing data from gene spots that were damaged, data were uploaded to the database program AMAD (http://www.microarrays.org/software.html), which normalized the data over the entire array. Red-to-green expression ratios (the ratio of means) were used only if the spot intensity was >150 U, which reduced variance due to weak fluorescence signal. We subsequently renormalized the data (ratio of means) to each microarray grid section, of which there are 16 per array, which decreased variance due to array position in the data set. Ultimately, expression data were handled and analyzed using the software programs FileMaker Pro (FileMaker, Santa Clara, CA), Excel (Microsoft, Redmond, WA), Cluster and TreeView (Eisen et al., 1998; http://rana.lbl.gov/EisenSoftware.htm). For performing average linkage clustering in the Cluster program, the similarity metric setting used was “Correlation (uncentered).”
To define regulated genes for our analyses, we used a criterion of two occurrences of induction or repression of greater than twofold (at any time during a given time course). Genes were considered to exhibit altered regulation in various mutants if they displayed a threefold difference in expression compared with wild-type strains at two or more time points. To determine the noise and sensitivity of our experimental and analysis procedures, we considered the expression ratios for every gene over seven different microarrays. Six wild-type RNA samples were harvested on different days before any osmolarity treatment and one sample that varied only slightly from the other six samples: wild-type treated with 0.0625 M KCl for 2.5 min. We found that the six zero time-point samples displayed variances of 0.05-0.08 when compared with each other. However, the sample treated with KCl displayed higher variances (0.16-0.19) compared with the six untreated samples. Thus, even in samples that differ only slightly, we can detect meaningful changes in gene expression. For reference, we compared wild type treated with 0.5 M KCl for 20 min (a condition in which many genes show regulation) to the six untreated wild-type samples, which yielded a variation of ∼1.85.
RESULTS
Response of Wild-Type Cells to High Osmolarity
We began investigating the S. cerevisiae response to high osmolarity by examining gene regulation in response to sorbitol and KCl, both of which are commonly used to study the osmotic stress response. We performed two time-course analyses of the response of wild-type yeast cells to high external osmolarity, sampling at 10 time points throughout 180 min of exposure. mRNA was isolated and used for two-color hybridization to DNA microarrays as described in MATERIALS AND METHODS (DeRisi et al., 1997).
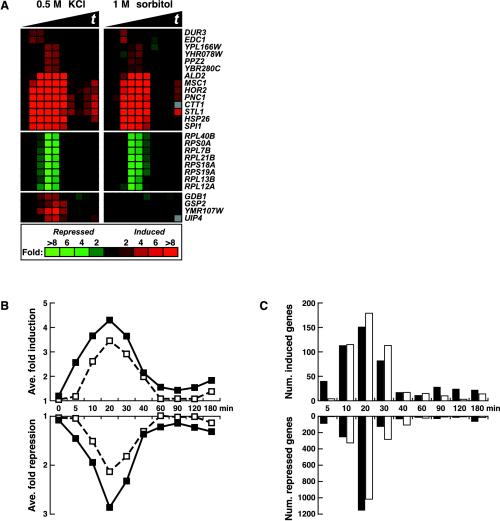
Based on hierarchical clustering of the data, 1 M sorbitol treatment altered the transcript levels of 1196 genes at least twofold (see MATERIALS AND METHODS); 0.5 M KCl treatment altered the levels of 2144 genes at least twofold. Merging these two data sets yielded a “hyperosmolarity-regulated” set of 2277 genes whose expression is altered by at least one solute. Almost all of the genes showed similar regulation by both 1 M sorbitol and 0.5 M KCl as demonstrated by the finding that >96% of the hyperosmolarity-regulated genes showed a positive Pearson correlation during response to KCl and sorbitol treatment (the mean Pearson r was 0.69 with a SD of 0.27). Expression of 488 genes increased and 1789 genes decreased; representative genes are displayed in Figure 2A.
Figure 2.
Identification of high osmolarity-regulated genes. DNA microarrays containing yeast open reading frames were used to assay gene expression in wild-type yeast (IH4506) treated with 0.5 M KCl or 1 M sorbitol. ORF microarrays were cohybridized with two samples: one derived from treated yeast and one derived from an untreated wild-type strain growing in normal osmolarity YEPD medium. Only genes that were regulated twofold or more in at least two time points in either the KCl or sorbitol time course were included. Osmolarity-regulated genes were defined as induced or repressed according to their position determined by hierarchical clustering (our unpublished data). (A) Example of the output of hierarchical clustering. Red color indicates increased mRNA abundance and green indicates decreased mRNA abundance (relative to the untreated reference culture). The solid black triangles in this and subsequent figures indicate increasing time of treatment, here from 0 to 180 min (0, 5, 10, 20, 30, 40, 60, 90, 120, 180 min). (B) The average fold change of mRNA levels is displayed as a plot for 488 osmoinduced and 1789 osmorepressed genes. Solid squares represent the average magnitude of regulation by 0.5 M KCl; open squares represent the average magnitude of regulation by 1 M sorbitol. (C) The number of induced and repressed genes is plotted according to their time point of maximal regulation for 0.5 M KCl (black bars) or 1 M sorbitol (open bars) treatments (genes as in B).
To compare the responses to the two different solutes, the average gene expression values were plotted for induced and repressed genes (Figure 2B). The changes in RNA levels in response to KCl and sorbitol seemed similar, with most changes taking place within the first 40 min of treatment. For the 488 osmoinduced genes, 82% underwent maximal changes in RNA levels within 40 min of KCl treatment, and 91% underwent maximal changes in RNA levels within 40 min of the sorbitol treatment (Figure 2C). For both KCl and sorbitol, the time at which the most genes showed maximal change was 20 min (Figure 2C). The response to high external osmolarity as assessed by gene expression was, therefore, rapid and occurred in the same time frame as other cellular events, such as presence of phosphorylated Hog1 protein in the nucleus (Ferrigno et al., 1998; Reiser et al., 1999).
Although 1 M sorbitol and 0.5 M KCl affected gene expression in a highly similar manner, there were several differences (Figure 2). Cells responded faster to KCl than to sorbitol, as can be observed by comparing the response to KCl or sorbitol at 5 min in Figure 2. Also, the magnitude of regulation was greater in response to KCl than to sorbitol. For example, 1 M sorbitol caused an average repression of 2.1-fold, whereas 0.5 M KCl caused a 2.9-fold repression at 20 min (Figure 2B). We found four genes that were regulated by only one of the solutes (Figure 2A, bottom). Because cells responded to KCl more vigorously than to sorbitol, KCl was used for subsequent experiments.
Response of a hog1 Mutant to KCl
To determine the contribution of the HOG pathway to the high osmolarity response, we first compared isogenic HOG1 and hog1Δ strains after exposure to 0.5 M KCl. 2144 genes exhibited altered regulation in the wild-type strain and 1809 genes in the mutant. Fifty-three percent (1369) of the genes were in common. In other words, approximately one-half of the genes whose expression was affected by high osmolarity in the wild-type strain exhibited a similar response in the absence of HOG1.
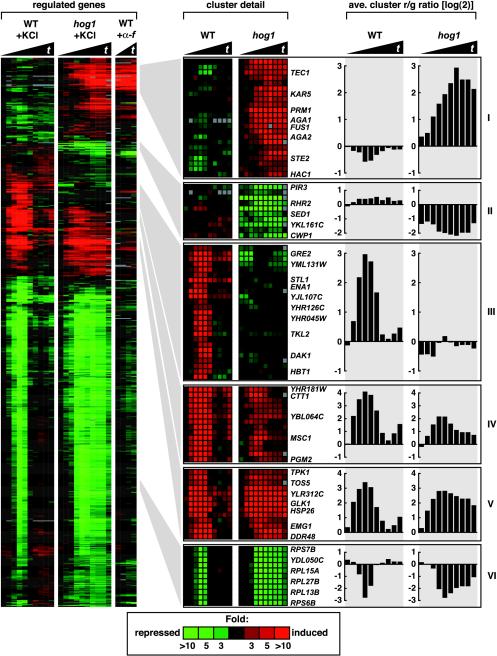
Further analysis of HOG1-dependent genes was accomplished by calculating the ratio of expression of a gene in wild-type cells to that in hog1 mutant cells, which defines the mutant effect ratio (Me). Genes with an Me of greater than threefold (up or down) for at least two time points defined a set of 579 genes that is strongly dependent on Hog1 for regulation. Clustering of these genes revealed six distinct gene expression patterns (Figure 3). Representative genes of all six clusters and plots of the average expression values for these selected genes are shown in Figure 3.
Figure 3.
Identification of 579 genes dependent on Hog1 for normal regulation. Genes are displayed which showed at least a twofold change in expression (at two or more time points) in either wild-type (IH4506) or hog1 (IH4553) strains treated with 0.5 M KCl and which showed at least two occurrences of a threefold difference (increased or decreased) between wild-type and hog1 strains. Six clusters were identified based on gene expression patterns and dependence on Hog1. The left-most three cluster diagrams represent gene expression values in the wild-type strain treated with 0.5 M KCl (0-180 min), the hog1 strain treated with 0.5 M KCl (0-180 min), and the wild-type strain treated with 38.5 μM α-factor (10-40 min). Samples were taken at intervals as in Figure 2 for the cultures treated with KCl and at 10-min intervals for the culture treated with α-factor. The two right-most cluster diagrams are expanded views of selected areas showing gene expression in wild-type and hog1 strains treated with 0.5 M KCl. Average gene expression plots of all genes within the expanded clusters shown on the far right further illustrate the various ways in which Hog1 influences gene expression.
We have previously demonstrated that high osmolarity inappropriately induces FUS1-lacZ expression and a mating response in hog1 and pbs2 mutants (O'Rourke and Herskowitz, 1998). Cluster I consists of genes that were inappropriately induced in the hog1 mutant. This cluster includes the pheromone-induced genes, FUS1, STE2, KAR5, AGA1, and PRM1 and several genes associated with filamentous growth, TEC1, PHD1, and PGU1 (shown previously for TEC1 by Davenport et al., 1999; and for several pheromoneinducible genes by Rep et al., 2000).
The second class of genes is a small cluster of nine ORFs that required HOG1 for basal expression and in some cases for very modest induction. Two genes encoding proteins with known or predicted biochemical roles are RHR2, which encodes a glycerol-3-phosphatase that produces glycerol and phosphate from glycerol 3-phosphate, and YKL161c, which codes for a MAPK. Three genes encoding cell wall proteins also required HOG1 for their basal expression: CWP1, SED1, and PIR3.
The third class contains 32 genes that exhibited very strong induction in wild-type cells but little in the hog1 mutant. Although the roles of these gene in adaptation to high osmolarity is unknown in most cases, some genes have known or predicted functions: ENA1 plays a role in ion homeostasis, STL1 codes for a putative hexose transporter, TKL2 codes for a transketolase, DAK1 codes for a putative dihydroxyacetone kinase, and THI4 codes for a putative enzyme involved in vitamin B1 biosynthesis. HBT1 is involved in polarized morphogenesis.
HOG Pathway-independent Induction of Genes
The fourth and fifth clusters of Figure 3 contain genes induced in both wild-type and hog1 mutants in response to high osmolarity. The response in wild-type cells was transient, peaking at 20 min and decreasing to near uninduced levels by 40-90 min. In cells lacking HOG1, some of these genes were still induced normally (especially genes in cluster V), although the down-regulation that occurred in wild-type cells was delayed or absent. Three of the genes in cluster V (HSP26, CVT17, and SSA1) encode proteins involved in protein stability or transit. Others code for proteins involved in glycogen utilization during stress conditions (GLK1, GSY1, SGA1, GDB1, and GLC3), stress-regulated small molecule production (HOR2, which encodes a DL-glycerol-3-phosphatase), and a phosphoglucomutase required for trehalose metabolism (PGM2). General stress-response genes such as CTT1, GRE1, HSP26, HSP12, and DDR48 are also in this group. Many genes in clusters IV and V are induced in response to diverse environmental stresses such as heat shock, protein-damaging agents, nitrogen starvation, and stationary phase adaptation (Gasch et al., 2000).
The final and largest group of genes (cluster VI) exhibited transient reductions in transcript levels in the wild-type strain in response to osmotic stress but greatly prolonged reductions in the hog1 mutant. Among genes of this group, 96 code for ribosomal protein subunits (RPS and RPL genes). Gasch et al. (2000) have observed that genes coding for ribosomal subunits are down-regulated during diverse stresses. Finding that ribosomal subunit-coding genes are down-regulated during osmotic stress is therefore not surprising. Our observations indicate that Hog1 plays a role in reestablishment of expression of these genes after stress. The defect in expression of these ribosomal genes in a hog1 mutant likely leads to global alterations in protein synthesis.
Roles of PBS2, STE11, and SSK1 in Response to 0.5 M KCl
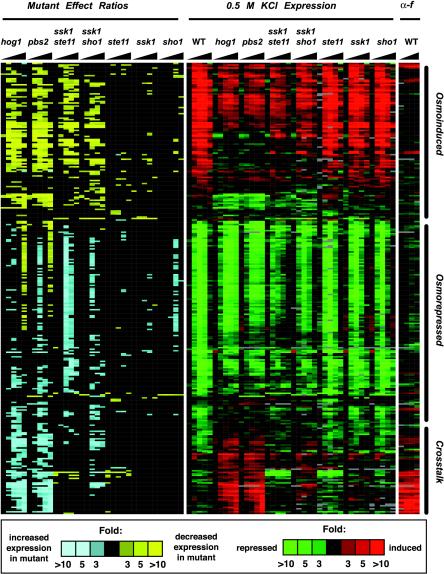
To evaluate how upstream components of the HOG pathway contribute to the pattern of gene regulation in response to high osmolarity, we carried out time-course analyses of strains deleted for PBS2, STE11, SSK1, or both STE11 and SSK1. Because the majority of gene regulation exhibited by wild-type cells took place within 40 min of KCl treatment (Figure 2), we assessed the response of mutant cells to 0.5 M KCl at 0, 10, 20, 30, and 40 min. We found that osmotic-regulated genes showed a variety of gene expression patterns in the absence of the HOG pathway components tested (Figure 4).
Figure 4.
Expression pattern of 296 genes that show altered expression in response to 0.5 M KCl in the absence of various HOG pathway components. Gene expression (green/red) is as in Figure 3 and Me ratio (yellow/blue) as described previously (O'Rourke and Herskowitz, 2002). Genes displayed here were those that satisfied two criteria: 1) an Me ratio of >3 or <0.333 for any two time points in any strain, and 2) induction or repression in at least two time points more than twofold in any individual strain treated with 0.5 M KCl. Gene expression values and corresponding Me ratios are displayed for samples taken at 0, 10, 20, 30, and 40 min. For clarity, the Me ratio is displayed in color only if it is >3 or <0.333. The number of genes that exhibited altered expression in each time series, as defined by the above-mentioned criteria are hog1, 145; pbs2, 162; ssk1 ste11, 133; ssk1 sho1, 75; ste11, 20; ssk1, 8; and sho1, 5. We compared expression of these 296 genes in four different wild-type time series and found that between one and three genes varied significantly between the different time series, which serves as a baseline for false-positive detection. Hierarchical clustering of the data defined three main clusters, denoted here as osmoinduced, osmorepressed, and cross-talk. Strains were wild-type (IH4506), hog1 (IH4553), pbs2 (IH4522), ssk1 ste11 (IH4531), ssk1 sho1 (IH4514), ste11 (IH4537), ssk1 (IH4508), and sho1 (IH4510).
The gene expression patterns of hog1 and pbs2 mutants were remarkably similar: of 196 genes that exhibited altered regulation within 40 min in either mutant, only four had significant differences between the two mutants (Figure 4; our unpublished data). Alterations in expression of LEU1 and YHR114w probably resulted from the fact that the PBS2 deletion was marked with LEU2, which seems to cause altered expression of these genes (also observed for other strains marked with LEU2; our unpublished observations). The other two genes (FIG1 and YBR012c) are α-factor inducible and displayed stronger induction in the pbs2 mutant than in the hog1 mutant. Because YBR012c was expressed very weakly, the difference between the hog1 and pbs2 mutants may not be significant. hog1 and pbs2 mutants thus exhibited an essentially identical response to osmotic stress.
Mutation of both SSK1 and STE11, which inactivates the two known inputs to Pbs2, resulted in some distinctive gene regulation patterns compared with the hog1 and pbs2 mutants (Figure 4). For example, among the 57 genes whose induction required HOG1 and PBS2 (see MATERIALS AND METHODS), 33 also showed reduced expression in the ssk1 ste11 mutant, indicating that Ste11, Ssk1, or both are necessary to activate Pbs2. In contrast, a number of genes whose induction was absolutely dependent on Hog1 and Pbs2 (STL1, ENA1, GRE2, RHR2, YJL107c, and others; Fig. i, web supplement) were still significantly induced in the ssk1 ste11 mutant. In addition, genes that displayed prolonged reductions at late time points in the hog1 and pbs2 mutants (Figure 3, cluster VI) did not show this response in the ssk1 ste11 double mutant. These observations suggest that there may be inputs in addition to the Ste11 and Ssk1 branches that activate Pbs2. Finally, unlike hog1 and pbs2 mutants, the ssk1 ste11 strain showed no detectable induction of pheromoneinducible genes. This was expected, as STE11 is required for signaling in the pheromone-response pathway (Fields et al., 1988; Roberts et al., 2000). Twelve pheromone-induced genes, including BAR1, SST2, STE2, MFA2, AGA2, GPA2, and NDJ1, showed reduced expression throughout the time course in both the ssk1 ste11 and ste11 mutants, indicating that the pheromone-response pathway plays an important role in maintaining a basal signal even when cells are not actively mating (as noted previously by Fields et al., 1988). Thus, blocking the two upstream branches of the HOG pathway, as in an ssk1 ste11 mutant, only partially phenocopies blocking the HOG pathway at the level of Hog1 or Pbs2.
Sho1 has been proposed to feed into Ste11 (Posas and Saito, 1997). To determine whether all of the actions of Ste11 in response to high osmolarity result from input from Sho1, we compared the high-osmolarity response of ssk1 sho1 and ssk1 ste11 strains. The ssk1 sho1 strain was qualitatively similar to the ssk1 ste11 strain except for two differences. First, the effect of the sho1Δ mutation was less severe than ste11Δ: the sho1 mutation allowed greater induction of individual osmoresponse genes than the ste11 mutation. Second, the ssk1 sho1 mutant exhibited induction of many of the genes that are inappropriately induced in the hog1 and pbs2 mutants, although the level of induction was lower than seen in hog1 mutants. Both of these observations support our hypothesis that an additional osmosensor works in parallel with Sho1 to promote Ste11 activation in wild-type cells and promote cross talk in mutants with a compromised HOG pathway (O'Rourke and Herskowitz, 1998, 2002).
Mutants defective in single components of the upstream HOG pathway (SHO1, STE11, or SSK1) resulted in few consistent perturbations of gene expression in response to 0.5 M KCl. Mutation of STE11 caused failure of basal and salt-induced expression of pheromone-inducible genes, as noted for the ssk1 ste11 strain. Deletion of SSK1 or SHO1 affected expression of few if any genes. Clearly, for response to 0.5 M KCl, either the SHO1 or SLN1 branch is sufficient for triggering the HOG pathway.
The Two Upstream HOG Pathway Branches Are Not Redundant In Vivo
The ability to assay expression of thousands of genes as reporters of the response to high osmolarity provided us with the opportunity to examine the response of cells to osmostress lower than that induced by 0.5 M KCl. We thus performed time-course analyses with wild-type and ssk1 ste11 double mutant strains by using KCl concentrations of 0.0625, 0.125, 0.25, 0.5, and 1.0 M (Table 1, supplemental data). We found that modest osmotic stress elicits changes in fewer genes than more severe osmotic stress (0.5 and 1.0 M). For example, 0.0625 M KCl altered the expression of only 279 genes, whereas 0.5 M KCl altered expression of 1828 genes in wild-type cells.
The two upstream branches of the HOG pathway are redundant for providing input to Pbs2 and Hog1 as assayed by growth on high-osmolarity medium (1.5 M sorbitol; Maeda et al., 1995) and induction of gene expression by 0.5 M KCl (Figure 4). We sought to determine whether the two branches are redundant for regulating gene induction at lower solute concentrations (0.0625 and 0.125 M KCl). Time-course analyses were performed on four strains: wild-type, and ssk1 ste11, ste11, and ssk1 mutants. For each KCl concentration, genes exhibiting strong alterations in regulation in any of the three mutant strains were selected and clustered together along with wild-type expression data.
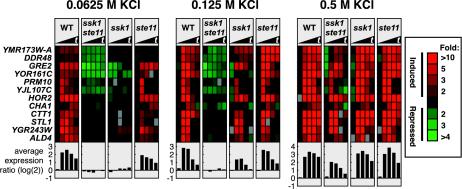
Figure 5 shows the results of this analysis for genes that were osmoinduced. There are several notable features. First, functional redundancy of the Sln1-Ssk1 and the Sho1-Ste11 branches was clearly exhibited at 0.5 M KCl: for a substantial cluster of genes (YNL194C, AFR1, etc.), the ssk1 and ste11 single-mutant strains exhibited induction similar to that of the wild-type strain. In contrast, little or no induction was observed for the ssk1 ste11 double mutant. Gene induction was strikingly different at lower KCl concentrations. At both 0.0625 and 0.125 M KCl, the ste11 single mutant exhibited an induction pattern similar to the wild-type pattern. In contrast, the ssk1 mutant was severely defective for induction even though it carried the wild-type STE11 allele. These observations indicate that at moderate concentrations of KCl, the Sln1-Ssk1 branch is functional, whereas the Sho1-Ste11 branch is not. Thus, these branches are not functionally redundant at these osmolarities. The analysis shown in Figure 5 also reveals a cluster of genes, which includes YGR243W, ALD2, and NCE103, that was induced in the ssk1 ste11 strain at 0.5 M KCl but not at the lower osmolarities. This seems to be a set of genes that is induced at moderate osmolarities via the Sln1-Ssk1 branch and at high osmolarity by any of three mechanisms, the Sln1-Ssk1 branch, the Sho1-Ste11 branch, or an SSK1- and SHO1-independent branch, presumably the general stress response system.
Figure 5.
Redundancy and nonredundancy of the Sho1-Ste11 and Sln1-Ssk1 branches at different osmolarities. Genes that exhibited severe induction defects in response to different concentrations of KCl are displayed. The genes shown exhibited alterations in regulation at two or more time points by a factor of at least threefold in any mutant strain and showed induction in the wild-type strain. For conciseness, only 12 of 98 genes are displayed. Samples were taken at 0, 5, 10, 20, and 30 min. The plots below the gene display show the average fold induction [in log(2) scale] of the genes shown. Wild-type (IH4506), ssk1 ste11 (IH4531), ssk1 (IH4508), and ste11 (IH4537) strains were used.
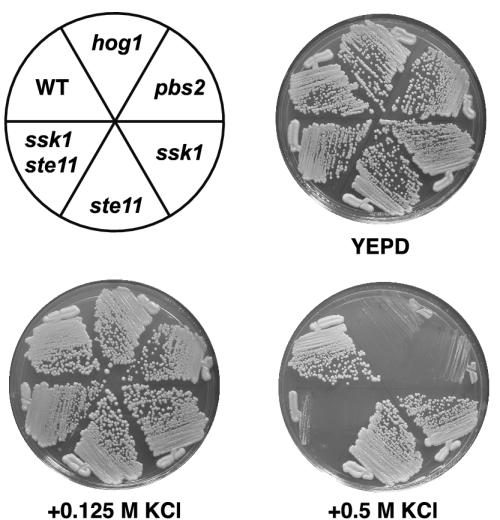
Even though the ssk1 single mutant strain is clearly defective for induction of many genes at the lowest two KCl concentrations, the ssk1 and ssk1 ste11 mutants did not exhibit a growth defect under these osmolarity concentrations (Figure 6). In contrast, the ssk1 ste11 strain exhibited a severe growth defect at 0.5 M KCl.
Figure 6.
Growth on high-osmolarity media. Strains were streaked onto the indicated media and photographed after 48 h of growth at 30°C. Strains as in Figure 4.
DISCUSSION
We have used genome-wide expression profiling to examine the response of wild-type and mutant yeast cells to a variety of osmotic conditions. Response of wild-type cells to 1 M sorbitol and 0.5 M KCl occurred with rapid kinetics and was transient (Figure 2). Response to KCl and sorbitol was very similar, though KCl induced a stronger response (Figure 2, B and C). A total of 2277 genes, nearly one-third of the yeast genome, exhibited significant regulation by KCl or sorbitol. These genes were identified by a time-course analysis in which expression levels were assessed at 10 time points (at 5, 10, 20, 30, 40, 60, 90, 120, and 180 min). We found that most genes exhibited maximal response at 20 min, which declined by 30 min and approached uninduced levels by 40 min (Figure 2D). Prior genomic studies of response to high osmolarity used fewer time points and provide a less complete picture of the response. For example, Posas et al. (2000) examined expression at 10 and 20 min of 0.4 and 0.8 M NaCl treatment; Rep et al. (2000) assayed response at 45 min of 0.7 M NaCl treatment and at 30 min after 0.5 M NaCl or 0.95 M sorbitol treatment; Yale and Bohnert (2001) assessed response at 10, 30, and 90 min after 1 M NaCl treatment; and Gasch et al. (2000) examined mRNA levels in response to 1 M sorbitol at 5, 15, 30, 45, 60, 90, and 120 min. Our studies indicate that a time course analysis up to 40 min provides a relatively complete picture of the response. In general, our observations on gene induction in wild-type cells agree with previous studies. However, some differences were observed, in particular, concerning the large group of genes coding for ribosomal proteins. We and others (Gasch et al., 2000; Rep et al., 2000; Causton et al., 2001) find that these genes are repressed, whereas Posas et al. (2000) and Yale and Bohnert (2001) find that this group of genes is induced by osmotic stress. The basis for this difference may result from differences in growth conditions.
Hog1 Plays Multiple Roles in the Response to High Osmolarity
We identified 579 genes whose RNA levels are dependent on Hog1 and showed that Hog1 controls three major aspects of the response to increased osmolarity: it determines the magnitude of gene induction, determines the duration of gene regulation, and limits activation of other MAPK cascades.
Only a relatively small subset of high osmolarity-induced genes (32; cluster III) have a strong requirement for HOG1 for induction. These genes, for example, STL1 and GRE2, are likely to be directly regulated by the HOG pathway because the kinetics of induction are rapid (beginning by 5 min) and because Hog1 and two transcription factors Hot1 and Sko1 (each of which bind Hog1) are associated with the promoters of STL1 and GRE2 (Alepuz et al., 2001; Proft and Struhl, 2002). In contrast, the ∼100 genes of cluster IV require HOG1 only for maximal induction. Rep et al. (2000) have shown that genes in this cluster (CTT1, ALD2, ALD3, HSP26, and SOL4) require the general stress response pathway (specifically Msn2 and Msn4) for their osmotic induction. Maximal induction of these genes thus requires both a HOG-dependent process and the general stress-response pathway.
Hog1 is required for restoration of gene expression after osmotic stress. The genes in clusters V and VI show essentially normal regulation at early times in the hog1 mutant, but mRNA levels fail to return to basal levels (Figure 3). Thus, Hog1 seems to play a role in down-regulating the response after cells have adapted to new osmotic conditions. Because only a minority of osmotically induced genes requires HOG1 for induction, it seems that Hog1 is involved in resetting expression levels of genes that are induced by the general stress response. Protein phosphatases activated by osmotic shock inactivate Hog1 after osmotic shock (Jacoby et al., 1997; Wurgler-Murphy et al., 1997; Mattison and Ota, 2000; Warmka et al., 2001) and are thereby involved in restoring gene expression to basal levels. These phosphatases may also be involved in resetting the general stress response pathway to basal levels.
The findings described here and by Rep et al. (2000) demonstrate that a large set of genes (cluster I; 88 genes) is misexpressed in hog1 mutants. The comprehensive identification of this group of genes confirms earlier work that demonstrates inappropriate activation of the mating and filamentation pathways in hog1 mutants (Hall et al., 1996; O'Rourke and Herskowitz, 1998; Davenport et al., 1999). We show elsewhere that expression of two-thirds of this gene set is mediated by Sho1 or Msb2 (O'Rourke and Herskowitz, 2002). Interestingly, some of these genes (e.g., SVS1 and PRY1) are not induced by mating pheromone, yet are induced by osmotic stress in a hog1 mutant.
Hog1 also maintains the basal expression of a group of genes whose expression is not regulated strongly by high osmolarity (including CWP1, SED1, and PIR3; Figure 3, cluster II). Correspondingly, HOG1 has known roles outside of the osmoresponse: HOG1 is required for pH-dependent induction of genes coding for cell wall structural proteins, including CWP1 (Kapteyn et al., 2001) and induction of various genes during heat stress (Winkler et al., 2002).
Pathway Profiling Reveals Additional Inputs for Pbs2 and Ste11
We have analyzed the induction profile in a variety of mutant strains to examine additional features of the HOG pathway and response to high osmolarity and have drawn several conclusions. First, it seems that the only role of the MAPKK Pbs2 is to regulate the MAPK Hog1, because mutants defective in PBS2 and HOG1 have nearly identical gene expression patterns (Figure 4; our unpublished data). Thus, the circuitry of the downstream portion of the HOG pathway is strictly linear.
Second, a comparison between the transcript profiles of pbs2 (or hog1) and ssk1 ste11 mutants indicates that additional inputs into Pbs2 may exist (Fig. i, web supplement). We find that STL1 and GRE2 are induced 8- to 38-fold in ssk1 ste11 and ssk1 sho1 mutants but exhibit little induction (<1.7-fold) when HOG1 or PBS2 is deleted. These observations indicate the existence of an input to Pbs2 in addition to Ste11 and SSK1. Such an input may be provided by a new MAPKKK that activates Pbs2 (e.g., Bck1) or by Ssk2/Ssk22 that is somehow activated independently of Ssk1. Van Wuytswinkel et al. (2000) have also argued for the existence of an input for Pbs2 that does not require Ste11, Ssk2, or Ssk22, which functions in response to high-solute conditions, e.g., 1.4 M KCl or NaCl (Van Wuytswinkel et al., 2000). This same input may be responsible for the induction that we observe in ste11 ssk1 mutants treated with 0.5 M KCl.
A third feature of the HOG pathway revealed by analysis of mutants is the existence of an additional osmosensing activity that functions in parallel with Sho1 to activate Ste11 (O'Rourke and Herskowitz, 1998, 2002). If Sho1 provided the sole input into Ste11, then sho1 mutants should exhibit the same phenotype as ste11 mutants. This was decidedly not the case: ste11 ssk1 strains exhibited a more severe phenotype than sho1 ssk1 strains with respect to the extent of induction of various genes (e.g., YLR042C) and inappropriate induction of pheromone-response genes. These differences indicate that there is another osmosensor in addition to Sho1 that can provide input to Ste11, a conclusion reached on other grounds by O'Rourke and Herskowitz (1998). We have recently identified this additional input, Msb2 (O'Rourke and Herskowitz, 2002).
Response to Different Osmolarities Reveals Redundancy and Nonredundancy between the Sho1 and Sln1 Branches
The Sho1 and Sln1 branches are redundant for promoting growth on high-osmolarity medium (Maeda et al., 1995), but they also display some functional differences. For example, these two branches activate Hog1 at different osmolarities and with different kinetics (Maeda et al., 1995; Van Wuytswinkel et al., 2000). In addition, the two branches differ in their roles in pathways other than response to high osmolarity. Sho1 is required for production of pseudohyphae (O'Rourke and Herskowitz, 1998), for signaling to Ste12 in response to protein glycosylation defects (Cullen et al., 2000), and for activating Hog1 in response to heat stress (Winkler et al., 2002). The Sln1 branch does not influence any of these responses but is specifically required for Hog1 activation in response to defects in GPI anchor synthesis (Toh-e and Oguchi, 2001).
We have taken advantage of the sensitivity of transcriptional profiling to show that the Sln1-Ssk1 branch governs the response to stress by moderately high osmolarity, e.g., 0.0625 M KCl or 0.125 M KCl (Figure 5) and thus that the Sho1-Ste11 and Sln1-Ssk1 branches are not equivalent for inducing gene expression in vivo (Figure 6). These results were surprising, because yeast mutants defective in HOG1, PBS2, and other components of the HOG pathway do not exhibit growth defects on media of moderate osmolarity (0.125 or 0.25 M KCl, Figure 6; our unpublished data). Presumably, the response to modest osmotic stress has biological consequences that are not captured in the simple laboratory tests that we have performed.
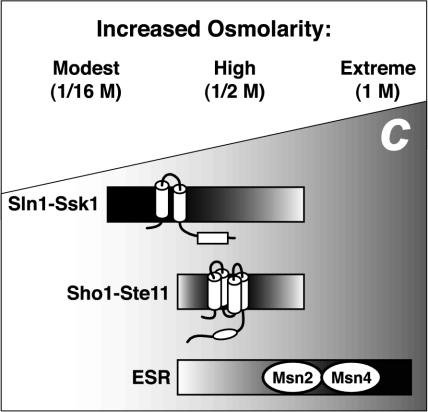
At higher osmolarities, both branches of the HOG pathway as well as the general stress response pathway are involved in the response. Previous work has shown that Hog1 phosphorylation in response to moderate osmolarity (0.1 and 0.2 M NaCl; Maeda et al., 1995) or severe osmotic stress (1.4 M KCl or NaCl; Van Wuytswinkel et al., 2000) is defective when the Sln1-Ssk1 branch is disabled. Thus, the Sln1-Ssk1 branch is active over a wide range of osmolarities for promoting Hog1 phosphorylation and gene induction. We likewise observed HOG1-dependent induction of genes at high osmolarity. We also observed HOG pathway-independent regulation at high osmolarities: for example, exposure of ssk1 ste11 mutants to high or extremely high osmolarity (0.5 or 1 M KCl) caused clear induction and repression of most osmoregulated genes. In summary, it seems that the SLN1-SSK1 branch of the HOG pathway is specialized for response to conditions of modest osmolarity, and other pathways such as the general stress response pathway apparently contribute, along with both branches of the HOG pathway, to osmoresponse during more extreme conditions (Figure 7).
Figure 7.
Different hyperosmotic conditions trigger different response pathways. C, concentration of solute shown increasing from left to right. The environmental stress response (ESR) pathway is preferentially used during extreme osmotic stress (Gasch et al., 2000). The Sln1-Ssk1 branch of the HOG pathway but not the Sho1-Ste11 branch is used during modest osmotic stress. At intermediate hyperosmotic stress, the ESR, Sln1-Ssk1, and Sho1-Ste11 pathways each contribute significantly to changes in gene expression.
Our results demonstrate that the bioassays originally used to monitor the cellular response to high osmolarity (in particular, osmosensitivity; Brewster et al., 1993) are not sufficiently sensitive to observe responses to more modest osmostresses. In this regard, it would be of interest to use transcript profiling to assay for differential functions of the Sho1 and Sln1 branches during response to other stimuli that activate these sensing pathways (O'Rourke and Hers-kowitz, 1998; Cullen et al., 2000; Toh-e and Oguchi, 2001; Winkler et al., 2002). In addition, screens could be performed on osmotically challenged yeast strains individually deleted for every nonessential gene using whole-genome expression profiling as a readout. This approach may identify knockout strains, which, like ssk1Δ, do not display growth defects on high-osmolarity medium but do exhibit a submaximal gene expression response.
Supplementary Material
Acknowledgments
We thank members of our laboratory and the laboratories of Joseph DeRisi and Wendell Lim for insightful discussion, and Joseph DeRisi, Brian Pulliam, and Jingchun Zhu for computer assistance. We also thank Joseph DeRisi for excellent suggestions for data analysis and Erin O'Shea for help in preparation of the manuscript. Kirsten Benjamin, Linda Huang, Hiten Madhani, and Jun Urano also provided useful comments on the manuscript. This work was supported by National Institutes of Health grant GM59466 (to I.H.). S.M.O. was supported by an National Institutes of Health training grant, the Markey Program in Biological Sciences, the Herbert W. Boyer Fund, and a UCSF Chancellor's Fellowship.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-07-0521. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-07-0521.
Online version of this article contains supplementary material. Online version is available at www.molbiolcell.org.
References
- Alepuz, P.M., Jovanovic, A., Reiser, V., and Ammerer, G. (2001). Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol. Cell 7, 767-777. [DOI] [PubMed] [Google Scholar]
- Bilsland-Marchesan, E., Arino, J., Saito, H., Sunnerhagen, P., and Posas, F. (2000). Rck2 kinase is a substrate for the osmotic stress-activated mitogen-activated protein kinase Hog1. Mol. Cell. Biol. 20, 3887-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster, J.L., de Valoir, T., Dwyer, N.D., Winter, E., and Gustin, M.C. (1993). An osmosensing signal transduction pathway in yeast. Science 259, 1760-1763. [DOI] [PubMed] [Google Scholar]
- Causton, H.C., Ren, B., Koh, S.S., Harbison, C.T., Kanin, E., Jennings, E.G., Lee, T.I., True, H.L., Lander, E.S., and Young, R.A. (2001). Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12, 323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, P.J., Schultz, J., Horecka, J., Stevenson, B.J., Jigami, Y., and Sprague, G.F., Jr. (2000). Defects in protein glycosylation cause SHO1-dependent activation of a STE12 signaling pathway in yeast. Genetics 155, 1005-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport, K.D., Williams, K.E., Ullmann, B.D., and Gustin, M.C. (1999). Activation of the Saccharomyces cerevisiae filamentation/invasion pathway by osmotic stress in high-osmolarity glycogen pathway mutants. Genetics 153, 1091-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nadal, E., Casadome, L., and Posas, F. (2003). Targeting the MEF2-like transcription factor Smp1 by the stress-activated Hog1 mitogen-activated protein kinase. Mol. Cell. Biol. 23, 229-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi, J.L., Iyer, V.R., and Brown, P.O. (1997). Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278, 680-686. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch, F. (2000). Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol. Rev. 24, 469-486. [DOI] [PubMed] [Google Scholar]
- Ferrigno, P., Posas, F., Koepp, D., Saito, H., and Silver, P.A. (1998). Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin beta homologs NMD5 and XPO1. EMBO J. 17, 5606-5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields, S., Chaleff, D.T., and Sprague, G.F., Jr. (1988). Yeast STE7, STE11, and STE12 genes are required for expression of cell-type-specific genes. Mol. Cell. Biol. 8, 551-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch, A.P., Spellman, P.T., Kao, C.M., Carmel-Harel, O., Eisen, M.B., Storz, G., Botstein, D., and Brown, P.O. (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin, M.C., Albertyn, J., Alexander, M., and Davenport, K. (1998). MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62, 1264-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, J.P., Cherkasova, V., Elion, E., Gustin, M.C., and Winter, E. (1996). The osmoregulatory pathway represses mating pathway activity in Saccharomyces cerevisiae: isolation of a FUS3 mutant that is insensitive to the repression mechanism. Mol. Cell. Biol. 16, 6715-6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby, T., Flanagan, H., Faykin, A., Seto, A.G., Mattison, C., and Ota, I. (1997). Two protein-tyrosine phosphatases inactivate the osmotic stress response pathway in yeast by targeting the mitogen-activated protein kinase, Hog1. J. Biol. Chem. 272, 17749-17755. [DOI] [PubMed] [Google Scholar]
- Kapteyn, J.C., ter Riet, B., Vink, E., Blad, S., De Nobel, H., Van Den Ende, H., and Klis, F.M. (2001). Low external pH induces HOG1-dependent changes in the organization of the Saccharomyces cerevisiae cell wall. Mol. Microbiol. 39, 469-479. [DOI] [PubMed] [Google Scholar]
- Maeda, T., Takekawa, M., and Saito, H. (1995). Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269, 554-558. [DOI] [PubMed] [Google Scholar]
- Mattison, C.P., and Ota, I.M. (2000). Two protein tyrosine phosphatases, Ptp2 and Ptp3, modulate the subcellular localization of the Hog1 MAP kinase in yeast. Genes Dev. 14, 1229-1235. [PMC free article] [PubMed] [Google Scholar]
- O'Rourke, S.M., and Herskowitz, I. (1998). The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12, 2874-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke, S.M., and Herskowitz, I. (2002). A third osmosensing branch in Saccharomyces cerevisiae requires the Msb2 protein and functions in parallel with the Sho1 branch. Mol. Cell. Biol. 22, 4739-4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke, S.M., Herskowitz, I., and O'Shea, E.K. (2002). Yeast go the whole HOG for the hyperosmotic response. Trends Genet. 18, 405-412. [DOI] [PubMed] [Google Scholar]
- Posas, F., Chambers, J.R., Heyman, J.A., Hoeffler, J.P., de Nadal, E., and Arino, J. (2000). The transcriptional response of yeast to saline stress. J. Biol. Chem. 275, 17249-17255. [DOI] [PubMed] [Google Scholar]
- Posas, F., and Saito, H. (1997). Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science 276, 1702-1705. [DOI] [PubMed] [Google Scholar]
- Posas, F., and Saito, H. (1998). Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 17, 1385-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas, F., Witten, E.A., and Saito, H. (1998). Requirement of STE50 for osmostress-induced activation of the STE11 mitogen-activated protein kinase kinase kinase in the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 18, 5788-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas, F., Wurgler-Murphy, S.M., Maeda, T., Witten, E.A., Thai, T.C., and Saito, H. (1996). Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86, 865-875. [DOI] [PubMed] [Google Scholar]
- Proft, M., Pascual-Ahuir, A., de Nadal, E., Arino, J., Serrano, R., and Posas, F. (2001). Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J. 20, 1123-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft, M., and Serrano, R. (1999). Repressors and upstream repressing sequences of the stress-regulated ENA1 gene in Saccharomyces cerevisiae: bZIP protein Sko1p confers HOG-dependent osmotic regulation. Mol. Cell. Biol. 19, 537-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft, M., and Struhl, K. (2002). Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell 9, 1307-1317. [DOI] [PubMed] [Google Scholar]
- Raitt, D.C., Posas, F., and Saito, H. (2000). Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 19, 4623-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramezani Rad, M., Jansen, G., Buhring, F., and Hollenberg, C.P. (1998). Ste50p is involved in regulating filamentous growth in the yeast Saccharomyces cerevisiae and associates with Ste11p. Mol. Gen. Genet. 259, 29-38. [DOI] [PubMed] [Google Scholar]
- Reed, R.H., Chudek, J.A., Foster, R., and Gadd, G.M. (1987). Osmotic significance of glycerol accumulation in exponentially growing yeasts. Appl. Environ. Microbiol. 53, 2119-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser, V., Ruis, H., and Ammerer, G. (1999). Kinase activity-dependent nuclear export opposes stress-induced nuclear accumulation and retention of Hog1 mitogen-activated protein kinase in the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell 10, 1147-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser, V., Salah, S.M., and Ammerer, G. (2000). Polarized localization of yeast Pbs2 depends on osmostress, the membrane protein Sho1 and Cdc42. Nat. Cell Biol. 2, 620-627. [DOI] [PubMed] [Google Scholar]
- Rep, M., Krantz, M., Thevelein, J.M., and Hohmann, S. (2000). The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J. Biol. Chem. 275, 8290-8300. [DOI] [PubMed] [Google Scholar]
- Rep, M., Reiser, V., Gartner, U., Thevelein, J.M., Hohmann, S., Ammerer, G., and Ruis, H. (1999). Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol. Cell. Biol. 19, 5474-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, C.J., et al. (2000). Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287, 873-880. [DOI] [PubMed] [Google Scholar]
- Rose, M.D., Winston, F., and Hieter, P. (1990). Methods in Yeast Genetics: A Laboratory Course Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Sakumoto, N., et al. (1999). A series of protein phosphatase gene disruptants in Saccharomyces cerevisiae. Yeast 15, 1669-1679. [DOI] [PubMed] [Google Scholar]
- Schiestl, R.H., and Gietz, R.D. (1989). High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16, 339-346. [DOI] [PubMed] [Google Scholar]
- Schüller, C., Brewster, J.L., Alexander, M.R., Gustin, M.C., and Ruis, H. (1994). The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene. EMBO J. 13, 4382-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker, D.D., et al. (2001). Experimental annotation of the human genome using microarray technology. Nature 409, 922-927. [DOI] [PubMed] [Google Scholar]
- Sprague, G.F., Jr. (1998). Control of MAP kinase signaling specificity or how not to go HOG wild. Genes Dev. 12, 2817-2820. [DOI] [PubMed] [Google Scholar]
- Stock, A.M., Robinson, V.L., and Goudreau, P.N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69, 183-215. [DOI] [PubMed] [Google Scholar]
- Toh-e, A., and Oguchi, T. (2001). Defects in glycosylphosphatidylinositol (GPI) anchor synthesis activate Hog1 kinase and confer copper-resistance in Saccharomyces cerevisiae. Genes Genet. Syst. 76, 393-410. [DOI] [PubMed] [Google Scholar]
- van Drogen, F., O'Rourke, S.M., Stucke, V.M., Jaquenoud, M., Neiman, A.M., and Peter, M. (2000). Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr. Biol. 10, 630-639. [DOI] [PubMed] [Google Scholar]
- Van Wuytswinkel, O., Reiser, V., Siderius, M., Kelders, M.C., Ammerer, G., Ruis, H., and Mager, W.H. (2000). Response of Saccharomyces cerevisiae to severe osmotic stress: evidence for a novel activation mechanism of the HOG MAP kinase pathway. Mol. Microbiol. 37, 382-397. [DOI] [PubMed] [Google Scholar]
- Warmka, J., Hanneman, J., Lee, J., Amin, D., and Ota, I. (2001). Ptc1, a type 2C Ser/Thr phosphatase, inactivates the HOG pathway by dephosphorylating the mitogen-activated protein kinase Hog1. Mol. Cell. Biol. 21, 51-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, A., Arkind, C., Mattison, C.P., Burkholder, A., Knoche, K., and Ota, I. (2002). Heat stress activates the yeast high-osmolarity glycerol mitogen-activated protein kinase pathway, and protein tyrosine phosphatases are essential under heat stress. Eukaryot. Cell 1, 163-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurgler-Murphy, S.M., Maeda, T., Witten, E.A., and Saito, H. (1997). Regulation of the Saccharomyces cerevisiae HOG1 mitogen-activated protein kinase by the PTP2 and PTP3 protein tyrosine phosphatases. Mol. Cell. Biol. 17, 1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yale, J., and Bohnert, H.J. (2001). Transcript expression in Saccharomyces cerevisiae at high salinity. J. Biol. Chem. 276, 15996-16007. [DOI] [PubMed] [Google Scholar]
- Zhan, X.L., and Guan, K.L. (1999). A specific protein-protein interaction accounts for the in vivo substrate selectivity of Ptp3 towards the Fus3 MAP kinase. Genes Dev. 13, 2811-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.