Abstract
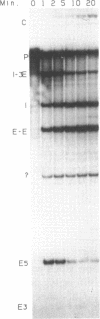
The group I intron from the chloroplast rRNA large subunit of Chlamydomonas reinhardtii (Cr.LSU) undergoes autocatalytic splicing in vitro. Cr.LSU displays a range of reactions typical of other group I introns. Under optimal conditions, the 5' cleavage step proceeds rapidly, but the exon-ligation step is relatively slow, and no pH dependent hydrolysis of the 3' splice site occurs. A requirement for high temperature and high [Mg2+] suggests involvement of additional splicing factors in vivo. The positions of three cyclization sites of the free intron have been mapped; two of these sites represent reactions analogous to 5'-splice site cleavage, whereas the third is an example of G-exchange. Cr.LSU contains an open reading frame (ORF) potentially encoding an 163 amino acid polypeptide. ORF function has been investigated by using chloroplast gene replacement via particle bombardment. We have shown that the ORF can be deleted from Cr.LSU without affecting splicing in vivo and it thus does not encode an essential splicing factor.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barfod E. T., Cech T. R. The conserved U.G pair in the 5' splice site duplex of a group I intron is required in the first but not the second step of self-splicing. Mol Cell Biol. 1989 Sep;9(9):3657–3666. doi: 10.1128/mcb.9.9.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Been M. D., Cech T. R. Selection of circularization sites in a group I IVS RNA requires multiple alignments of an internal template-like sequence. Cell. 1987 Sep 11;50(6):951–961. doi: 10.1016/0092-8674(87)90522-8. [DOI] [PubMed] [Google Scholar]
- Been M. D., Cech T. R. Sites of circularization of the Tetrahymena rRNA IVS are determined by sequence and influenced by position and secondary structure. Nucleic Acids Res. 1985 Dec 9;13(23):8389–8408. doi: 10.1093/nar/13.23.8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Been M. D., Perrotta A. T. Group I intron self-splicing with adenosine: evidence for a single nucleoside-binding site. Science. 1991 Apr 19;252(5004):434–437. doi: 10.1126/science.2017681. [DOI] [PubMed] [Google Scholar]
- Belfort M. Phage T4 introns: self-splicing and mobility. Annu Rev Genet. 1990;24:363–385. doi: 10.1146/annurev.ge.24.120190.002051. [DOI] [PubMed] [Google Scholar]
- Boynton J. E., Gillham N. W., Harris E. H., Hosler J. P., Johnson A. M., Jones A. R., Randolph-Anderson B. L., Robertson D., Klein T. M., Shark K. B. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science. 1988 Jun 10;240(4858):1534–1538. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]
- Burke J. M., Belfort M., Cech T. R., Davies R. W., Schweyen R. J., Shub D. A., Szostak J. W., Tabak H. F. Structural conventions for group I introns. Nucleic Acids Res. 1987 Sep 25;15(18):7217–7221. doi: 10.1093/nar/15.18.7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J. M., Irvine K. D., Kaneko K. J., Kerker B. J., Oettgen A. B., Tierney W. M., Williamson C. L., Zaug A. J., Cech T. R. Role of conserved sequence elements 9L and 2 in self-splicing of the Tetrahymena ribosomal RNA precursor. Cell. 1986 Apr 25;45(2):167–176. doi: 10.1016/0092-8674(86)90380-6. [DOI] [PubMed] [Google Scholar]
- Cech T. R. Conserved sequences and structures of group I introns: building an active site for RNA catalysis--a review. Gene. 1988 Dec 20;73(2):259–271. doi: 10.1016/0378-1119(88)90492-1. [DOI] [PubMed] [Google Scholar]
- Cech T. R. Self-splicing of group I introns. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- Doudna J. A., Cormack B. P., Szostak J. W. RNA structure, not sequence, determines the 5' splice-site specificity of a group I intron. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7402–7406. doi: 10.1073/pnas.86.19.7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B. Group I introns as mobile genetic elements: facts and mechanistic speculations--a review. Gene. 1989 Oct 15;82(1):91–114. doi: 10.1016/0378-1119(89)90034-6. [DOI] [PubMed] [Google Scholar]
- Durocher V., Gauthier A., Bellemare G., Lemieux C. An optional group I intron between the chloroplast small subunit rRNA genes of Chlamydomonas moewusii and C. eugametos. Curr Genet. 1989 Apr;15(4):277–282. doi: 10.1007/BF00447043. [DOI] [PubMed] [Google Scholar]
- Erickson J. M., Rahire M., Rochaix J. D. Chlamydomonas reinhardii gene for the 32 000 mol. wt. protein of photosystem II contains four large introns and is located entirely within the chloroplast inverted repeat. EMBO J. 1984 Dec 1;3(12):2753–2762. doi: 10.1002/j.1460-2075.1984.tb02206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriga G., Lambowitz A. M. RNA splicing in neurospora mitochondria: self-splicing of a mitochondrial intron in vitro. Cell. 1984 Dec;39(3 Pt 2):631–641. doi: 10.1016/0092-8674(84)90470-7. [DOI] [PubMed] [Google Scholar]
- Gauthier A., Turmel M., Lemieux C. A group I intron in the chloroplast large subunit rRNA gene of Chlamydomonas eugametos encodes a double-strand endonuclease that cleaves the homing site of this intron. Curr Genet. 1991 Jan;19(1):43–47. doi: 10.1007/BF00362086. [DOI] [PubMed] [Google Scholar]
- Harris E. H., Burkhart B. D., Gillham N. W., Boynton J. E. Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: correlation of genetic and physical maps of the chloroplast genome. Genetics. 1989 Oct;123(2):281–292. doi: 10.1093/genetics/123.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrin D. L., Chen Y. F., Schmidt G. W. RNA splicing in Chlamydomonas chloroplasts. Self-splicing of 23 S preRNA. J Biol Chem. 1990 Dec 5;265(34):21134–21140. [PubMed] [Google Scholar]
- Inoue T., Sullivan F. X., Cech T. R. New reactions of the ribosomal RNA precursor of Tetrahymena and the mechanism of self-splicing. J Mol Biol. 1986 May 5;189(1):143–165. doi: 10.1016/0022-2836(86)90387-6. [DOI] [PubMed] [Google Scholar]
- Kay P. S., Menzel P., Inoue T. Two guanosine binding sites exist in group I self-splicing IVS RNAs. EMBO J. 1988 Nov;7(11):3531–3537. doi: 10.1002/j.1460-2075.1988.tb03229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchino Y., Nishimura S. Enzymatic RNA sequencing. Methods Enzymol. 1989;180:154–163. doi: 10.1016/0076-6879(89)80099-0. [DOI] [PubMed] [Google Scholar]
- Kuhsel M. G., Strickland R., Palmer J. D. An ancient group I intron shared by eubacteria and chloroplasts. Science. 1990 Dec 14;250(4987):1570–1573. doi: 10.1126/science.2125748. [DOI] [PubMed] [Google Scholar]
- Latham J. A., Zaug A. J., Cech T. R. Self-splicing and enzymatic cleavage of RNA by a group I intervening sequence. Methods Enzymol. 1990;181:558–569. doi: 10.1016/0076-6879(90)81151-j. [DOI] [PubMed] [Google Scholar]
- Lemieux C., Boulanger J., Otis C., Turmel M. Nucleotide sequence of the chloroplast large subunit rRNA gene from Chlamydomonas reinhardtii. Nucleic Acids Res. 1989 Oct 11;17(19):7997–7997. doi: 10.1093/nar/17.19.7997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Ellington A. D., Couture S., Szostak J. W. Phylogenetic and genetic evidence for base-triples in the catalytic domain of group I introns. Nature. 1990 Oct 11;347(6293):578–580. doi: 10.1038/347578a0. [DOI] [PubMed] [Google Scholar]
- Michel F., Netter P., Xu M. Q., Shub D. A. Mechanism of 3' splice site selection by the catalytic core of the sunY intron of bacteriophage T4: the role of a novel base-pairing interaction in group I introns. Genes Dev. 1990 May;4(5):777–788. doi: 10.1101/gad.4.5.777. [DOI] [PubMed] [Google Scholar]
- Michel F., Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990 Dec 5;216(3):585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Newman S. M., Boynton J. E., Gillham N. W., Randolph-Anderson B. L., Johnson A. M., Harris E. H. Transformation of chloroplast ribosomal RNA genes in Chlamydomonas: molecular and genetic characterization of integration events. Genetics. 1990 Dec;126(4):875–888. doi: 10.1093/genetics/126.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oard J. H., Paige D. F., Simmonds J. A., Gradziel T. M. Transient gene expression in maize, rice, and wheat cells using an airgun apparatus. Plant Physiol. 1990 Feb;92(2):334–339. doi: 10.1104/pp.92.2.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partono S., Lewin A. S. Autocatalytic activities of intron 5 of the cob gene of yeast mitochondria. Mol Cell Biol. 1988 Jun;8(6):2562–2571. doi: 10.1128/mcb.8.6.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partono S., Lewin A. S. Splicing of COB intron 5 requires pairing between the internal guide sequence and both flanking exons. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8192–8196. doi: 10.1073/pnas.87.21.8192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partono S., Lewin A. S. The rate and specificity of a group I ribozyme are inversely affected by choice of monovalent salt. Nucleic Acids Res. 1991 Feb 11;19(3):605–609. doi: 10.1093/nar/19.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman P. S., Butow R. A. Mobile introns and intron-encoded proteins. Science. 1989 Dec 1;246(4934):1106–1109. doi: 10.1126/science.2479980. [DOI] [PubMed] [Google Scholar]
- Price J. V., Engberg J., Cech T. R. 5' exon requirement for self-splicing of the Tetrahymena thermophila pre-ribosomal RNA and identification of a cryptic 5' splice site in the 3' exon. J Mol Biol. 1987 Jul 5;196(1):49–60. doi: 10.1016/0022-2836(87)90510-9. [DOI] [PubMed] [Google Scholar]
- Rochaix J. D., Rahire M., Michel F. The chloroplast ribosomal intron of Chlamydomonas reinhardii codes for a polypeptide related to mitochondrial maturases. Nucleic Acids Res. 1985 Feb 11;13(3):975–984. doi: 10.1093/nar/13.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder R., von Ahsen U., Belfort M. Effects of mutations of the bulged nucleotide in the conserved P7 pairing element of the phage T4 td intron on ribozyme function. Biochemistry. 1991 Apr 2;30(13):3295–3303. doi: 10.1021/bi00227a018. [DOI] [PubMed] [Google Scholar]
- Tabak H. F., Van der Horst G., Osinga K. A., Arnberg A. C. Splicing of large ribosomal precursor RNA and processing of intron RNA in yeast mitochondria. Cell. 1984 Dec;39(3 Pt 2):623–629. doi: 10.1016/0092-8674(84)90469-0. [DOI] [PubMed] [Google Scholar]
- Tabak H. F., Van der Horst G., Winter A. J., Smit J., Van der Veen R., Kwakman J. H., Grivell L. A., Arnberg A. C. Reactions mediated by yeast mitochondrial group I and II introns. Cold Spring Harb Symp Quant Biol. 1987;52:213–221. doi: 10.1101/sqb.1987.052.01.026. [DOI] [PubMed] [Google Scholar]
- Turmel M., Boulanger J., Lemieux C. Two group I introns with long internal open reading frames in the chloroplast psbA gene of Chlamydomonas moewusii. Nucleic Acids Res. 1989 May 25;17(10):3875–3887. doi: 10.1093/nar/17.10.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M., Boulanger J., Schnare M. N., Gray M. W., Lemieux C. Six group I introns and three internal transcribed spacers in the chloroplast large subunit ribosomal RNA gene of the green alga Chlamydomonas eugametos. J Mol Biol. 1991 Mar 20;218(2):293–311. doi: 10.1016/0022-2836(91)90713-g. [DOI] [PubMed] [Google Scholar]
- Walstrum S. A., Uhlenbeck O. C. The self-splicing RNA of Tetrahymena is trapped in a less active conformation by gel purification. Biochemistry. 1990 Nov 20;29(46):10573–10576. doi: 10.1021/bi00498a022. [DOI] [PubMed] [Google Scholar]
- Xu M. Q., Kathe S. D., Goodrich-Blair H., Nierzwicki-Bauer S. A., Shub D. A. Bacterial origin of a chloroplast intron: conserved self-splicing group I introns in cyanobacteria. Science. 1990 Dec 14;250(4987):1566–1570. doi: 10.1126/science.2125747. [DOI] [PubMed] [Google Scholar]
- Yarus M., Illangesekare M., Christian E. Selection of small molecules by the Tetrahymena catalytic center. Nucleic Acids Res. 1991 Mar 25;19(6):1297–1304. doi: 10.1093/nar/19.6.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug A. J., Grabowski P. J., Cech T. R. Autocatalytic cyclization of an excised intervening sequence RNA is a cleavage-ligation reaction. Nature. 1983 Feb 17;301(5901):578–583. doi: 10.1038/301578a0. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Kent J. R., Cech T. R. Reactions of the intervening sequence of the Tetrahymena ribosomal ribonucleic acid precursor: pH dependence of cyclization and site-specific hydrolysis. Biochemistry. 1985 Oct 22;24(22):6211–6218. doi: 10.1021/bi00343a027. [DOI] [PubMed] [Google Scholar]
- van der Horst G., Tabak H. F. New RNA-mediated reactions by yeast mitochondrial group I introns. EMBO J. 1987 Jul;6(7):2139–2144. doi: 10.1002/j.1460-2075.1987.tb02481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]