Abstract
OBJECTIVE
Pigment epithelium–derived factor (PEDF) is an adipocyte-secreted factor involved in the development of insulin resistance in obesity. Previous studies have identified PEDF as a regulator of triacylglycerol metabolism in the liver that may act through adipose triglyceride lipase (ATGL). We used ATGL−/− mice to determine the role of PEDF in regulating lipid and glucose metabolism.
RESEARCH DESIGN AND METHODS
Recombinant PEDF was administered to ATGL−/− and wild-type mice, and whole-body energy metabolism was studied by indirect calorimetry. Adipose tissue lipolysis and skeletal muscle fatty acid metabolism was determined in isolated tissue preparations. Muscle lipids were assessed by electrospray ionization–tandem mass spectrometry. Whole-body insulin sensitivity and skeletal muscle glucose uptake were assessed.
RESULTS
PEDF impaired the capacity to adjust substrate selection, resulting in a delayed diurnal decline in the respiratory exchange ratio, and suppressed daily fatty acid oxidation. PEDF enhanced adipocyte lipolysis and triacylglycerol lipase activity in skeletal muscle. Muscle fatty acid uptake and storage were unaffected, whereas fatty acid oxidation was impaired. These changes in lipid metabolism were abrogated in ATGL−/− mice and were not attributable to hypothalamic actions. ATGL−/− mice were also refractory to PEDF-mediated insulin resistance, but this was not related to changes in lipid species in skeletal muscle.
CONCLUSIONS
The results are the first direct demonstration that 1) PEDF influences systemic fatty acid metabolism by promoting lipolysis in an ATGL-dependent manner and reducing fatty acid oxidation and 2) ATGL is required for the negative effects of PEDF on insulin action.
Adipose tissue biology is markedly affected by obesity, and its endocrine role has been extensively investigated. Studies using proteomic approaches estimate that ∼90–260 individual proteins are released by adipocytes (1–4). Several adipose-secreted factors that are elevated in obesity are implicated in the pathogenesis of metabolic dysfunction and insulin resistance, including tumor necrosis factor-α (5), interleukin-6 (6), resistin (7), and retinol-binding protein 4 (8). These data support a major role for adipose tissue in regulating whole-body fatty acid metabolism and insulin action.
Pigment epithelium–derived factor (PEDF, SerpinF1) is upregulated in individuals with the metabolic syndrome (9,10) and patients with type 2 diabetes (11,12). Although PEDF is best known for its antiangiogenic and neuroprotective functions (13), recent work has implicated PEDF in the development of obesity-related insulin resistance (3,14). PEDF induced proinflammatory signaling, increased adipocyte lipolysis, and promoted lipid accumulation in muscle and liver that was associated with insulin resistance (3).
PEDF is thought to exert its biologic actions by binding to a cell surface receptor (15,16). A recently identified cell surface receptor that possesses phospholipase activity was reported in retinal pigment epithelial cells (17). Surprisingly, this putative PEDF receptor was reported to be adipose triglyceride lipase (ATGL), a highly conserved triacylglycerol lipase that is critical for the maintenance of lipid and glucose homeostasis (18–23). Others have shown that recombinant PEDF is transported into cells and colocalizes with ATGL at lipid droplets, and coimmunoprecipitation studies indicate that ATGL interacts with PEDF (24). PEDF-deficient mice have hepatic steatosis, and some evidence supports the premise that the PEDF–ATGL nexus may be important in conveying PEDF’s modulation of lipid metabolism (24).
In the current study, we used pharmacologic and genetic models to examine the role of PEDF in systemic fatty acid metabolism. We tested the hypothesis that ATGL is required for the metabolic actions of PEDF.
RESEARCH DESIGN AND METHODS
Cell culture.
L6 myoblasts and 3T3-L1 adipocytes were maintained as described in the Supplementary Methods. Recombinant PEDF was purified from a HEK293 cell line stably transfected with human PEDF (25).
Animal maintenance and experimental protocols.
Experimental procedures were approved by the School of Biomedical Sciences Animal Ethics Committee (Monash University) and conformed to National Health & Medical Research Council (Australia) guidelines regarding the care and use of experimental animals. ATGL and hormone-sensitive lipase (HSL)−/− mice were generated as described (18,26). Knockout and wild-type (Wt) mice were generated by breeding heterozygous mice for the deleted allele. Mice were housed under controlled temperature (∼22°C) and a 12-h light/dark cycle and had free access to standard mouse chow and water. Female or male mice were aged 9–12 weeks.
For the refeeding studies, mice were fasted for 16 h and killed, or fasted for 24 h then provided access to food for 2 h before being killed. For in vivo PEDF administration experiments, mice were allowed ad libitum access to food or were fasted for 8 h, injected with 0.9% saline or PEDF, then maintained until being killed 8 h later. For ex vivo metabolic analysis, mice were fasted for 4 h and assays were performed at 1100. For the intracerebroventricular (ICV) experiments, male C57BL/6 mice were anesthetized under isoflurane and placed in a stereotaxic device. A guide cannula (Plastics One, Roanoke, VA) was implanted into the left lateral ventricle (anteroposterior −0.3 mm, lateral +1.0 mm to bregma and dorsoventral −2.5 mm below skull). The support plate of the cannula was attached to the skull, and the mice were allowed 4 days of recovery.
Metabolic assessment.
Whole-body metabolic monitoring was performed in a Comprehensive Laboratory Animal Monitoring System (Columbus Instruments, Columbus, OH). Mice were housed individually in closed chambers for 60 h for assessment of oxygen uptake, carbon dioxide production, activity, and food intake. Mice were injected with PEDF (50 μg i.p.), or for ICV experiments, mice were injected with a 2.5-μL volume of artificial cerebrospinal fluid (aCSF) or PEDF (0.365 μg/μL) before the onset of the dark phase. All compounds were injected using a 28-gauge stainless steel injector placed in and projecting 0.5 mm below the tip of the cannula.
Lipolysis.
3T3-L1 adipocytes or isolated epididymal fat were incubated in Krebs buffer, 2% BSA, and glucose (8 mmol/L) for 2 h. PEDF (100 nmol/L) or isoproterenol (1 μmol/L) were added as indicated. Glycerol (Sigma, St. Louis, MO) and free fatty acid (FFA; Wako, Richmond, VA) were analyzed using commercial assays.
Fatty acid metabolism.
Fatty acid metabolism was assessed in myotubes or isolated muscles as described previously (27).
Mitochondrial isolation and respiration.
Mitochondrial isolation and respiration were performed as described in the Supplementary Methods.
Triacylglycerol hydrolase activity.
Cell lysates were incubated in a substrate consisting of 5 mmol/L triolein, 14 × 106 dpm [9, 10-3H] triolein, 0.6 mg phospholipid (phosphatidylcholine/phosphatidylinositol 3:1 w/w), 0.1 mol/L potassium phosphate, and 20% BSA, and activity was determined as the release of 3H-FFA (28).
Glucose uptake.
Myotubes or isolated extensor digitorum longus muscles were pretreated with 100 nmol/L PEDF or an equal volume of PBS for 2 h. 2-Deoxy-d-glucose uptake was measured as described (3).
Insulin tolerance test.
Mice were injected with PEDF (50 µg i.p.) or 0.9% saline and 2 h later with insulin (0.5 units/kg; Actrapid, Novo Nordisk, Bagsværd, Denmark). Tail blood was collected at 15-min intervals for 90 min, and blood glucose was determined using a glucometer (Accu-Chek, Roche Diagnostics, Mannheim, Germany).
Cell fractionation, immunofluorescence analysis, and immunoblot analysis.
Methods described in Supplementary Methods.
Quantitative RT-PCR.
RNA was extracted in Qiazol reagent, followed by isolation using an RNeasy Tissue Kit (Qiagen, Doncaster, Victoria, Australia). RNA quantity was determined at 260 nm (NanoDrop p2000 Spectrometer, Biolab, Clayton, Australia), reverse transcribed (Invitrogen, Mt. Waverley, Victoria, Australia), and gene products were determined by real-time quantitative RT-PCR (ep realplex Mastercycler, Eppendorf, Hamburg, Germany) using TaqMan Assays-on-Demand (Applied Biosystems, Scoresby, Victoria, Australia). 18S was used as a reference gene and did not vary between groups. The mRNA levels were determined by a comparative CT method.
Electrospray ionization–tandem mass spectrometry of muscle lipids.
ATGL−/− and Wt littermates were injected with recombinant PEDF or saline at 0700 h and allowed access to food and water. Mice were killed at 1600 h, and the vastus lateralis muscle was removed. A 60-mg portion was homogenized in PBS, and 50 μg protein (10–20 μL) was extracted with chloroform/methanol (2:1; 20 volumes) after the addition of internal standards. Analysis was performed by electrospray ionization–tandem mass spectrometry using a PE Sciex API 4000 Q/TRAP mass spectrometer with a turbo-ionspray source and Analyst 1.5 data system. Quantification of individual lipid species was performed using scheduled multiple-reaction monitoring in positive ion mode. For detailed methods see Supplementary Methods.
Statistical analysis.
Statistical analysis was performed using unpaired Student t test. A two-way ANOVA with repeated measures was applied where appropriate, and a Student-Newman-Keuls post hoc analysis was performed. Statistical significance was set a priori at P < 0.05.
RESULTS
PEDF enhances basal adipose tissue lipolysis in an ATGL-dependent manner.
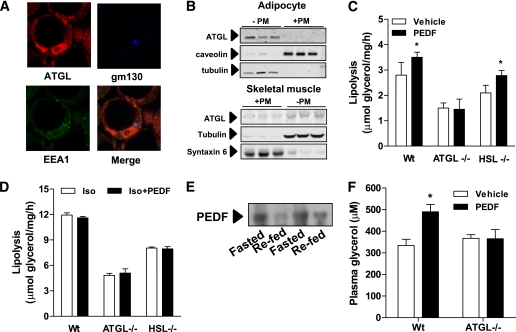
We previously demonstrated that PEDF increases basal lipolysis in 3T3-L1 adipocytes, however, the mechanism remained unresolved (3). ATGL is a critical mediator of triacylglycerol lipolysis in multiple tissues (19,21,23) and appears to interact with PEDF (24). ATGL was recently proposed to be a receptor for PEDF (17). This is a curious observation because ATGL has previously been localized to lipid droplets or the cytoplasm of adipocytes (19,23,29,30). We also verified the localization of ATGL to lipid droplets in adipocytes (Fig. 1A); however, we failed to observe any detectable ATGL in purified plasma membrane fractions isolated from adipocytes (Fig. 1B), and very little was detected in myotubes (Fig. 1B).
FIG. 1.
ATGL is the major target of PEDF-mediated lipolysis in adipose tissue. A: Immunofluorescence microscopy shows distribution of ATGL in the cytoplasm and around lipid droplets in 3T3-L1 adipocytes. Notably, ATGL is not observed at the plasma membrane (PM) or with endosomal (early endosome antigen 1 [EEA1]: green) or Golgi markers (gm130: blue). B: ATGL colocalizes with the cytoplasmic marker tubulin, but not with membrane markers in a subcellular fractionation. Western blot shows location of various subcellular markers in a fractionation from 3T3-L1 adipocytes (top) and L6 myotubes (below). +PM, plasma membrane–containing fraction. C: Epididymal fat pads were excised from lean ATGL−/−, HSL−/−, or Wt littermates, and lipolysis was assessed as glycerol release into the buffer. PEDF: 100 nmol/L (n = 4 per group). *P < 0.05 vs. vehicle within the same genotype. D: PEDF (100 nmol/L) does not affect β-adrenergic–stimulated lipolysis in epididymal fat pads. Fat pads were incubated for 2 h in isoproterenol (1 μmol/L), and glycerol release was determined (n = 4 per group). E: Immunoblot of plasma PEDF in mice fasted for 16 h or 2 h after refeeding. F: In vivo lipolysis is increased by PEDF. Lean ATGL−/− or Wt mice were injected with PEDF intraperitoneally, blood was obtained after 30 min, and glycerol was assessed in the plasma (n = 6 mice per group). *P < 0.05 vs. vehicle. Data in graphs are mean ± SEM. (A high-quality digital representation of this figure is available in the online issue.)
We next sought to determine whether ATGL is required for PEDF’s stimulatory effects on lipolysis. PEDF increased basal lipolysis by ∼25% in isolated adipose tissue explants (Fig. 1C, Supplementary Fig. 1A) but did not affect β-adrenergic stimulated lipolysis (Fig. 1D, Supplementary Fig. 1B). This stimulatory effect of PEDF on basal lipolysis was not evident in explants from ATGL−/− mice (Fig. 1C) and may be explained by a direct interaction between ATGL and PEDF as previously shown (17,24). It is unlikely that the other key lipolytic enzyme, HSL, is important for this process because adipose tissue from HSL−/− mice exhibited PEDF-stimulated lipolysis (Fig. 1C). The PEDF-induced glycerol release in HSL−/− explants suggests activation of another diacylglycerol lipase(s), as observed in HSL−/− mice with β-adrenergic stimulation (31,32), and/or reflects an increased availability of diacylglycerol substrate for HSL in Wt mice. Interestingly, plasma PEDF is elevated during fasting and reduced with refeeding in mice (Fig. 1E), which is temporally consistent with lipolysis rates and therefore a prolipolytic role for PEDF.
Next, we assessed the in vivo relevance of these observations. An acute PEDF injection into Wt mice increased plasma glycerol; however, these effects were abolished in ATGL−/− mice (Fig. 1F). Collectively, these in vitro and whole-animal studies demonstrate that ATGL is required for PEDF’s lipolytic effects.
PEDF alters whole-body energy metabolism by modulating fatty acid oxidation.
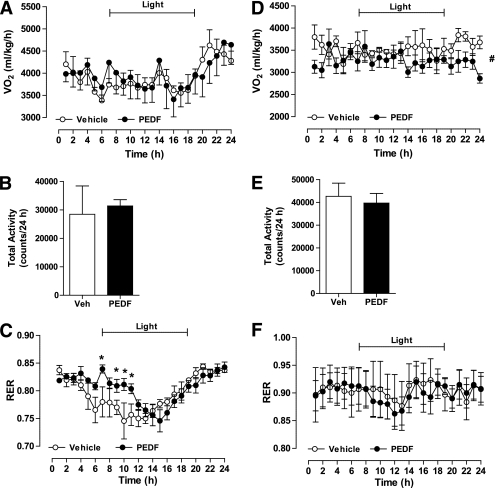
Having established that PEDF increases basal lipolysis and thereby fatty acid availability, we next assessed PEDF’s effects on whole-body energy homeostasis. PEDF administration did not affect total energy expenditure (Fig. 2A), daily physical activity (Fig. 2B), or food intake (data not shown) in Wt mice. Whole-body substrate oxidation was calculated by indirect calorimetry, and PEDF administration was shown to delay the diurnal switch from carbohydrate to fat oxidation in the fasted state (Fig. 2C). We repeated these experiments in ATGL−/− mice and observed a mild decrease in 24-h oxygen consumption (Fig. 2D) and no change in daily physical activity (Fig. 2E). Whole-body carbohydrate oxidation was elevated in ATGL−/− mice, as reported previously (18,33); however, the PEDF-induced alterations in substrate metabolism were not evident (Fig. 2F).
FIG. 2.
PEDF alters whole-body fatty acid metabolism. A–C: Lean C57Bl/6 mice were injected with recombinant PEDF or sterile saline and placed in a metabolic monitoring station. Oxygen uptake (VO2; A) and total activity (B) were similar between groups. C: The respiratory exchange ratio (RER) was assessed during the light and dark cycles and was increased during the early phase of the light cycle in PEDF-treated mice (n = 6 mice per group). *P < 0.05 vs. vehicle at the corresponding time. D–F: VO2, activity, and the RER were assessed in ATGL−/− mice after recombinant PEDF or sterile saline administration (n = 4 per group). #P < 0.05, main effect for treatment. All data are presented as means ± SEM.
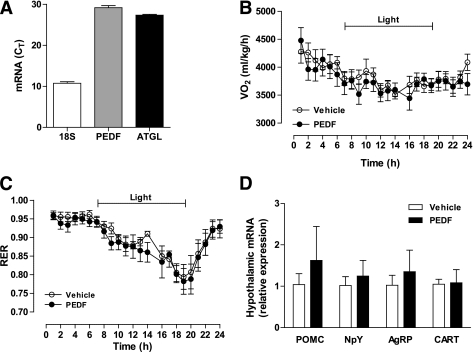
Intracerebroventricular PEDF does not affect energy expenditure or substrate partitioning.
Hypothalamic sensing of nutrients and hormones regulates whole-body energy homeostasis, affecting both feeding and energy expenditure (34). Accordingly, we examined whether PEDF was exerting metabolic effects by centrally mediated pathways. We first established the presence of PEDF and ATGL transcripts in the hypothalamus (Fig. 3A). To determine the central effects of PEDF on energy metabolism, intracerebroventricular PEDF or aCSF was administered in lean mice. Whole-body energy expenditure (Fig. 3B), substrate partitioning (Fig. 3C), food intake, and activity (not shown) were not altered with the ICV PEDF injection. ICV PEDF did not influence the expression of hypothalamic neuropeptides that modulate feeding and energy metabolism (Fig. 3D).
FIG. 3.
PEDF’s metabolic effects do not depend on central actions. A: The hypothalamus of C57Bl/6 mice was excised, and 18S, PEDF, and ATGL mRNA expression were assessed by qRT-PCR (n = 8). B: C57Bl/6 mice were injected with 0.9 μg PEDF or aCSF and placed in a metabolic monitoring station for assessment of oxygen uptake (VO2). C: RER was also assessed (n = 8 for PEDF and n = 11 for vehicle). D: Mice were injected with 0.9 μg PEDF or aCSF, and the hypothalami were excised after 6 h to assess mRNA of hypothalamic neuropeptides that modulate feeding and energy metabolism. Pro-opiomelanocortin (POMC), agouti-related peptide (AgRP), neuropeptide Y (NPY), and cocaine- and amphetamine-regulated transcript (CART) were measured (n = 8 per group). All data are presented as means ± SEM.
PEDF influences fatty acid metabolism via direct effects on peripheral tissues
Cell culture studies.
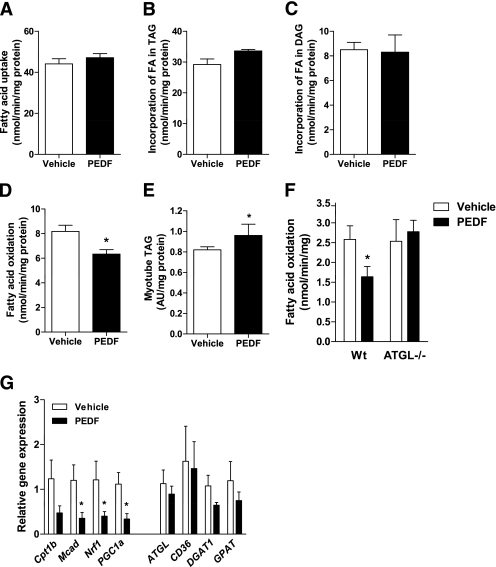
Because skeletal muscle is a major site for whole-body substrate metabolism, these whole-body metabolic effects raise the possibility that PEDF exerts negative effects on skeletal muscle by interfering with fatty acid metabolism. We first examined fatty acid metabolism in cultured myotubes. PEDF did not affect fatty acid uptake into myotubes (Fig. 4A) or the incorporation of extracellular fatty acids into triacylglycerol (P = 0.07, Fig. 4B) and diacylglycerol (Fig. 4C), indicating that PEDF does not acutely affect fat storage in muscle. PEDF increased intracellular muscle TG hydrolase activity and the oxidation of fatty acids derived from intramyocellular triacylglycerol (Supplementary Fig. 2A and B). Concomitantly, PEDF reduced exogenous fatty acid oxidation by 23% (representing those fatty acids derived from outside the cell, Fig. 4D). Notably, the rate of exogenous fatty acid oxidation was 90% higher than TG-derived fatty acid oxidation; hence, total fatty acid oxidation was reduced by 18% in PEDF-treated myotubes. The net reduction in overall fatty acid oxidation was due to reduced “complete oxidation” (14CO2 production: ↓40 ± 7% in PEDF, n = 12–15, P < 0.05), suggestive of defective mitochondrial fatty acid oxidation and not impaired mitochondrial fatty acid transport.
FIG. 4.
PEDF modulates fatty acid (FA) metabolism in skeletal muscle. L6 myotubes were treated with 100 nmol/L PEDF or saline (vehicle) for 2 h. Total FA uptake (A), incorporation of FA into triacylglycerol (TAG; B) or diacylglycerol (DAG; C), and FA oxidation (D) were assessed using 0.5 mmol/L [1-14C] palmitate (n = 6–12 for each group). E: Myotube TAG content is shown after 6-h treatment with 0.5 mmol/L oleate in the incubation media (n = 4–6 per group). All data are presented as means ± SEM. *P < 0.05 vs. vehicle. F: PEDF (100 nmol/L) reduces FA oxidation in intact skeletal muscle. Soleus muscles were removed from ATGL−/− and Wt littermates before assessment of FA oxidation using 0.5 mmol/L [1-14C] oleate ex vivo. *P < 0.05 vs. vehicle within the same genotype (n = 6 per group). G: Genes associated with FA oxidation and mitochondrial biogenesis (left) and TAG storage and degradation (right) were assessed in vastus lateralis of mice treated with saline or PEDF for 5 days (n = 6 for each group). *P < 0.05 vs. vehicle.
Intramyocellular triacylglycerol levels were increased in myotubes (Fig. 4E), demonstrating that the incorporation of new fatty acids into the triacylglycerol pool exceeded the capacity for PEDF-mediated intramyocellular triacylglycerol hydrolysis and whole-cell fatty acid oxidation. In support of this conclusion, when myotubes were incubated in the absence of free fatty acids, PEDF treatment resulted in intramyocellular triacylglycerol depletion (Supplementary Fig. 2C).
Animal studies.
PEDF also blunted fatty acid oxidation in isolated soleus muscle of Wt mice (Fig. 4F). Studies in mice also revealed effects of nutritional status. When PEDF was injected into fasting mice, plasma FFA levels were increased and muscle TG was unchanged (Supplementary Fig. 3A and B), whereas PEDF administration in mice fed ad libitum did not affect plasma FFA, and skeletal muscle TG was decreased (Supplementary Fig. 3D and E). Studies in isolated soleus muscle from ATGL−/− revealed no effects of PEDF on fatty acid oxidation (Fig. 4F), TG hydrolase activity, or esterification into triacylglycerol and diacylglycerol (Supplementary Fig. 4A–C). These data indicated that PEDF interacts with ATGL to generate a signal that disrupts mitochondrial function. To test this notion, we examined whether PEDF alters respiration in mitochondria isolated from Wt mice. Surprisingly, PEDF had no effect on mitochondrial respiration in the absence or presence of ADP, oligomycin, or carbonyl cyanide p-[trifluoromethoxy] phenylhydrazone (FCCP; Supplementary Fig. 5). Although PEDF appears to bind endothelial cell-surface F1Fo-ATP synthase and reduce its ATP synthesis activity (35), the addition of PEDF to isolated mitochondria in vitro did not affect respiration (data not shown).
Prolonged PEDF administration affects lipid metabolism in skeletal muscle.
We next examined the effects of prolonged PEDF administration on skeletal muscle fatty acid metabolism. C57Bl/6 mice were continuously infused with recombinant PEDF or saline for 5 days (3) and muscles were excised. Fatty acid uptake and fatty acid storage into triacylglycerol were not affected by PEDF treatment (data not shown), whereas fatty acid oxidation was reduced by 22% (saline: 3.7 ± 0.2 vs. PEDF: 2.9 ± 0.3 nmol/mg/min, P = 0.05). Further studies revealed that PEDF downregulates the expression of genes involved in fatty acid β-oxidation and oxidative phosphorylation (Fig. 4G). Genes involved in fatty acid uptake and triacylglycerol metabolism were unaffected. Collectively, PEDF modulates whole-body energy homeostasis by impairing skeletal muscle fatty acid metabolism by both acute biochemical mechanisms and longer-term transcriptional suppression of β-oxidation and oxidative phosphorylation genes.
PEDF has mild effects on liver triacylglycerol metabolism.
Dysfunctional liver lipid metabolism leads to steatosis and insulin resistance. Forced ATGL expression in obese mice (36,37) reduces liver triacylglycerol, and livers of PEDF−/− mice store more triacylglycerol (24). Here, we showed that acute PEDF administration in vivo increased triacylglycerol hydrolase activity in liver lysates from Wt but not ATGL−/− mice (Supplementary Fig. 6); however, this did not alter TG content in fasted or ad libitum–fed mice (Supplementary Fig. 3C and F). The absence of a functional effect may reflect the low hepatic ATGL abundance (27).
ATGL is required for PEDF’s inhibitory effect on skeletal muscle glucose metabolism.
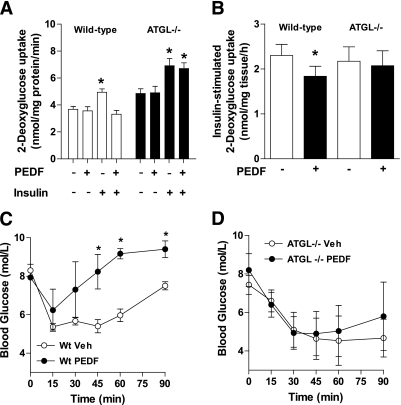
To test whether ATGL is required for PEDF’s inhibitory effects on insulin-mediated glucose disposal (3,14), myocytes were generated from skeletal muscle satellite cells of ATGL−/− and Wt littermate mice and differentiated into multinucleated myotubes. As expected, insulin increased glucose uptake in both Wt and ATGL−/− myotubes (Fig. 5A). PEDF reduced insulin-mediated glucose uptake in Wt myotubes, but this effect was completely abrogated in ATGL−/− myotubes, thereby demonstrating a role for ATGL in PEDF’s inhibition of insulin-mediated glucose metabolism (Fig. 5A). These effects were also demonstrated in soleus muscle from Wt and ATGL−/− mice ex vivo (Fig. 5B). We next performed an insulin tolerance test in vivo and observed a pronounced reduction in systemic insulin sensitivity in Wt mice with PEDF treatment (Fig. 5C), but no effect of PEDF to blunt insulin sensitivity in ATGL−/− mice (Fig. 5D).
FIG. 5.
PEDF causes skeletal muscle insulin resistance in an ATGL-dependent manner. A: 2-Deoxy-d-glucose (2-DG) uptake experiments in primary myotubes. Wt (□) and ATGL−/− (■) myotubes were pretreated with saline or PEDF (100 nmol/L) for 2 h. The media was removed, and basal and insulin-stimulated 2-DG uptake was determined (n = 6 for each group where each experiment was performed in triplicate on two occasions). *P < 0.05 vs. vehicle within the same genotype. Values are means ± SEM. B: Extensor digitorum longus muscles were removed from ATGL−/− (■) and Wt littermates (□) before assessment of insulin-stimulated glucose uptake (n = 4–6 for each group where each experiment was performed on two occasions). *P < 0.05 vs. –PEDF within the same genotype. Values are means ± SEM. Insulin tolerance tests were performed in Wt (C) and ATGL−/− mice (D). PEDF was injected intraperitoneally 2 h before insulin administration. PEDF (n = 6–7 per treatment). *P < 0.05 vs. corresponding time point within the same genotype.
Altogether, these results demonstrate that PEDF induces insulin resistance in skeletal muscle via an ATGL-dependent process. Because ATGL catalyzes multiple functions in lipid metabolism (21), we performed an unbiased lipidomics analysis of skeletal muscle to determine whether PEDF action produces a lipid that may interfere with insulin signal transduction. PEDF reduced total triacylglycerol in Wt mice, but no effects were noted in ATGL−/− mice. Aside from sphingomyelin, which was increased by 17%, there were no changes in the total contents of other lipids (Table 1). There were some minor changes in several lipid species (Supplementary Table 1).
TABLE 1.
Effect of PEDF on muscle lipids in Wt and ATGL−/− mice
| Lipid type | Wt |
ATGL−/− |
||
|---|---|---|---|---|
| Vehicle | PEDF | Vehicle | PEDF | |
| Triacylglycerol | 202,356 ± 50,054 | 72,565 ± 8,131* | 243,282 ± 17,744 | 210,888 ± 52,728 |
| Diacylglycerol | 53,860 ± 4,521 | 42,232 ± 2,304 | 75,273 ± 26,727 | 54,336 ± 24,595 |
| Phosphatidylglycerol | 1,597 ± 181 | 2,052 ± 77 | 1,789 ± 75 | 1,873 ± 101 |
| Phosphatidylinositol | 20,493 ± 4,134 | 22,446 ± 2,594 | 21,346 ± 2,273 | 20,742 ± 1,775 |
| Phosphatidylethanolamine | 34,203 ± 3,803 | 40,704 ± 1,193 | 37,618 ± 952 | 38,991 ± 1,299 |
| Phosphatidylserine | 8,156 ± 463 | 7,630 ± 413 | 8,749 ± 992 | 7,744 ± 385 |
| Cholesterol ester | 3,738 ± 1,348 | 4,606 ± 1,008 | 4,670 ± 1,113 | 3,928 ± 620 |
| Sphingomyelin | 261,548 ± 3,995 | 305,516 ± 15,202† | 355,098 ± 22,736 | 320,042 ± 14,868 |
| Ceramide | 1,924 ± 123 | 1,850 ± 111 | 1,874 ± 147 | 1,809 ± 60 |
| Dihydroceramide | 253 ± 18 | 372 ± 75 | 302 ± 35 | 266 ± 44 |
| Dihexosylceramide | 205 ± 45 | 209 ± 42 | 250 ± 24 | 208 ± 24 |
| Monohexosylceramide | 797 ± 118 | 635 ± 96 | 893 ± 216 | 624 ± 57 |
| GM3 ganglioside | 152 ± 7 | 120 ± 12 | 135 ± 13 | 156 ± 13 |
| Lysophosphatidylcholine | 5,212 ± 1,897 | 3,110 ± 303 | 2,907 ± 320 | 3,135 ± 277 |
Units are expressed as pmol/μmol phosphatidylcholine. Wt, Wt + PEDF, ATGL−/− n = 4; ATGL−/− + PEDF n = 8. Values are mean ± SD.
*P < 0.05 vs. ATGL−/− PEDF.
†P < 0.05 vs. vehicle of the same genotype.
DISCUSSION
Obesity is associated with defective fatty acid metabolism in adipose tissue, liver, and muscle. We previously identified PEDF as an adipocyte-secreted factor that is elevated in obesity and causes insulin resistance (3). Here, we used genetic approaches in cell culture models and in vivo functional experiments to reveal several important metabolic functions of PEDF: 1) PEDF increases adipose tissue lipolysis in an ATGL-dependent manner; 2) PEDF alters systemic fat metabolism by reducing skeletal muscle fatty acid oxidation and suppressing gene expression of β-oxidation/oxidative metabolism proteins; and 3) ATGL is implicit in PEDF-mediated insulin resistance.
A unique feature of PEDF is the absence of an active c-reactive loop that renders the protein noninhibitory. Thus, an important question posed by these experiments is how does PEDF exert its metabolic actions? PEDF is a growth factor that presumably binds to cell surface receptor(s) and triggers intracellular signaling (17,38). Although one group identified ATGL as a lipase-linked membrane-bound receptor for PEDF (17), we and others have been unable to detect ATGL at the plasma membrane in adipocytes (19,22,30). PEDF likely exerts some biologic effects via a receptor at the plasma membrane (38). We cannot exclude the possibility that PEDF acts by modifying the activity of another bioactive molecule, but it is notable that serpin activity for this protein remains undetected.
PEDF leads to the rapid activation of the Jun NH2-terminal kinase and extracellular signal–related kinase signaling pathways, consistent with a PEDF receptor–mediated event (3). It should be noted that the latter was retained in muscle of ATGL−/− mice, again making it unlikely that ATGL is the PEDF receptor. It is also possible that PEDF is transported across the plasma membrane to exert its biologic actions. Chung et al. (24) incubated HepG2 cells with recombinant His-tagged PEDF and demonstrated uptake of PEDF into cells and trafficking to intracellular lipid droplets. Others have shown high-affinity binding of recombinant PEDF to ATGL (17). Thus, PEDF appears to be exerting its biologic effects by independent mechanisms: 1) activating plasma membrane receptors and downstream signaling and 2) via the interaction of PEDF with ATGL, perhaps after sarcolemmal transport of PEDF. This study was designed to examine the metabolic effects of PEDF and the requirement of ATGL for these processes.
ATGL exerts a critical function in the degradation of triacylglycerol stored in intracellular lipid droplets (19,21–23,39) and possesses transacylase activity (21). By using genetic ATGL and HSL ablation, we extended our previous observation that PEDF induces adipocyte lipolysis (3) by demonstrating that PEDF-mediated lipolysis is dependent on ATGL. The finding of a greater PEDF-induced FFA release in Wt mice (42%) compared with glycerol release (20%) is also consistent with PEDF stimulating ATGL, which does not possess diacylglycerol lipase activity (19).
A previous study showed that triacylglycerol content in PEDF−/− hepatocytes was elevated and the addition of recombinant PEDF reduced triacylglycerol content (24). These authors used a nonspecific suicide inhibitor, (bromomethylene)-3-(1-naphthalenyl)-2H-tetrahydropyran-2-one, that blocks all calcium-independent phospholipase A2 lipase activity, including ATGL, and reported an attenuation of PEDF’s apparent lipolytic effects. We showed that PEDF can increase TG hydrolase activity in the liver, but this is not sufficient to alter triacylglycerol content. This is likely to reflect the relatively low abundance of ATGL in the liver compared with the adipocyte (19). Our studies in mice also revealed an increase in plasma PEDF levels during fasting, which is temporally associated with increases in adipocyte lipolysis. Thus, PEDF is a lipolytic hormone that mobilizes fatty acids from triacylglycerol stores. As PEDF is elevated in obesity and type 2 diabetes (9,12), it may contribute to the increased systemic mobilization of fatty acids frequently observed in these conditions (40,41).
An imbalance between fatty acid delivery, storage, and oxidation contributes to skeletal muscle lipid accumulation, which is mechanistically linked to the development of insulin resistance (42). Data presented here suggest that some of PEDF’s in vivo metabolic effects depend on nutritional status. Total energy expenditure was not altered by PEDF, but substrate utilization was affected: PEDF reduced daily fatty acid oxidation by ∼5% (P = 0.02), an effect that was mediated by suppressing the normal diurnal substrate switching from carbohydrate oxidation in the fed state (dark) to fatty acid oxidation in the fasted state (light). Impaired substrate switching, or “metabolic inflexibility,” refers to an impaired capacity to adjust substrate selection in response to a changing nutrient supply and is a common feature of obesity and its related diseases, including type 2 diabetes (43).
Skeletal muscle is a major regulator of whole-body fatty acid disposal and oxidative enzyme activity, and tissue fatty acid oxidation is reduced in obesity and type 2 diabetes (44–48). Studies in cultured myotubes and isolated muscles confirmed the whole-body metabolic analysis and revealed that PEDF reduces fatty acid oxidation. PEDF appears to be inducing concurrent functions in muscle that leads to a “lipotoxic” environment (3). PEDF increased muscle triacylglycerol lipase activity and the oxidation of the liberated triacylglycerol-derived fatty acids. The increased intramyocellular flux competed with fatty acids derived from extracellular sources, which accounted for ∼25% of the reduction in fatty acid oxidation from extracellular sources, suggesting that a mechanism other than substrate flux reduces total fatty acid oxidation. The net effect of this acute dysregulation in vitro is muscle lipid storage, demonstrating that the flux and oxidation of fatty acids derived from intracellular triacylglycerol is less than the uptake and oxidation of exogenous fatty acid. Our studies examining the effect of nutritional status (fast vs. fed) on PEDF actions in mice support this conclusion.
We have also shown that fatty acid oxidation is reduced with 5-day PEDF treatment, and subsequent transcript profiling in skeletal muscle revealed that PEDF decreased the expression of genes associated with fatty acid β-oxidation and mitochondrial biogenesis. This result was somewhat unexpected, because stimulation of ATGL would be expected to produce fatty acid ligands that activate peroxisome proliferator–activated receptors and increase the expression of genes involved in mitochondrial fat oxidation (49). PEDF likely targets several transcription factors or coactivators, given its known pleiotropic actions (13). Collectively, PEDF modulates whole-body energy homeostasis by impairing skeletal muscle fatty acid metabolism acutely via the suppression of fatty acid oxidation and, in the longer-term, by inducing transcriptional suppression of several genes involved with β-oxidation and oxidative phosphorylation.
The accumulation of ectopic lipid contributes to insulin resistance, and interventions that stimulate fatty acid oxidation can decrease lipid accumulation in muscle that associates with insulin sensitization (50–52). We previously showed that PEDF caused insulin resistance in lean, healthy mice in association with lipid metabolite accumulation (3). Here, we extended those studies by demonstrating that ATGL is required for PEDF-induced insulin resistance in muscle. Studies in primary muscle cells and isolated intact muscles demonstrate that this effect is intrinsic to muscle and is not affected by other known negative regulators of insulin action such as altered circulating lipids, hormones, or proinflammatory cytokines. These studies supported the premise that PEDF-mediated activation of ATGL produces lipid ligands that inhibit insulin action in muscle. However, our detailed lipidomics analysis did not reveal this lipid signal, if one exists. Notably, we did not measure FFA species, and others have shown that specific fatty acids (e.g., C16:1n7-palmitoleate) can modulate systemic metabolism (53).
Finally, impaired mitochondrial function has been linked with insulin resistance development (46,48), and PEDF inhibits endothelial cell surface ATP synthesis activity (35). Our studies do not support a role for PEDF in acute mitochondrial dysfunction; however, we cannot exclude the possibility that PEDF is producing a metabolite that can influence mitochondrial function in vivo. Further studies are required to address this possibility.
In summary, these studies have extended the understanding of PEDF’s biologic targets and metabolic actions. These results show that aspects of defective lipid metabolism in obesity can be recapitulated with PEDF. Specifically, PEDF enhances basal adipose tissue lipolysis via ATGL, reduces skeletal muscle fatty acid oxidation, and requires ATGL to induce insulin resistance. The interaction between PEDF and ATGL contributes to the metabolic dysregulation that results in lipid deposition and insulin resistance in obesity and type 2 diabetes, conditions characterized by increased PEDF levels (3,9,11,12).
ACKNOWLEDGMENTS
These studies were supported by research grants and a fellowship from the National Health and Medical Research Council (NHMRC) of Australia, the Diabetes Australia Research Trust, the William Buckland Foundation (ANZ Trustees Program), and a Monash Fellowship (to M.J.W.). M.L.B. is an Australian Postgraduate Award holder. E.J.D. has received an RPB Career Development Award and is supported by the Juvenile Diabetes Research Foundation.
M.L.B. and Z.B.A. researched data and reviewed and edited the manuscript. E.J.D. and R.Z. provided reagents and reviewed and edited the manuscript. P.J.M. researched data and reviewed and edited the manuscript. M.J.W. researched data and wrote the manuscript.
The authors thank Maria Matzaris, Romana Stark, Seamus Crowe, Joanne Pagnon, Andrew Hoy (Monash University), and Lindsay Wu (Garvan Institute) for technical assistance.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db10-0845/-/DC1.
REFERENCES
- 1.Zvonic S, Lefevre M, Kilroy G, et al. Secretome of primary cultures of human adipose-derived stem cells: modulation of serpins by adipogenesis. Mol Cell Proteomics 2007;6:18–28 [DOI] [PubMed] [Google Scholar]
- 2.Alvarez-Llamas G, Szalowska E, de Vries MP, et al. Characterization of the human visceral adipose tissue secretome. Mol Cell Proteomics 2007;6:589–600 [DOI] [PubMed] [Google Scholar]
- 3.Crowe S, Wu LE, Economou C, et al. Pigment epithelium-derived factor contributes to insulin resistance in obesity. Cell Metab 2009;10:40–47 [DOI] [PubMed] [Google Scholar]
- 4.Wang P, Mariman E, Keijer J, et al. Profiling of the secreted proteins during 3T3-L1 adipocyte differentiation leads to the identification of novel adipokines. Cell Mol Life Sci 2004;61:2405–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science 1993;259:87–91 [DOI] [PubMed] [Google Scholar]
- 6.Fried SK, Bunkin DA, Greenberg AS. Omental and subcutaneous adipose tissues of obese subjects release interleukin-6: depot difference and regulation by glucocorticoid. J Clin Endocrinol Metab 1998;83:847–850 [DOI] [PubMed] [Google Scholar]
- 7.Steppan CM, Bailey ST, Bhat S, et al. The hormone resistin links obesity to diabetes. Nature 2001;409:307–312 [DOI] [PubMed] [Google Scholar]
- 8.Yang Q, Graham TE, Mody N, et al. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature 2005;436:356–362 [DOI] [PubMed] [Google Scholar]
- 9.Yamagishi S, Adachi H, Abe A, et al. Elevated serum levels of pigment epithelium-derived factor in the metabolic syndrome. J Clin Endocrinol Metab 2006;91:2447–2450 [DOI] [PubMed] [Google Scholar]
- 10.Wang P, Smit E, Brouwers MC, et al. Plasma pigment epithelium-derived factor is positively associated with obesity in Caucasian subjects, in particular with the visceral fat depot. Eur J Endocrinol 2008;159:713–718 [DOI] [PubMed] [Google Scholar]
- 11.Ogata N, Matsuoka M, Matsuyama K, et al. Plasma concentration of pigment epithelium-derived factor in patients with diabetic retinopathy. J Clin Endocrinol Metab 2007;92:1176–1179 [DOI] [PubMed] [Google Scholar]
- 12.Jenkins A, Zhang SX, Gosmanova A, et al. Increased serum pigment epithelium derived factor levels in type 2 diabetes patients. Diabetes Res Clin Pract 2008;82:e5–e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tombran-Tink J, Barnstable CJ. PEDF: a multifaceted neurotrophic factor. Nat Rev Neurosci 2003;4:628–636 [DOI] [PubMed] [Google Scholar]
- 14.Famulla S, Lamers D, Hartwig S, et al. Pigment epithelium-derived factor is one of the most abundant proteins secreted by human adipocytes and induces insulin resistance and inflammatory signaling in muscle and fat cells. Int J Obes (Lond). 12 October 2010 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 15.Bilak MM, Becerra SP, Vincent AM, Moss BH, Aymerich MS, Kuncl RW. Identification of the neuroprotective molecular region of pigment epithelium-derived factor and its binding sites on motor neurons. J Neurosci 2002;22:9378–9386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filleur S, Volz K, Nelius T, et al. Two functional epitopes of pigment epithelial-derived factor block angiogenesis and induce differentiation in prostate cancer. Cancer Res 2005;65:5144–5152 [DOI] [PubMed] [Google Scholar]
- 17.Notari L, Baladron V, Aroca-Aguilar JD, et al. Identification of a lipase-linked cell membrane receptor for pigment epithelium-derived factor. J Biol Chem 2006;281:38022–38037 [DOI] [PubMed] [Google Scholar]
- 18.Haemmerle G, Lass A, Zimmermann R, et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science 2006;312:734–737 [DOI] [PubMed] [Google Scholar]
- 19.Zimmermann R, Strauss JG, Haemmerle G, et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 2004;306:1383–1386 [DOI] [PubMed] [Google Scholar]
- 20.Grönke S, Mildner A, Fellert S, et al. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab 2005;1:323–330 [DOI] [PubMed] [Google Scholar]
- 21.Jenkins CM, Mancuso DJ, Yan W, Sims HF, Gibson B, Gross RW. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem 2004;279:48968–48975 [DOI] [PubMed] [Google Scholar]
- 22.Smirnova E, Goldberg EB, Makarova KS, Lin L, Brown WJ, Jackson CL. ATGL has a key role in lipid droplet/adiposome degradation in mammalian cells. EMBO Rep 2006;7:106–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villena JA, Roy S, Sarkadi-Nagy E, Kim KH, Sul HS. Desnutrin, an adipocyte gene encoding a novel patatin domain-containing protein, is induced by fasting and glucocorticoids: ectopic expression of desnutrin increases triglyceride hydrolysis. J Biol Chem 2004;279:47066–47075 [DOI] [PubMed] [Google Scholar]
- 24.Chung C, Doll JA, Gattu AK, et al. Anti-angiogenic pigment epithelium-derived factor regulates hepatocyte triglyceride content through adipose triglyceride lipase (ATGL). J Hepatol 2008;48:471–478 [DOI] [PubMed] [Google Scholar]
- 25.Duh EJ, Yang HS, Suzuma I, et al. Pigment epithelium-derived factor suppresses ischemia-induced retinal neovascularization and VEGF-induced migration and growth. Invest Ophthalmol Vis Sci 2002;43:821–829 [PubMed] [Google Scholar]
- 26.Haemmerle G, Zimmermann R, Hayn M, et al. Hormone-sensitive lipase deficiency in mice causes diglyceride accumulation in adipose tissue, muscle, and testis. J Biol Chem 2002;277:4806–4815 [DOI] [PubMed] [Google Scholar]
- 27.Turpin SM, Hoy AJ, Brown RD, et al. Adipose triacylglycerol lipase is a major regulator of hepatic lipid metabolism but not insulin sensitivity in mice. Diabetologia 2011;54:146–156 [DOI] [PubMed] [Google Scholar]
- 28.Watt MJ, Heigenhauser GJ, Spriet LL. Effects of dynamic exercise intensity on the activation of hormone-sensitive lipase in human skeletal muscle. J Physiol 2003;547:301–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartz R, Zehmer JK, Zhu M, et al. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J Proteome Res 2007;6:3256–3265 [DOI] [PubMed] [Google Scholar]
- 30.Granneman JG, Moore HP, Granneman RL, Greenberg AS, Obin MS, Zhu Z. Analysis of lipolytic protein trafficking and interactions in adipocytes. J Biol Chem 2007;282:5726–5735 [DOI] [PubMed] [Google Scholar]
- 31.Fortier M, Soni K, Laurin N, et al. Human hormone-sensitive lipase (HSL): expression in white fat corrects the white adipose phenotype of HSL-deficient mice. J Lipid Res 2005;46:1860–1867 [DOI] [PubMed] [Google Scholar]
- 32.Osuga J, Ishibashi S, Oka T, et al. Targeted disruption of hormone-sensitive lipase results in male sterility and adipocyte hypertrophy, but not in obesity. Proc Natl Acad Sci USA 2000;97:787–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huijsman E, van de Par C, Economou C, et al. Adipose triacylglycerol lipase deletion alters whole body energy metabolism and impairs exercise performance in mice. Am J Physiol Endocrinol Metab 2009;297:E505–E513 [DOI] [PubMed] [Google Scholar]
- 34.Woods SC, D’Alessio DA. Central control of body weight and appetite. J Clin Endocrinol Metab 2008;93(Suppl. 1):S37–S50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Notari L, Arakaki N, Mueller D, Meier S, Amaral J, Becerra SP. Pigment epithelium-derived factor binds to cell-surface F(1)-ATP synthase. FEBS J 2010;277:2192–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reid BN, Ables GP, Otlivanchik OA, et al. Hepatic overexpression of hormone-sensitive lipase and adipose triglyceride lipase promotes fatty acid oxidation, stimulates direct release of free fatty acids, and ameliorates steatosis. J Biol Chem 2008;283:13087–13099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turpin SM, Hoy AJ, Brown RD, et al. Adipose triacylglycerol lipase is a major regulator of hepatic lipid metabolism but not insulin sensitivity in mice. Diabetologia 2011;54:146–156 [DOI] [PubMed] [Google Scholar]
- 38.Filleur S, Nelius T, de Riese W, Kennedy RC. Characterization of PEDF: a multi-functional serpin family protein. J Cell Biochem 2009;106:769–775 [DOI] [PubMed] [Google Scholar]
- 39.Kershaw EE, Hamm JK, Verhagen LA, Peroni O, Katic M, Flier JS. Adipose triglyceride lipase: function, regulation by insulin, and comparison with adiponutrin. Diabetes 2006;55:148–157 [PMC free article] [PubMed] [Google Scholar]
- 40.Horowitz JF, Coppack SW, Paramore D, Cryer PE, Zhao G, Klein S. Effect of short-term fasting on lipid kinetics in lean and obese women. Am J Physiol 1999;276:E278–E284 [DOI] [PubMed] [Google Scholar]
- 41.Nurjhan N, Consoli A, Gerich J. Increased lipolysis and its consequences on gluconeogenesis in non-insulin-dependent diabetes mellitus. J Clin Invest 1992;89:169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Savage DB, Petersen KF, Shulman GI. Disordered lipid metabolism and the pathogenesis of insulin resistance. Physiol Rev 2007;87:507–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelley DE, Mandarino LJ. Fuel selection in human skeletal muscle in insulin resistance: a reexamination. Diabetes 2000;49:677–683 [DOI] [PubMed] [Google Scholar]
- 44.Sparks LM, Xie H, Koza RA, et al. A high-fat diet coordinately downregulates genes required for mitochondrial oxidative phosphorylation in skeletal muscle. Diabetes 2005;54:1926–1933 [DOI] [PubMed] [Google Scholar]
- 45.Holloway GP, Thrush AB, Heigenhauser GJ, et al. Skeletal muscle mitochondrial FAT/CD36 content and palmitate oxidation are not decreased in obese women. Am J Physiol Endocrinol Metab 2007;292:E1782–E1789 [DOI] [PubMed] [Google Scholar]
- 46.Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes 2002;51:2944–2950 [DOI] [PubMed] [Google Scholar]
- 47.Kim JY, Hickner RC, Cortright RL, Dohm GL, Houmard JA. Lipid oxidation is reduced in obese human skeletal muscle. Am J Physiol Endocrinol Metab 2000;279:E1039–E1044 [DOI] [PubMed] [Google Scholar]
- 48.Ritov VB, Menshikova EV, He J, Ferrell RE, Goodpaster BH, Kelley DE. Deficiency of subsarcolemmal mitochondria in obesity and type 2 diabetes. Diabetes 2005;54:8–14 [DOI] [PubMed] [Google Scholar]
- 49.Ahmadian M, Duncan RE, Varady KA, et al. Adipose overexpression of desnutrin promotes fatty acid use and attenuates diet-induced obesity. Diabetes 2009;58:855–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bruce CR, Hoy AJ, Turner N, et al. Overexpression of carnitine palmitoyltransferase-1 in skeletal muscle is sufficient to enhance fatty acid oxidation and improve high-fat diet-induced insulin resistance. Diabetes 2009;58:550–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bruce CR, Thrush AB, Mertz VA, et al. Endurance training in obese humans improves glucose tolerance and mitochondrial fatty acid oxidation and alters muscle lipid content. Am J Physiol Endocrinol Metab 2006;291:E99–E107 [DOI] [PubMed] [Google Scholar]
- 52.Choi CS, Savage DB, Abu-Elheiga L, et al. Continuous fat oxidation in acetyl-CoA carboxylase 2 knockout mice increases total energy expenditure, reduces fat mass, and improves insulin sensitivity. Proc Natl Acad Sci USA 2007;104:16480–16485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao H, Gerhold K, Mayers JR, Wiest MM, Watkins SM, Hotamisligil GS. Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell 2008;134:933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]