Abstract
Heat shock triggers the assembly of nuclear stress bodies that contain heat shock factor 1 and a subset of RNA processing factors. These structures are formed on the pericentromeric heterochromatic regions of specific human chromosomes, among which chromosome 9. In this article we show that these heterochromatic domains are characterized by an epigenetic status typical of euchromatic regions. Similarly to transcriptionally competent portions of the genome, stress bodies are, in fact, enriched in acetylated histone H4. Acetylation peaks at 6 h of recovery from heat shock. Moreover, heterochromatin markers, such as HP1 and histone H3 methylated on lysine 9, are excluded from these nuclear districts. In addition, heat shock triggers the transient accumulation of RNA molecules, heterogeneous in size, containing the subclass of satellite III sequences found in the pericentromeric heterochromatin of chromosome 9. This is the first report of a transcriptional activation of a constitutive heterochromatic portion of the genome in response to stress stimuli.
INTRODUCTION
The genomic material in eukaryotic nuclei can be roughly partitioned in two cytologically distinct entities termed eu- and hetero-chromatin. Heterochromatin was originally defined as that portion of the genome that remains condensed and intensely stained with DNA intercalating dyes throughout the cell cycle. It represents a significant fraction of most eukaryotic genomes and is generally associated with telomeres and pericentric regions of chromosomes. Contrary to euchromatin, heterochromatic regions consist predominantly of repetitive DNA, including satellite sequences and middle repetitive sequences related to transposable elements and retroviruses. Although not devoid of genes, these regions are typically gene-poor (Hennig, 1999).
Establishment of heterochromatin depends on two basic elements: the histone modification code (Strahl and Allis, 2000) and the interaction of nonhistone chromosomal proteins (Nielsen et al., 2001). The covalent modifications of histone tails by deacetylase and methyl-transferase activities act in concert to establish the “histone code” essential for assembly of silenced chromatin. In general, the histone tails in heterochromatin are relatively hypo-acetylated, whereas histone H3 and H4 tails found in euchromatin are generally acetylated. Concerning the nonhistone chromosomal proteins, one of the best characterized is heterochromatin protein 1 (HP1), which was originally identified as a protein primarily concentrated in the pericentric heterochromatin. HP1 homologues exist in evolutionary distant organisms such as Schizosaccharomyces pombe (Swi6p) and Homo sapiens (HP1α, HP1β, and HP1γ; Eissenberg and Elgin, 2000) and are characterized by an amino-terminal chromo domain that mediates the interaction with chromatin (Nakayama et al., 2001). Methylation of lysine 9 (K9) of histone H3 creates a binding site for recruitment of Swi6/HP1 (Maison et al., 2002; Peters et al., 2002).
The factors underlying the selection of a specific chromosomal locus for the assembly of heterochromatin are still elusive; however, repetitive DNA rather than specific DNA sequences are likely involved. Also the function of heterochromatin is not yet completely understood. In addition to serve important roles in chromosome mechanics, particularly in the centromere function (Hennig, 1999), heterochromatic structures are involved in the inactivation of genes normally resident in euchromatic domains. This is suggested by two key observations. First, inactivation of X-chromosome in mammals, a mechanism that leaves the inactive X as a visibly stained structure, the Barr body (Brockdorff, 2002). The second indication came from the analysis of position effect variegation (PEV) in Drosophila, that is the mosaic pattern of expression exhibited by genes placed near centric heterochromatin by chromosomal rearrangements or transposition events.
Stress treatments such as heat shock or exposure to heavy metals drastically affect the function and the structure of the eukaryotic cell. Among the numerous effects triggered by heat shock on the structure of the cell nuclei, one of the most intriguing is the transitory appearance of novel nuclear districts, termed stress bodies (Weighardt et al., 1999). Stress bodies are exclusively detectable in human cells (Denegri et al., 2002) and consist of clusters of perichromatin granules, i.e., highly packed forms of ribonucleoprotein complexes (Chiodi et al., 2000). They are sites of accumulation of heat shock factor 1 (HSF1; Jolly et al., 1999) and of a subset of RNA processing factors, including two proteins of the heterogeneous nuclear ribonucleoprotein complexes (hnRNP HAP and hnRNP M), two members of the SR family of splicing factors (SF2/ASF and SRp30c), and the RNA-binding protein Sam68 (Denegri et al., 2001). We have recently shown that the assembly of stress bodies depends on the presence of pericentromeric heterochromatic regions on human chromosomes 9, 12, and 15. These heterochromatic regions most likely act as recruiting centers as suggested by their colocalization with stress bodies (Denegri et al., 2002). Moreover, the subclass of satellite III elements in the heterochromatic pericentromeric q1.2 band of chromosome 9 contains sequence motifs similar to the HSF1-binding site. It has been proposed that the interaction between HSF1 and satellite III sequences directs the recruitment of HSF1 to stress bodies (Jolly et al., 2002).
In this article we show that after heat shock the heterochromatic regions associated with stress bodies are characterized by an higher-order organization typical of euchromatic portions of the genome. Moreover, heat shock triggers the production of satellite III transcripts, highly heterogeneous in size, that are undetectable in unstressed cells. To our knowledge this is the first indication that environmental stresses such as heat shock can elicit a transient alteration of the higher-order structure of specific heterochromatic regions and induce the transcriptional activation of silent portions of the genome.
MATERIALS AND METHODS
Cell Culture and Cell Treatments
HeLa cells were grown in DMEM medium (Sigma, St. Louis, MO), 10% fetal calf serum (Sigma), 50 μg/ml gentamicin, and 2 mM l-glutamine. For heat shock experiments, cells grown in monolayers were incubated 1 h at 42°C in complete medium made with 40 mM HEPES, pH 7.0, and allowed to recover at 37°C as indicated in the text.
When required, HeLa cells were treated with different concentrations of trichostatin A (TSA; Sigma) dissolved in ethanol at a concentration of 1 mg/ml.
Indirect Immunofluorescence
HeLa cells grown on coverslips were washed once with phosphate-buffered saline (PBS), fixed for 7 min in 4% formaldehyde, and subsequently permeabilized in 0.5% Triton X-100 for 7 min on ice. Primary antibodies were diluted to working concentration in PBS containing 5% skimmed milk (Difco, Detroit, MI) and then added to the coverslips. Primary antibodies used were as follows: affinity-purified rabbit anti-HAP polyclonal antibody (Weighardt et al., 1999), mouse anti-HAP monoclonal antibody (mAb) 16C3 (Weighardt et al., 1999), rabbit antihyperacetylated histone H4 (Upstate, Lake Placid, NY), rabbit acetyl-histone H4 antibody set (Ac K5; Ac K8; Ac K12; Upstate), rat anti-HSF1 mAb 10H8 (NeoMarker, Freemont, CA), rat anti-HP1β mAb MAC353 and rat anti-HP1γ mAb MAC385 (Serotec, Cergy Saint-Christophe, France) and mouse anti-HP1α mAb 3446 (Chemicon, International, Temecula, CA), mouse anti-SF2/ASF mAb-96 (Zymed, San Francisco, CA), and rabbit anti-4x-methil-histone H3(Lys9) polyclonal antibody (kindly provided by Dr. Thomas Jenuwein, Research Institute for Molecular Pathology, Vienna). After 1hat37°C in a humid chamber, coverslips were washed three times with PBS. Secondary antibodies used were rhodamine-conjugated anti-rabbit, anti-mouse, or anti-rat immunoglobulin (IgG) goat antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) and fluorescein isothiocyanate (FITC)-conjugated anti-rabbit or anti-mouse IgG goat antibodies (Jackson ImmunoResearch Laboratories). Secondary antibodies were diluted at the final concentration recommended by the supplier in PBS-made 5% skimmed milk and were added to coverslips. After 1 h at 37°C in a humid chamber, coverslips were washed three times with PBS, rinsed, and mounted in 90% glycerol in PBS. Confocal microscopy was performed with a TCS-NT digital scanning confocal microscope (Leica, Deerfield, IL) equipped with a 63 ×/NA = 1.32 oil immersion objective. We used the 488-nm laser line for excitation of FITC (detected at 500 nm < FITC <540 nm) and the 543-nm laser line for the rhodamine fluorescence (detected at >590 nm). The pinhole diameter was kept at 1 μm. Images were exported to Adobe Photoshop (Adobe Systems, Mountain View, CA).
RNA Extraction and Northern Blotting
Total RNAs were isolated from HeLa cells using the guanidinium thiocyanate solubilization method (Chirgwin et al., 1979). Northern blotting was performed as described (Sambrook and Russel, 2001). Prehybridization and hybridization solutions: 4× SSC (1× SSC: 0.15 M NaCl, 0.015 M Na citrate), 4× Denhardt's solution, 50% formamide, 0.5% SDS, 7.5 mM Na4P2O7, 12.5 mM NaH2PO4, and 0.3 mg/ml tRNA. Probes were labeled with the Mega-prime DNA labeling System (Amersham, Buckinghamshire, UK). After an overnight hybridization, membranes were washed twice in 0.2× SSC, 1% SDS at 60°C. Probes used were as follows: pHuR98 (ATCC, Manassas, VA), containing a monomer of the subclass of satellite III sequences on chromosome 9, pAL1 pBR12, pMC15 pDMX1 specific for the alphoid DNA on chromosomes 1, 12, 15, and X. In addition we used the β-actin cDNA (Clontech Laboratories, Palo Alto, CA) as a control of loaded material.
In Situ Hybridization to RNA
Cells were grown on coverslips and heat-shocked as described above. After 3 h of recovery at 37°C, cells were fixed in 4% paraformaldehyde in PBS for 15 min, washed thoroughly in PBS, and permeabilized in 0.5% Triton X-100 on ice for 5 min. Cells were then washed in PBS and 2× SSC and incubated overnight at 42°C in hybridization buffer (1 μg/ml tRNA [Sigma-Aldrich], 2× SSC, 10% dextran sulfate [Sigma-Aldrich], 5× Denhardt's solution, 25% formamide) containing 5 ng/ml 5′ biotinylated oligonucleotide probe. The following morning, cells were washed in 2× SSC and the biotinylated probe was detected with fluorescein-avidin (Vector Laboratories, Burlingame, CA). The signal was amplified by incubation with antiavidin D antibody (Vector Laboratories) followed by fluorescein-avidin. Cells were counterstained with anti-HAP rabbit antibodies and rhodamine-conjugated anti-rabbit antibodies. The oligonucleotide probes used were as follows: Direct (5′-GGA ATG GCA TGG ATT GGA AT-3′), and Reverse (5′-ATT CCA ATC CAT GCC ATT CC-3′). Both oligonucleotides were 5′ biotinylated and are complementary to nucleotides 97-106 of the satellite III sequence of human chromosome 9 (accession number: X06137). Cells were also hybridized to a control oligo (5′-CCGGGAAGCTAGAGTAAGTAG-3′) complementary to a pBR322 sequence.
RESULTS
Trichostatin A Prevents the Formation of Nuclear Stress Bodies
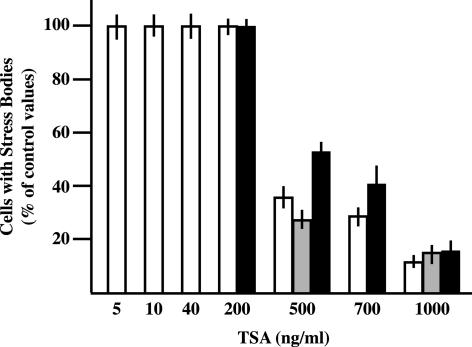
We have previously reported that nuclear stress bodies form on heterochromatic regions of specific human chromosomes (Denegri et al., 2002). This raises the possibility that the chromatin packaging status could be directly involved in this process. We decided to investigate this possibility by means of TSA, an inhibitor of histone deacetylases (Yoshida et al., 1995). HeLa cells were incubated 6 h with increasing concentrations of TSA and, after drug withdrawal, were heat-shocked 1 h at 42°C. After 1 h of recovery at 37°C, cells were fixed and stained with antibodies directed against hnRNP HAP, HSF1, or splicing factor SF2/ASF, three components of stress bodies (Denegri et al., 2001). The results of this analysis are reported in Figure 1. We observed that doses higher than 500 ng/ml drastically reduced the fraction of cells in which HAP and SF2/ASF were recruited to stress bodies. This fraction dropped to ∼10% of the control value when cells were treated with 1000 ng/ml TSA. At this high concentration a full effect was already detectable after 1 h of incubation (unpublished data). Although the distribution of HSF1 was less drastically perturbed, this factor also displayed an homogenous distribution in most of the cell nuclei at the highest TSA dose. It is worth noticing that these treatments did not increase the fraction of apoptotic cells measured by DAPI staining after 48 h of recovery (unpublished data). The effect of the drug was completely reversible and an interval of 3 h between TSA withdrawal and heat shock was sufficient to rescue the ability of the cells to form stress bodies.
Figure 1.
TSA inhibits the assembly of stress bodies. HeLa cells were treated for 6 h with increasing concentrations of TSA. After removal of the drug, cells were heat-shocked 1 h at 42°C and then allowed to recover 1 h at 37°C before fixation with 4% formaldehyde. Cells were then independently stained with (1) rabbit polyclonal antibodies against hnRNP HAP, (2) mAb-96 against SF2/ASF, or (3) rat mAb 10H8 against HSF1. Primary antibodies were revealed with FITC-conjugated anti-rabbit, anti-mouse, or anti-rat goat antibodies. The fraction of cells with stress bodies was measured in three independent experiments counting 500 cells per experiments. This fraction was expressed as a percentage of the value measured in cells treated only with the solvent of TSA (ethanol). White bars, hnRNP HAP; gray bars, SF2/ASF; black bars, HSF1.
Stress Bodies Contain Acetylated Histone H4
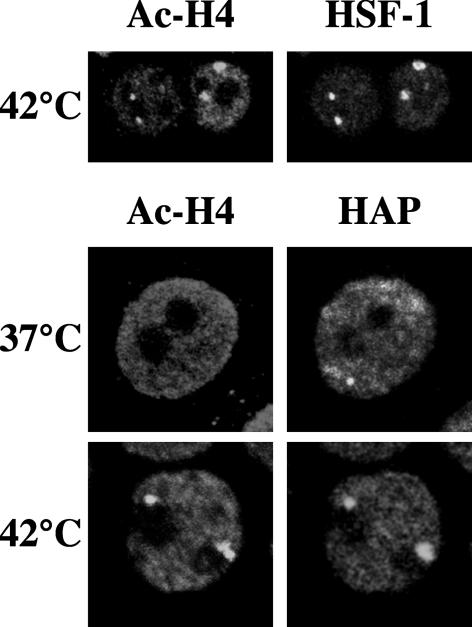
The result reported above prompted us to investigate in more detail the relationship between chromatin and stress bodies. A standard marker of heterochromatin domains, from yeast to mammals, is the presence of underacetylated histone H4 (Turner, 2002). On the other hand, TSA, an inhibitor of histone deacetylases that induces a drastic increase in the level of acetylated histones, prevents the assembly of stress bodies. A reasonable prediction is that stress bodies might correspond to nuclear districts particularly poor of acetylated histones. We decided to verify this prediction by assessing the relative distribution of acetylated histone H4 (Ac-H4), hnRNP HAP, and HSF1 in unstressed and heat-shocked HeLa cells. Surprisingly, immunofluorescence analysis showed a perfect colocalization of Ac-H4 with hnRNP HAP and HSF1 in stress bodies that, contrary to our expectations, were the nuclear districts most intensely decorated by the anti-(Ac-H4) antibodies (Figure 2). This staining pattern suggests that stress bodies are assembled on large chromatin territories characterized by a high density of Ac-H4. Notably, anti-(Ac-H4) antibodies do not decorate structures similar to stress bodies in unstressed cells (Figure 2), indicating that acetylation of histone H4 bound to these chromosomal domains is specifically triggered by heat shock.
Figure 2.
Stress bodies contain acetylated histone H4. HeLa cells were heat-shocked 1 h at 42°C and then allowed to recover 1 h at 37°C. Cells were then fixed in 4% formaldehyde and costained with a rabbit polyclonal antibody against the hyper-acetylated histone H4 and with either anti-HAP 16C3 mAb or the rat anti-HSF1 10H8 mAb. The distribution of hnRNP HAP and Ac-H4 in unstressed cells was also determined (37°C). Primary antibodies were revealed with FITC-conjugated anti-rabbit and with rhodamine-conjugated anti-rat or anti-mouse goat antibodies. Cells were then analyzed by confocal laser microscopy. Images of the same cells are shown.
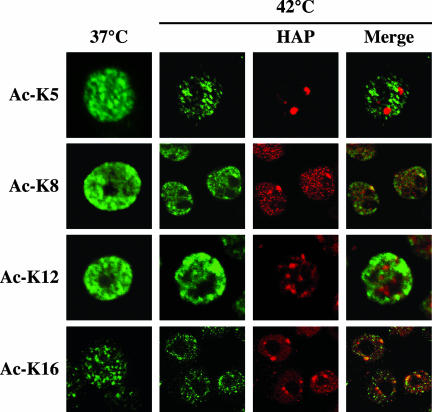
Histone H4 can be acetylated in vivo on four lysine residues (K5, K8, K12, and K16) all located in the N-terminal tail. Lysine-specific H4 acetylation has been linked to the regulation of mammalian chromatin (Johnson et al., 1998). However the details of this control mechanism are still mostly undefined. A few evidences have recently linked acetylation of K8 and K16 with transcription (Johnson et al., 1998; Agalioti et al., 2002). Acetylation of K5 and K12, on the other hand, identifies newly synthesized histone H4 (Sobel et al., 1995). It is generally assumed that deposition of histone H4 acetylated on K5 and K12 during DNA replication is followed by rapid deacetylation to preserve the underacetylated pattern of histone H4, a critical parameter for the maintenance of the silenced state of heterochromatin (Taddei et al., 1999). To decipher the pattern of histone H4 modification in stress bodies, we stained unstressed and heat-shocked HeLa cells with commercially available antibodies able to distinguish between the four acetylated lysines. The immunofluorescence analysis in Figure 3 shows that stress bodies lie in nuclear districts devoid of Ac-K5 and Ac-K12. On the contrary, they are recognized by antibodies specific for Ac-K8 and Ac-K16. The association with Ac-K8 is particularly evident and, in fact, stress bodies are the nuclear regions most intensely stained by antibodies against this acetylated lysine. In this respect, the staining pattern observed with anti-Ac-K8 is highly similar to that obtained with antibodies directed to the hyper-acetylated form of histone H4. Collectively, these results indicate that the chromatin regions associated to stress bodies are enriched in histone H4 isoforms characterized by acetylation of K8 and K16, a posttranslational modification that distinguishes transcriptionally competent portions of the genome.
Figure 3.
Pattern of acetylation of histone H4 associated to stress bodies. Unstressed (37°C) or heat-shocked HeLa cells (1 h at 42°C followed by 1 h of recovery at 37°C) (42°C) were stained with rabbit polyclonal antibodies directed against different acetylated lysine residues of histone H4 as indicated. Primary antibodies were revealed with a FITC-conjugated anti-rabbit goat antibody. To reveal the localization of stress bodies, heat-shocked cells were costained with anti-HAP mAb 16C3 and with a rhodamine-conjugated anti-mouse goat antibody. Confocal laser microscopy images of representative cells are shown.
Stress Bodies Lack Distinguishing Features of Heterochromatin
Although formed by arrays of satellite sequences and cytologically defined as heterochromatin, chromosomal domains associated to stress bodies are characterized by the presence of Ac-H4, a marker of euchromatic portions of the genome. We decided to explore this aspect more in detail by determining whether some features typical of heterochromatin domains were still present in these regions.
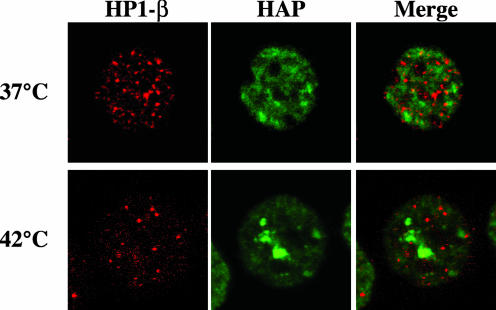
Pericentromeric heterochromatin is assembled in a specific higher-order structure, characterized by the presence of hypo-acetylated histones and of nonhistone proteins such as HP1. In accordance with the notion that HP1 and Ac-H4 label, respectively, hetero- and euchromatin domains, antibodies directed against HP1β failed to recognize stress bodies stained by anti-hnRNP HAP antibodies (Figure 4). An identical pattern was observed with antibodies against the other two members of the HP1 family, HP1α and γ (Supplementary Figure 1). Moreover, no colocalization of HP1β with hnRNP HAP was detectable in unstressed cells (Figure 4). The absence of colocalization with stress bodies was unlikely to originate from massive degradation of HP1 in response to heat shock since the steady state level of these proteins remained constant throughout the experiment (Supplementary Figure 2).
Figure 4.
Stress bodies are not major sites of accumulation of HP1β. HeLa cells either unstressed (37°C) or heat-shocked 1 h at 42°C followed by 1 h at 37°C (42°C) were fixed in 4% formaldehyde. Cells were costained with a rabbit polyclonal antibody against hnRNP HAP, to reveal stress bodies, and with the rat mAb MAC353 specific for HP1β. Primary antibodies were revealed with FITC-conjugated anti-rabbit and rhodamine-conjugated anti-rat goat antibodies. Confocal laser microscopy images of the same fields are shown.
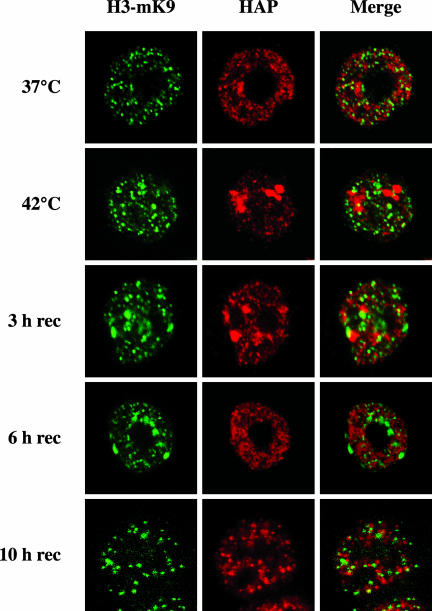
Another marker of heterochromatin is histone H3 methylated on K9 (H3-mK9). Two antibodies against H3-mK9 are available. The first one was raised against a linear K9-dimethylated peptide and detects any silenced chromatin domains with no particular preference for pericentromeric heterochromatin. The second antibody was raised against a branched peptide presenting four fingers of the K9-dimethylated TARSK consensus sequence of the H3 terminus (Maison et al., 2002). It has recently been shown that the structure recognized by the latter antibody is part of the binding site of HP1 and is sensitive to TSA treatment (Maison et al., 2002). In unstressed cells hnRNP HAP and the antigen recognized by antibody against the branched peptide occupy mutually exclusive nuclear sites (Figure 5), consistently with the fact that methylated histone H3 marks trascriptionally silent portions of the genome, whereas hnRNP HAP labels ribonucleoprotein complexes. No colocalization between the two antigens was also observed after 1 h of heat-shock at 42°C. However, during recovery from stress a more complex pattern was detectable in a subset of cells. After 3 h of recovery, in ∼10% of the cells, methylated H3 accumulated in large structures adjacent to stress bodies. These structures were more evident, always in subset of cells, after an additional 3 h of recovery when hnRNP HAP, as previously described (Weighardt et al., 1999), was already redistributed throughout the nuclear volume. Finally, these structures were no longer visible after 10 h of recovery. The relevance of this finding for the dynamics of stress bodies is still matter of investigation.
Figure 5.
Stress bodies are not major sites of accumulation of histone H3 methylated on K9 (H3-mK9). HeLa cells either unstressed (37°C) heat-shocked 1 h at 42°C (42°C) or allowed to recover for the indicated time intervals were fixed in 4% formaldehyde. Cells were stained with the anti-HAP mAb 16C3 and with the rabbit antibody raised against the branched α-methH3-K9 peptide. Primary antibodies were revealed with FITC-conjugated anti-mouse and rhodamine-conjugated anti-rabbit goat antibodies. Confocal laser microscopy images of the same fields are shown.
Kinetics of Histone H4 Acetylation in Stress Bodies
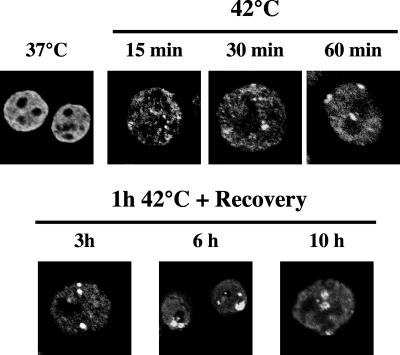
We have previously described the kinetics of the recruitment of HSF1 and hnRNP HAP to stress bodies. Although HSF1 is already detectable in these structures after a very short incubation at 42°C, the recruitment of HAP is temporally delayed and peaks after 3 h of recovery at 37°C (Weighardt et al., 1999). To determine the kinetics of H4 acetylation, HeLa cells were heat-shocked 1 h at 42°C and allowed to recover for increasing time intervals at 37°C. Cells were then analyzed by indirect immunofluorescence with antibodies against Ac-H4. As shown in Figure 6, an altered distribution of Ac-H4 was already observed after 15 min at 42°C. In these cells a number of small dots was detectable, some of which appeared to be clustered in the proximity of the nucleoli. Structures comparable to stress bodies were clearly visible after 30 min at 42°C, but the intensity of the signals and the size of these structures kept growing and peaked at 6 h of recovery. At longer times (10 h of recovery) the fraction of cells displaying stress bodies started to diminish, and stress bodies had a fuzzier appearance. A normal distribution of Ac-H4 was detectable after 20 h of recovery (unpublished data). Altogether this analysis indicates that heat shock induces a transient modification of the higher-order structure of the heterochromatic regions associated with stress bodies.
Figure 6.
Kinetics of histone H4 acetylation in stress bodies. HeLa cells were heat-shocked for 15, 30, or 60 min at 42°C. After 1 h of heat shock, cells were also allowed to recover at 37°C for the indicated time intervals. Cells were then stained with a rabbit polyclonal antibody against the hyper-acetylated form of histone H4 and with a FITC-conjugated anti-rabbit goat antibody. Confocal laser images of representative cells are shown. The distribution of histone H4 in unstressed HeLa cells is shown as a control (37°C).
Heat Shock Induces Transcription of Satellite III DNA Sequences
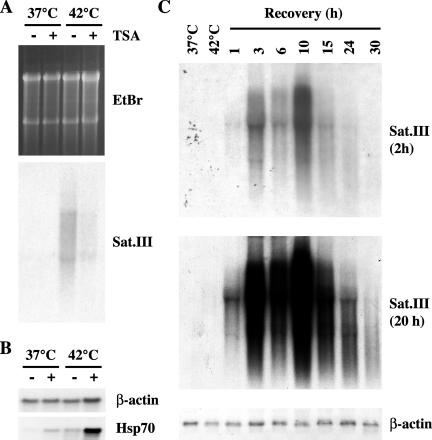
After heat shock the heterochromatic domains associated to stress bodies have a higher-order structure competent for transcription. An open possibility is that heat shock can trigger the transcriptional activation of these heterochromatic regions. We decided to verify this hypothesis in the case of the large pericentromeric heterochromatic q1.2 band of human chromosome 9, one of the recruiting centers for stress bodies. This band is mainly composed of arrays of satellite III sequences that were used as probes in Northern blot analysis of total RNAs prepared from unstressed and from heat-shocked HeLa cells (1 h at 42°C followed by 1 h of recovery at 37°C). In addition, we studied whether pretreatment with TSA, under conditions that prevent the formation of stress bodies (1000 ng/ml), could affect the transcription of this region of the genome. As shown in Figure 7A, pHuR98 detected a continuum of transcripts, ranging in size from >5 kb to less that 2 kb, that were exclusively present in heat-shocked cells. No hybridization signal was visible in unstressed cells even after overexposure of the membrane (unpublished data). Notably, the production of these transcripts was almost completely abrogated by TSA.
Figure 7.
Heat shock induces the production of satellite III transcripts. (A) HeLa cells were treated for 6 h with TSA (1 μg/ml; +) or with an equal volume of ethanol (-). After washing out the drug, cells were heatshocked 1 h at 42°C and allowed to recover 1 h at 37°C. Total RNAs were extracted and 10 μg were analyzed in Northern blotting with the pHuR98 probe specific for the subclass of satellite III sequences in the heterochromatic q1.2 band of human chromosome 9. The hybridization signal (overnight exposure) is shown along with an image of the gel stained with ethidium bromide to reveal the loaded material. (B) The same total RNAs described in A were hybridized to β-actin and hsp70 cDNA probes. (C) Total RNAs were prepared from unstressed cells (37°C) or from cells heat-shocked 1 h at 42°C (42°C) and then allowed to recover at 37°C for the indicated times. RNAs, 10 μg, were hybridized with the pHuR98 probe. Two exposures of the same filter are shown. As a control the filter was also hybridized to the β-actin cDNA.
Then we asked whether satellite III transcripts accumulated in the cells in parallel with the presence of Ac-H4 in stress bodies and we analyzed in Northern blot total RNAs from HeLa cells heat-shocked 1 h at 42°C and then allowed to recover at 37°C for increasing time intervals. The result of this analysis is shown in Figure 7C. Satellite III transcripts were barely detectable at the end of the incubation at 42°C. Their level drastically increased during the recovery at 37°C and peaked at 10 h of recovery when the intensity of the hybridization signal was 250-fold higher than that detectable at the end of heat shock. Satellite III transcripts were still visible after 30 h of recovery. Intriguingly two bands of ∼5000 and 2000 base pairs were detectable over the smear, with the longest being prevalent at early times and the other increasing later on. Selection on an oligo dT column showed that these transcripts were poly-adenylated (unpublished data).
The effect of heat shock seems to be specific for satellite III sequences. Indeed, in Northern blot analysis of the same RNA preparations we failed to observe hybridization signals with α-satellite probes specific for the pericentromeric heterochromatin of human chromosome 12 and 15 that also colocalize with stress bodies (Denegri et al., 2002; see DISCUSSION). Finally no, signal was observed with α-satellite probes for the heterochromatic regions of human chromosomes 1, and X or with a probe for β-satellite sequences distributed on several human chromosomes (unpublished data).
Satellite III Transcripts Colocalize with hnRNP HAP in Stress Bodies
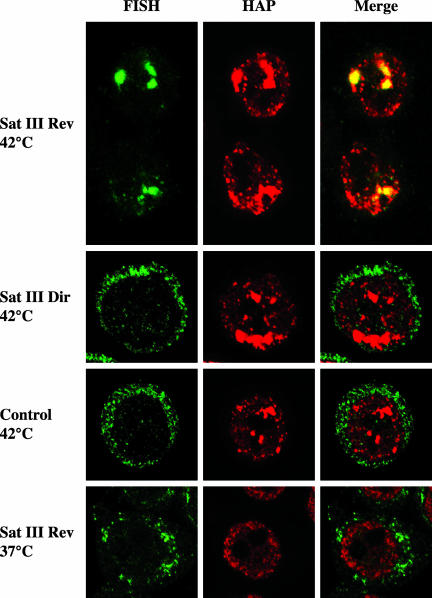
We have previously shown that stress bodies contain RNA molecules whose nature is still undefined (Chiodi et al., 2000). The results in the previous section raise the possibility that these RNAs are in fact satellite III transcripts. To verify this possibility, we investigated the distribution of satellite III transcripts by in situ hybridization. Heat-shocked HeLa cells were allowed to recover 3 h at 37°C and then independently hybridized to two biotinylated 20-mers each specific for one strand of the satellite III sequence on human chromosome 9. As shown in Figure 8, the Reverse 20-mer, complementary to the upper strand of the satellite sequence, efficiently stained the very same structures recognized by anti-hnRNP HAP antibodies. On the contrary, no signal was observed with the Direct oligo, complementary to the bottom strand, or with a control oligonucleotide (Figure 8). Hybridization of the Reverse oligo was specific and, in fact, no signal was detectable in unstressed cells (Figure 8) or in cells pretreated with RNAse (unpublished data). Notably, all the stress bodies were stained by the Reverse 20-mer, indicating the presence of satellite III transcripts in all of these structures.
Figure 8.
Satellite III transcripts colocalize with hnRNP HAP in stress bodies. HeLa cells were heat-shocked 1 h at 42°C and then allowed to recover 3 h at 37°C. Cells were analyzed by in situ hybridization with oligonucleotides complementary to either the upper strand (Sat III Rev 42°C) or the lower strand (Sat III Dir 42°C) of the satellite III sequence. Heat-shocked cells were also hybridized to a control oligo complementary to a pBR322 sequence (Control 42°C). Finally the Sat III Rev 20-mer was hybridized to unstressed HeLa cells (Sat III Rev 37°C). The distribution of the hybridized oligos was revealed with fluorescein-avidin. The distribution of hnRNP HAP in the same cells was revealed with rabbit polyclonal anti-HAP and rhodamine-conjugated goat anti-rabbit antibodies. Confocal laser microscopy images of the same fields are shown.
DISCUSSION
In this article we have studied the epigenetic status of the heterochromatic domains on which nuclear stress bodies are assembled. In heat-shocked cells these chromosomal domains display distinctive euchromatic features: they are associated with acetylated histone H4, a marker of transcription competent portions of the genome, but not with HP1 and histone H3-mK9, which are involved in heterochromatin silencing. Histone H4 associated to stress bodies is acetylated on K8 and K16, a pattern that has been linked to transcriptional activation. In fact, genes transcribed by RNA polymerase I and II are enriched in H4 acetylated on K16 (Johnson et al., 1998). In addition acetylation of K8 has been shown to mediate the recruitment of the SWI/SNF complex on the IFN-β gene and to be required for gene activation (Agalioti et al., 2002).
Accumulation of acetylated histone H4 in stress bodies is accompanied by the transcriptional activation of the heterochromatic regions associated with these structures. Indeed, heat shock triggers the production of transcripts, highly heterogeneous in size, composed of satellite III sequences specific of the pericentromeric heterochromatic band q1.2 of the human chromosome 9. We have previously reported that α-satellite sequences of pericentromeric heterochromatin of human chromosomes 12 and 15 colocalize with stress bodies. Although we have not detected transcripts containing α-satellite elements from these regions, it is still possible that portions of these domains, other than α-satellite sequences, are transcribed in response to heat shock. This possibility is supported by the observation that all the stress bodies contain satellite III transcripts. It is worth noticing that heat shock does not indiscriminately induce transcription of satellite DNA sequences. In fact, in addition to α-satellite sequences of chromosome 12 and 15, we failed to detect transcription of α-satellite sequences of human chromosomes X and 1 and of β-satellite sequences scattered on different chromosomes. It has been previously shown that HSF1 binds to chromosome 9 satellite III in vitro. It is conceivable that binding of HSF1 directs acetylation of histone H4, production of satellite III transcripts, and recruitment of the subset of RNA processing factors found in these structures. In this model stress bodies, even though assembled on different chromosomes, would identify sites of transcription of satellite III elements.
In summary, our analysis provides the first indication that environmental stress can trigger a transient alteration of the epigenetic program and of the higher-order structure of specific heterochromatic regions leading to the transcription of otherwise silent portions of the genome.
Functional Considerations
In most eukaryotes, the bulk of the heterochromatin is found in peri- and centromeric regions. Although this distribution certainly reflects the contribution in the centromere activity, the analysis of the phenomenon of position effect variegation (PEV) has unraveled a link between heterochromatin and transcriptional silencing. PEV was initially recognized as a process in which a gene, upon translocation in proximity of heterochromatin, is silenced in a proportion of cells in which it would normally be expressed (Dillon and Festenstein, 2002). In addition to being the result of pathological processes such as chromosomal rearrangements, relocation of euchromatic genes close to heterochromatic masses has been suggested to be part of a physiological mechanism to control the gene expression program (Cockell and Gasser, 1999). DNA-binding proteins such as Ikaros, a regulator of both B- and T-lymphocyte development, are likely mediators of this effect. It has been proposed that the interaction of Ikaros with genes that are destined for inactivation mediates their recruitment to centromeric foci, where they may then assemble into heterochromatin (Cobb et al., 2000). In this model heterochromatin would represent a “closed” chromatin configuration that limits the accessibility of regulatory DNA sequences to trans-acting factors. In this perspective, our observation that in heat-shock cells large heterochromatic regions are characterized by an epigenetic status typical of euchromatic portions of the human genome could be relevant for the expression program of specific genes. However, it is unlikely a role in the transcriptional regulation of heatshock genes as suggested by the observation that genes of heat-shock proteins do not colocalize with the pericentric heterochromatin of chromosome 9 in unstressed cells or more in general with stress bodies in heat-shocked cells (Jolly et al., 1999), and the same holds true for the HSF1 gene located on human chromosome 8.
Transcription of Satellite III Sequences
An intriguing result of our analysis is the identification of transcripts arising from the pericentromeric heterochromatic band of human chromosome 9 in response to heat shock. The kinetics of accumulation and disappearance of these poly-adenylated transcripts parallels the presence of Ac-H4 on the pericentromeric bands associated with stress bodies, suggesting a tight link between the two events. The function of these transcripts is still matter of speculation. However, their size heterogeneity is compatible with the idea that these molecules are in fact composed of a variable number of tandem repeats of satellite III sequences arguing against a possible coding capacity. We hypothesize that these transcripts could serve some structural function in the organization of stress bodies, namely in the recruitment of the subset of RNA-binding proteins found in these structures. This possibility is supported by two observations. First, the upper strand satellite III sequence, which is transcribed in response to heat shock, contains possible binding sites for SF2/ASF, one of the proteins found in stress bodies. The second indication is the ability of TSA to prevent the formation of stress bodies. The mechanism through which TSA prevents the formation of stress bodies is still unknown. However, it could be related to the ability of the drug to induce activation of heat-shock genes (see Figure 7B), an event that is known to confer resistance to successive stress treatments. We have previously observed that the formation of stress bodies is triggered by a subset of stress agents, among which are heat shock and cadmium sulfate (Denegri et al., 2001). These structures do not form in response to other stresses such as H2O2 and hyper-osmolarity, which induce the production of heat shock proteins. It is not surprising, therefore, that TSA, although triggering transcription of heat-shock genes, is unable to induce the formation of stress bodies. Our finding that TSA also inhibits the production of satellite III transcripts is consistent with the idea that these molecules are indeed the recruiters of RNA-binding proteins to stress bodies. In this model human stress bodies can be assimilated to the ω-speckles observed in Drosophila cells after heat shock. In Drosophila thermal stress increases the production of noncoding hsrω-n transcripts that direct the recruitment of a subset of RNA-processing factors in correspondence of the 93D locus where the hsrω gene is located (Prasanth et al., 2000). The peculiarity of the human system is the fact that, contrary to hsrω-n transcripts, satellite III transcripts are exclusively detectable in stressed cells. Although it is highly probable that similar mechanisms operate during the formation of ω-speckles in Drosophila and of stress bodies in human cells, some points will deserve further investigation. For instance, we have previously reported that stress bodies contain RNA molecules synthesized both before and after heat shock (Chiodi et al., 2000). An intriguing possibility is that ribonucleoprotein complexes assembled in other nuclear districts are recruited to stress bodies by an interaction with satellite III RNAs.
An alternative and appealing possibility is that satellite III transcripts could participate to the mechanism that reestablishes silencing of the heterochromatic q1.2 band of chromosome 9 disrupted by heat shock. Indeed, several experimental evidences suggest the involvement of RNA molecules in heterochromatin formation. For instance, the interaction of HP1 with heterochromatic domain depends on as yet unidentified RNA molecules (Maison et al., 2002; Muchardt et al., 2002) and the large noncoding X-IST transcript is a key player in the process of X chromosome inactivation in mammal females (Boumil and Lee, 2001). More recently it has been shown that RNA molecules play also a role in the establishment of heterochromatin domains in the yeast S. pombe (Volpe et al., 2002). In this case double-stranded RNA molecules arising from centromeric repeats have been suggested to direct formation and maintenance of heterochromatin through RNA interference.
In conclusion, the characterization of stress bodies carried out here has unveiled novel roles of heterochromatin in the organization of the nuclear function.
Supplementary Material
Acknowledgments
This work was supported by a grant of the “Associazione Italiana per la Ricerca sul Cancro” (AIRC) to G.B., by the program MIUR/FIRB “Post-genoma” (contract number RBNEOKXC9_001 and RBNE015MPB_003) to S.R. and G.B. respectively, by the program MIUR-CNR “Biomolecole per la salute umana” L.95/95, and by a grant from Progetto CNR-MIUR “Genomica Funzionale” L.449/97 to G.B.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-07-0487. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-07-0487.
Online version of this article contains supplementary figure material. Online version is available at www.molbiolcell.org.
References
- Agalioti, T., Chen, G., and Thanos, D. (2002). Deciphering the transcriptional histone acetylation code for a human gene. Cell 111, 381-392. [DOI] [PubMed] [Google Scholar]
- Boumil, R.M., and Lee, J.T. (2001). Forty years of decoding the silence in X-chromosome inactivation. Hum. Mol. Genet. 10, 2225-2232. [DOI] [PubMed] [Google Scholar]
- Brockdorff, N. (2002). X-chromosome inactivation: closing in on proteins that bind Xist RNA. Trends Genet 18, 352-358. [DOI] [PubMed] [Google Scholar]
- Chiodi, I., Biggiogera, M., Denegri, M., Corioni, M., Weighardt, F., Cobianchi, F., Riva, S., and Biamonti, G. (2000). Structure and dynamics of hnRNP-labelled nuclear bodies induced by stress treatments. J. Cell Sci. 113, 4043-4053. [DOI] [PubMed] [Google Scholar]
- Chirgwin, J.M., Przybyla, A.E., MacDonald, R.J., and Rutter, W.J. (1979). Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry 18, 5294-5299. [DOI] [PubMed] [Google Scholar]
- Cobb, B.S., Morales-Alcelay, S., Kleiger, G., Brown, K.E., Fisher, A.G., and Smale, S.T. (2000). Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding. Genes Dev. 14, 2146-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell, M., and Gasser, S.M. (1999). Nuclear compartments and gene regulation. Curr. Opin. Genet. Dev. 9, 199-205. [DOI] [PubMed] [Google Scholar]
- Denegri, M., Chiodi, I., Corioni, M., Cobianchi, F., Riva, S., and Biamonti, G. (2001). Stress-induced nuclear bodies are sites of accumulation of pre-mRNA processing factors. Mol. Biol. Cell 12, 3502-3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denegri, M., Moralli, D., Rocchi, M., Biggiogera, M., Raimondi, E., Cobianchi, F., De Carli, L., Riva, S., and Biamonti, G. (2002). Human chromosomes 9, 12, and 15 contain the nucleation sites of stress-induced nuclear bodies. Mol. Biol. Cell 13, 2069-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon, N., and Festenstein, R. (2002). Unravelling heterochromatin: competition between positive and negative factors regulates accessibility. Trends Genet. 18, 252-258. [DOI] [PubMed] [Google Scholar]
- Eissenberg, J.C., and Elgin, S.C. (2000). The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev. 10, 204-210. [DOI] [PubMed] [Google Scholar]
- Hennig, W. (1999). Heterochromatin. Chromosoma 108, 1-9. [DOI] [PubMed] [Google Scholar]
- Johnson, C.A., O'Neill, L.P., Mitchell, A., and Turner, B.M. (1998). Distinctive patterns of histone H4 acetylation are associated with defined sequence elements within both heterochromatic and euchromatic regions of the human genome. Nucleic Acids Res. 26, 994-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly, C., Konecny, L., Grady, D.L., Kutskova, Y.A., Cotto, J.J., Morimoto, R.I., and Vourc'h, C. (2002). In vivo binding of active heat shock transcription factor 1 to human chromosome 9 heterochromatin during stress. J. Cell Biol. 156, 775-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly, C., Usson, Y., and Morimoto, R.I. (1999). Rapid and reversible relocalization of heat shock factor 1 within seconds to nuclear stress granules. Proc. Natl. Acad. Sci. USA 96, 6769-6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison, C., Bailly, D., Peters, A.H., Quivy, J.P., Roche, D., Taddei, A., Lachner, M., Jenuwein, T., and Almouzni, G. (2002). Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30, 329-334. [DOI] [PubMed] [Google Scholar]
- Muchardt, C., Guilleme, M., Seeler, J.S., Trouche, D., Dejean, A., and Yaniv, M. (2002). Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep. 3, 975-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama, J., Rice, J.C., Strahl, B.D., Allis, C.D., and Grewal, S.I. (2001). Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110-113. [DOI] [PubMed] [Google Scholar]
- Nielsen, A.L., Oulad-Abdelghani, M., Ortiz, J.A., Remboutsika, E., Chambon, P., and Losson, R. (2001). Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell 7, 729-739. [DOI] [PubMed] [Google Scholar]
- Peters, A.H., Mermoud, J.E., O'Carroll, D., Pagani, M., Schweizer, D., Brockdorff, N., and Jenuwein, T. (2002). Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat. Genet. 30, 77-80. [DOI] [PubMed] [Google Scholar]
- Prasanth, K.V., Rajendra, T.K., Lal, A.K., and Lakhotia, S.C. (2000). Omega speckles - a novel class of nuclear speckles containing hnRNPs associated with noncoding hsr-omega RNA in Drosophila. J. Cell Sci. 113, 3485-3497. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and Russel, D.W. (2001). Molecular Cloning, 3rd edition, CSHL Press.
- Sobel, R.E., Cook, R.G., Perry, C.A., Annunziato, A.T., and Allis, C.D. (1995). Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl. Acad. Sci. USA 92, 1237-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl, B.D., and Allis, C.D. (2000). The language of covalent histone modifications. Nature 403, 41-45. [DOI] [PubMed] [Google Scholar]
- Taddei, A., Roche, D., Sibarita, J.B., Turner, B.M., and Almouzni, G. (1999). Duplication and maintenance of heterochromatin domains. J. Cell Biol. 147, 1153-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, B.M. (2002). Cellular memory and the histone code. Cell 111, 285-291. [DOI] [PubMed] [Google Scholar]
- Volpe, T.A., Kidner, C., Hall, I.M., Teng, G., Grewal, S.I., and Martienssen, R.A. (2002). Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833-1837. [DOI] [PubMed] [Google Scholar]
- Weighardt, F., Cobianchi, F., Cartegni, L., Chiodi, I., Villa, A., Riva, S., and Biamonti, G. (1999). A novel hnRNP protein (HAP/SAF-B) enters a subset of hnRNP complexes and relocates in nuclear granules in response to heat shock. J. Cell Sci. 112, 1465-1476. [DOI] [PubMed] [Google Scholar]
- Yoshida, M., Horinouchi, S., and Beppu, T. (1995). Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays 17, 423-430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.