Abstract
OBJECTIVE
TR4 is a nuclear receptor without clear pathophysiological roles. We investigated the roles of hepatic TR4 in the regulation of lipogenesis and insulin sensitivity in vivo and in vitro.
RESEARCH DESIGN AND METHODS
TR4 activity and phosphorylation assays were carried out using hepatocytes and various TR4 wild-type and mutant constructs. Liver tissues from TR4 knockout, C57BL/6, and db/db mice were examined to investigate TR4 target gene stearoyl-CoA desaturase (SCD) 1 regulation.
RESULTS
TR4 transactivation is inhibited via phosphorylation by metformin-induced AMP-activated protein kinase (AMPK) at the amino acid serine 351, which results in the suppression of SCD1 gene expression. Additional mechanistic dissection finds TR4-transactivated SCD1 promoter activity via direct binding to the TR4-responsive element located at −243 to −255 on the promoter region. The pathophysiological consequences of the metformin→AMPK→TR4→SCD1 pathway are examined via TR4 knockout mice and primary hepatocytes with either knockdown or overexpression of TR4. The results show that the suppression of SCD1 via loss of TR4 resulted in reduced fat mass and increased insulin sensitivity with increased β-oxidation and decreased lipogenic gene expression.
CONCLUSIONS
The pathway from metformin→AMPK→TR4→SCD1→insulin sensitivity suggests that TR4 may function as an important modulator to control lipid metabolism, which sheds light on the use of small molecules to modulate TR4 activity as a new alternative approach to battle the metabolic syndrome.
Metabolic syndrome, which includes obesity, dyslipidemia, and the proinflammatory state, linked well with insulin resistance, which can be regarded as a disease with dysregulation of not only glucose homeostasis but also lipid metabolism (1).
Stearoyl-CoA desaturase (SCD) is a family of rate-limiting enzymes involved in the biosynthesis of monounsaturated fatty acids from saturated fatty acids, and its activity has been implicated in the metabolic syndrome (2). SCD1 knockout (SCD1−/−) mice showed impaired triglyceride and cholesterol ester biosynthesis with increased insulin sensitivity (3). Early studies showed that the gene expression of SCD1 was well regulated by sterol regulatory element–binding proteins (SREBPs), which are important transcription factors for the regulation of fatty acids and cholesterol metabolism (4). Insulin and glucagon act oppositely in the transcriptional regulation of hepatic SREBP-1c, with insulin inducing and glucagon repressing SREBP-1c expression (5). Constitutively expressing SREBP-1c in mice leads to increased SCD1 expression and lipogenesis, whereas knocking out SREBP-1c (SREBP-1c−/−) results in a decreased SCD1 expression (6,7).
AMP-activated protein kinase (AMPK) functions as a sensor of cellular energy that can be activated by glucose deprivation, high AMP-to-ATP ratios, and the antidiabetes drug metformin (8). Several mechanisms of AMPK action on lipid and glucose metabolism have been studied: AMPK regulates the expression of lipid synthesis genes by modulating the activities of transcription factors and coactivators in liver and other peripheral tissues (9).
TR4 is a member of the nuclear receptor superfamily. Based on the anatomical profiling of nuclear receptor expression, TR4 was classified in the central nervous system, circadian, and basal metabolism group (10). Recent studies also indicate the rhythmic expression of TR4 in four metabolic tissues (liver, white adipose tissue, brown adipose tissue, and muscle) over light and dark cycles (11). Using TR4 knockout (TR4−/−) mice (12,13) as model, we identified a novel pathway, metformin→AMPK→TR4→SCD1→insulin sensitivity, in liver that proved that TR4 may function as a regulator in lipid metabolism. Small molecules, such as metformin, AICAR, or AMPK inhibitor compound C (CpdC), are able to modulate TR4 transactivation to control the metabolic syndrome.
RESEARCH DESIGN AND METHODS
Animal use and care.
All animal procedures were approved by the Animal Care and Use Committee of the University of Rochester. TR4−/− male mice used in this study were generated from heterozygous breeding pairs provided by Lexicon Genetics and genotyped as previously described (13). In the fructose-feeding study, 8-week-old male TR4+/+, TR4−/−, and C57BL/6 mice were fed for 6 weeks with a fructose diet consisting of 60% fructose by weight (Dyets). Leptin receptor–deficient db/db mice were purchased from The Jackson Laboratory (Bar Harbor, ME).
Analytical procedures.
Blood was collected from fed or overnight-fasted animals, and 5–10 μL of plasma samples were used for measuring concentrations of insulin (Crystal Chem, Inc., Downers Grove, IL), free fatty acids (Wako Diagnostics, Richmond, VA), triglycerides (Sigma-Aldrich, St. Louis, MO), and glucose (LifeScan, Milpitas, CA), according to the manufacturers’ protocols. For determination of tissue triglyceride content, tissue pieces (100 mg) were quick frozen in liquid nitrogen and homogenized on ice. Triglycerides were extracted from homogenized tissues with chloroform/methanol and resuspended in t-butyl alcohol. Resuspended samples were then mixed with Triton X-100/methanol (2:1, vol/vol) to completely dissolve the lipid suspension, and an aliquot of this solution was used for the triglyceride assay kit (Sigma-Aldrich). For determination of mitochondrial β-oxidation, the assay uses the incubation with [14C]palmitate with or without potassium cyanide followed by trapping released 14CO2 by base, as described previously (14).
Protein dephosphorylation.
Cell lysates were prepared, and protein concentrations were quantified by the Bradford method (Bio-Rad, Hercules, CA). The lysates were suspended in 1 × NE Buffer (1 µg/10 µL; New England Biolabs, Ipswich, MA), and 1 unit/µg protein lysates of calf intestinal alkaline phosphatase (CIP; New England Biolabs) was added and incubated at 37°C for 60 min.
Western blot analysis.
Protein extracts from cell and liver tissue samples were prepared by homogenizing the tissue in modified lysis buffer. Protein samples (60 μg) were tested for TR4, SCD1, or AMPK phosphorylation levels by Western blot analyses. Western blotting was performed with anti-TR4, anti-SCD1 (Alpha Diagnostic International, San Antonio, TX), anti–phospho-AMPK (Cell signaling), or anti-α1AMPK antibodies (Cell Signaling Technology, Danvers, MA) (15).
Determination of gene expression.
Total RNA was isolated from different tissues using TRIzol reagent (Invitrogen, Carlsbad, CA), and cDNA was synthesized using SuperScript II and random hexamer primers (Invitrogen). Quantitative RT-PCR was performed using SYBR green supermix reagent with the iCycler real-time PCR amplifier (Bio-Rad). The results are given as percentage over control after normalization of mRNA to 18S rRNA expression. A list of primer sequences for real-time PCR is available in Supplementary Table 1.
Reagents, plasmids, and luciferase assay.
We used 500 μmol/L metformin (Sigma-Aldrich), 1 mmol/L AICAR (Calbiochem, San Diego, CA), and 10 μmol/L CpdC (Dorsomorphin; Calbiochem). The plasmid pCMX-TR4 has been described previously (15). The human SCD1 5′ promoter region consisting of −487 to +67 bp was amplified by PCR from HepG2 genomic DNA and cloned into pGL3 reporter vector (Promega, Madison, WI) to generate pGL-SCD-Luc. Deleted pGL-SCD1 (−237/+67)-Luc containing −237 to +67 bp of SCD1 5′ promoter region was generated by PCR amplification from pGL-SCD1-Luc and subcloning into pGL3 (Promega). The DR1 × 3-Luc plasmid contains three copies of the TR4- response element (TR4RE) direct repeat (DR) (15). Hepa1–6, HepG2, and COS-1 cells were maintained in Dulbecco’s modified Eagle’s medium containing 10% FCS. Transfections were performed by using SuperFect (Qiagen, Valencia, CA). Relative luciferase activities were measured in the luciferase reporter assay system (Promega). The plasmid pRL-TK (Promega) for internal control was cotransfected in all transfection experiments.
Glucose and insulin tolerance tests.
For the glucose tolerance test, mice were fasted overnight, followed by an intraperitoneal d-glucose injection (2 g/kg body wt). Blood glucose was measured by tail bleeding at 0, 15, 30, 60, and 120 min after the injection. For the insulin tolerance test, mice were injected intraperitoneally with human insulin (Sigma-Aldrich) at 1 unit/kg body wt, and blood glucose was measured at 0, 15, 30, and 60 min after the administration.
Histological analysis.
Tissues were fixed in fresh 4% buffered paraformaldehyde and then dehydrated through a series of graded alcohols before being embedded in paraffin. Tissue sections were stained with hematoxylin and examined under light microscopy. To investigate the lipid amount in both TR4+/+ and TR4−/− livers, Oil Red O staining was performed (16).
Chromatin immunoprecipitation assay.
Chromatin immunoprecipitation (ChIP) assays were performed in HepG2 cells, as described in previous studies (12). Immunoprecipitations were performed at 4°C overnight, with 2 µg TR4 antibody number 15. The list of primers used can be found in Supplementary Table 1.
Statistical analysis.
Experiments were carried out at least in duplicate and repeated three times. The result was expressed as means ± SD. The P values calculated from the Student t test and one-way ANOVA <0.05 are interpreted as statistically significant.
RESULTS
AMPK phosphorylates hepatic TR4 at the serine 351 site.
Using peptide mapping and motif scan, we found that the amino acid serine 351 (Ser351) phosphorylation site, which is conserved in human, mouse, and rat TR4, is a potential target of AMPK (Fig. 1A). We used SDS-PAGE analysis to examine the TR4 phosphorylation status in mouse hepatocyte Hepa1–6 cells and found that endogenous TR4 displays three bands. The addition of CIP to cytosolic extracts reduced the number of TR4 bands to the fastest moving component (Fig. 1B), suggesting that TR4 is a phosphoprotein and a potential target for AMPK.
FIG. 1.
AMPK inhibits TR4 transactivation through TR4 protein phosphorylation. A: Mouse, human, and rat TR4 amino acid sequence of the putative AMPK phosphorylation site. B: TR4 phosphorylation status by immunoblot with TR4 antibodies. Hepa1–6 cell lysates were treated with CIP or with buffer as the control. C: TR4-wt, TR4-S351A, and TR4-S351E protein were translated in the transcription/translation system containing [35S]methionine, as described in Supplemental Methods. D: The effect of TR4-wt and mutants (S351A and S351E) on the transcriptional activity of the TR4 target gene PEPCK-Luc reporter plasmid in Hepa1–6 cells (**P < 0.01; *P < 0.05 vs. vector control). E: Autoradiogram of the TR4 protein phosphorylated in vitro by AMPK. TR4-wt and S351 mutants were incubated with purified rat liver AMPK and [γ-32P]ATP, in the presence or absence of AMP, as indicated. Lane 1: 35S-labeled TR4 protein. F: DR1 × 3-Luc reporter vector was cotransfected with TR4 in Hepa1–6 cells. After overnight recovery, the transfected cells were treated with AICAR or CpdC for 24 h, and luciferase activity was measured (*P < 0.05 vs. lane 1). G: DR1 × 3-Luc reporter vector was cotransfected with TR4 and AMPK RNAi (target sequence: AMPK-a1 5′-GAATCCTGTGACAAGCACA-3′) in Hepa1–6 cells. The transfected cells were harvested after 48 h, and luciferase activity was measured (*P < 0.05 vs. lane 1).
We then mutated the Ser351 phosphorylation site of TR4 from serine to alanine (S351A) to mimic dephosphorylated TR4 and from serine to glutamic acid (S351E) to mimic phosphorylated TR4, and then we re-evaluated these modifications on TR4 transactivation. Using a luciferase reporter assay utilizing the TR4 target gene (12), PEPCK promoter (PEPCK-Luc), with in vitro transcription/translation system equally expressed TR4-wt, S351A, and S351E (Fig. 1C), we found higher TR4 transactivation for the S351A protein and lower TR4 transactivation for the S351E protein compared with TR4-wt (Fig. 1D) in Hepa1–6 cells, suggesting that phosphorylation of TR4 at S351 resulted in suppression of TR4 transactivation in vitro.
An in vitro phosphorylation assay further demonstrated that TR4 can be phosphorylated by AMPK in the presence of AMP (TR4-wt could be phosphorylated at Ser351) (Fig. 1E, lane 4 vs. 3) and that mutation of Ser351 completely abolished phosphorylation of TR4 in the presence or absence of AMP (Fig. 1E, lanes 5–8). This result suggested that AMPK is an upstream signal capable of modulating TR4 protein via phosphorylation of TR4 at Ser351.
We next tested whether modulating AMPK activity, via its activator AICAR or inhibitor CpdC, resulted in the modulation of TR4 transactivation. Inactivation of AMPK via CpdC, leading to dephosphorylation of AMPK (17), resulted in enhanced TR4 transactivation on the DR1 × 3-Luc reporter, which contains TR4RE (Fig. 1F). In contrast, activation of AMPK by AICAR resulted in suppression of TR4 transactivation (Fig. 1F). Furthermore, reducing endogenous AMPK in hepatocytes by AMPK-RNA interference (RNAi) (a gift from Dr. J.P. Bolanos, Department of Biochemistry and Molecular Biology, University of Salamanca, Salamanca, Spain) induced TR4 transactivation (Fig. 1G). Together, results from Fig. 1A–G demonstrate that hepatic TR4 is a target of AMPK and that changing AMPK activity may modulate TR4 transactivation via reversible phosphorylation at the Ser351 site.
Metformin induces AMPK-mediated TR4 phosphorylation with decreased TR4 transactivation.
Metformin has been used widely as an antidiabetes agent, and it activates AMPK. In vitro studies (18,19) suggest that AMPK inhibits some metabolic gene expressions, for example PEPCK and SCD1. The underlying mechanisms that link metformin, AMPK, and TR4, and how they influence each other, remain unclear. Incubation of Hepa1–6 cells with metformin resulted in increased phosphorylation of AMPK at amino acid threonine 172, without changing AMPK protein expression (Fig. 2A), which might then result in increased phosphorylation of TR4 (Fig. 2B). The addition of CIP to metformin-treated Hepa1–6 cell lysates resulted in the disappearance of metformin-induced TR4 phosphorylation, suggesting that metformin, like AICAR, can also activate AMPK-mediated TR4 phosphorylation in Hepa1–6 cells.
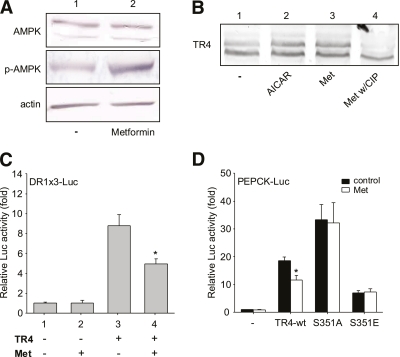
FIG. 2.
Effect of metformin on TR4 activity. A: AMPK and phosphor-AMPK in Hepa1–6 cells treated with metformin. B: TR4 phosphorylation status by Western blot. Hepa1–6 cells were treated with AICAR and metformin, and cell lysates were harvested. Cell lysates were then treated with CIP. C: DR1 × 3-Luc reporter vector was cotransfected with TR4 in Hepa1–6 cells. After overnight recovery, the transfected cells were treated with metformin and luciferase activity was measured (*P < 0.05 vs. lane 3). D: PEPCK-Luc reporter vector was cotransfected with TR4-wt, S351A, or S351E in Hepa1–6 cells and treated with or without metformin (*P < 0.05 vs. control). (A high-quality color representation of this figure is available in the online issue.)
The consequence of metformin-induced AMPK-mediated TR4 phosphorylation resulted in decreased TR4 transactivation using either DR1 × 3-Luc (Fig. 2C) or PEPCK promoter-Luc (Fig. 2D). In contrast, metformin failed to repress the transactivation of TR4 mutants (TR4-S351A and S351E) (Fig. 2D). Together, results from Fig. 2A–D suggest that metformin can modulate TR4 transactivation via induction of AMPK-mediated TR4 phosphorylation.
TR4 and SREBP-1c induce SCD1 gene expression independently.
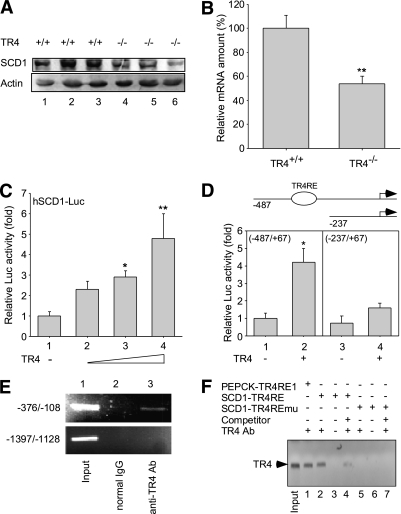
We noted first that SCD1 expression was markedly reduced in the liver of TR4−/− mice (Fig. 3A and B). The promoter-Luc assay in HepG2 cells confirmed that TR4 induced hSCD1-Luc reporter activity in a dose-dependent manner (Fig. 3C), suggesting that TR4 can induce SCD1 gene expression at the transcriptional level. Using a series of deletion mutations with or without putative TR4RE (a DR site with GGGGCAGGGGCA for the binding of TR4), we found that TR4 has minimal activity with the deletion mutant of SCD1 promoter (SCD1 [−237]) that lost the TR4RE site (Fig. 3D). ChIP assays further confirmed that TR4 bound directly on the SCD1 promoter (−108 to −376) in HepG2 cells in vivo (Fig. 3E).
FIG. 3.
SCD1 expression and effect of TR4 on the transcriptional activity of the SCD1 promoter. A: Hepatic SCD1 protein levels of TR4+/+ and TR4−/− male mice. The Western blots are representative of 10 mice per group. B: Quantitative PCR using total RNAs extracted from liver of TR4+/+ and TR4−/− mice was performed and quantified (**P < 0.01 vs. TR4+/+; n = 6). C: Hep1–6 cells were cultured and transiently transfected with SCD1 reporter without or with increasing amounts of TR4-expressing wild-type plasmid (*P < 0.05; **P < 0.01 vs. lane 1). D: SCD1 promoter full-length (−487) or deletion (−237) mutant were cotransfected with TR4 (*P < 0.05 vs. lane 1). E: ChIP assay using TR4-specific antisera. Lane 1: Input control. Lane 2: Control IP with normal mouse IgG. Lane 3: PCR product was obtained from immunoprecipitates using TR4 antibodies. F: DNA pull-down assay using biotin-labeled oligonucleotides containing the PEPCK-TR4RE1, SCD1-TR4RE, and SCD1-TR4REmu. Protein extracts were incubated with biotin-labeled double-stranded oligonucleotides containing the putative TR4RE, and the bound proteins were pulled down with streptavidin-agarose beads and analyzed by immunoblotting with antibodies against TR4. A total of 25 µg of protein extract was loaded in the “input” lane as a control (lane 1); 100 µg of proteins were used in each pull-down (lanes 1–3, 5, and 6). Non–biotin-labeled oligonucleotides were used as competition probes (lanes 4 and 7).
To determine the binding of TR4 to TR4RE in vitro, we used the nonradioactive biotin–labeled probes to perform the DNA pull-down assay. As shown in Fig. 3F, the input control was an indication of TR4 protein; biotin-labeled probes containing SCD1-TR4RE were able to bind efficiently to TR4 (Fig. 3F, lane 2). However, TR4 failed to bind to the TR4RE mutant (TR4REmu) (Fig. 3F, lane 5). Thus, results from Fig. 3A–F indicate that TR4 can induce SCD1 gene expression via direct binding to the TR4RE located on the SCD1 5′ promoter.
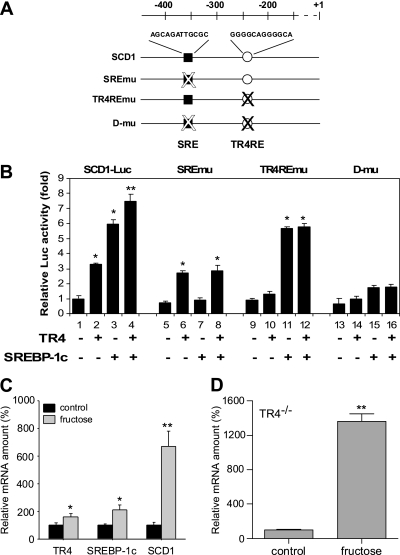
Analysis of the promoter of SCD1 found other conserved elements, including the sterol regulatory element (SRE) (20,21), might be able to mediate SCD1 induction via SREBP-1c (Fig. 4A). SCD1 promoter activity assay in Hepa1–6 cells demonstrated that SREBP-1c, like TR4, could also activate SCD1 transactivation, and that the addition of both TR4 and SREBP-1c resulted in the additive transactivation effects (Fig. 4B, lanes 1–4). Although SREBP-1c activation was eliminated following the mutation of the SRE (pSCD1-SREmu) reporter (20), TR4 effects persisted using pSCD1-SREmu (Fig. 4B, lanes 5–8). In contrast, TR4 activation was eliminated following mutation of the TR4RE (pSCD1-TR4REmu), but the SREBP-1c effect persisted (Fig. 4B, lanes 9–12). Double mutations of the SRE and TR4RE (pSCD1-d-mu) resulted in no SCD1 induction in the presence of TR4 and SREBP-1c (Fig. 4B, lanes 13–16), suggesting that TR4 and SREBP-1c activate SCD1 independently.
FIG. 4.
TR4 and SREBP-1c effects on SCD1 gene regulation. A: Schematic representation of the proximal promoters of the hSCD1. The sequences that correspond to a SRE (■), as well as TR4RE (○) are indicated. A mutated motif is indicated (with an X). B: Effect of mutation of the SRE and TR4RE on the transcriptional regulation of promoter-reporter genes. Hep1–6 cells were transiently transfected with pSCD1-(−487), pSCD1-SREmu, pSCD1-TR4REmu, or pSCD1-d-mu, as indicated (*P < 0.05; **P < 0.01 vs. control). C: Hepatic TR4, SREBP-1c, and SCD1 gene expression in wild-type mice fed with the fructose diet for 6 weeks. The relative gene expression amount was quantified by quantitative PCR (*P < 0.05; **P < 0.01 vs. control diet; n = 7). D: Hepatic SCD1 mRNA expression upon control diet and long-term fructose feeding in TR4−/− mice (**P < 0.01 vs. control diet; n = 7 per group).
Previous studies have shown that the SCD1 gene is regulated at the transcriptional level by a number of dietary factors, such as glucose and fructose, via a SREBP-1c–dependent mechanism (7,22), with less SCD1 expression in normal diet–fed SREBP-1c−/− mice (7,23). However, long-term fructose feeding results in increased SCD1 expression in both SREBP-1c−/− and SREBP-1c+/+ mice, with even higher SCD1 induction found in SREBP-1c−/− mice (7). These data suggest that fructose-induced SCD1 gene expression acts through a mechanism that is independent of SREBP-1c. In a similar series of experiments to determine whether fructose-induced SCD1 gene expression is TR4-dependent, we found, in wild-type mice fed with a fructose diet, that TR4, as well as SCD1 and SREBP-1c gene expression, were induced (Fig. 4C). Increased SCD1 gene expression was also induced by fructose in both TR4+/+ and TR4−/− mice. It is noteworthy that TR4−/− mice fed with fructose had significantly higher SCD1 gene expression compared with TR4+/+ mice (Fig. 4C and D), and this result is similar to that obtained in fructose-fed SREBP-1c−/− mice (7,23). These data suggest that fructose induced SCD1 gene expression in a TR4-independent manner. Together, results from Fig. 4 indicate that TR4 and SREBP-1c induce hepatic SCD1 gene expression independently.
TR4 induces SCD1 gene expression in the primary hepatocytes from wild-type and db/db mice.
To confirm the above results in Hepa1–6 cells showing that TR4 modulates SCD1 gene expression, we performed transductions to deliver TR4 shRNA into Hepa1–6 cells and primary hepatocytes. Using vector-based TR4 RNA interference (pRetro-shTR4a) to knockdown TR4 expression transiently, we evaluated the effect of TR4 silencing on SCD1 reporter activity in Hepa1–6 cells. As shown in Fig. 5A, TR4-mediated SCD1 promoter transcriptional activation was significantly reduced in the TR4 knockdown cells (Fig. 5A, lane 4). We also used mice primary hepatocytes with knockdown of TR4 (via RNAi) (Fig. 5B). As shown in Fig. 5C, the addition of lentiviral vector with TR4-RNAi, but not TR4-scramble-RNAi, resulted in the suppression of SCD1 gene expression. These results are consistent with the above in vivo data.
FIG. 5.
Inhibition of TR4 reduces SCD1 promoter activity and gene expression. A: Effect of pRetro-shTR4a (TR4 RNAi) or control pRetro-shNS (scramble RNAi) on TR4-mediated SCD1-Luc activity (*P < 0.05 vs. lane 3) in Hepa1–6 cells. The TR4 (B) and SCD1 (C) mRNA were quantified in TR4+/+ mice primary hepatocytes cultured and transiently transduced by lentiviral vector carrying pLVTHM-shTR4 (TR4 RNAi) and scramble RNAi (*P < 0.05; **P < 0.01 vs. scramble RNAi). D: Quantitative PCR using total RNAs extracted from liver of wild-type (WT) and db/db mice was performed and quantified (*P < 0.05; **P < 0.01 vs. WT; n = 5). E: Hepatic SCD1 and TR4 gene expression in db/db mice with or without metformin injection (*P < 0.05 vs. control; n = 5). F: Effect of TR4 RNAi and metformin on SCD1 mRNA amounts from primary hepatocytes (*P < 0.05). NS, nonsignificant.
We additionally used db/db mice that show induction of SCD1 in liver (24) to confirm our finding of hepatic SCD1 induction by TR4. As expected, we found increased expression of both TR4 and SCD1 in livers of db/db mice compared with wild-type mice (Fig. 5D). Injection of metformin into db/db mice resulted in the decreased hepatic SCD1 gene expression (Fig. 5E) that may be a result of the decreased TR4 transactivation (Supplementary Fig. 1A). We further confirmed these results via using primary hepatocytes from db/db mice treated with metformin in the presence or absence of TR4-RNAi. As shown in Fig. 5F, treatment with metformin resulted in the reduction of SCD1 gene expression in db/db mice (Fig. 5F, lanes 1 and 2), and silencing TR4 abolished the inhibition of SCD1 expression by metformin (Fig. 5F, lanes 3 and 4), suggesting the role of TR4 in the metformin suppression of SCD1.
Potential pathophysiological consequences of the newly identified metformin→AMPK→TR4→SCD1 pathway.
Early studies demonstrated that altered SCD1 gene expression resulted in changes in lipid oxidative and lipogenic pathways. To dissect the consequences of TR4-induced SCD1 expression, we compared gene profiles involved in hepatic fatty acid oxidation and lipogenesis in TR4+/+ and TR4−/− mice. TR4−/− liver showed enhanced mRNA levels of genes involved in lipid β-oxidation, carnitine palmitoyltransferase-1, acyl-CoA oxidases, and peroxisome proliferator–activated receptor-α (Supplementary Fig. 2A). As expected, enhanced β-oxidation gene expression may then result in the higher rates of isolated hepatic mitochondrial β-oxidation in TR4−/− mice compared with TR4+/+ mice, using radioisotope-labeled tracer experiments with [U-14C]palmitate (Supplementary Fig. 2B).
We also examined the genes involved in hepatic lipogenic pathways. Acetyl-CoA carboxylase and fatty acid synthase were reduced in TR4−/− mice liver (Supplementary Fig. 2C). It is noteworthy that TR4−/− mice showed no difference in SREBP-1c- and carbohydrate-responsive element–binding protein (ChREBP) expression, suggesting that the reduction of SCD1 we observed is not a result of the loss of SREBP-1c or ChREBP in TR4−/− mice (Supplementary Fig. 2C).
Increased β-oxidation and reduced lipogenic gene expression via TR4-modulated SCD1 expression in TR4−/− mice may result in the lipoatrophy. Although TR4−/− mice had smaller sizes and lower body weights than TR4+/+ mice, TR4+/+ and TR4−/− mice had a similar basal daily food intake normalized to body weight. Epididymal and retroperitoneal fat masses of TR4−/− mice were markedly reduced when these tissues were normalized by body weight compared with those of TR4+/+ mice (Fig. 6A). In contrast to the dramatic reduction of white adipose tissue, other organs, such as liver, heart, kidney, and testis, were similar to those of TR4+/+ mice (Fig. 6A). We also found that TR4−/− mice had obvious reduction in fat-pad size (Fig. 6B). Histological analysis showed that adipocytes of epididymal fat pads from TR4−/− mice were much smaller and more heterogeneous in size than those of TR4+/+ mice (Fig. 6C). Additional histological analysis showed significantly less lipid accumulation in liver from TR4−/− mice via using Oil Red O staining (Fig. 6C). Liver triglyceride level of TR4−/− mice consistently was reduced to ~50% of that of TR4+/+ mice. However, skeletal muscle triglyceride levels were similar to that of TR4+/+ mice (Fig. 6D).
FIG. 6.
TR4 deficiency leads to reduction of lipid content. A: Comparison of weights in different tissues from 10-week-old TR4−/− and TR4+/+ male mice. The weight of each tissue was normalized by body weight. Each bar represents the means ± SD (n = 5). E.Fat, epididymal fat; R.Fat, retroperitoneal fat (*P < 0.05 vs. TR4+/+). B: Epididymal fat pads isolated from TR4+/+ and TR4−/− mice. C: Histological analyses of epididymal fat (upper panel) and liver (lower panel). Sections were stained with hematoxylin and eosin or/and Oil Red O (liver) (×400). D: Triglyceride contents in liver and skeletal muscle (*P < 0.05 vs. TR4+/+; n = 7 per group). (A high-quality color representation of this figure is available in the online issue.)
In addition to reduced fat mass, loss of the SCD1 gene might also result in enhanced systemic insulin sensitivity (25,26). We measured blood glucose and insulin concentrations and found that TR4−/− mice had 10% reduction in fed and 20% reduction in fasting blood glucose concentrations, compared with those of TR4+/+ mice (Fig. 7A). Serum insulin levels were lower in fed TR4−/− mice, and fasting caused a decrease of insulin levels in both TR4+/+ and TR4−/− mice (Fig. 7B). Intraperitoneal glucose injection showed reduced blood glucose in TR4−/− mice at all time periods compared with TR4+/+ mice (Fig. 7C). Furthermore, blood glucose levels of TR4−/− mice were returned to normal levels at 60 min after glucose injection. However, blood glucose levels of TR4+/+ mice were still higher than normal glucose levels until 120 min after glucose injection. Increased insulin sensitivity was further confirmed in insulin tolerance tests, showing an enhanced glucose-lowering effect in TR4−/− mice compared with that of TR4+/+ mice (Fig. 7D).
FIG. 7.
TR4−/− male mice show increased insulin sensitivity. Plasma concentrations of glucose (A) and insulin (B) in animals fed or fasted overnight. Values are given as means ± SD (*P < 0.05; **P < 0.01 vs. TR4+/+; n = 5–7 per group). C: Glucose tolerance tests. Each point represents the means ± SD (n = 5). D: Insulin tolerance tests. Each point represents the means ± SD (*P < 0.05; **P < 0.01; ***P < 0.001 vs. TR4+/+; n = 5–6 per group). E: Muscle tissue subjected to immunoprecipitation with anti-IRβ, or anti–IRS-1 antibody, followed by immunoblot analysis using antiphosphotyrosine antibody. The Western blots are representative of five separate experiments with independent tissue lysate preparations.
To further examine whether increased systemic insulin sensitivity is accompanied with increased insulin signaling, we also examined insulin signaling in skeletal muscles of TR4−/− and TR4+/+ mice. The TR4−/− mice showed increased basal tyrosine phosphorylation of insulin receptor-β (IRβ) in skeletal muscle compared with TR4+/+ mice before infusion of insulin (Fig. 7E), indicating that TR4−/− mice are more sensitive to insulin. Both TR4+/+ and TR4−/− mice showed insulin-induced tyrosine phosphorylation of IRβ in skeletal muscle with, as expected, a much higher extent in TR4−/− mice. To further confirm increased insulin signaling in TR4−/− mice, we measured tyrosine phosphorylation of insulin receptor substrate (IRS)-1. Consistent with phosphorylation of IRβ, TR4−/− mice showed enhanced basal phosphorylation of IRS-1. Insulin infusion further phosphorylates IRS-1 in skeletal muscle of both TR4+/+ and TR4−/− mice with approximately onefold higher phosphorylation of IRS-1 in TR4−/− mice (Fig. 7E; Supplementary Fig. 1C).
DISCUSSION
Transactivation of nuclear receptors can be activated by ligands or via modulation through post-translational modifications, such as phosphorylation, acetylation, and sumoylation, that result in the alteration of biological function (27). Here, we found, for the first time, that transactivation of TR4 could be altered by a small molecule, such as metformin. Metformin has been shown to activate AMPK via an LKB1-dependent mechanism (28). The activated AMPK may phosphorylate TR4 and inhibit TR4 transactivation activity and subsequently downregulate TR4 target gene expression. These data suggest that TR4 activity can be altered by phosphorylation. It also provides a platform in which small molecules can change TR4 phosphorylation status in the absence of an identified ligand(s) and potentially modulate TR4 target gene expression, which show the clinical application of the physiological functions of TR4.
A previous study (29) showed that TR4 transactivation could also be modulated by mitogen-activated protein kinase phosphorylation that resulted in the recruitment of corepressors and coactivators, with hyperphosphorylated TR4 showing lower activity and hypophosphorylated TR4 showing higher activity. The same phenomena were observed in AMPK phosphorylation on TR4 proteins, that TR4 transactivation could be negatively regulated by AMPK-mediated phosphorylation. However, TR4 behavior after phosphorylation remains unclear. The AMPK phosphorylation site, Ser351, is located on the hinge region, which is important for nuclear receptor nuclear localization, suggesting that TR4 nuclear/cytosolic translocation may be regulated by AMPK phosphorylation. The impact of AMPK phosphorylation of TR4 on the DNA-binding ability, protein stability, and coregulator(s) recruitment, however, remains unclear.
The data from the SREBP-1c−/− and TR4−/− fructose-feeding experiments indicate that the lipogenic effects of fructose may be mediated by TR4 or SREBP-1c, whereas other transcription factors may also participate in the regulation independently. Two possible candidates mediating the SREBP-1c- or TR4-independent mechanisms are ChREBP and liver X receptor (LXR). ChREBP has been known as a transcription factor mediating glucose sensing and lipogenesis (30). However, the role of ChREBP on SCD1 regulation is yet to be determined. LXR, the major transcription factor that activates SREPB-1c and ChREBP transcription, integrates hepatic carbohydrate and lipid metabolism (31). In a recent study (32), LXR was also identified as a transcription factor regulating SCD1 gene expression directly. Through the LXR-response element on SCD1 promoter, LXR could induce SCD1 gene expression and increase the hepatic triglyceride-to-monounsaturated fatty acid-to-saturated fatty acid ratio. It has been demonstrated that the SCD1 induction in SREBP-1c−/− or TR4−/− mice fed with fructose is through the SREBP-1c- or TR4-independent pathway. It is possible that the SCD1 regulation in the absence of SREBP-1c is mediated by TR4, ChREBP, and LXR. In contrast, the induced SCD1 gene expression in TR4−/− mice may be mediated by SREBP-1c, ChREBP, and LXR. However, there is no study about the fructose or high-carbohydrate effect of SCD1 gene expression on LXR−/− or ChREBP−/− mice. Therefore, the roles of ChREBP and LXR in fructose-fed SREBP-1c−/− or TR4−/− mice remain unclear.
Here, we showed that TR4-deficient mice have reduced fat-pad size, enhanced lipid oxidation, and insulin sensitivity as a result of reduced SCD1 gene expression in liver. These metabolic changes may protect TR4−/− mice from a variety of dietary and genetic conditions that promote obesity and insulin resistance. In support of this notion, SCD1−/− mice were found to be resistant to diet-induced obesity, have reduced adiposity, and have increased insulin sensitivity. Furthermore, the regulation of SCD1 gene expression is an important component of the metabolic response. The TR4−/− mouse model recapitulates the phenotypes observed in standard diet–fed SCD1-deficient mice. In agreement with our TR4−/− model, Kang et al. (33) also observed that systemic loss of TR4 results in less weight gain, lower hepatic triglyceride levels, reduced lipid accumulation in adipose tissue, and greatly decreased lipogenic gene expression. Taken together, our findings support the idea that SCD1 may play important roles in the TR4−/− mice lipid metabolism. These roles strengthen the importance of TR4 regulation on SCD1 gene expression and the pathophysiological significance of this newly identified pathway (Fig. 8).
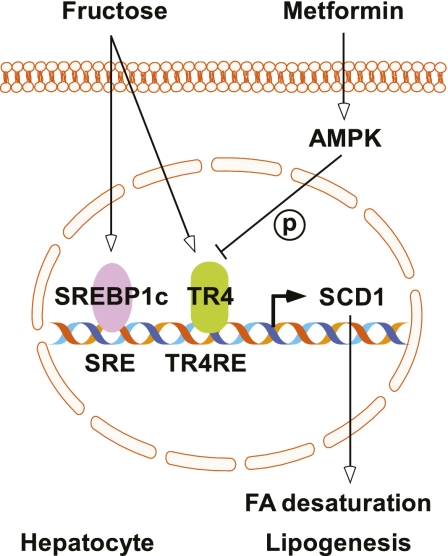
FIG. 8.
TR4 acts as a key regulator of SCD1 gene expression and lipid metabolism in hepatocytes. TR4 gene expression was induced by fructose, and its activity was inhibited by AMPK-mediated phosphorylation. The binding of TR4 to the SCD1 promoter through TR4RE motif activates SCD1 expression and stimulates lipogenesis. (A high-quality color representation of this figure is available in the online issue.)
TR4 may function as a regulator to modulate many nuclear receptor–mediated pathways, such as retinoid X receptor, retinoid acid receptor, and androgen receptor (34). Knockout of TR4 in mice leads to growth retardation (13), as well as imbalance in glucose and lipid metabolism (12,15). Recently, studies (35,36) suggested that TR4 might exert its function by interacting with certain fatty acids and lipids. In general, saturated fatty acids and mono- and polyunsaturated fatty acids all show different degrees of activation of TR4. Although fatty acids are able to interact with TR4 directly, it remains possible that fatty acids may regulate TR4 activity via phosphorylation or other posttranslational modification. Because SCD1 gene expression also can be regulated by saturated and unsaturated fatty acids, it suggests that fatty acid treatment may induce TR4 activation and TR4 target gene, SCD1, via a TR4-dependent pathway. The SCD1 gene expression regulated by saturated and unsaturated fatty acids may also be involved in the TR4-independent pathway.
Together, the newly identified pathways from metformin to AMPK to TR4 in the present studies provide the first evidence showing that hepatic TR4 transactivation can be modulated by a small molecule, such as metformin, AICAR, or CpdC, which may lead to the development of a platform for using TR4 as a target to battle TR4-related diseases, such as metabolic syndrome.
ACKNOWLEDGMENTS
This work was supported by the George Whipple Professorship Endowment at the University of Rochester, the National Institutes of Health Grants CA-127300 and DK-73414, and Department of Health Clinical Trial and Research Center of Excellence Grant DOH99-TD-B-111-004 (to China Medical University, Taichung, Taiwan).
No potential conflicts of interest relevant to this article were reported.
E.K. and N.-C.L. researched data and wrote the manuscript. I-C.Y. and H.-Y.L. researched data. Y.-F.L. and J.D.S. contributed to the discussion and reviewed and edited the manuscript. L.-M.C. contributed to the discussion. C.C. wrote the manuscript and contributed to the discussion.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db10-0393/-/DC1.
REFERENCES
- 1.Potenza MV, Mechanick JI. The metabolic syndrome: definition, global impact, and pathophysiology. Nutr Clin Pract 2009;24:560–577 [DOI] [PubMed] [Google Scholar]
- 2.Flowers MT, Ntambi JM. Role of stearoyl-coenzyme A desaturase in regulating lipid metabolism. Curr Opin Lipidol 2008;19:248–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ntambi JM, Miyazaki M. Recent insights into stearoyl-CoA desaturase-1. Curr Opin Lipidol 2003;14:255–261 [DOI] [PubMed] [Google Scholar]
- 4.Sampath H, Ntambi JM. Stearoyl-coenzyme A desaturase 1, sterol regulatory element binding protein-1c and peroxisome proliferator-activated receptor-alpha: independent and interactive roles in the regulation of lipid metabolism. Curr Opin Clin Nutr Metab Care 2006;9:84–88 [DOI] [PubMed] [Google Scholar]
- 5.Azzout-Marniche D, Bécard D, Guichard C, Foretz M, Ferré P, Foufelle F. Insulin effects on sterol regulatory-element-binding protein-1c (SREBP-1c) transcriptional activity in rat hepatocytes. Biochem J 2000;350:389–393 [PMC free article] [PubMed] [Google Scholar]
- 6.Kim HJ, Miyazaki M, Man WC, Ntambi JM. Sterol regulatory element-binding proteins (SREBPs) as regulators of lipid metabolism: polyunsaturated fatty acids oppose cholesterol-mediated induction of SREBP-1 maturation. Ann N Y Acad Sci 2002;967:34–42 [DOI] [PubMed] [Google Scholar]
- 7.Miyazaki M, Dobrzyn A, Man WC, et al. Stearoyl-CoA desaturase 1 gene expression is necessary for fructose-mediated induction of lipogenic gene expression by sterol regulatory element-binding protein-1c-dependent and -independent mechanisms. J Biol Chem 2004;279:25164–25171 [DOI] [PubMed] [Google Scholar]
- 8.Wong AK, Howie J, Petrie JR, Lang CC. AMP-activated protein kinase pathway: a potential therapeutic target in cardiometabolic disease. Clin Sci (Lond) 2009;116:607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGee SL, Hargreaves M. AMPK and transcriptional regulation. Front Biosci 2008;13:3022–3033 [DOI] [PubMed] [Google Scholar]
- 10.Freedman LP. Anatomy of the steroid receptor zinc finger region. Endocr Rev 1992;13:129–145 [DOI] [PubMed] [Google Scholar]
- 11.Yang X, Downes M, Yu RT, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell 2006;126:801–810 [DOI] [PubMed] [Google Scholar]
- 12.Liu NC, Lin WJ, Kim E, et al. Loss of TR4 orphan nuclear receptor reduces phosphoenolpyruvate carboxykinase–mediated gluconeogenesis. Diabetes 2007;56:2901–2909 [DOI] [PubMed] [Google Scholar]
- 13.Collins LL, Lee YF, Heinlein CA, et al. Growth retardation and abnormal maternal behavior in mice lacking testicular orphan nuclear receptor 4. Proc Natl Acad Sci USA 2004;101:15058–15063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Gouill E, Jimenez M, Binnert C, et al. Endothelial nitric oxide synthase (eNOS) knockout mice have defective mitochondrial β-oxidation. Diabetes 2007;56:2690–2696 [DOI] [PubMed] [Google Scholar]
- 15.Kim E, Xie S, Yeh SD, et al. Disruption of TR4 orphan nuclear receptor reduces the expression of liver apolipoprotein E/C-I/C-II gene cluster. J Biol Chem 2003;278:46919–46926 [DOI] [PubMed] [Google Scholar]
- 16.Yang JS, Kim JT, Jeon J, et al. Changes in hepatic gene expression upon oral administration of taurine-conjugated ursodeoxycholic acid in ob/ob mice. PLoS ONE 2010;5:e13858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fediuc S, Gaidhu MP, Ceddia RB. Regulation of AMP-activated protein kinase and acetyl-CoA carboxylase phosphorylation by palmitate in skeletal muscle cells. J Lipid Res 2006;47:412–420 [DOI] [PubMed] [Google Scholar]
- 18.Horike N, Sakoda H, Kushiyama A, et al. AMP-activated protein kinase activation increases phosphorylation of glycogen synthase kinase 3beta and thereby reduces cAMP-responsive element transcriptional activity and phosphoenolpyruvate carboxykinase C gene expression in the liver. J Biol Chem 2008;283:33902–33910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim YD, Park KG, Lee YS, et al. Metformin inhibits hepatic gluconeogenesis through AMP-activated protein kinase–dependent regulation of the orphan nuclear receptor SHP. Diabetes 2008;57:306–314 [DOI] [PubMed] [Google Scholar]
- 20.Tabor DE, Kim JB, Spiegelman BM, Edwards PA. Identification of conserved cis-elements and transcription factors required for sterol-regulated transcription of stearoyl-CoA desaturase 1 and 2. J Biol Chem 1999;274:20603–20610 [DOI] [PubMed] [Google Scholar]
- 21.Bené H, Lasky D, Ntambi JM. Cloning and characterization of the human stearoyl-CoA desaturase gene promoter: transcriptional activation by sterol regulatory element binding protein and repression by polyunsaturated fatty acids and cholesterol. Biochem Biophys Res Commun 2001;284:1194–1198 [DOI] [PubMed] [Google Scholar]
- 22.Hasty AH, Shimano H, Yahagi N, et al. Sterol regulatory element-binding protein-1 is regulated by glucose at the transcriptional level. J Biol Chem 2000;275:31069–31077 [DOI] [PubMed] [Google Scholar]
- 23.Biddinger SB, Almind K, Miyazaki M, Kokkotou E, Ntambi JM, Kahn CR. Effects of diet and genetic background on sterol regulatory element–binding protein-1c, stearoyl-CoA desaturase 1, and the development of the metabolic syndrome. Diabetes 2005;54:1314–1323 [DOI] [PubMed] [Google Scholar]
- 24.Loffler M, Bilban M, Reimers M, Waldhäusl W, Stulnig TM. Blood glucose-lowering nuclear receptor agonists only partially normalize hepatic gene expression in db/db mice. J Pharmacol Exp Ther 2006;316:797–804 [DOI] [PubMed] [Google Scholar]
- 25.Rahman SM, Dobrzyn A, Dobrzyn P, Lee SH, Miyazaki M, Ntambi JM. Stearoyl-CoA desaturase 1 deficiency elevates insulin-signaling components and down-regulates protein-tyrosine phosphatase 1B in muscle. Proc Natl Acad Sci USA 2003;100:11110–11115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahman SM, Dobrzyn A, Lee SH, Dobrzyn P, Miyazaki M, Ntambi JM. Stearoyl-CoA desaturase 1 deficiency increases insulin signaling and glycogen accumulation in brown adipose tissue. Am J Physiol Endocrinol Metab 2005;288:E381–E387 [DOI] [PubMed] [Google Scholar]
- 27.Faus H, Haendler B. Post-translational modifications of steroid receptors. Biomed Pharmacother 2006;60:520–528 [DOI] [PubMed] [Google Scholar]
- 28.Towler MC, Hardie DG. AMP-activated protein kinase in metabolic control and insulin signaling. Circ Res 2007;100:328–341 [DOI] [PubMed] [Google Scholar]
- 29.Huq MD, Gupta P, Tsai NP, Wei LN. Modulation of testicular receptor 4 activity by mitogen-activated protein kinase-mediated phosphorylation. Mol Cell Proteomics 2006;5:2072–2082 [DOI] [PubMed] [Google Scholar]
- 30.Dentin R, Denechaud PD, Benhamed F, Girard J, Postic C. Hepatic gene regulation by glucose and polyunsaturated fatty acids: a role for ChREBP. J Nutr 2006;136:1145–1149 [DOI] [PubMed] [Google Scholar]
- 31.Pégorier JP, Le May C, Girard J. Control of gene expression by fatty acids. J Nutr 2004;134:2444S–2449S [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Zhang X, Chen L, et al. Liver X receptor agonist TO-901317 upregulates SCD1 expression in renal proximal straight tubule. Am J Physiol Renal Physiol 2006;290:F1065–F1073 [DOI] [PubMed] [Google Scholar]
- 33.Kang HS, Okamoto K, Kim YS, et al. Nuclear orphan receptor TAK1/TR4-deficient mice are protected against obesity-linked inflammation, hepatic steatosis, and insulin resistance. Diabetes 2011;60:177–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee YF, Lee HJ, Chang C. Recent advances in the TR2 and TR4 orphan receptors of the nuclear receptor superfamily. J Steroid Biochem Mol Biol 2002;81:291–308 [DOI] [PubMed] [Google Scholar]
- 35.Tsai NP, Huq M, Gupta P, Yamamoto K, Kagechika H, Wei LN. Activation of testicular orphan receptor 4 by fatty acids. Biochim Biophys Acta 2009;1789:734–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie S, Lee YF, Kim E, et al. TR4 nuclear receptor functions as a fatty acid sensor to modulate CD36 expression and foam cell formation. Proc Natl Acad Sci USA 2009;106:13353–13358 [DOI] [PMC free article] [PubMed] [Google Scholar]