Abstract
OBJECTIVE
Previous animal studies suggest a functional relationship between metabolism, type 2 diabetes, and the amplitude of daily rhythms in white adipose tissue (WAT). However, data interpretation is confounded by differences in genetic background and diet or limited sampling points. We have taken the novel approach of analyzing serial human WAT biopsies across a 24-h cycle in controlled laboratory conditions.
RESEARCH DESIGN AND METHODS
Lean (n = 8), overweight/obese (n = 11), or overweight/obese type 2 diabetic (n = 8) volunteers followed a strict sleep–wake and dietary regimen for 1 week prior to the laboratory study. They were then maintained in controlled light–dark conditions in a semirecumbent posture and fed hourly during wake periods. Subcutaneous WAT biopsies were collected every 6 h over 24 h, and gene expression was measured by quantitative PCR.
RESULTS
Lean individuals exhibited significant (P < 0.05) temporal changes of core clock (PER1, PER2, PER3, CRY2, BMAL1, and DBP) and metabolic (REVERBα, RIP140, and PGC1α) genes. The BMAL1 rhythm was in approximate antiphase with the other clock genes. It is noteworthy that there was no significant effect (P > 0.05) of increased body weight or type 2 diabetes on rhythmic gene expression.
CONCLUSIONS
The robust nature of these rhythms and their relative phasing indicate that WAT now can be considered as a peripheral tissue suitable for the study of in vivo human rhythms. Comparison of data between subject groups clearly indicates that obesity and type 2 diabetes are not related to the amplitude of rhythmic WAT gene expression in humans maintained under controlled conditions.
Circadian clocks regulate daily changes in a wide range of physiological and behavioral functions, including the timing of sleep. Disruption of circadian biology or sleep has profound metabolic consequences (1), which has led to the development of a major research effort to understand the links between these processes. By understanding the temporal regulation of metabolic physiology, it is anticipated that common behaviors and clinical interventions can be optimized to reduce the impact of modern society on the incidence of metabolic diseases, such as diabetes.
There are clear links between circadian clocks and metabolism. In murine models, transgenic disruption of key clock–related genes induces marked metabolic phenotypes (2–5). In humans, chronic desynchrony of internal circadian time with the external environment (e.g., during shift work) correlates with increased incidence of obesity, type 2 diabetes, and cardiometabolic diseases (6). Moreover, consistent with the transgenic murine data, recent work has linked polymorphisms of human circadian clock genes with metabolic dysfunction (7–9).
The mammalian circadian timing system consists of a “master” clock within the suprachiasmatic nuclei (SCN) of the hypothalamus and a series of “peripheral” clocks that are located in extra-SCN regions of the brain as well as in almost all other tissues (10). The environmental light–dark cycle synchronizes the circadian system by acting through the SCN clock, which then maintains temporal order of the peripheral clocks via multiple rhythmic outputs (10). These SCN-driven pathways include neuronal (e.g., sympathetic tone), endocrine (e.g., melatonin and cortisol secretion), and behavioral (e.g., feeding) activity.
Murine white adipose tissue (WAT), as with many other tissues, contains endogenous circadian timing properties (11,12), and ~10–20% of murine WAT transcriptome is estimated to exhibit a 24-h variation (13,14). It is noteworthy that mice that are genetically susceptible to obesity and type 2 diabetes (15) or fed a high-fat diet (16) exhibit reduced amplitude rhythms of gene expression in WAT, leading to the hypothesis that the robustness of WAT clocks is functionally associated with metabolic phenotype.
This hypothesis has not been rigorously tested in humans. The principal reason for this important gap in our knowledge stems from the difficulty in obtaining multiple serial biopsies of tissue from human volunteers. To date, the literature on rhythmic gene expression in human WAT has primarily relied on either analysis of single-time–point biopsies maintained in culture (17) or the relative expression of genes at a single time point (18,19). It is clear that neither of these methods is ideal for the assessment of 24-h rhythms in vivo.
In this study, serial human biopsies across a 24-h cycle were analyzed using a carefully controlled protocol designed to minimize the effect of confounding factors on rhythmic physiology. We studied three defined groups of human volunteers to investigate daily WAT rhythms; lean individuals were compared with subjects who were overweight/obese or overweight/obese with type 2 diabetes.
RESEARCH DESIGN AND METHODS
All aspects of the study were conducted in accordance with the Declaration of Helsinki and received a favorable ethical opinion from the Surrey Research Ethics Committee and the University of Surrey Ethics Committee.
Volunteers completed questionnaires to assess general health and ensure normal sleep and diurnal preference. They then attended a prescreening session at the University of Surrey, typically within 1 month of the laboratory session and during the early afternoon. BMI and waist circumference were measured, and fasting blood samples were provided for measurement of plasma HbA1c, insulin and glucose concentrations, and homeostasis model assessment of insulin resistance (HOMA-IR; calculated using HOMA calculator version 2.2 software [Diabetes Trial Unit; University of Oxford, Oxford, U.K.]).
Twenty-seven male volunteers were recruited (8 lean, 11 overweight, and 8 overweight with type 2 diabetes). Of the patients with type 2 diabetes, average time since diagnosis was 6.9 ± 2.3 years; three type 2 diabetic patients were controlled by diet and exercise, and the other five patients were treated with combinations of metformin, statins, ramipril, and lisinopril. One participant within the lean group was a smoker and was required to refrain from smoking for 1 week prior to the study. None of the participants had undertaken shift work within 5 years of the study or crossed any time zones within 1 month of the study.
For 1 week prior to the laboratory study, volunteers were required to maintain prescribed daily feeding times and sleep activity (sleep 2230–0630 h), which were monitored using wrist actigraphy (AWL; Cambridge Neurotechnology, Cambridge, U.K.), sleep diaries, food diaries, and recorded messages on a laboratory time-stamped answer telephone. Participants also were required to refrain from eating fatty or sugary foods and drinking alcohol or caffeine throughout the week. For the final 3 days of this baseline week, food was provided by the research team to enable control of timed behavior together with both the quantity and quality of nutrient intake. During these 3 days, the daily caloric content of the supplied food was basal metabolic rate times 1.5 with ~35% of energy from fat.
Laboratory study.
All experimental procedures were carried out at the Surrey Clinical Research Centre. Volunteers arrived in the afternoon of day 0 for a night of adaptation. Throughout the 2-day study, they were required to maintain a semirecumbent posture. They were required to remain awake with lights on between 0630 and 2230 h (440–825 lux in direction of gaze) and allowed to sleep with lights off between 2230 and 0630 h (0 lux). During the waking period, participants were fed with hourly nutritional drinks (Fortisip; Nutricia, Schiphol, the Netherlands) and were allowed to drink water ad libitum. Daily energy intake was basal metabolic rate times 1.1, divided equally over the waking hours.
Four subcutaneous WAT biopsies were taken under a local anesthetic (lidocaine) from four different sites of the upper buttock region of each participant at 6-hourly intervals for 24 h, beginning at 1030 h on day 1. The sampling order of the four biopsy sites was randomized between groups and subjects. This WAT depot is known to exhibit metabolic activity (20).
Analysis of gene expression.
WAT biopsies were washed with saline and snap-frozen in liquid nitrogen before storage at −80°C. Total RNA was extracted using TRIzol, according to the manufacturer’s instructions. cDNA was synthesized, and expression was measured for genes integral to the circadian clock (PER1, PER2, PER3, CRY2, BMAL1, and DBP) and involved in metabolic activity (REVERBα, RIP140, and PGC1α) by quantitative TaqMan real-time PCR, as described previously (21) (Table 1). Standard curves (R2 > 0.99) were generated using human genomic DNA (Promega, Southampton, U.K.), and expression of all genes was normalized to GAPDH, as in previous studies of adipose clock gene expression (15,16).
TABLE 1.
TaqMan PCR primer probe sets
| Gene | Upper primer (5′ to 3′ end) | Lower primer (5′ to 3′ end) | TaqMan probe (5′ to 3′) | GenBank accession number | Region amplified (bp) | Product size (bp) |
|---|---|---|---|---|---|---|
| PER1 | CCCTTTGGTGACCCCAATG | GCCCCATAAGGATAGCTGGAT | TGGCCTTGGTGCTCCCTAACTATCTGTTC | NM_002616 | 2,974–3,051 | 78 |
| PER2 | GTCCAGCCCCCACCTTTC | GGGAAGGAATAACTGGGTAGCA | CTGCCCCTTTGGCGCCTGTCAT | NM_022817 | 2,908–2,978 | 71 |
| PER3 | CCTGGACCCTGAACATGCA | TGTGAGCCCCACGTGTTTAA | CCAGCCCCTTTGACCTCGGAAGA | NM_016831 | 2,136–2,200 | 65 |
| CRY2 | AGTGGGCTGAGGGCAAGAC | CAGCCCTCCTGCCTCAGTT | TTCCCTTGGATTGATGCCATCATGACC | NM_021117 | 1,128–1,197 | 70 |
| DBP | CGTGGAGGTGTTGATGACCTT | TCGTGGCCAGGAATGCTT | AACCCGACCCAGCTGATCTTGCC | NM_001352 | 982–1,050 | 69 |
| BMAL1 | TGCCTCGTCGCAATTGG | ACCCTGATTTCCCCGTTCA | CGACTGCATTCTCATGTAGTTCCACAACCA | NM_001178 | 1,342–1,409 | 68 |
| REVERBα | CTTCAATGCCAACCATGCAT | CCTGATTTTCCCAGCGATGT | AGGTAGCCCTCCAGCCACCACCC | NM_021724 | 1,565–1,629 | 65 |
| RIP140 | GGCAAACAGGATAGCACATTACTG | CCTGATTTGTTGTGACAGAGCAA | TTTGCTTCAGTCATTCAGCTCTAGGCTGCA | NM_003489 | 974–1,060 | 87 |
| PGC1α | CCTGCTCGGAGCTTCTCAAAT | TTTCTGTTCTCTGTGGGTTTGGT | TCTGACCACAAACGATGACCCTCCTCA | NM_013261 | 737–809 | 73 |
| GAPDH | CAAGGTCATCCATGACAACTTTG | GGGCCATCCACAGTCTTCTG | ACCACAGTCCATGCCATCACTGCCA | NM_002046 | 585–674 | 90 |
Statistical analysis.
Prescreen data were analyzed using one-way ANOVA with Tukey’s post hoc test. Correlation analyses were conducted using linear regression, and the gene expression time course from biopsies was performed using one-way or two-way repeated-measures ANOVA (factors time and group) with Tukey’s post hoc test.
RESULTS
Prescreen data are shown in Table 2. Despite efforts to age-match the participants, there was a significant (P < 0.05) difference in age, with post hoc analysis revealing a significant difference only between the overweight and type 2 diabetic groups. However, subsequent analysis using age as a covariant revealed that age, per se, did not alter gene expression (data not shown). There were significant (P < 0.05) differences in BMI, waist circumference, fasting plasma glucose and insulin concentrations, HbA1c levels, and HOMA-IR, with highest values occurring in participants with type 2 diabetes. There was no significant difference in plasma insulin, glucose, HbA1c, or HOMA-IR between the lean and overweight/obese groups, indicating that our overweight/obese participants were insulin-sensitive.
TABLE 2.
Participant prelaboratory data
| Variable | Lean subjects | Overweight/obese subjects | Type 2 diabetic subjects |
|---|---|---|---|
| n | 8 | 11 | 8 |
| Age (years) | 53.6 ± 6.0 | 49.9 ± 7.5 | 58.0 ± 4.5* |
| BMI (kg/m2) | 23.2 ± 1.4 | 30.2 ± 2.3† | 31.2 ± 3.2† |
| Waist circumference (cm) | 88.9 ± 6.5 | 105 ± 4.3† | 110.9 ± 10.6† |
| Fasting glucose (mmol/L) | 4.2 ± 0.7 | 4.8 ± 0.7 | 6.7 ± 1.3*† |
| Fasting insulin (pmol/L) | 28.1 ± 16.8 | 39.2 ± 17.4 | 93.6 ± 78.7*† |
| HbA1c (mmol/L) | 5.4 ± 0.4 | 5.4 ± 0.5 | 6.8 ± 0.8*† |
| HOMA-IR | 0.5 ± 0.1 | 0.7 ± 0.1 | 1.9 ± 0.5*† |
Data are means ± SE.
*P < 0.05 vs. overweight/obese group.
†P < 0.05 vs. lean group.
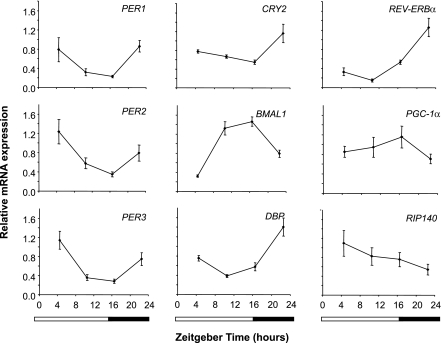
In lean individuals, there was a significant (P < 0.05) effect of time on the expression of multiple genes involved in circadian (PER1, PER2, PER3, CRY2, BMAL1, and DBP) and metabolic functions (REVERBα, RIP140, and PGC1α) (Fig. 1). Minimal expression of REVERBα and DBP occurred around the middle to end of the afternoon. In keeping with circadian gene expression in other tissues (10), the phase of PER1, PER2, PER3, and CRY2 was slightly delayed compared with REVERBα and DBP, with minimal expression occurring around the evening light–dark transition. The BMAL1 rhythm was in approximate antiphase to the other circadian genes, with maximal expression around the evening light–dark transition.
FIG. 1.
Robust 24-h changes of gene expression in human WAT from lean individuals. Data are presented as means ± SE of n = 8–11 values, normalized to glyceraldehyde 3-phosphate dehydrogenase. There was a significant effect of time (P < 0.05, one-way repeated-measures ANOVA) on the expression of each gene. Horizontal bars represent wake (white) and sleep (black) periods during the laboratory study. Zeitgeber time 0 represents the time of lights on (equivalent to clock time 0630 h).
Analysis by two-way repeated-measures ANOVA of the data from all of the experimental groups confirmed a significant (P < 0.05) effect of time on gene expression (Fig. 2). However, it is surprising that we did not observe a significant effect of experimental group on the expression of any gene. The only gene to exhibit a significant (P < 0.01) time × group interaction was BMAL1. Tukey’s post hoc analysis revealed that BMAL1 expression 16.5 h after lights on (equivalent to clock time 2300 h) was significantly (P < 0.05) lower in the overweight/obese participants than in the lean or type 2 diabetic groups.
FIG. 2.
Comparison of 24-h gene expression profiles in human WAT from individuals who are lean (solid line, ♦), overweight/obese (dashed line, ■), or overweight/obese with type 2 diabetes (dotted line, ▲). Data are presented as means ± SE of n = 8–11 values, normalized to glyceraldehyde 3-phosphate dehydrogenase. For all genes, there was a significant (P < 0.05) effect of time but no subject group effect on gene expression (two-way repeated-measures ANOVA). With the exception of BMAL1 (P < 0.01), there was no significant interaction between time and subject group on gene expression. Horizontal bars represent wake (white) and sleep (black) periods during the laboratory study. Zeitgeber time 0 represents the time of lights on (equivalent to clock time 0630 h).
DISCUSSION
Consistent with a previous study (22), our data reveal robust diurnal rhythmicity within human WAT in vivo. To date, limited data have been published concerning clock gene expression in this tissue. Because of difficulties in acquiring human tissue, some studies have measured clock gene expression in WAT collected at a single time point (18,19), although it is clearly difficult to infer rhythmical changes from such a temporally limited dataset. An elegant compromise has been to culture explants of tissue taken at a single time point and then measure gene expression in fragments of the biopsies over a 24-h time course (17). Although such studies highlight endogenous WAT rhythms, they do not reflect gene expression in vivo. Therefore, we measured gene expression in human tissue explants harvested across a whole 24-h cycle to provide in vivo human WAT rhythms.
Animal models suggest that obesity and type 2 diabetes reduce rhythm amplitude in murine WAT because lean C57BL/6J mice exhibit higher amplitude WAT rhythms than obese/diabetic KK and KK-Ay mice (15). Other data revealed reduced amplitude rhythms in mice that became obese as a result of a high-fat diet (16). However, Ando et al. (15) compared mice with differing genetic backgrounds, which complicate interpretation of their data, and it is possible that dietary intervention could directly regulate gene expression rather than obesity per se.
In contrast to the above studies, we observed minimal differences between clock gene rhythms in human WAT. Interpretation of the minor difference in BMAL1 expression is not clear. Although murine Bmal1 has been implicated in the control of adipogenesis in vitro (23), juvenile Bmal1−/− mice develop adipose depots comparably to their wild-type littermates (24). Furthermore, the similarity between gene expression profiles in lean individuals and those who were overweight with type 2 diabetes suggests that BMI is not the cause of the time × group interaction for BMAL1 in the overweight, nondiabetic individuals.
The similarity of gene expression profiles between our experimental groups may partly result from a lack of extreme phenotypic differences. Although there was not a large disparity in BMI between our groups, all participants fell into the current clinical guidelines for lean/healthy, obese, and type 2 diabetic individuals. Therefore, there was no effect of obesity or type 2 diabetes, per se, on human WAT rhythmicity. It remains possible that severely obese individuals may exhibit reduced amplitude WAT rhythmicity; however, interpretation of data from such individuals is complicated by various confounding factors, such as comorbidities associated with type 2 diabetes.
Another difference between our study and previous work (25) is the high level of glycemic control in our participants. We specifically aimed to investigate the relationship between WAT rhythmicity, body weight, and presence of type 2 diabetes. Therefore, we eliminated as many additional factors as possible. Diurnal adipose gene expression may be modulated by the level of glycemic control and/or drugs taken by diabetic individuals. Alternatively, the association between metabolic state and WAT rhythmicity might vary between adipose depots. However, because of limitations of sampling and experimental group size, we could not directly address these possibilities in the current study.
The persistence of 24-h rhythms in WAT from patients with type 2 diabetes now suggests that the link between human circadian and metabolic physiology occurs outside of WAT, at least in the earlier stages of the disease.
ACKNOWLEDGMENTS
This work was funded by Diabetes UK (grant 08/0003607), the Biotechnology and Biological Sciences Research Council (grant BB/D526853/1), and Stockgrand, Ltd., U.K. No other potential conflicts of interest relevant to this article were reported.
D.T.O. researched data and reviewed and edited the manuscript. S.M., S.B., and J.W. researched data. P.T., D.J.S., and M.D.R. reviewed and edited the manuscript. J.D.J. wrote the manuscript.
The authors thank the clinical and administrative staff of the Surrey Clinical Research Centre. The authors also thank Dr. Caroline Bodinham (Faculty of Health and Medical Sciences, University of Surrey, U.K.) for assistance with the prescreen assays.
REFERENCES
- 1.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell 2008;134:728–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raspé E, Duez H, Mansén A, et al. Identification of Rev-erbalpha as a physiological repressor of apoC-III gene transcription. J Lipid Res 2002;43:2172–2179 [DOI] [PubMed] [Google Scholar]
- 3.Turek FW, Joshu C, Kohsaka A, et al. Obesity and metabolic syndrome in circadian clock mutant mice. Science 2005;308:1043–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oishi K, Atsumi G, Sugiyama S, et al. Disrupted fat absorption attenuates obesity induced by a high-fat diet in Clock mutant mice. FEBS Lett 2006;580:127–130 [DOI] [PubMed] [Google Scholar]
- 5.Green CB, Douris N, Kojima S, et al. Loss of nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc Natl Acad Sci USA 2007;104:9888–9893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lowden A, Moreno C, Holmbäck U, Lennernäs M, Tucker P. Eating and shift work: effects on habits, metabolism and performance. Scand J Work Environ Health 2010;36:150–162 [DOI] [PubMed] [Google Scholar]
- 7.Scott EM, Carter AM, Grant PJ. Association between polymorphisms in the Clock gene, obesity and the metabolic syndrome in man. Int J Obes (Lond) 2008;32:658–662 [DOI] [PubMed] [Google Scholar]
- 8.Sookoian S, Gemma C, Gianotti TF, Burgueño A, Castaño G, Pirola CJ. Genetic variants of Clock transcription factor are associated with individual susceptibility to obesity. Am J Clin Nutr 2008;87:1606–1615 [DOI] [PubMed] [Google Scholar]
- 9.Monteleone P, Tortorella A, Docimo L, et al. Investigation of 3111T/C polymorphism of the CLOCK gene in obese individuals with or without binge eating disorder: association with higher body mass index. Neurosci Lett 2008;435:30–33 [DOI] [PubMed] [Google Scholar]
- 10.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol 2010;72:517–549 [DOI] [PubMed] [Google Scholar]
- 11.Johnston JD, Frost G, Otway DT. Adipose tissue, adipocytes and the circadian timing system. Obes Rev 2009;10(Suppl. 2):52–60 [DOI] [PubMed] [Google Scholar]
- 12.Gimble JM, Floyd ZE. Fat circadian biology. J Appl Physiol 2009;107:1629–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zvonic S, Ptitsyn AA, Conrad SA, et al. Characterization of peripheral circadian clocks in adipose tissues. Diabetes 2006;55:962–970 [DOI] [PubMed] [Google Scholar]
- 14.Ptitsyn AA, Zvonic S, Conrad SA, Scott LK, Mynatt RL, Gimble JM. Circadian clocks are resounding in peripheral tissues. PLOS Comput Biol 2006;2:e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ando H, Yanagihara H, Hayashi Y, et al. Rhythmic messenger ribonucleic acid expression of clock genes and adipocytokines in mouse visceral adipose tissue. Endocrinology 2005;146:5631–5636 [DOI] [PubMed] [Google Scholar]
- 16.Kohsaka A, Laposky AD, Ramsey KM, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab 2007;6:414–421 [DOI] [PubMed] [Google Scholar]
- 17.Gómez-Santos C, Gómez-Abellán P, Madrid JA, et al. Circadian rhythm of clock genes in human adipose explants. Obesity (Silver Spring) 2009;17:1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gómez-Abellán P, Hernández-Morante JJ, Luján JA, Madrid JA, Garaulet M. Clock genes are implicated in the human metabolic syndrome. Int J Obes (Lond) 2008;32:121–128 [DOI] [PubMed] [Google Scholar]
- 19.Wu X, Xie H, Yu G, et al. Expression profile of mRNAs encoding core circadian regulatory proteins in human subcutaneous adipose tissue: correlation with age and body mass index. Int J Obes (Lond) 2009;33:971–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan S, Minihane AM, Talmud PJ, et al. Dietary long-chain n-3 PUFAs increase LPL gene expression in adipose tissue of subjects with an atherogenic lipoprotein phenotype. J Lipid Res 2002;43:979–985 [PubMed] [Google Scholar]
- 21.Otway DT, Frost G, Johnston JD. Circadian rhythmicity in murine pre-adipocyte and adipocyte cells. Chronobiol Int 2009;26:1340–1354 [DOI] [PubMed] [Google Scholar]
- 22.Loboda A, Kraft WK, Fine B, et al. Diurnal variation of the human adipose transcriptome and the link to metabolic disease. BMC Med Genomics 2009;2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimba S, Ishii N, Ohta Y, et al. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc Natl Acad Sci USA 2005;102:12071–12076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev 2006;20:1868–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ando H, Takamura T, Matsuzawa-Nagata N, et al. Clock gene expression in peripheral leucocytes of patients with type 2 diabetes. Diabetologia 2009;52:329–335 [DOI] [PubMed] [Google Scholar]