Abstract
Mus81 is a highly conserved endonuclease with homology to the XPF subunit of the XPF-ERCC1 complex. In yeast Mus81 associates with a second subunit, Eme1 or Mms4, which is essential for endonuclease activity in vitro and for in vivo function. Human Mus81 binds to a homolog of fission yeast Eme1 in vitro and in vivo. We show that recombinant Mus81-Eme1 cleaves replication forks, 3′ flap substrates, and Holliday junctions in vitro. By use of differentially tagged versions of Mus81 and Eme1, we find that Mus81 associates with Mus81 and that Eme1 associates with Eme1. Thus, complexes containing two or more Mus81-Eme1 units could function to coordinate substrate cleavage in vivo. Down-regulation of Mus81 by RNA interference reduces mitotic recombination in human somatic cells. The recombination defect is rescued by expression of a bacterial Holliday junction resolvase. These data provide direct evidence for a role of Mus81-Eme1 in mitotic recombination in higher eukaryotes and support the hypothesis that Mus81-Eme1 resolves Holliday junctions in vivo.
INTRODUCTION
Maintaining genomic integrity is an essential component of the process of cell duplication and division. Cells are especially vulnerable to DNA damage during the process of DNA replication. Replication forks that encounter relatively innocuous forms of damage such as a single strand break or a modified base can increase the damage by generating a double strand break or by fixing a mutation. To cope with the need to complete replication in the presence of DNA damage, organisms have evolved with a number of mechanisms for replicating through or around damaged sites in ways that cause minimal genome instability. One option involves the use of bypass or translesion polymerases that act at sites where the template has been damaged in such a way that it cannot be deciphered by the replicative polymerases (Friedberg et al., 2002). A second mechanism, involves regression of the fork and annealing of the nascent strands to form a Holliday junction structure (Higgins et al., 1976). The leading strand can thus be extended, using the nascent lagging strand as a template. Subsequent migration of the Holliday junction back toward the nascent end restores the replication fork, allowing replication to continue. Use of such a template-switching mechanism does not directly repair the damage, but it does allow replication to proceed downstream of the damaged site and thus circumvents the problems associated with incomplete replication. Stalled replication forks can also be repaired through a recombination repair process in which a Holliday junction formed by the regressed fork is resolved into an intact duplex and a free double strand end. The double strand end is repaired by homologous recombination, a process involving resection of one strand of the free end, invasion of the intact linear duplex, and the creation of new replication fork (Haber, 1999). The importance of homologous recombination in replication restart is well established in prokaryotes. Evidence that this is also an important pathway in eukaryotes is accumulating (Cox et al., 2000; Michel, 2000; McGlynn and Lloyd, 2002). Two activities that resolve Holliday junctions into linear duplex have been identified in human somatic cell extracts (Chen et al., 2001; Constantinou et al., 2002). One, known as Resolvase A, introduces symmetrical related nicks into strands of like polarity, and associates with branch migration activities (Constantinou et al., 2001, 2002). The identities of the proteins that comprise this activity are not known. The second human activity that resolves Holliday junctions is associated with Mus81 (Chen et al., 2001; Constantinou et al., 2002). Mus81-associated endonuclease cleaves Holliday junction structures into linear duplexes but does not cut at symmetrically related sites and the products are not ligatable in vitro (Chen et al., 2001). Mus81 is a component of a highly conserved endonuclease with homology to the XPF subunit of the XPF-ERCC1 complex (Boddy et al., 2000; Interthal and Heyer, 2000; Chen et al., 2001; Mullen et al., 2001). In yeast, Mus81 mutants are hypersensitive to DNA-damaging agents that impede DNA replication, and Mus81 mutation is lethal in combination with disruption of the RecQ like helicases Rqh1 or Sgs1 (Boddy et al., 2000; Mullen et al., 2001). In humans, mutations in the RecQ like helicases BLM and WRN are associated with genomic instability and increased incidence of cancer (Hickson, 2003). BLM and WRN have both been demonstrated to branch-migrate Holliday junctions in vitro (Constantinou et al., 2000; Karow et al., 2000). Thus, the genetic interactions and enzymatic properties of Mus81 and the RecQ helicases suggest that they act independently at stalled replication forks to promote replication restart.
Mus81 associates with a second protein, known as Eme1 in fission yeast and Mms4 in budding yeast. Eme1/Mms4 is essential for Mus81 endonuclease activity in vitro and for in vivo function (Boddy et al., 2001; Kaliraman et al., 2001; Mullen et al., 2001; Doe et al., 2002). Based on the phenotypes of fission yeast mutants of Mus81 and Eme1, i.e., sensitivity to agents that impede DNA replication, recombination-dependent failure to complete meiosis, and evidence that Mus81 containing complexes resolve Holliday junctions into linear duplex in vitro, we proposed that Mus81 and Eme1 are subunits of a nuclear Holliday junction resolvase (Boddy et al., 2001). A similar function was proposed for the human enzyme (Chen et al., 2001). On the other hand, in specific strains of budding yeast, Mus81 and Mms4 mutants do not have a profound meiotic recombination defect, suggesting that Mus81-Mms4 is not a major meiotic Holliday junction resolvase in budding yeast (De Los Santos et al., 2001, 2003). Furthermore, highly purified recombinant Mus81-Mms4 or Mus81-Eme1 and partially purified Mus81-complexes from human cells extracts have greater activity with 3′ flap and replication fork structures than with Holliday junctions (Kaliraman et al., 2001; Constantinou et al., 2002; Whitby et al., 2003). The preference Mus81 shows for these substrates led to the suggestion that replication forks or 3′ flaps, and not Holliday junctions, are the relevant in vivo substrate of Mus81 (Kaliraman et al., 2001; Constantinou et al., 2002; Doe et al., 2002; Bastin-Shanower et al., 2003).
To gain more insight into the function of Mus81 in human cells we have identified human homologs of Eme1 that bind Mus81 in vitro and in vivo. The sequence we designated Eme1A was independently identified and shown to interact with Mus81 by two other groups (Ciccia et al., 2003; Ogrunc and Sancar, 2003). We find that recombinant Mus81-Eme1 efficiently cleaves replication forks and 3′ flap substrates in vitro. Mus81-Eme1 generates a lower, but readily detectable, yield of linear duplex product from Holliday junction structures. Resolution of Holliday junctions requires coordinate cleavage on opposing strands of the junction. By use of differentially tagged versions of Mus81 and Eme1, we find that Mus81 associates with Mus81, and that Eme1 associates with Eme1. Thus, complexes containing two or more Mus81-Eme1 units could function to coordinate substrate cleavage in vivo. Down-regulation of Mus81 by RNA interference reduces mitotic recombination in human somatic cells. The recombination defect is rescued by expression of a bacterial Holliday junction resolvase. These data provide direct evidence for a role of Mus81-Eme1 in mitotic recombination in higher eukaryotes and support the hypothesis that Mus81-Eme1 resolves Holliday junctions in vivo.
MATERIALS AND METHODS
Cell Culture and Mitotic Recombination Assays
HeLa cells, 293 (human embryonic kidney cells), and GM847L22 (Prince et al., 2001) were grown in DMEM, supplemented with 10% enriched calf serum, 100 μg/ml penicillin and streptomycin. For routine culture, GM847L22 were maintained in presence of 400 μg/ml G418. Sf9 cells were grown in Excell-401 media (JRH Biosciences, Lenexa, KS) with 50 μg/ml penicillin and streptomycin. To assay mitotic recombination 5 × 105 cells were plated in G418 free medium 16 h before transfection. Thymidine (2 mM) was added to the culture medium, and cells were grown for 16 h. Cells were cultured in the normal growth medium for a further 24 h. Cells were fixed with 2% formaldehyde in phosphate-buffered saline (PBS) for 10 min, washed with PBS twice, and assayed for β-galactosidase activity by incubation in PBS containing 1 mg/ml X-Gal, 4 mM potassium ferrocyanide, 4 mM potassium ferricyanide, 2 mM MgCl2 at 37°C overnight (Herzing and Meyn, 1993). The number of blue cells was scored using 20× objective on an inverted light microscope. Statistical significances were calculated using a Student's t test.
Expression of Recombinant Proteins and RNA Interference (RNAi)
3HaMus81 wild-type and endonuclease dead (Chen et al., 2001) was cloned into pCDNA3 (Invitrogen, Carlsbad, CA) by using EcoRI and XhoI sites. Eme1 was FLAG tagged at the C′ terminus by using the oligonucleotide 5′GCCCGCTCGAGTCACTTGTCATCGTCGTCCTTGTAGTCAGCACTATCTAAAGA-3′ and inserted in pCDNA3 by using EcoRI and XhoI. Mus81 was FLAG tagged at the C′ terminus by using the oligonucleotide 5′-CTCGAGTCACTTGTCATCGTCGTCCTTGTAGTCGGTCAAGGGGCCGTAGC-3′. 3HaEme1 was made by cloning Eme1 into pcDNA-3Ha (Melchionna et al., 2000) by using NdeI and XhoI. Human cells were transfected using Effectene (QIAGEN, Valenica, CA) or FuGENE 6 (Roche Diagnostics, Indianapolis, IN)) according to the manufacturer's instructions. For expression in Sf9 cells, Gst-Mus81 (Chen et al., 2001) was cloned into pFastBac (Invitrogen) by using EcoRI and HindIII, and Eme1-FLAG was cloned using EcoRI and XhoI. The Bac-to-Bac system (Invitrogen) was used to generate recombinant viruses. All constructs were verified by sequencing. Two 19-nucleotide regions (corresponding to 178-197 and 292-311 of Mus81 open reading frame; Chen et al., 2001) were selected and cloned into pSuper and used as described in Brummelkamp et al., (2002). Polymerase chain reaction (PCR) was carried out on HeLa cell cDNA library (BD Biosciences Clonetech, Palo Alto, CA) by using sequences present in all three forms of Eme1 (5′-CGGAATTCACCATGGCTCTAAAGAAGTCATCACC-3′ and 5′-GCCCGCTCGAGTCAGTCAGCACTATCTAAAGAGAG-3′). The PCR products were cloned into pTopo (Invitrogen). Restriction enzyme analysis of six clones gave a pattern corresponding to Eme1B. Sequencing of two clones verified that the transcript corresponding to Eme1B is expressed in HeLa cells. NLSRusA-2Ha wild-type and inactive was cloned into pCDNA3 for expression in human cells by using pRep1-RusA and pRep1-RusA-D70N (Boddy et al., 2001) as starting constructs.
Gene Cloning
Three clones corresponding to the sequence AK0055926 were retrieved from American Type Culture Collection (Manassas, VA) collection of human expressed tagged sequences. ATCC clone 2899969 (coding for Eme1A) came from a cervical carcinoma library, ATCC clone 3905809 (coding for Eme1B) came form a uterine tumor library, and ATCC clone 814686 (coding for Eme1C) came form normal B-cell library. The three clones were sequenced directly. Where a difference between the clones was observed multiple reads from both directions was used to confirm specific nucleotides.
Nuclease Assays and Western Analysis
Nuclease assays were carried out as described previously (Chen et al., 2001; Constantinou et al., 2002). Antibody to the Ha-epitope was from Babco (Richmond, CA). Antibody to the FLAG-epitope (FLAG-M2) was from Sigma-Aldrich (St. Louis, MO). Antibody to Mus81 was described in Chen et al. (2001). Cells lysates, immune-precipitates, and immune-blots analysis was carried out as in Chen et al. (2001).
Immune-Fluorescence Microscopy
Conditions for growing XPA cells, localized UV-irradiation, and for developing immune-fluorescence images were as described in Gao et al. (2003).
RESULTS
Identification of a Human Homolog of Eme1
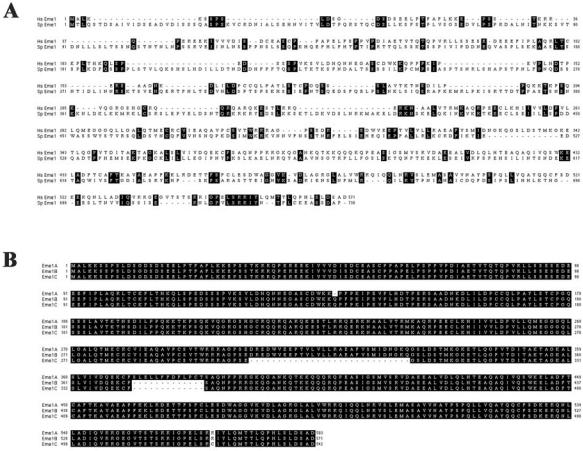
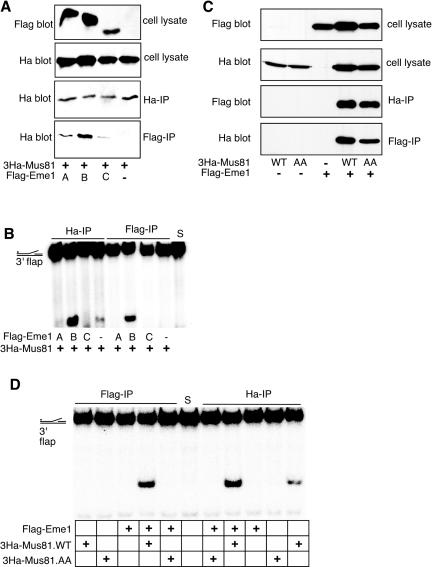
Reiterative PSI-BLAST searches (www.ncbi.nlm.nih.gov/blast/Blast) were used to identify mammalian homologs of Schizosaccharomyces pombe Eme1 (SpEme1). In the first iteration, an uncharacterized open reading frame (AL356173) from Neurospora crassa was identified as having significant similarity to spEme1. The same open reading frame has previously been noted to have homology to the Mus81 binding partner in budding yeast Mms4 (Mullen et al., 2001). Reiteration of the search identified several highly related, overlapping human sequences with significant similarity to SpEme1 (Figure 1A). Three clones corresponding to the sequence AK0055926 were retrieved from American Type Culture Collection and were sequenced to confirm their identity. Surprisingly, the three clones were found to differ slightly and were designated Eme1A, Eme1B, and Eme1C (Figure 1B). The identification of a single genomic locus (17q21.33) in the National Center for Biotechnology Information human genome database suggested that the three variants derive from a single gene. The absence of specific sequences (corresponding to amino acids 303-331 and 372-384 in Eme1A) suggested that the different forms might be due to incomplete splicing. However, the absence of a single codon (at position 138) in the longest sequence, and the substitution of an arginine for a cysteine (at position 525) in the shortest one, was not compatible with this interpretation. To determine which, if any, of the three forms of human Eme1 binds to Mus81, epitope-tagged constructs of all three variants were tested for the ability to interact with Mus81 and to support Mus81-associated endonuclease activity. HeLa cells were transiently transfected with 3HaMus81 and FLAG-tagged Eme1. As shown in Figure 2A, 3HaMus81 was detected in immune-precipitates of all three forms of Eme1. However, the amount of 3HaMus81 associated with FLAG-Eme1B was reproducibly higher than FLAG-Eme1A or C. Human Mus81 has robust activity against a 3′ flap substrate in vitro (Constantinou et al., 2002); therefore, Mus81 and Eme1 immune-complexes were assayed for associated endonuclease activity by using this substrate (Figure 2B). The activity of 3HaMus81 was greatly increased in samples that had been cotransfected with FLAG-Eme1B, but not with FLAG-Eme1A or FLAG-Eme1C. Likewise, when the different forms of Eme1 were immune-precipitated using the FLAG antibody, the B form had readily detectable activity. Longer exposures reveal endonuclease activity in both FLAG-Eme1A and FLAG-Eme1C immune-precipitates (our unpublished data); however, Eme1B-associated endonuclease activity was consistently higher.
Figure 1.
Identification of a human homolog of SpEme1. (A) A human cDNA encoding an open reading frame of 571 amino acids with similarity to spEme1 (Boddy et al., 2001) was identified by the PSI-BLAST algorithm. Alignment of Eme1 homologs was generated using the Clustal W program. Identities are shown in reverse. (B) Three sequence variants of human Eme1 were obtained from American Type Culture Collection. Eme1A (ATCC clone 2899969) encodes a protein of 583 amino acids, Eme1B (ATCC clone 3905809) encodes a protein of 571 amino acids, and Eme1C (ATCC clone 814686) encodes a protein of 542 amino acids.
Figure 2.
Eme1 interacts with Mus81 and has Mus81-dependent endonuclease activity. HeLa cells were transiently transfected with 3HaMus81 in the presence or absence of FLAG-Eme1. Forty-eight hours after transfection, lysates and immune-precipitates were probed for the presence of 3Ha-Mus81 and FLAG-Eme1. (A) 3HaMus81 was detected in FLAG immune-precipitates from cells that express FLAG-Eme1A, B, or C. (B) Ha and FLAG immune-precipitates were assayed for associated endonuclease activity by using a 3′ flap substrate. Coexpression of 3HaMus81 and FLAG-Eme1B resulted in highest activity. (C) FLAG-Eme1B was detected in Ha immune-precipitates from cells that express wild-type 3Ha-Mus81 (WT) and an endonuclease dead version of Mus81 (AA). 3HaMus81 (wild-type and an endonuclease dead version) were detected in a FLAG immune-precipitate from cells that express FLAG-Eme1B (D) FLAG-Eme1B immune-precipitates from cells that were cotransfected with wild-type but not endonuclease dead 3HaMus81 cleave 3′ flap structures. 3HaMus81WT immune-precipitates have associated endonuclease activity that was increased when cells are cotransfected with FLAG-Eme1. S, substrate alone. The black dot represents radioactive phosphate labeling of the substrate.
To determine which forms of Eme1 are expressed in HeLa cells, oligonucleotide primers common for all three variants were used to amplify sequences from a HeLa cell cDNA library. The products of PCR were cloned and restriction enzyme analysis of six clones gave a pattern corresponding to Eme1B. Sequencing of 2 clones verified that the transcript corresponding to Eme1B is expressed in HeLa cells. Although this analysis does not exclude the possibility that the A or C form of Eme1 is expressed in other cell types, or at low levels in HeLa cells, transcripts corresponding to the B form of the protein were readily detectable, therefore we conducted further analysis of Eme1 by using the B form of the protein.
We first asked whether the endonuclease activity detected in an Eme1 immune-precipitate was dependent on Mus81. As shown in Figure 2C, FLAG-Eme1 associated both with wild-type Mus81 and with a mutant version of Mus81 that lacks associated endonuclease activity (Chen et al., 2001). Endonuclease activity was detected in a FLAG-Eme1 immune-precipitate from cells that had been simultaneously transfected with wild-type 3HaMus81, but not in cells that had been transfected with FLAG-Eme1 alone (Figure 2D). Thus, Eme1 associated endonuclease activity is dependent on coexpression of wild-type Mus81. As reported previously (Chen et al., 2001), Ha-immune-precipitates from cells that had been transfected with 3HaMus81 had detectable endonuclease activity in the absence of transfected Eme1 (Figure 2D).
Eme1 Localizes in Nucleoli and to Regions of Damaged DNA
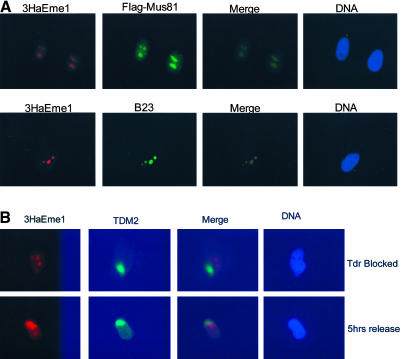
Recent studies of human Mus81 showed that it localizes to nucleoli and to regions of damaged DNA in S-phase cells (Gao et al., 2003). If Eme1 is acting in complex with Mus81, it should show the same distinctive pattern of subcellular localization. This was tested using indirect-immune-fluorescence of cells that had been transfected with vectors encoding 3HaEme1 and FLAG-Mus81. As previously seen, Mus81 was retained in the nucleoli of cells that were subjected to in situ extraction before fixation (Gao et al., 2003). 3HaEme1 was found to exactly colocalize with Mus81 in two to three regions of the nucleus (Figure 3A). Costaining of 3HaEme1 and the nucleolar marker nucleophosmin/B23 confirmed that the Eme1 is retained within nucleoli (Figure 3A). To determine whether Eme1 is recruited to regions of damaged DNA in a cell cycle-dependent manner, cells were synchronized at the G1/S boundary by double-thymidine block. Arrested cells, or cells that had been released for 5 h, were UV-irradiated through polycarbonate filters containing 10-μm-diameter pores and 15 min later, samples were fixed and stained for regions of damage and for Eme1. To prevent repair of UV-photoproducts by NER, simian virus 40 (SV40)-transformed fibroblasts with a defect in XPA were used in this analysis (Mone et al., 2001). When thymidine-arrested cells were UV irradiated, regions of damage were clearly visualized (Figure 3B) by using the cyclobutane pyrimidine dimer-specific monoclonal antibody TDM2 (Mizuno et al., 1991; Mori et al., 1991). 3HaEme1 was found in the nucleoli of thymidine-arrested cells, but there was no evidence that Eme1 was recruited to regions of damaged DNA (1 of 25 cases). By contrast, 5 h after thymidine release when the majority of cells were replicating DNA, Eme1 was found both in nucleoli and within regions of damage (25 of 30 cases). Thus, the pattern of Eme1 localization in damaged and undamaged cells is identical to that recently reported for Mus81 (Gao et al., 2003).
Figure 3.
Eme1 colocalizes with Mus81 in nucleoli and is recruited to regions of damage in S-phase cells. HeLa cells that were transfected with vectors encoding 3HaEme1 and FLAG-Mus81 were grown on coverslips for 24 h and subjected to in situ extraction before fixation. (A) Samples were processed for immune-fluorescence by using antibodies to the Ha epitope (red), to the FLAG epitope (green) or nucleophosmin/B23 (green). Samples were costained with 4,6-diamidino-2-phenylindole to reveal DNA. (B) XPA cells were transfected with 3HaEme1 and FLAG-Mus81, grown on coverslips, and synchronized at the G1/S boundary by double-thymidine (Tdr) block. A UV-opaque filter containing pores of ∼10 μm in diameter was placed over cells that were exposed to 60 mJm-2 254-nm light and then cultured for 15 min before in situ fraction and fixation. Cells were stained for Eme1 (red) and with TDM-2 (green) to detect the regions of UV-induced cyclobutane pyrimidine dimer. Top, typical cell that was in thymidine at the time of irradiation. Bottom, typical cell 5 h after release from the thymidine block.
Activity of Recombinant Mus81-Eme1
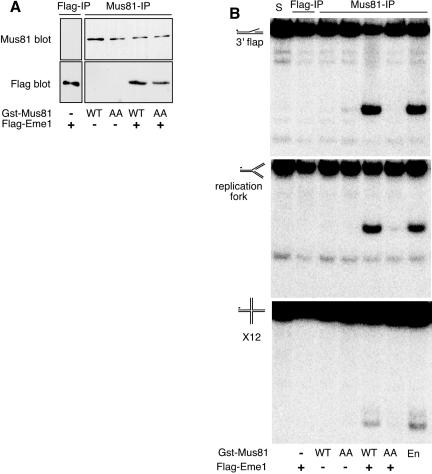
Eme1 is essential for Mus81 activity and function in fission yeast (Boddy et al., 2001). Likewise, Mms4 is essential for Mus81 activity and function in budding yeast (Kaliraman et al., 2001; Mullen et al., 2001). To test whether Eme1 is essential for the activity of human Mus81, insect cells were infected with baculoviruses encoding Gst-Mus81, FLAG-Eme1, or both (Figure 4A). Immune-precipitated Gst-Mus81 and FLAG-Eme1 were assayed using a 3′ flap, a replication fork, or a Holliday junction structure (X12). Gst-Mus81 alone had no detectable activity on any of these substrates. Likewise, immune-precipitated FLAG-Eme1B had no detectable endonuclease activity (Figure 4B). In contrast, when Gst-Mus81 and FLAG-Eme1 were coexpressed immune-precipitates of Mus81 readily cleaved the 3′ flap and replication fork structures. A lower but detectable and specific activity was also observed using the X12 substrate (Figure 4B). Thus, the endonuclease activity of Mus81 depends on Eme1 and vice versa. A similar pattern of activity was seen in immune-precipitates of FLAG-Eme1 and after purification of recombinant Mus81-Eme1 by GSH-Sepharose chromatography (our unpublished data). The data obtained by immune-precipitation of recombinant Mus81-Eme1 is shown because it allows direct comparison with the activity of immune-precipitated endogenous Mus81. Like endogenous Mus81, recombinant Mus81-Eme1 has a lower activity against a Holliday junction substrate. As was previously seen for endogenous and transfected Mus81, the reaction product was linear duplex (Chen et al., 2001). No cleavage was detected when a mutant form of Mus81 (Mus81AA) was used.
Figure 4.
Endonuclease activity of recombinant Mus81-Eme1. Gst-Mus81 or FLAG-Eme1B was immune precipitated from Sf9 cells infected with viruses encoding the indicated proteins. (A) Mus81 immune-precipitates were probed for the presence of Gst-Mus81 and FLAG-Eme1B. (B) Recombinant Mus81-Eme1 cleaves 3′ flap, replication fork, and Holliday junction (X12) structures in vitro. The activity associated with Mus81 immune-precipitates from HeLa cells is shown for comparison (En). S, substrate alone. The black dot represents radioactive phosphate labeling of the substrates.
Association of Mus81-Eme1 Catalytic Units
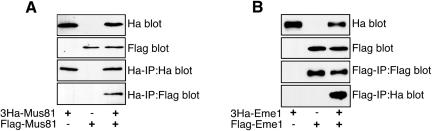
The generation of linear duplex products requires coordinate cleavage of two opposing strands of the X12 substrate and implies that the enzyme exist in complexes containing two catalytic units. To determine whether Mus81-Eme1 exists in the higher order complexes needed to coordinate cleavage of Holliday junctions, human cells were simultaneously transfected with plasmids encoding two different forms of Mus81, 3HaMus81, and FLAG-Mus81. As shown in Figure 5A, FLAG-Mus81 was detected in an immune-precipitate of 3HaMus81. Control samples in which cells were transfected with one construct show that there is no cross-reactivity between the immune-precipitating antibody. Likewise, when 3HaEme1 was cotransfected with FLAG-Eme1, 3HaEme1 was detected in immune-precipitates of FLAG-Eme1 (Figure 5B). This analysis does not distinguish the number of Mus81, or Eme1, molecules that coprecipitate with each other; however, it does demonstrate that at least two molecules of Mus81, and of Eme1, associate in vivo.
Figure 5.
Mus81 and Eme1 self-associate. 293 cells were transfected with 3HaMus81, FLAG-Mus81, or both. Forty hours after transfection, lysates and Ha immune-precipitates were probed for the presence of 3HaMus81 and FLAGMus81. (A) FLAG-Mus81 was detected in Ha immune-precipitates from cells that express 3HaMus81. (B) 3Ha-Eme1 was detected in FLAG immune-precipitates from cells that express FLAG-Eme1.
RNA Interference with Mus81 Expression Reduces Viability
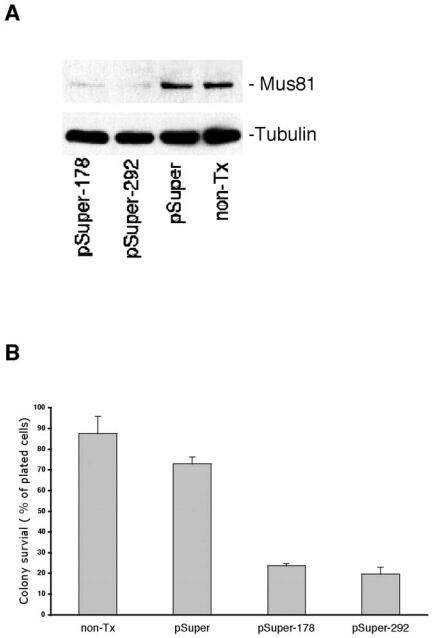
The role of Mus81 in human cells was investigated using RNAi to suppress Mus81 expression (Brummelkamp et al., 2002). As shown in Figure 6A, Mus81 protein levels were substantially reduced in cells that were transfected with pSuper vectors containing 19-nucleotide sequences that target two regions of Mus81 mRNA (pSuper-178 and pSuper-292) (Brummelkamp et al., 2002). No loss of Mus81 was seen in cells transfected with control vector (pSuper). The effect Mus81-RNAi had on cell viability was determined by assaying colony survival. As shown in Figure 6B, suppression of Mus81 expression was associated with a significant reduction in the ability of cells to form viable colonies. Loss of viability seemed to be specifically associated with reduced Mus81 expression because it was seen after transfection with two different constructs that cause reduced Mus81 expression and was not seen after transfection of control RNAi vectors. Flow cytometric analysis did not reveal a significant change in the cell cycle distribution 48 or 72 h after transfection with pSuper-178 or pSuper-292. Experiments to determine the specific cause of cell death have not been informative.
Figure 6.
Suppression of Mus81 by RNAi results in reduced viability. (A) Transfection with pSuper-178, pSuper-292, but not empty pSuper results in reduced Mus81 protein. NonTx, untransfected cells. (B) HeLa cells were transfected with pSuper-178, pSuper-292, or pSuper and plated at low density. Colonies were counted after 10 d of growth and staining with Geimsa. Triplicate plates were counted for each point. Results are the average of three independent transfections.
RNAi Suppression of Mus81 Reduces Mitotic Recombination
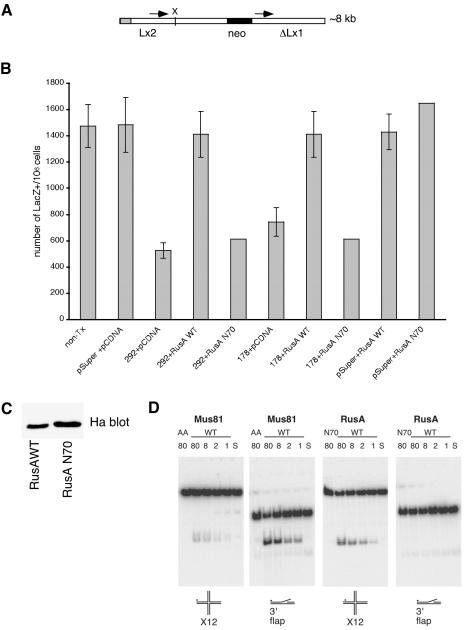
A Mus81 meiotic defect that depends on recombination has been reported both in fission yeast and budding yeast (Interthal and Heyer, 2000; Boddy et al., 2001; Kaliraman et al., 2001). To determine whether Mus81 is needed for mitotic recombination in human cells, we took advantage of an SV40-transformed human fibroblast line, GM847L22 (Prince et al., 2001), which contains a single integrated copy of the mitotic recombination reporter plasmid pLrec (Herzing and Meyn, 1993; Meyn, 1993). A schematic of the Lrec cassette is shown in Figure 7A. It contains two direct repeats of genetically inactive β-galactosidase (LacZ) genes and can give rise to LacZ+ cells by gene conversion, unequal sister chromatid exchange, or by intrachromosomal recombination (Herzing and Meyn, 1993; Meyn, 1993; Prince et al., 2001). This system has previously been used to demonstrate that cells from ataxia telangiectasia patients have increased mitotic recombination rates (Meyn, 1993) and that loss of the Werner syndrome protein (WRN) is associated with decreased productive mitotic recombination (Prince et al., 2001; Saintigny et al., 2002). A feature of this reporter is that β-galactosidase activity can be scored directly in single cells, thus it is compatible with transient down-regulation through use of RNAi. Furthermore, it does not depend on long-term viability of the culture. After transfection with plasmids that suppress Mus81 expression (pSuper-178, pSuper-292) or control plasmids (pSuper or pCDNA), GM847L22 cells were grown in the presence of thymidine to increase the incidence of recombination (Lundin et al., 2002). After a further 24 h growth in normal medium, cells were stained for β-galactosidase activity and the frequency of recombination was scored. Untransfected cultures generated 1470 ± 180 recombinants per million cells (Figure 7B). A similar number of LacZ+ cells were seen after transfection of control vectors. The number of recombinants was reduced fourfold (p = 0.0003) and twofold (p = 0.0006) in cells that had been transfected with the Mus81-RNAi plasmids, pSuper-292, and pSuper-178, respectively. These data suggested that suppression of Mus81 reduces mitotic recombination but could also indicate that Mus81-RNAi interfered with β-galactosidase expression. To determine whether Mus81-RNAi-transfected cells were capable of expressing active β-galactosidase, cells were transfected with a plasmid carrying a single intact copy of the β-galactosidase and with pSuper, pSuper-178, or pSuper-292 plasmids. Cells were processed and stained for β-galactosidase activity exactly as described above. A similar percentage (84 ± 2%) of β-galactosidase positive cells was present in all cases. Therefore, we interpret these data to indicate that down-regulation of Mus81 suppresses recombination between the two inactive LacZ alleles rather than expression of β-galactosidase activity per se.
Figure 7.
Suppression of Mus81 results in reduced mitotic recombination and is rescued by expression of a bacterial Holliday junction resolvase, RusA. (A) pLrec contains a direct repeat of two inactive LacZ genes separated by the neomycin resistance gene (black box). Expression is under the control of the SV40 promoter (gray box). Base pairs (693) of identical sequence in the two LacZ alleles are indicated by arrows. Lx2 is inactive due to an insertion at a site indicated by X. ΔLx1 lacks 727 base pairs of 5′ sequence (Herzing and Meyn, 1993). The cell-line GM847L22 contains a single intact copy of pLrec (Prince et al., 2001). (B) Incidence of LacZ+ cells. Cells (5 × 105) were plated in G418-free medium 16 h before transfection with the indicated plasmid. The amount of DNA transfected was kept constant by use of empty vector. Thymidine (2 mM) was added to the culture medium and cells were grown for 16 h. Cells were cultured in normal growth medium for a further 24 h, before staining for β-galactosidase activity. Duplicate dishes were used to monitor cell number and expression of Mus81 and RusA. Error bars represent data from four separate experiments. (C) Ha immune-blots of cell lysates indicate that the active and mutant forms of RusA were expressed at comparable levels. (D) Immune-precipitates of wild-type RusA were active on X12 structures but not 3′ flaps structures in vitro. The 80, 8, 2, and 1 indicate the relative proportion of the enzyme added to a constant amount of substrate. Mutant versions of RusA, RusAN70A, or Mus81-Eme1, Mus81AA, did not cleave either structure. Wild-type Mus81-Eme1 converted both X12 and 3′ flap structures into linear duplex products. Quantification of product yield was calculated using Image Quant (Amersham Biosciences, Piscataway, NJ) software. S, substrate alone.
RusA Rescues Mus81-dependent Recombination
RusA rescues the meiotic defect and hypersensitivity to agents that cause replication fork stalling of Mus81 mutants (Boddy et al., 2001; Doe et al., 2002). We reasoned that if suppression of Mus81 in human cells results in the accumulation of Holliday junctions, the reduction in recombination would be rescued by expression of active RusA (Bolt and Lloyd, 2002). As shown in Figure 7B, expression of wild-type Ha-tagged RusA did not significantly affect the incidence of recombination in cells that were transfected with empty vector (p = 0.65), suggesting that at this level of expression, RusA does not drive increased recombination in human cells. In contrast, when active, nuclear RusA (RusAWT) was cotransfected with plasmids encoding Mus81-RNAi it increased the incidence of recombination to the levels seen in untransfected control cultures (Figure 7B). An endonuclease dead version of RusA (RusAN70) did not significantly increase the number of recombinants in Mus81-RNAi transfected cells. Immune blotting showed that the wild-type and mutant forms of RusA were equally expressed (Figure 7C). Ha-immune-precipitates and endonuclease activity assays confirmed that wild-type, but not mutant RusA, was active on a Holliday junction (X12) substrate (Figure 7D). The observation that RusA rescues Mus81-dependent recombination suggests that the two enzymes might be acting on a common substrate in vivo. We therefore tested the ability of RusA and Mus81-Eme1 to cleave an X12 or 3′ flap structure in vitro. Both enzymes were recovered from HeLa cells by using the Ha-antibody. As shown in Figure 7D, Mus81-Eme1 had readily detectable activity on both substrates. Mus81-Eme1 showed 10- to 12-fold more activity on the 3′ flap compared with the X12 across the range of enzyme amount used. Likewise, RusA cleaved the X12 structure, even after a 80-fold dilution of the immune-precipitate RusA detectably cleaved the X12 structure. By contrast, incubating equal amounts of RusA with a 3′ flap structure yielded no detectable product. Even with detector sensitivity set 10-fold higher no product was detected in the samples in which RusA was incubated with a 3′ flap structure. This analysis suggests that if RusA cleaves 3′ flaps in vitro, it does so at <1000-fold the efficiency it acts on the X12 structure.
DISCUSSION
To better understand the function of Mus81 in human cells, we set out to identify a human homolog of the Mus81 binding partner Eme1 or Mms4. A human sequence with limited homology to fission yeast Eme1 was identified by PSI-BLAST. While this work was in progress, two groups reported identification of the sequence that we refer to as Eme1A. Ogrunc and Sancar (2003) designated this protein HsMms4 and showed that it binds to Mus81 in vivo and that immune complexes of transfected Mus81-Mms4 (Eme1A) have endonuclease activity. Ciccia et al. (2003) also identified Eme1A, and by use of bacterial expression they showed that the two proteins associate to form an endonuclease complex. Our analysis of the in vitro substrate specificity of recombinant Mus81-Eme1 closely parallels that of Ciccia et al. (2003) and indicates that Mus81-Eme1 cleaves 3′ flaps more efficiently than Holliday junction structures.
In this study, three variants of Eme1 were retrieved from the human expressed sequence tag collection at American Type Culture Collection. Because the sequences included both potential polymorphic variations and splice variants, all three were tested for the ability to interact with Mus81 and to support Mus81-associated endonuclease activity. Even though the same promoter drove expression, Eme1B was present at slightly higher levels than the other two proteins. We found that the three variants associate with and activate Mus81 to differing degrees. The higher endonuclease activity in Eme1B containing immune-precipitates seemed to result mainly from increased association between 3HaMus81, but it is also possible that Eme1B stimulated Mus81 activity more than Eme1A or Eme1C. The 13-amino acid sequence insert in Eme1A is not present in a homologous murine sequence (accession no. CB182498). Furthermore, the presence of consensus splice donor and acceptor sites flanking this insert suggest that the construct corresponding to Eme1A was the result of incomplete splicing. Nevertheless, Eme1A can bind to and activate Mus81 (Ciccia et al., 2003; Ogrunc and Sancar, 2003). The likely source of Eme1C is more difficult to predict. The clone was derived from normal tissue. Relative to the other two sequences, it lacks residues 303-331 and contains a cysteine in place of arginine in the carboxy terminus of the protein. This substitution is particularly intriguing as it is present within a block of significant homology with fission yeast Eme1 (Figure 1A). Determining whether the three variants of Eme1 are differentially expressed, or whether the single site substitutions are polymorphic variants in the human populations, and whether they are associated with human disease will be of interest.
As reported previously, we found that immune-precipitates of 3HaMus81 have associated endonuclease activity in the absence of transfected Eme1 (Chen et al., 2001). By contrast, immune-precipitates of Eme1 were not active unless Mus81 was cotransfected. Given that Mus81 expressed in Sf9 cells is not active in the absence of Eme1, we assume the activity in a Mus81-immune-precipitate reflects the ability of Mus81 to associate with endogenous human Eme1. This idea is supported by the substantial increase in Mus81-associated endonuclease activity detected on transfection of Eme1. The reason transfected Eme1 fails to form an active complex with endogenous Mus81 is not clear. Transfection of Eme1A or C leads to a reproducible reduction in the activity associated with endogenous Mus81 (Figure 2B), suggesting that transfected Eme1A or C can disrupt the endogenous Mus81 containing complex.
The hypothesis that Mus81 is a component of a Holliday junction resolvase in vivo has been challenged on the basis of its ability to preferentially cleave branched Y-structures and 3′ flaps in vitro. Ciccia et al. (2003) showed that recombinant human Mus81-Eme1 has ∼12-fold less activity when assayed using a mobile Holliday junction (X26) structure compared with a 3′ flap or a replication fork. Cleavage of a fixed Holliday junction (X0) was ∼75-fold less than a 3′ flap (Ciccia et al., 2003). Our analysis of recombinant Mus81-Eme1 is in agreement with this and with previous studies in showing that Mus81-Eme1 preferentially cleaves 3′ flaps and replication forks in vitro (Kaliraman et al., 2001; Constantinou et al., 2002; Bastin-Shanower et al., 2003; Whitby et al., 2003). However, we contend that the ability to cleave other branched structures in vitro does not disqualify the Mus81-Eme1 complex from resolving Holliday junctions in vivo. The data presented here and that presented in Ciccia et al. (2003) clearly demonstrate that recombinant human Mus81-Eme1 is capable of resolving Holliday junction into linear duplexes in vitro. Mus81-Eme1 does not behave like the prokaryotic Holliday junction resolvase RuvC, either by displaying strong sequence selectivity, nor by catalyzing symmetrical cleavage of junctions (Chen et al., 2001; Constantinou et al., 2002). Nevertheless, Mus81-Eme1 is capable of catalyzing the biologically critical process of converting a four-way DNA junction into two linear duplexes in vitro, and thus it might do so in vivo. Partially purified preparations of human Mus81 have robust Holliday junction activity that is dependent on the Mus81 protein (e.g., Figure 2 of Constantinou et al., 2001). Comparison of this activity with the relatively weak activity of recombinant Mus81-Eme1 (this work; Ciccia et al., 2003), suggests that cofactors may improve the Holliday junction resolution activity of Mus81-Eme1 in extracts (Boddy et al., 2001). The identification of human Eme1 should facilitate characterization of ancillary factors that enhance Mus81-Eme1 function.
Mus81 and Eme1 colocalize in the nucleoli of human cells and both proteins are recruited to regions of damaged DNA, specifically in S phase (Gao et al., 2003; Figure 3). The recruitment of Mus81 and Eme1 to localized region of UV damage, specifically in S-phase cells, is consistent with the hypothesis that the Mus81-Eme1 complex functions in recombination repair in human cells.
Sequence and structural analysis of junction resolving enzymes from different sources suggests that they have evolved independently from several different ancestors (Aravind et al., 2000; Lilley and White, 2001). Despite these differences, all characterized Holliday junction resolving enzymes bind junction DNA in dimeric form (Lilley and White, 2001). The association of two catalytic units enables paired cleavage of opposed strands on the four-way junction. By use of differentially tagged version of Mus81 and Eme1, we find that at least two molecules of Mus81 and of Eme1 associate. Further analysis will be needed to determine whether the ability of Mus81-Eme1 to resolve Holliday junctions into linear duplex is dependent on the correct coordination of two active Mus81-Eme1 heterodimers in vivo.
The mitotic phenotypes associated with loss of Mus81-function in fission yeast and budding yeast are very similar. Disruption of Mus81 results in increased sensitivity to genotoxic agents that impede DNA replication and Mus81 mutation is lethal in combination with disruption of the RecQ homologs. Nevertheless, both yeast are viable in the absence of Mus81 (Boddy et al., 2000; Interthal and Heyer, 2000). As a first step in directly determining the role of Mus81-Eme1 in human cells, we have used RNAi to suppress expression of Mus81. This analysis has allowed us to draw two conclusions about the function of Mus81 in human cells. The first is that down-regulation of Mus81 results in a significant loss of cell viability. Suppression of Mus81 protein expression was maximal between 48-72 h after transfection; however, at this time no distinct morphological or cell cycle defect could be determined. At later time points, the number of detached, dead cells in culture increased in line with the reduced number of Mus81-RNAi-treated cells that were able to form viable colonies. Analysis of Mus81-RNAi-transfected cultures failed to define a clear cause of cell death. Mus81-Eme1 is required to repair problems arising during DNA replication in yeast. In humans, Mus81-Eme1 is recruited to sites of DNA damage, specifically in cells that are replicating DNA (Gao et al., 2003). It is possible that replication problems arise frequently in human cells, and that Mus81 is essential even in absence of extraneous replicational stress. It will be of interest to determine whether germ-line disruption of Mus81 or Eme1 has the same effect as acute loss in somatic cells.
A second finding of these studies is that inhibition of Mus81 results in reduced mitotic recombination in human cells. The possibility that the reduced recombination rate was an indirect consequence of the loss in viability associated with Mus81-RNAi was considered. We found that Mus81-RNAi-treated cells expressed active β-galactosidase at the same frequency and at the same level as control cells when a single intact copy of the LacZ open-reading frame was present on a plasmid. The rate of recombination was calculated as the incidence of Lac+ cells per million live cells at the time of harvest; therefore, Mus81-RNAi cells that were lost before this point were not included in the analysis. By the criteria of trypan blue exclusion, cell cycle profile and β-galactosidase expression the remaining cells were viable and capable of expressing active β-galactosidase at the time of analysis. It will be important to determine whether Mus81-deficient cells die because they are unable to process intermediates that arise specifically during DNA replication. Mus81 protein is not detectable in quiescent cells. The increase in abundance seen on serum stimulation and reentry into replicative cycle suggest that Mus81 is required specifically in cells that are actively dividing (Gao et al., 2003).
Evidence that Mus81-Eme1 is active on 3′ flap and replication fork structures as well as Holliday junctions in vitro predict a number of possible roles for Mus81-Eme1 in recombination repair. A role in directly cleaving replication forks has been proposed (Kaliraman et al., 2001; Constantinou et al., 2002; Doe et al., 2002). However, two recently emerged lines of evidence suggest that Mus81 does not act directly on replication forks in vivo. First, the extreme sensitivity of yeast strains that lack Mus81 to camptothecin, a topoisomerase inhibitor that is thought to cause fork collapse, strongly suggests that Mus81 is required after replication fork collapse (Doe et al., 2002; Bastin-Shanower et al., 2003) and is not consistent with the hypothesis that Mus81 activity is required to cleave stalled forks. Second, mutations in genes that act early in recombination (Rad51, Rad52, and Rad54) suppress the synthetic lethality of Mus81-Sgs1 strains (Fabre et al., 2002; Bastin-Shanower et al., 2003). If Mus81-Eme1 acts directly on replication forks its growth defects would not be rescued by disruption of these genes.
Does the observation that expression of RusA rescues the Mus81-dependent recombination defect differentiate the role of Mus81 in the remaining pathways? The observation that Mus81-Eme1 efficiently cleaves 3′ flaps in vitro suggests a role in trimming flaps that might arise after extension of a 3′ end during the process of synthesis-dependent strand annealing (SDSA), in which a strand of the sister chromatid is used as a template for extension of a free 3′ end (Kaliraman et al., 2001; Constantinou et al., 2002; Bastin-Shanower et al., 2003). SDSA is an attractive model for double strand break repair. Because it can be accomplished without forming a Holliday junction, it could account for mitotic recombination without crossover (Paques and Haber, 1999). However, models of SDSA are based on the presence of two double strand ends generated by a break. In the case of replication restart, cleavage of the regressed fork would generate only one double strand end. Thus, replication fork recapture inevitably results in the formation of at least one Holliday junction (McGlynn and Lloyd, 2002). If Mus81 deficiency resulted in a failure to cleave a 3′ flap generated by SDSA, it would not lead to Holliday junction accumulation, and thus one would not expect expression of a Holliday junction resolvase to rescue a defect in SDSA. The ability of RusA to rescue Mus81-dependent recombination suggests that Holliday junctions accumulate in Mus81-deficient cells. Clearly, this interpretation is dependent on knowing the specificity of RusA. The ability of RusA to act alone and to retain activity when expressed fused to nuclear localization signals in yeast and human cells has been exploited in several studies (Doe et al., 2000; Boddy et al., 2001; Doe et al., 2002; Saintigny et al., 2002; Bastin-Shanower et al., 2003; Odagiri et al., 2003). Extensive analysis of RusA has shown that its nuclease activity is highly specific for Holliday junctions and that it is unlikely to cleave other structures in vivo (Bolt and Lloyd, 2002; Doe et al., 2002). Consistent with these reports, we find that HeLa cell expressed RusA did not detectably cleave a 3′ flap structures, whereas the protein had robust activity on an X12 structure. The data suggest that even when expressed in a heterologous system, RusA retains its preference for symmetrical structures. Although we cannot know for certain all the targets of RusA in vivo, the available data strongly support the hypothesis that it acts only on Holliday junctions. Thus, the rescue of Mus81-dependent recombination by a Holliday junction resolvase, coupled with biochemical evidence that Mus81-Eme1 resolves Holliday junction in vitro, suggests that Mus81-Eme1 resolves Holliday junctions in human cells.
Acknowledgments
We are grateful to Ray Monnat for providing GM847L22 and advice on use of the cells, to Reuven Agami for providing pSuper, and to members of the Scripps Cell Cycle group for encouragement and advice. P.-H.L.G. and M.N.B. are Research Special Fellows of the Leukemia and Lymphoma Society. This work was funded by a National Cancer Institute grant awarded to C.H.M.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-08-0580. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-08-0580.
References
- Aravind, L., Makarova, K.S., and Koonin, E.V. (2000). Survey and summary: Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 28, 3417-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin-Shanower, S.A., Fricke, W.M., Mullen, J.R., and Brill, S.J. (2003). The mechanism of mus81-mms4 cleavage site selection distinguishes it from the homologous endonuclease rad1-rad10. Mol. Cell Biol. 23, 3487-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy, M.N., Gaillard, P.H., McDonald, W.H., Shanahan, P., Yates, J.R., and Russell, P. (2001). Mus81-eme1 are essential components of a Holliday junction resolvase. Cell 107, 537-548. [DOI] [PubMed] [Google Scholar]
- Boddy, M.N., Lopez-Girona, A., Shanahan, P., Interthal, H., Heyer, W.D., and Russell, P. (2000). Damage tolerance protein mus81 associates with the FHA1 domain of checkpoint kinase cds1. Mol. Cell Biol. 20, 8758-8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolt, E.L., and Lloyd, R.G. (2002). Substrate specificity of RusA resolvase reveals the DNA structures targeted by RuvAB and RecG in vivo. Mol. Cell 10, 187-198. [DOI] [PubMed] [Google Scholar]
- Brummelkamp, T.R., Bernards, R., and Agami, R. (2002). A system for stable expression of short interfering RNAs in mammalian cells. Science 296, 550-553. [DOI] [PubMed] [Google Scholar]
- Chen, X.B., Melchionna, R., Denis, C.M., Gaillard, P.H., Blasina, A., Van de Weyer, I., Boddy, M.N., Russell, P., Vialard, J., and McGowan, C.H. (2001). Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell 8, 1117-1127. [DOI] [PubMed] [Google Scholar]
- Ciccia, A., Constantinou, A., and West, S.C. (2003). Identification and characterization of the human Mus81/Eme1 endonuclease. J. Biol. Chem. 278, 25172-25178. [DOI] [PubMed] [Google Scholar]
- Constantinou, A., Chen, X.B., McGowan, C.H., and West, S.C. (2002). Holliday junction resolution in human cells: two junction endonucleases with distinct substrate specificities. EMBO J. 21, 5577-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinou, A., Davies, A.A., and West, S.C. (2001). Branch migration and Holliday junction resolution catalyzed by activities from mammalian cells. Cell 104, 259-268. [DOI] [PubMed] [Google Scholar]
- Constantinou, A., Tarsounas, M., Karow, J.K., Brosh, R.M., Bohr, V.A., Hickson, I.D., and West, S.C. (2000). Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 1, 80-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, M.M., Goodman, M.F., Kreuzer, K.N., Sherratt, D.J., Sandler, S.J., and Marians, K.J. (2000). The importance of repairing stalled replication forks. Nature 404, 37-41. [DOI] [PubMed] [Google Scholar]
- De Los Santos, T., Hunter, N., Lee, C., Larkin, B., Loidl, J., and Hollingsworth, N.M. (2003). The mus81/mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164, 81-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Los Santos, T., Loidl, J., Larkin, B., and Hollingsworth, N.M. (2001). A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics 159, 1511-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe, C.L., Ahn, J.S., Dixon, J., and Whitby, M.C. (2002). Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 277, 32753-32759. [DOI] [PubMed] [Google Scholar]
- Doe, C.L., Dixon, J., Osman, F., and Whitby, M.C. (2000). Partial suppression of the fission yeast rqh1(-) phenotype by expression of a bacterial Holliday junction resolvase. EMBO J. 19, 2751-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre, F., Chan, A., Heyer, W.D., and Gangloff, S. (2002). Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. USA 99, 16887-16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg, E.C., Wagner, R., and Radman, M. (2002). Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science 296, 1627-1630. [DOI] [PubMed] [Google Scholar]
- Gao, H., Chen, X.-B., and McGowan, C.H. (2003). Mus81 endonuclease localizes to nucleoli and to regions of DNA damage in human S-phase cells. Mol. Biol. Cell 14, 4826-4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber, J.E. (1999). DNA recombination: the replication connection. Trends Biochem. Sci. 24, 271-275. [DOI] [PubMed] [Google Scholar]
- Herzing, L.B., and Meyn, M.S. (1993). Novel lacZ-based recombination vectors for mammalian cells. Gene 137, 163-169. [DOI] [PubMed] [Google Scholar]
- Hickson, I.D. (2003). RecQ helicases: caretakers of the genome. Nat. Rev. Cancer 3, 169-178. [DOI] [PubMed] [Google Scholar]
- Higgins, N.P., Kato, K., and Strauss, B. (1976). A model for replication repair in mammalian cells. J. Mol. Biol. 101, 417-425. [DOI] [PubMed] [Google Scholar]
- Interthal, H., and Heyer, W.D. (2000). MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet. 263, 812-827. [DOI] [PubMed] [Google Scholar]
- Kaliraman, V., Mullen, J.R., Fricke, W.M., Bastin-Shanower, S.A., and Brill, S.J. (2001). Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 15, 2730-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow, J.K., Constantinou, A., Li, J.L., West, S.C., and Hickson, I.D. (2000). The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl. Acad. Sci. USA 97, 6504-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley, D., and White, M. (2001). The junction-resolving enzymes. Nat. Rev. Mol. Cell Biol. 2, 433-443. [DOI] [PubMed] [Google Scholar]
- Lundin, C., Erixon, K., Arnaudeau, C., Schultz, N., Jenssen, D., Meuth, M., and Helleday, T. (2002). Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol. Cell Biol. 22, 5869-5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn, P., and Lloyd, R.G. (2002). Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell Biol. 3, 859-870. [DOI] [PubMed] [Google Scholar]
- Melchionna, R., Chen, X.B., Blasina, A., and McGowan, C.H. (2000). Threonine 68 is required for radiation-induced phosphorylation and activation of cds1. Nat. Cell Biol. 2, 762-765. [DOI] [PubMed] [Google Scholar]
- Meyn, M.S. (1993). High spontaneous intrachromosomal recombination rates in ataxia-telangiectasia. Science 260, 1327-1330. [DOI] [PubMed] [Google Scholar]
- Michel, B. (2000). Replication fork arrest and DNA recombination. Trends Biochem. Sci. 25, 173-178. [DOI] [PubMed] [Google Scholar]
- Mizuno, T., Matsunaga, T., Ihara, M., and Nikaido, O. (1991). Establishment of a monoclonal antibody recognizing cyclobutane-type thymine dimers in DNA: a comparative study with 64M-1 antibody specific for (6-4)photoproducts. Mutat. Res. 254, 175-184. [DOI] [PubMed] [Google Scholar]
- Mone, M.J., Volker, M., Nikaido, O., Mullenders, L.H., van Zeeland, A.A., Verschure, P.J., Manders, E.M., and van Driel, R. (2001). Local UV-induced DNA damage in cell nuclei results in local transcription inhibition. EMBO Rep. 2, 1013-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, T., Nakane, M., Hattori, T., Matsunaga, T., Ihara, M., and Nikaido, O. (1991). Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or (6-4)photoproduct from the same mouse immunized with ultraviolet-irradiated DNA. Photochem. Photobiol. 54, 225-232. [DOI] [PubMed] [Google Scholar]
- Mullen, J.R., Kaliraman, V., Ibrahim, S.S., and Brill, S.J. (2001). Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157, 103-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odagiri, N., Seki, M., Onoda, F., Yoshimura, A., Watanabe, S., and Enomoto, T. (2003). Budding yeast mms4 is epistatic with rad52 and the function of Mms4 can be replaced by a bacterial Holliday junction resolvase. DNA Repair (Amst) 2, 347-358. [DOI] [PubMed] [Google Scholar]
- Ogrunc, M., and Sancar, A. (2003). Identification and characterization of human MUS81-MMS4 structure specific endonuclease. J. Biol. Chem. 278, 21715-21720. [DOI] [PubMed] [Google Scholar]
- Paques, F., and Haber, J.E. (1999). Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince, P.R., Emond, M.J., and Monnat, R.J., Jr. (2001). Loss of Werner syndrome protein function promotes aberrant mitotic recombination. Genes Dev. 15, 933-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintigny, Y., Makienko, K., Swanson, C., Emond, M.J., and Monnat, R.J., Jr. (2002). Homologous recombination resolution defect in Werner syndrome. Mol. Cell Biol. 22, 6971-6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby, M.C., Osman, F., and Dixon, J. (2003). Cleavage of model replication forks by fission yeast mus81-eme1 and budding yeast mus81-mms4. J. Biol. Chem. 278, 6928-6935. [DOI] [PubMed] [Google Scholar]