Limbal stromal niche cells heterogeneously express embryonic stem cell markers. They were isolated and expanded on different Matrigel substrate by maintaining their original phenotype. Their phenotype is critical to the prevention of differentiation of limbal epithelial progenitors.
Abstract
Purpose.
Limbal stromal niche cells heterogeneously express embryonic stem cell (SC) markers. This study was conducted to isolate and expand them and to prove that their phenotype is critical for supporting SCs.
Methods.
Human limbus was isolated by dispase or collagenase. Single cells were seeded on coated, 2D, or 3D Matrigel and were serially passaged in modified embryonic SC medium (MESCM), supplemented hormonal epithelial medium (SHEM), or Dulbecco's modified Eagle's medium plus 10% fetal bovine serum (DF) before they were seeded in 3D Matrigel. Sphere growth was achieved by mixing expanded single cells with dispase-isolated epithelial cells in 3D Matrigel. Expression of SC markers was analyzed by qRT-PCR, immunofluorescence staining, and Western blot; SC clonal growth was measured on 3T3 feeder layers.
Results.
Collagenase, but not dispase, isolated subjacent mesenchymal cells, of which the expression of Oct4, Sox2, Nanog, Rex1, SSEA4, N-cadherin, and CD34 was promoted in MESCM more than SHEM or DF. Reunion of PCK+ and Vim+ cells generated spheres in 3D Matrigel, but spindle cells emerged on 2D or coated Matrigel. Serial passages on coated Matrigel resulted in rapid expansion of spindle cells, of which the expression of ESC markers had declined but could be regained after reseeding in 3D Matrigel in MESCM but not in SHEM or DF. Resultant epithelial spheres mixed with spindle cells expanded in MESCM expressed more p63α, less CK12, and more holoclones than those mixed with spindle cells expanded in DF.
Conclusions.
Limbal stromal niche cells expressing SC markers can be isolated and expanded to prevent differentiation and maintain clonal growth of limbal epithelial progenitors.
The corneal epithelium, like other epithelial tissues, undergoes constant renewal by a population of adult lineage-committed epithelial stem cells (SCs) anatomically located in limbal palisades of Vogt.1,2 Although the mechanism remains elusive, the quiescence, self-renewal, and fate of limbal epithelial SCs are conceivably controlled in this unique niche.3 As a first step toward addressing these issues, it is important to isolate putative limbal niche cells (NCs). To that end, we have recently reported that the traditional method of isolating an intact human limbal epithelial sheet using dispase, which cleaves the basement membrane,4 fails to remove the entire limbal basal progenitors and only removes few subjacent mesenchymal cells (MCs).5 In contrast, collagenase that cleaves stromal interstitial, but not basement membrane, collagens isolates a cluster of cells consisting of not only entire limbal epithelial progenitors but also abundant subjacent stromal MCs.5 Interestingly, the latter cells are as small as 5 μm in diameter and heterogeneously express some embryonic SC markers including Oct4, Sox2, Nanog, Rex1, and SSEA4 as well as other SC markers such as Nestin, N-cadherin, and CD34.5 Because disruption of the physical close association between limbal basal epithelial cells and these MCs by trypsin and EDTA (T/E) results in the loss or marked reduction of epithelial clonal growth in three different in vitro assays,5 we speculated that these stromal MCs may serve as NCs. Even if it were true, we still do not know whether their phenotype of expressing these SC markers is critical for their niche function.
Although the presence of NCs is implicated in the previous study,5 characterization of these NCs depends on successful isolation and expansion. In this regard, Dravida et al.6 isolated SSEA4+ cells by magnetic cell sorting from limbal explant outgrowth, cultured them on a substrate coated with Matrigel (BD Biosciences, Franklin Lakes, NJ) in a modified embryonic SC medium containing bFGF and LIF (hereafter termed MESCM), and successfully expanded multipotent fibroblastlike cells expressing Oct4, Sox2, Nanog, Rex1, SSEA4, and TDGF1 for more than 20 passages. Herein, we report our modified method of isolating and expanding these NCs and provide strong evidence that the expression of SC markers is the hallmark for them to prevent differentiation and maintain clonal growth of limbal epithelial progenitors.
Materials and Methods
All materials used for cell isolation and culturing are listed in Supplementary Table S1, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-8441/-/DCSupplemental.
Isolation of Limbal Epithelial Sheets and Clusters
Human corneoscleral rims from donors younger than 60 years and <5 days from death to culturing were obtained from the Florida Lions Eye Bank (Miami, FL) and were managed in accordance with the Declaration of Helsinki. The isolation of limbal epithelial sheets4 or clusters5 by either dispase or collagenase, respectively, was consistent with what we have reported. In short, after corneoscleral tissue was rinsed three times with Hanks' balanced salt solution containing 50 μg/mL gentamicin and 1.25 μg/mL amphotericin B, the remaining sclera, conjunctiva, iris, trabecular meshwork, and corneal endothelium were removed. Then the tissues were cut into 12 one-clock-hour segments, from which each limbal segment was obtained by incisions made at 1 mm within and beyond the anatomic limbus. An intact epithelial sheet was isolated by digestion of each limbal segment at 4°C for 16 hours with 10 mg/mL neural protease (Dispase II; Roche Applied Science, Indianapolis, IN) in MESCM made of DMEM/F-12 (1:1) supplemented with 10% knockout serum, 5 μg/mL insulin, 5 μg/mL transferrin, 5 ng/mL sodium selenite, 4 ng/mL bFGF, 10 ng/mL hLIF, 50 μg/mL gentamicin, and 1.25 μg/mL amphotericin B. In parallel, other limbal segments were digested at 37°C for 18 hours with 1 mg/mL collagenase A in MESCM, SHEM, or DF to generate limbal clusters. SHEM consists of DMEM/F-12 (1:1) supplemented with 5% fetal bovine serum (FBS), 0.5% dimethyl sulfoxide, 2 ng/mL hEGF, 5 μg/mL insulin, 5 μg/mL transferrin, 5 ng/mL selenium, 0.5 μg/mL hydrocortisone, 1 nM cholera toxin, 50 μg/mL gentamicin, and 1.25 μg/mL amphotericin B. DF is made of DMEM containing 10% FBS, 50 μg/mL gentamicin, and 1.25 μg/mL amphotericin B. Limbal epithelial sheets and clusters were further digested with 0.25% trypsin and 1 mM EDTA (T/E) at 37°C for 15 minutes to yield single cells.
Coated, 2D, and 3D Matrigel Culture and Treatment
Matrigel with different thicknesses—that is, coated, thin (2D), and thick (3D) gel—were prepared by adding the plastic dish with 5% diluted Matrigel, 50 μL 50% diluted Matrigel, and 200 μL of 50% diluted Matrigel (all in DMEM) per square centimeter, respectively, by incubation at 37°C for 1 hour before use. On 3D Matrigel, dispase and collagenase-isolated cells were seeded at a density of 5 × 104/cm2 in MESCM. In parallel, on coated and 2D Matrigel, 5 × 104/cm2 or 1 × 105/cm2 collagenase-isolated cells were seeded in MESCM, SHEM, or DF. At passage (P)0, cells on day 5 cultured in coated, 2D, and 3D Matrigel in MESCM were added with 10 μM of 5-ethynyl-2′-deoxyuridine (EdU) for 24 hours. Spheres in 3D gel at different time points were harvested by digestion in 10 mg/mL neural protease (Dispase II; Roche Applied Science) at 37°C for 2 hours, of which some were rendered into single cells by T/E. On 80% confluence on coated Matrigel, single cells were continuously passaged at a density of 5 × 103 cells/cm2. At P4, the expanded cells were also reseeded in 3D Matrigel at a density of 5 × 104 cells/cm2 in three different media for 6 days. Afterward, P4 expanded cells from 3D Matrigel were prelabeled with red fluorescent nanocrystals (Qtracker cell labeling kits; Invitrogen, Carlsbad, CA), mixed at a 1:4 ratio with dispase-isolated epithelial cells, seeded at a density of 5 × 104/cm2 in 3D Matrigel containing MESCM, and cultured for 10 days. The extent of total expansion was measured by the number of population doubling from P1 to P4 using the formula: number of cell doublings (NCD) = log10(y/x)/log102, where y is the final density of the cells and x is the initial seeding density of the cells.
3T3 Clonal Cultures
The epithelial progenitor status of the sphere growth was determined by a clonal assay on 3T3 fibroblast feeder layers in SHEM. The feeder layer was prepared by treating 80% subconfluent 3T3 fibroblasts with 4 μg/mL mitomycin C at 37°C for 2 hours in DMEM containing 10% newborn calf serum before seeding at the density of 2 × 104 cells/cm2. Single cells obtained from day 10 spheres were then seeded on mitomycin C–treated 3T3 feeder layers, at a density of 100 cells/cm2 for 2 weeks. Resultant clonal growth was assessed by rhodamine B staining, and colony-forming efficiency (CFE) was measured by calculating the percentage of the clone number divided by the total number of cells seeded. Clone morphology was subdivided into holoclone, meroclone, and paraclone based on the criteria for skin keratinocytes.7
Immunofluorescence Staining
Limbal epithelial sheets or clusters obtained by dispase or collagenase digestion, respectively, were cryosectioned to 6 μm. Spheres, EdU-labeled cells, and the P4 isolated mesenchymal cells were prepared for cytospin using a centrifuge system (Cytofuge; StatSpin, Inc., Norwood, MA) at 1000 rpm for 8 minutes. For immunofluorescence staining, 4% formaldehyde-fixed samples were permeated with 0.2% Triton X-100 in PBS for 15 minutes and were blocked with 2% BSA in PBS for 1 hour at room temperature before they were incubated in the primary antibody overnight at 4°C. Corresponding secondary antibodies were then incubated for 1 hour using appropriate isotype-matched, nonspecific IgG antibodies as controls. EdU-labeled cells were detected by fixation in 4% formaldehyde for 15 minutes, followed by 0.2% Triton X-100 in PBS for 15 minutes, blocking with 2% BSA in PBS for 1 hour, and incubation in reaction cocktails (Click-iT; Invitrogen) for 30 minutes before they were subjected to PCK immunostaining. Nuclear counterstaining was achieved by Hoechst 33342 before they were analyzed with a confocal microscope (LSM 700; Zeiss, Thornwood, NY). Detailed information about primary and secondary antibodies and agents used for immunostaining is listed in Supplementary Table S2, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-8441/-/DCSupplemental.
Quantitative Real-Time Polymerase Chain Reaction
Total RNA was extracted from limbal clusters freshly isolated by collagenase on day 0 and cells on coated and 3D gel at different passages with an RNA isolation kit (RNeasy Mini; Qiagen, Valencia, CA). Total RNA (1–2 μg) was reverse transcribed to cDNA with a high-capacity cDNA transcription kit (Applied Biosystems, Foster City, CA). qRT-PCR was carried out in a 20-μL solution containing cDNA, gene expression assay (TaqMan; Invitrogen) (Supplementary Table S3, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-8441/-/DCSupplemental), and PCR master mix (Universal; Applied Biosystems). The results were normalized by internal control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Relative gene expression data were analyzed by the comparative CT method (ΔΔCT).
Immunoblot Analysis
Proteins from day 10 spheres were extracted by RIPA buffer supplemented with proteinase inhibitors and phosphatase. The protein concentration was determined by a BCA protein assay (Pierce, Rockford, IL). Equal amounts of proteins in total cell extracts were separated by 10% SDS-PAGE and transferred to nitrocellulose membranes that were then blocked with 5% (wt/vol) fat-free milk in TBST (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.05% (vol/vol) Tween-20), followed by sequential incubation with specific primary antibodies and their respective secondary antibodies using β-actin as the loading control. Immunoreactive bands were visualized by a chemiluminescence reagent (Western Lighting; Pierce). Antibodies used are listed in Supplementary Table S2, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-8441/-/DCSupplemental.
Statistical Analysis
The significance of the differences between groups was determined by Student's unpaired t-test. Test results were reported as two-tailed P values, where P < 0.05 was considered statistically significant.
Results
Collagenase Isolates More Subjacent Mesenchymal Cells
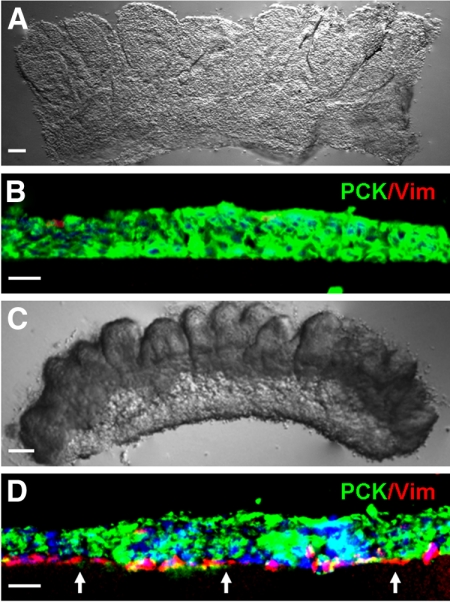
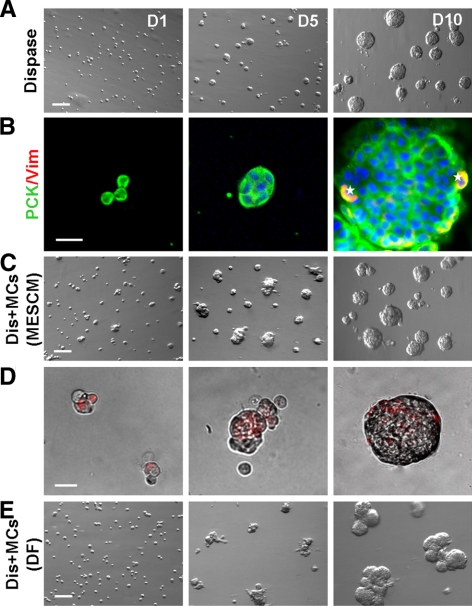
Anatomically, the limbal niche is located at the limbal palisades of Vogt, in which the epithelial-mesenchymal interface is undulated and consists of intermittent projections.8,9 Several studies suggest that limbal epithelial SCs may lie deep in the stroma in cryptlike structures.9–12 These anatomic features in the limbus suggest that limbal epithelial SCs might closely interact with cells in the underlying limbal stroma. As reported4 and commonly practiced, digestion with dispase removed an intact human limbal epithelial sheet (Fig. 1A) that consisted nearly exclusively of PCK+ cells (Fig. 1B). Nonetheless, as recently reported,5 digestion with collagenase resulted in a cluster of cells (Fig. 1C) that consisted of not only entire PCK+ epithelial cells but also many subjacent PCK−/Vim+ cells (Fig. 1D). These results indicated that collagenase, but not dispase, could isolate both limbal progenitors and closely associated stromal MCs.
Figure 1.
Collagenase but not dispase isolates more subjacent Vim+ cells. Dispase digestion of the one-clock-hour limbal segment at 4°C for 16 hours removed the entire PCK+ epithelial sheet (A), which consisted predominantly of PCK+ (green) cells (B). In contrast, collagenase digestion at 37°C for 18 hours isolated a cluster (C), which contained significantly more closely associated PCK-/Vim+ (red) cells (D, arrows). Nuclear counterstaining by Hoechst 33342. Scale bars: 100 μm (A, C); 20 μm (B, D).
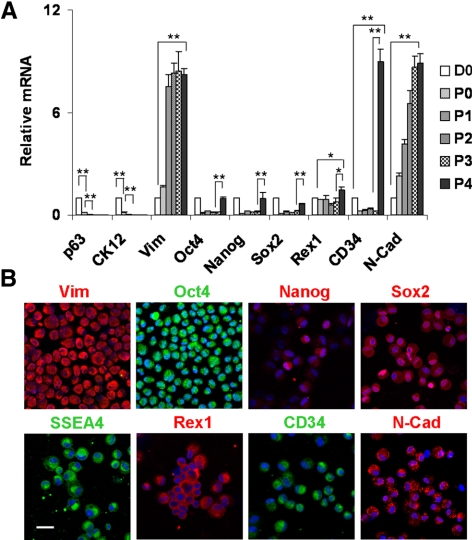
Clusters Isolated by Collagenase in MESCM Express Most ESC Markers
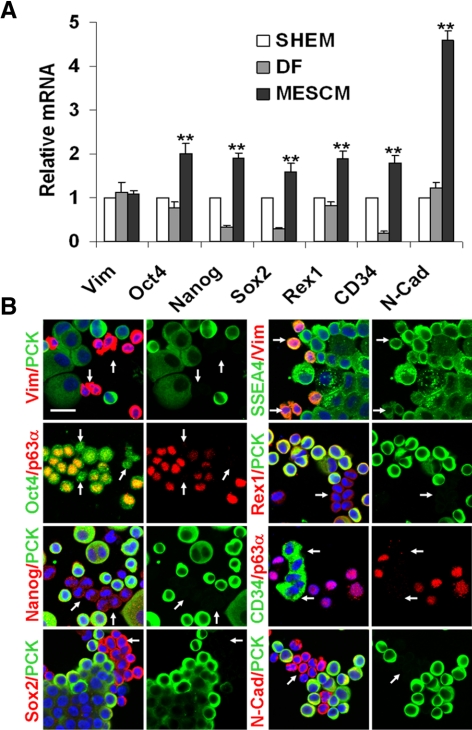
Previously, we isolated the limbal clusters by collagenase digestion in SHEM, which contains FBS.5 We reported that MCs in such collagenase-isolated limbal clusters are as small as 5 μm in diameter and heterogeneously express various SC markers, including Oct4, Sox2, Nanog, Rex1, SSEA4, Nestin, N-cadherin, and CD34.5 To prepare further isolation of these putative NCs, we digested limbal segments with collagenase in MESCM and compared the expression of the markers with that in SHEM or DF. qRT-PCR showed that the transcript level of Vim was not different among these three media (Fig. 2A), suggesting that they resulted in similar numbers of MCs. However, the transcript levels of Oct4, Nanog, Sox2, Rex1, CD34, and N-cadherin in MESCM were all significantly higher than those in SHEM and DF (n = 3, all P < 0.01; Fig. 2A), suggesting that expression of these markers by collagenase-isolated clusters was better maintained in MESCM. As a comparison, except for that of Oct4, Rex1, and N-cadherin, the expression of all other markers was notably reduced in DF (Fig. 2A). Our previous study showed that all small PCK+ epithelial cells were p63α+ but Vim−.5 Thus, we performed double immunostaining in PCK−, p63α−, or Vim+ MCs with the SC markers. Results showed that these small nonepithelial MCs indeed heterogeneously expressed Oct4, Nanog, SSEA4, Sox2, Rex1, CD34, and N-cadherin (Fig. 2B). Some of these SC markers were also expressed in some small epithelial cells. Collectively, these findings indicated that expression of these SC markers by small PCK−/p63α−/Vim+ cells was best maintained during collagenase digestion in MESCM.
Figure 2.
Collagenase-isolated clusters expressed more SC markers when digested in MESCM. qRT-PCR showed that collagenase-isolated clusters in MESCM expressed significantly more Oct4, Nanog, Sox2, Rex1, CD34, and N-cadherin (N-Cad) transcripts than those digested in SHEM and DF (A, n = 3, **P < 0.01). Double immunostaining between PCK (green), p63α (red), or Vim (red) and other markers revealed that small (PCK−/p63α−/Vim+) nonepithelial cells were Oct4+ (green), Nanog+ (red), Sox2+ (red), SSEA4+ (green), Rex1+ (red), CD34+ (green), and N-Cad+ (red) (D, arrows). Scale bar, 20 μm.
Different Growths on Three Matrigel Substrates
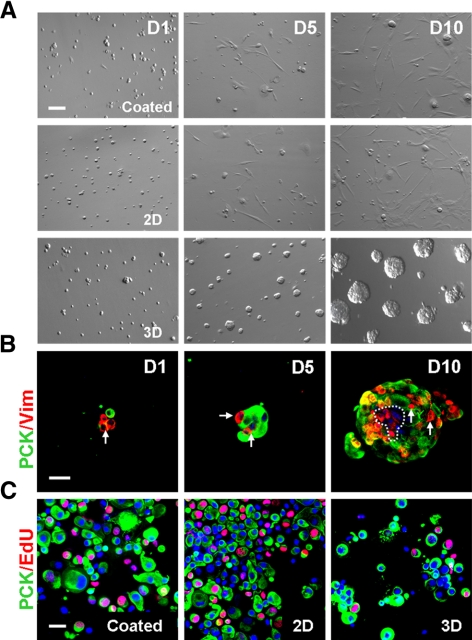
Our previous study showed that disruption of the close association between PCK+ epithelial progenitors and Vim+ MCs diminished epithelial clonal growth in three different assays, suggesting that the latter might serve as NCs.5 We speculated that such close association between PCK+ and Vim+ cells in collagenase-isolated clusters might be attained by preservation of the basement membrane. We then reasoned that single PCK+ and Vim+ cells generated by T/E, which disrupted their close association in collagenase-isolated clusters, might be reunited in the basement membrane substrate prepared by Matrigel. Indeed, spheres emerged in 3D Matrigel when cultured in MESCM, whereas predominant spindle cells without spheres occurred in coated and 2D Matrigel (Fig. 3A). Double immunostaining showed that spheres formed in 3D Matrigel consisted of both PCK+ cells and Vim+ cells on day 1, and both cells increased in number on days 5 and 10 (Fig. 3B). The proliferative activity measured by nuclear EdU labeling on day 5 for 24 hours was higher in coated and 2D Matrigel than in 3D Matrigel (Fig. 3C). The labeling index was 25.6% ± 3.2% and 27.3% ± 2.6% in PCK+ cells and 13.6% ± 1.5% and 12.9% ± 2.4% in PCK− cells in coated and 2D Matrigel, respectively. They were significantly higher than 12.5% ± 2.0% in PCK+ cells and 2.6% ± 1.2% in PCK− cells in 3D Matrigel (n = 5; all P < 0.01). These results suggested that cell proliferation was higher on coated and 2D Matrigel, where spindle cells emerged, than in 3D Matrigel, where sphere growth formed by the reunion of single PCK+ and Vim+ cells.
Figure 3.
Different growth by collagenase-isolated cells in coated, 2D, and 3D Matrigel. Single cells from collagenase-isolated limbal clusters (Fig. 1) were seeded in coated, 2D, and 3D Matrigel at 5 × 104/cm2 in MESCM. Spheres emerged in 3D Matrigel, whereas predominant spindle cells were found in coated and 2D Matrigel (A). The sphere in 3D Matrigel was formed by the reunion of single PCK+ (green) cells and Vim+ (red) cells, both of which increased in cell numbers in 10 days (B). Double staining between PCK (green) and EdU (red) confirmed that EdU+ nuclei were high in coated and 2D Matrigel but low in 3D Matrigel (C). Scale bar, 20 μm.
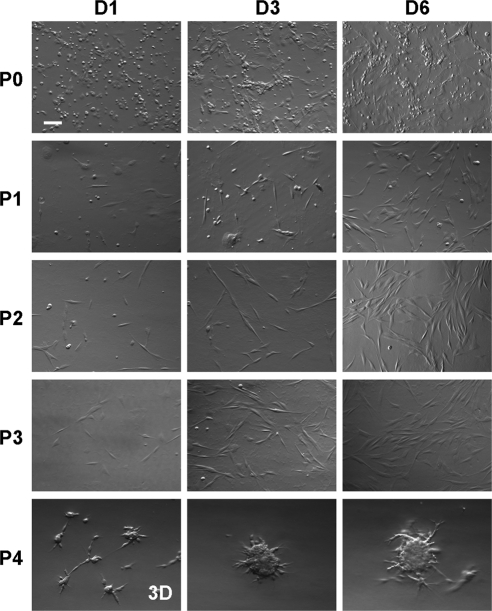
Spindle Cells Proliferate and Dominate on Coated Matrigel after Serial Passages
Because spheres formed in 3D Matrigel contained both PCK+ and Vim+ cells and Vim+ cells therein grew more slowly than PCK+ cells when judged by the EdU-labeling index (Fig. 3), 3D Matrigel was not an ideal substrate for isolating and expanding Vim+ MCs. In contrast, spindle cells emerged among small, round cells on coated Matrigel and rapidly increased in number on further passages (Fig. 4). Although some small, round cells were noted in P0, spindle cells dominated from P2 onward (Fig. 4). When reseeded in 3D Matrigel, single P4 cells began to form aggregates with stellate borders as early as day 1, increased in size, but then ceased to grow on day 6 (Fig. 4). These changes in proliferative activity were also reflected by the population doubling time, which was 40 and 39 hours for spindle cells at P2 and P3 in coated Matrigel but was significantly lengthened to 881 hours when reseeded in 3D Matrigel at P4 (Table 1).
Figure 4.
Expansion of spindle cells on coated Matrigel by serial passages. Single cells derived from collagenase-isolated limbal clusters were seeded at 1 × 105/cm2 on coated Matrigel in MESCM. Spindle cells emerged among small round cells in P0, rapidly proliferated, and became dominant after P2. These spindle cells in P4 could still form spheres when reseeded in 3D Matrigel at a density of 5 × 104/cm2. Scale bar, 100 μm.
Table 1.
Population Doubling of Cultured MCs from Collagenase-Isolated Clusters
| Passage | Seeding Density (×105/cm2) | Culture Time (d) | Final Density (×105/cm2) | Number of Cell Doublings | Cumulative Number of Cell Doublings | Population Doubling Time (h) |
|---|---|---|---|---|---|---|
| 0 | 1 | 6 | 1.05 | 0.07 | 0.07 | 2045.8 |
| 1 | 0.05 | 6 | 0.09 | 0.85 | 0.92 | 169.8 |
| 2 | 0.05 | 6 | 0.6 | 3.58 | 4.50 | 40.2 |
| 3 | 0.05 | 6 | 0.66 | 3.72 | 8.23 | 38.7 |
| 4 | 0.5 | 6 | 0.56 | 0.16 | 8.39 | 880.7 |
Results of seeding density, culture time, and final density from P0 to P4, from which population doubling and doubling time were calculated.
Reversibility of Phenotype of Spindle Cells Expanded in MESCM
Compared with that of the D0 cluster immediately isolated by collagenase, qRT-PCR revealed a rapid disappearance of p63 (i.e., an epithelial progenitor marker13) and CK12 (i.e., a corneal epithelial differentiation marker14,15) by P2 cells (Fig. 5A), suggesting that coated Matrigel successfully eliminated all epithelial cells by successive passages. From P0 to P3, there was a significant decline in expression of Oct4, Nanog, Sox2, and CD34 transcripts but a steady significant increase of expression of Vim and N-cadherin transcripts (Fig. 5A; all P < 0.01; n = 3). On reseeding in 3D Matrigel at P4, the transcript levels of Oct4, Nanog, Sox2 Rex1, and CD34 were significantly increased compared with P3 cells (all P < 0.01; n = 3), whereas those of CD34, Rex1, and N-cadherin were significantly higher than those of D0 clusters (all P < 0.01; n = 3). When reseeded in 3D Matrigel, these spindle cells at P4 indeed re-expressed Oct4, Nanog, SSEA4, Sox2, Rex1, CD34, and N-cadherin (Fig. 5B).
Figure 5.
Phenotypic characterization of expanded mesenchymal cells. Compared with D0 clusters immediately isolated by collagenase, qRT-PCR revealed the rapid disappearance of p63 and CK12 transcripts by P2, a significant decline of Oct4, Nanog, Sox2, and CD34 (n = 3, **P < 0.01), a steady increase of Vim and N-Cad (n = 3, **P < 0.01), and no change in Rex1 from on coated Matrigel from P0 to P3 (A). On reseeding in 3D Matrigel at P4, Oct4, Nanog, Sox2, Rex1, and CD34 transcripts were significantly increased (n = 3, *P < 0.05 and **P < 0.01, compared with P3 cells) (A). All cells derived from P4 aggregates were Vim+ and heterogeneously expressed SC markers (B). Scale bar, 20 μm.
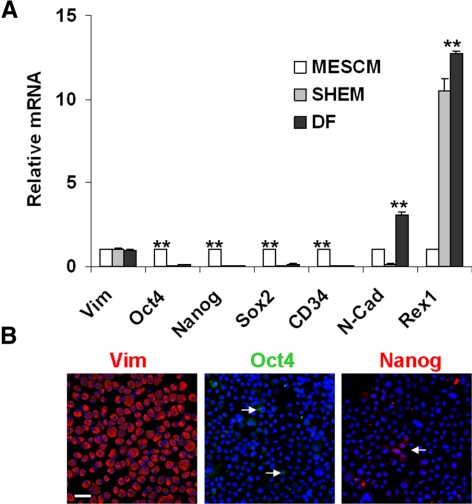
As a comparison, we also isolated and expanded spindle MCs on coated Matrigel in SHEM and DF. On reseeding in 3D Matrigel at P4, they also formed similar aggregates (not shown). However, qRT-PCR showed that these cells did not regain expression of these SC markers (Fig. 6A). Immunostaining confirmed the lack of such expression (Fig. 6B). Collectively, these data showed that the phenotype of expressing embryonic SC markers was regained by spindle cells expanded by continuous passages on coated Matrigel only in MESCM but not in SHEM or DF.
Figure 6.
Unique recovery of SC markers by MESCM. Vim+ spindle cells were continuously expanded in MESCM, SHEM, or DF on coated Matrigel up to P3. On reseeding in 3D Matrigel on P4, qRT-PCR showed re-expression of Oct4, Nanog, Sox2, and CD34 by cells cultured in MESCM, but not in SHEM and DF (n = 3, **P < 0.01) (A). When the P4 cells were harvested from 3D Matrigel in DF, immunostaining showed that all cells were Vim+ and very few were Oct4+ and Nanog+ (B, arrows). Scale bar, 20 μm.
Spheres Formed by Reunion between Dispase-Isolated Epithelial Cells and MCs Isolated and Expanded in Different Media
Figure 3 showed that reunion between PCK+ epithelial cells and Vim+ MCs obtained from collagenase-isolated clusters led to sphere growth. We found that PCK+ epithelial cells obtained from dispase-isolated limbal epithelial sheets, which contained few Vim+ cells (Fig. 1B), could also yield similar sphere growth in 3D Matrigel containing MESCM (Fig. 7A). Double immunostaining shows that these spheres consisted of predominantly PCK+ epithelial cells, of which few coexpressed Vim on day 10 (Fig. 7B). Thus, we mixed dispase-isolated epithelial cells with MCs that had been expanded on coated Matrigel up to P4, followed by seeding in 3D Matrigel in different media at the ratio of 4:1 to match the finding that 20% of collagenase-isolated clusters is made of PCK−/Vim+ MCs.5 More and relatively larger spheres were generated by MCs expanded in MESCM (Figs. 7A, 7C). These spheres consisted of epithelial cells and MCs prelabeled by red nanocrystals (Qdot; Life Technologies Corporation, Carlsbad, CA) (Fig. 7D). In contrast, spheres generated by mixing with MCs expanded in DF tended to adhere to one another on day 10 (Fig. 7E), which also consisted of both epithelial cells and MCs prelabeled by red nanocrystals (Qdot; Life Technologies) (not shown).
Figure 7.
Reunion of dispase-isolated epithelial cells and expanded MCs. In 3D Matrigel containing MESCM, dispase-isolated epithelial cells (dispase) formed spheres (A), which consisted of PCK+ epithelial cells (B); few also coexpressed Vim (B, yellow, marked by stars). When single dispase-isolated epithelial cells (Dis) were mixed with P4 MCs expanded in 3D Matrigel in MESCM or DF, they also formed spheres (C and E, respectively). Such spheres consisted of epithelial cells and MCs prelabeled by nanocrystals (red) (D). Spheres formed by MCs isolated in DF tended to adhere to one another on day 10 (E). Scale bars: 100 μm (A, C, E); 20 μm (B, D).
Maintenance of Limbal Epithelial Progenitor Status by Expanded NCs
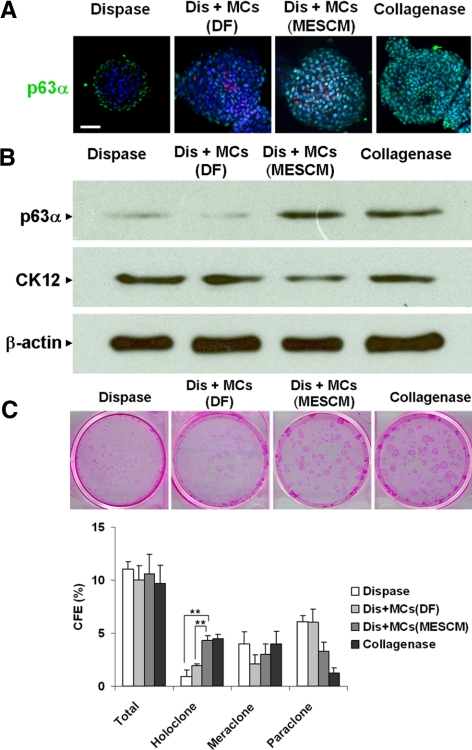
Although similar spheres were formed by dispase-isolated epithelial cells with or without mixing with expanded MCs (Fig. 7), immunofluorescence staining of p63a showed that dispase+MCs (MESCM) had more p63α expression than dispase+MCs (DF) (Fig. 8A). Western blot analysis followed by densitometry confirmed that spheres formed by collagenase-isolated limbal clusters (Fig. 3) expressed 3.5-fold p63α and 0.6-fold CK12 compared with those formed by dispase-isolated limbal epithelial sheets (Fig. 8B). Compared with spheres formed by dispase-isolated limbal epithelial cells, the addition of MCs expanded in MESCM resulted in spheres expressing 3.9-fold more p63α and 0.5-fold less CK12 (i.e., to a level similar to levels formed by collagenase-isolated clusters). In contrast, the addition of MCs expanded in DF resulted in spheres expressing 0.7-fold p63α and 0.7-fold CK12 (Fig. 8B). As reported,5 spheres from collagenase-isolated limbal clusters generated more holoclones than dispase-isolated limbal epithelial sheets on growth-arrested 3T3 feeder layers, presumably because of inclusion of the entire limbal basal epithelial progenitors (Fig. 8C). Compared with this baseline finding, spheres generated by mixing dispase-isolated epithelial cells with MCs expanded in MESCM had significantly more holoclone than those mixed with MCs isolated in DF (Fig. 8C). Collectively, these findings suggest that reunion with MCs expanded in MESCM prevents corneal epithelial differentiation and promotes clonal growth of limbal epithelial progenitors, similar to reunion with native NCs just isolated from the in vivo state.
Figure 8.
Maintenance of limbal epithelial progenitor status by MCs Expanded in MESCM but not DF. D10 spheres in 3D Matrigel were formed by dispase-isolated limbal epithelial cells alone (dispase) or were mixed with MCs expanded on coated Matrigel in DF (Dis+MCs [DF]) or in MESCM (Dis+MCs [MESCM]) or by collagenase-isolated clusters (Collagenase). Immunofluorescence staining of p63α demonstrated that Dis+MCs (MESCM) had more p63α (green) expression than Dis+MCs (DF) (these MCs were prelabeled by nanocrystals [red]) (A). Western blot analysis confirmed that dispase+MCs (MESCM) had more p63α but less CK12 than dispase+MCs (DF) using β-actin as the loading control (B). Spheres generated by Dis+MCs (MESCM) had significantly more holoclone than those by Dis+MCs (DF) using dispase and collagenase as controls (C, n = 3, **P < 0.01). Scale bar, 20 μm.
Discussion
In the locale of limbal palisades of Vogt, the basement membrane is undulated and fenestrated8,10 and limbal epithelial SCs lie deep in the stroma, as suggested by cryptlike structures disclosed by serial histologic sectioning10–12 and ultrastructural analyses.9 We have provided the first evidence supporting close physical contact between limbal epithelial SCs and MCs that lie immediately subjacent in the limbal stroma.5 As reported,5 digestion with collagenase, but not dispase, effectively isolated not only the entire limbal epithelial cells but also subjacent MCs (Fig. 1). Furthermore, qRT-PCR confirmed that limbal clusters isolated by the digestion of collagenase in SHEM expressed Oct4, Nanog, SSEA4, Sox2, Rex1, N-cadherin, and CD34 (Fig. 2). Among them N-cadherin,16 nestin,17 and Oct418 have been found in human limbal basal epithelial cells. Although further proof is needed that small PCK+/p63a+ epithelial cells that also express these SC markers (Fig. 2B) might represent genuine SCs, the present study provides strong evidence supporting the notion that small PCK−/p63α−/Vim+ cells that express these SC markers represent limbal native NCs.
The best way to isolate these NCs was to use collagenase digestion in MESCM because qRT-PCR and double immunostaining disclosed that expression of the SC markers was better maintained in MESCM than in SHEM or DF (Fig. 2). To isolate similar cells, Dravida et al.6 used SSEA4 magnetic beads to isolate a subset from a mixture of cells derived from limbal explants outgrowth. In contrast, we achieved successful isolation by seeding collagenase-isolated limbal clusters on coated Matrigel to rapidly eliminate PCK+ cells, as evidenced by the disappearance of p63 and CK12 (Fig. 5) and the emergence of spindle cells (Fig. 4) at P2. We chose MESCM because Dravida et al.6 successfully used it to expand the cells into multipotent fibroblastlike cells expressing Oct4, Sox2, Nanog, Rex1, SSEA4, and TDGF1. In agreement with their finding,6 we also found that serial passages on coated Matrigel in MESCM resulted in rapid expansion, as shown by a high EdU-labeling index (Fig. 3C) and a short doubling time around 40 hours (Table 1). The difference in the proliferation of Vim+ cells in 3D versus 2D and coated Matrigel was attributed primarily to the physical property of Matrigel because the same composition was used. We speculate that such a difference might be caused by the difference in matrix stiffness or rigidity, which may activate such mitotic signaling mediated by ERK and RhoA.19 However, the expression of SC markers rapidly declined compared with that by just isolated only by collagenase (Fig. 5). Cells expanded in our study might not have been the same as those expanded by Dravida et al.6 because ours expressed CD34 whereas theirs did not. For the first time, we disclosed that such a loss was transient because the expression of SC markers could be regained and promoted by reseeding in 3D Matrigel containing MESCM (Fig. 5). Unlike Dravida et al.,6 we did not include the remaining limbal stroma; therefore, we concluded that these NCs expressing SC markers were immediately subjacent to limbal basal epithelial cells.
In contrast to dominant Vim+ spindle cells that arose from coated or 2D Matrigel, spheres emerged in 3D Matrigel containing MESCM because of reunion of PCK+ epithelial cells and Vim+ MCs (Fig. 3). Our recent report20 showed that such reunion is mediated by the SDF-1 preferentially expressed by limbal epithelial cells and CXCR4 expressed by stromal MCs. Herein, using single cells obtained from dispase-isolated epithelial sheets, which were largely devoid of MCs (Fig. 1), we observed similar spheres nearly exclusively made of PCK+ cells in 3D Matrigel (Fig. 7). One should not confuse these spheres with the “neurospheres” and “mammospheres” generated from nonadherent precursor cells in Matrigel–free cultures containing the neural SC medium.21–23 Because of the dramatic outcome in yielding either spheres or spindle cells in different Matrigel substrates, future studies are needed to determine how matrix rigidity might influence the NC morphology and phenotype.19
Re-expression of SC markers by MCs expanded on coated Matrigel appeared to be crucial for endowing them with the NC phenotype. This notion was shown by studying spheres generated by dispase-isolated limbal epithelial cells. Successful isolation of entire limbal basal progenitor cells, including SCs in collagenase-isolated limbal clusters, generate more holoclones than dispase-isolated limbal epithelial sheets on 3T3 fibroblast feeder layers.5 Herein, we further showed that spheres formed from collagenase-isolated clusters in 3D Matrigel expressed more p63α, an epithelial progenitor marker,13,24 and less CK12, a marker for corneal epithelial differentiation,14,15 and generated more holoclones on 3T3 fibroblast feeder layers than the dispase-isolated limbal epithelial sheet (Fig. 8). Such a dramatic outcome is attributed to the reunion between PCK+ cells and Vim+ cells because disruption of the reunion before sphere growth by AMD3100, which specifically blocks the chemoattraction between SDF-1 and CXCR4, results in corneal epithelial differentiation and loss of holoclones.20 We further demonstrated that mere reunion with MCs was not sufficient because spheres formed by MCs expanded on coated Matrigel in DF, though able to partake in the reunion, did not yield the same result (Fig. 8). The failure of the latter MCs to prevent corneal differentiation and maintain holoclones of limbal epithelial progenitors was correlated with their irreversible loss of expression of SC markers during expansion in either DF or SHEM (Fig. 7). Thus, caution should be exercised in the choice of media during isolation and expansion of limbal NCs to preserve their unique phenotype of expressing these SC markers.
In conclusion, limbal stromal niche cells expressing SC markers can be isolated and expanded to prevent the differentiation of limbal epithelial progenitors. The in vitro model of sphere cultures in 3D Matrigel can be used to investigate how limbal NCs might regulate limbal epithelial SC quiescence, self-renewal, and fate in the future.
Supplementary Material
Acknowledgments
The authors thank Angela Y. Tseng for assistance with the preparation of this manuscript.
Footnotes
Supported by National Institutes of Health, National Eye Institute Grant RO1 EY06819 (SCGT).
Disclosure: H.-T. Xie, None; S.-Y. Chen, TissueTech, Inc. (E); G.-G. Li, None; S.C.G. Tseng, TissueTech, Inc. (I), P
References
- 1. Schermer A, Galvin S, Sun T-T. Differentiation-related expression of a major 64K corneal keratin in vivo and in culture suggests limbal location of corneal epithelial stem cells. J Cell Biol. 1986;103:49–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lavker RM, Tseng SC, Sun TT. Corneal epithelial stem cells at the limbus: looking at some old problems from a new angle. Exp Eye Res. 2004;78:433–446 [DOI] [PubMed] [Google Scholar]
- 3. Li W, Hayashida Y, Chen YT, Tseng SC. Niche regulation of corneal epithelial stem cells at the limbus. Cell Res. 2007;17:26–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Espana EM, Romano AC, Kawakita T, Di Pascuale M, Smiddy R, Tseng SC. Novel enzymatic isolation of an entire viable human limbal epithelial sheet. Invest Ophthalmol Vis Sci. 2003;44:4275–4281 [DOI] [PubMed] [Google Scholar]
- 5. Chen SY, Hayashida Y, Chen MY, Xie HT, Tseng SC. A new isolation method of human limbal progenitor cells by maintaining close association with their niche cells. Tissue Eng Part C Methods. 2011;17:537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dravida S, Pal R, Khanna A, Tipnis SP, Ravindran G, Khan F. The transdifferentiation potential of limbal fibroblast-like cells. Brain Res Dev Brain Res. 2005;160:239–251 [DOI] [PubMed] [Google Scholar]
- 7. Barrandon Y, Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci U S A. 1987;84:2302–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gipson IK. The epithelial basement membrane zone of the limbus. Eye. 1989;3(part 2):132–140 [DOI] [PubMed] [Google Scholar]
- 9. Shortt AJ, Secker GA, Munro PM, Khaw PT, Tuft SJ, Daniels JT. Characterization of the limbal epithelial stem cell niche: novel imaging techniques permit in vivo observation and targeted biopsy of limbal epithelial stem cells. Stem Cells. 2007;25:1402–1409 [DOI] [PubMed] [Google Scholar]
- 10. Dua HS, Shanmuganathan VA, Powell-Richards A, Tiqhe PJ, Joseph A. Limbal epithelial crypts: a novel anatomical structure and a putative limbal stem cell niche. Br J Ophthalmol. 2005;89:529–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shanmuganathan VA, Foster T, Kulkarni BB, et al. Morphological characteristics of the limbal epithelial crypt. Br J Ophthalmol. 2007;91:514–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yeung AM, Schlotzer-Schrehardt U, Kulkarni B, Tint NL, Hopkinson A, Dua HS. Limbal epithelial crypt: a model for corneal epithelial maintenance and novel limbal regional variations. Arch Ophthalmol. 2008;126:665–669 [DOI] [PubMed] [Google Scholar]
- 13. Pellegrini G, Dellambra E, Golisano O, et al. p63 identifies keratinocyte stem cells. Proc Natl Acad Sci U S A. 2001;98:3156–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen WY, Mui MM, Kao WW, Liu CY, Tseng SC. Conjunctival epithelial cells do not transdifferentiate in organotypic cultures: expression of K12 keratin is restricted to corneal epithelium. Curr Eye Res. 1994;13:765–778 [DOI] [PubMed] [Google Scholar]
- 15. Liu C-Y, Zhu G, Converse R, et al. Characterization and chromosomal localization of the cornea-specific murine keratin gene Krt1.12. J Biol Chem. 1994;269:24627–24636 [PubMed] [Google Scholar]
- 16. Hayashi R, Yamato M, Sugiyama H, et al. N-Cadherin is expressed by putative stem/progenitor cells and melanocytes in the human limbal epithelial stem cell niche. Stem Cells. 2007;25:289–296 [DOI] [PubMed] [Google Scholar]
- 17. Umemoto T, Yamato M, Nishida K, Yang J, Tano Y, Okano T. Limbal epithelial side-population cells have stem cell-like properties, including quiescent state. Stem Cells. 2006;24:86–94 [DOI] [PubMed] [Google Scholar]
- 18. Zhou SY, Zhang C, Baradaran E, Chuck RS. Human corneal basal epithelial cells express an embryonic stem cell marker OCT4. Curr Eye Res. 2010;35:978–985 [DOI] [PubMed] [Google Scholar]
- 19. Paszek MJ, Zahir N, Johnson KR, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254 [DOI] [PubMed] [Google Scholar]
- 20. Xie HT, Chen SY, Li GG, Tseng SC. Limbal epithelial stem/progenitor cells attract stromal niche cells by SDF-1/CXCR4 signaling to prevent differentiation. Stem Cells. 2011;29:1874–1885 [DOI] [PubMed] [Google Scholar]
- 21. Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710 [DOI] [PubMed] [Google Scholar]
- 22. Reynolds BA, Weiss S. Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev Biol. 1996;175:1–13 [DOI] [PubMed] [Google Scholar]
- 23. Dontu G, Abdallah WM, Foley JM, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Di Iorio E, Barbaro V, Ruzza A, Ponzin D, Pellegrini G, De Luca M. Isoforms of DeltaNp63 and the migration of ocular limbal cells in human corneal regeneration. Proc Natl Acad Sci U S A. 2005;102:9523–9528 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.