Abstract
The luteinizing hormone receptor (LHR) is a member of a subfamily of G protein-coupled receptors that is characterized by its alternative splicing. In a previous study, we identified a splice site mutation of intron 6 (IVS6-3C>A) in a patient suffering from Leydig cell hypoplasia, which leads to aberrant splicing of LHR mRNA. In vitro expression analysis confirmed that this mutation results in the skipping of exon 7 in the mature mRNA of the LHR gene. In this study, we determined the impact of IVS6-3C>A on the RNA secondary structure and function of LHR-Del7. The three-dimensional structure of the leucine-rich repeats in LHR was predicted by molecular modeling. Radioactive ligand-binding assays verified that LHR-Del7 has no binding affinity for hCG. Furthermore, we detected negligible cAMP production in cells transfected with LHR-Del7. Cells co-expressing LHR-WT and LHR-Del7 were able to generate cAMP in response to hCG, but there was no significant difference between cells transfected with LHR-WT/vector and LHR-WT/LHR-Del7, although the variant was able to localize to cell surface, similar to wild-type receptor. These results indicated that LHR-Del7 does not have a dominant negative effect on LHR-WT cell surface expression, and although the pathological splicing variant LHR-Del7 was able to localize to cell membranes it failed to bind hCG and had no effect on wild-type LHR.
Keywords: function study, splicing variant, LHR
Introduction
The luteinizing hormone receptor (LHR), together with the FSH and thyroid-stimulating hormone receptors, belongs to a subfamily of G protein-coupled receptors (GPCRs) that is responsible for transducing extracellular signals into intracellular responses by activating the G protein cascade (Chesler et al., 2007). The LHR has a large extracellular hormone-binding domain that contains a number of leucine-rich repeats (LRRs), seven membrane-spanning α helices and a short C-terminal intracellular domain (Ascoli et al., 2002). The LHR gene is localized on human chromosome 2p21 and contains 11 exons. The first 10 exons encode the extracellular domain (ECD), while the last exon encodes a small portion of the ECD, the transmembrane domain and the cytoplasmic C-terminal domain. The activating mutations of LHR resulting in constitutively elevated basal levels of cAMP production cause familial male-limited precocious puberty, whereas loss-of-function mutations of LHR lead to Leydig cell hypoplasia (LCH), a 46,XY disorder of sexual development.
In a previous study, we identified a splice site mutation in intron 6 (IVS6-3C>A) leading to aberrant splicing of LHR mRNA as a molecular cause of LCH. In vitro expression analysis of an allelic minigene showed that the mutation IVS6-3C>A decreased the activity of the 3′ splice acceptor site, leading to a skipping of exon 7 in the mature mRNA of the LHR gene (Qiao et al., 2009). The groups of Laue et al. (1996) and Gromoll et al. (2000) have detected two additional deletions in the ECD of LHR (i.e. exons 8 and 10) in patients suffering from hypospadias or micropenis. The deletion of exon 8 abolished hormone binding, whereas LHR lacking exon 10 is only responsive to hCG. Furthermore, a variety of LHR splice variants have been identified in humans as well as in several other species (Loosfelt et al., 1989; Aatsinki et al., 1992; Bacich et al., 1994; Kawate and Okuda, 1998; Saint-Dizier et al., 2003; Dickinson et al., 2009). It has been suggested that alternative splicing mechanisms resulting in exon skipping is a hallmark of GPCRs; splicing variants of GnRH and the D3 dopamine receptor were found to regulate the function of the wild-type receptors by dimerizing with them (Grosse et al., 1997; Elmhurst et al., 2000). These findings prompted us to investigate whether LHR-Del7 could interact with LHR-WT or even modulate its function.
In this study, we have delineated the impact of the IVS6-3C>A mutation on RNA secondary structure and the function of LHR deleted exon 7. We also studied the effect of the splicing variant LHR-Del7 co-expressed with LHR-WT to determine whether the splicing variant can act as a negative regulator of the wild-type receptor.
Materials and Methods
Plasmid construction
The pCDNA 3.0-LHR-WT plasmid was generously provided by Dr Aaron J.W. Hsueh. The construction of pCDNA3.1-LHR and EGFPN2-LHR was described previously (Qiao et al., 2009). Mutations were introduced into pCDNA 3.0-LHR-WT using two rounds of PCR mutagenesis with primers as described below. The following primers were used, with the restriction endonuclease sites indicated in bold and underlined: LHR HX FOR: 5′-ATCTCAAGCTTTCAGAGGACTTAATGAG-3′ (HindIII); LHR HX REV: 5′-CATCTTTCTAGAGTGATGACGGTGAG-3′(XbaI); LHR-Del7 FOR: 5′-GAATCTGTAACACTGGAGCTAAAGGAAAAC-3′; LHR-Del7 REV: 5′-TTTTCCTTTAG CTCCAGTGTTACAGATTCAT-3′. The primer sets LHR HX FOR/LHR-Del7 REV and LHR-Del7 FOR/LHR HX REV were used separately in the first round of PCR amplification. The resulting fragments were mixed and the final mutant fragment was obtained using the LHR HX FOR and LHR HX REV primers.
To construct the mutant human LHR expression vector, the re-amplified fragment was digested with HindIII and XbaI and the resulting fragment was used to replace the wild-type sequences in pcDNA3.1-LHR and EGFPN2-LHR, generating pcDNA3.1-LHR-Del7 and EGFPN2-LHR-Del7. In addition, we also constructed pcDNA3.1-LHR-Del7-3FLAG for functional study. Plasmids were isolated and purified with anion exchange columns (Qiagen, Hilden, Germany).
RNA structure and protein steric conformation
RNA secondary structure was predicted with the program RNA structure version 4.6 (Mathews et al., 2004) (http://rna.urmc.rochester.edu/rnastructure.) The nucleotide sequence (150 bp) covered the entirety of exon 7, plus 37 bp 5′and 44 bp 3′ of the exon. The maximum energy difference allowed by the program was 10% and the maximum number of structures allowed was 20. A three-dimensional computer model of the LHR-Del7 protein was predicted with the program NOC (version 3.0; http://noch.sourceforge.net). Using the crystal structure of the hormone-binding domain of human follicle-stimulating hormone receptor (Fan and Hendrickson, 2005; Vischer et al., 2006) (PDB code 1XWD, chain C) as a template, the ECD (residues 50-263) of LHR (access number: P22888) was modeled by homology using SWISS-MODEL (http://swissmodel.expasy.org/) (Arnold et al., 2006) at a resolution of 2.92 Å.
Transfection of 293T cells and cAMP assays
293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 100 pg penicillin/streptomycin and 2 mM glutamine. Twenty-four hours before transfection, cells were split in a 100-mm culture dish at 50–60% confluence in DMEM without antibiotics. Cells were transfected with 10 μg construct plasmid DNA and 500 ng β-galactosidase plasmid DNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Forty-eight hours after transfection, cell pellets were resuspended in DMEM supplemented with 0.2% bovine serum albumin (Calbiochem, La Jolla, CA, USA). A 250 µ1 cell suspension containing 100 000 cells was pipetted into a 24-well tissue culture plate (Cellstar, Greiner, Frickenhausen, Germany) and pre-incubated at 37°C in an atmosphere containing 5% CO2 for 15 min. Increasing doses of hCG (0–1000 ng/ml) were added to each well, and cells were incubated for 1 h in a 500 μl volume in the presence of 0.25 mM 3-isobutyl-1-methylxanthine (Sigma, St. Louis, MO, USA). All measurements were done in triplicate. The total cAMP in each well was measured with a specific ELISA kit (R&D Systems, Minneapolis, MN, USA). β-galactosidase activity was assayed to normalize the transfection efficiency with a specific kit (Beyotime, Haimen, China).
Confocal microscopic analysis of LHR-Del7 in 293T cells
293T cells were transferred to a 35-mm dish with a 20-mm bottom well made of No. 1 glass (Shenyou Biotechnology Co., Hangzhou, China) and transfected with 4 μg pEGFPN2-LHR-WT, pEGFPN2-LHR-Del7 as well as with pEGFPN2-LHR-WT and pCDNA3.1-LHR-Del7 at different ratios (1:1, 1:2, 1:5) using Lipofectamine 2000. The living cells were observed directly at 48 h after transfection with a confocal laser-scanning microscope (LSM5 Pascal, Zeiss, Oberkochen, Germany).
Western blot analysis
The plasmids pEGFPN2-LHR-WT and pCDNA3.1-LHR-Del7-3FLAG were co-transfected into 293T cells at different ratios (1:1, 1:2, 1:5) with Lipofectamine 2000 (Invitrogen). Cells were then collected, and the protein was extracted in RIPA lysis buffer containing phenylmethylsulphonyl fluoride 48 h after transfection. Proteins were separated on a 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel and then transferred to a nitrocellulose membrane. Anti-FLAG antibody (Sigma) and anti-GFP (Cell signal) were used to detect LHR-Del7 and LHR-WT, respectively. Anti-β-actin (Abcam) was used as a loading control. A western blotting luminal reagent (Thermo) was used to detect the signals. All experiments were repeated at least twice.
Immunofluorescence
293T cells grown on poly-l-lysine pretreated coverslips were transfected with pEGFPN2-LHR-WT and pCDNA3.1-LHR-Del7-3FLAG at a ratio of 1:1. Forty-eight hours after transfection, cells were fixed by 4% paraformaldehyde and anti-FLAG antibody (sigma) was used as the primary antibody. Then cells were incubated with anti-murine immunoglobulin G (Alexa Fluor 568 mouse, Invitrogen) for 1 h at 37°C. After a final washing step, slides were mounted with (Prolong gold antifade reagent) (Invitrogen) and analyzed in a confocal fluorescence system (LSM5 Pascal).
Receptor-binding assay
Receptor-binding assays were performed in both intact cells and detergent-solubilized extracts as described previously (Qiao et al., 2009). Intact cells (2 × 106) and detergent-solubilized receptors were incubated overnight at room temperature and 4°C, respectively, with increasing amounts of unlabeled hCG (0.1–l00 ng/tube) in the presence of [125I]-hCG (70 000 cpm/0.5 ng, Perkin-Elmer, MA, USA). Non-specific binding was determined in samples containing an excess of unlabeled hCG (100 IU, Sigma, St. Louis, MO, USA). All experiments were done in triplicate. Non-linear regression curve-fitting was performed with GraphPad Prism v. 5.0 (GraphPad Software Inc., San Diego, CA, USA).
Statistical analysis
The statistical significance of differences between groups was determined by the two-tailed-test. P< 0.05 was considered significant.
Results
Aberrant splicing of LHR mRNA
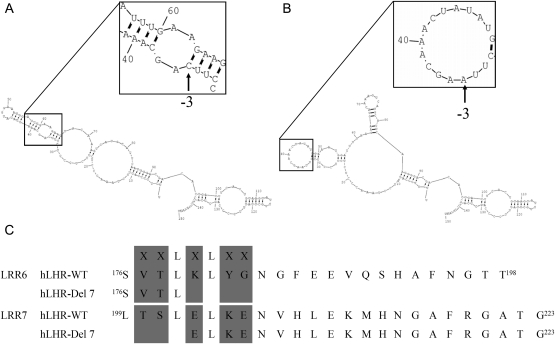
According to in vitro experiments, the splice site mutation in intron 6 (IVS6-3C>A) causes an aberrant splicing product of LHR. Using an RNA structure prediction program, we investigated whether the secondary RNA structure might be altered in this splicing variant. We analyzed the secondary structure of the wild-type LHR and then introduced the C>A mutation at the −3 acceptor splice site of intron 6. The native human LHR intron 6 −3 acceptor splice site is localized in a short stem (Fig. 1A inset), which is necessary for the recognition of the 3′ splicing site; however, in the mutated LHR pre-mRNA, the −3 acceptor splice site of intron 6 is situated in a loop (Fig. 1B inset) that contains no complementary sequences. The global secondary structure and the number of stems and loops are greatly changed after this mutation. However, the overall free energy of the molecule does not change dramatically (wild type, −22.1; Del7, −21.6). Compared with wild-type LHR, the LHR-Del7 protein lacks 23 amino acids that comprise part of the hydrophobic core of LRR6 and LRR7 (Fig. 1C).
Figure 1.
Prediction of secondary RNA structure of LHR. (A) Secondary RNA structure of wild-type LHR. The nucleotide sequence (150 bp) covering 37 bp 5′ and 44 bp 3′ of the exon was analyzed by the program RNAstructure. Regions covering the 3′ acceptor splice site were given in the inset at a higher magnification. IVS6-3 acceptor splice sites are indicated by arrows. (B) Secondary RNA structure of mutant LHR. The free energy (△G) calculated by RNAstructure program was not significantly different than that of the wild-type transcript. (C) LRR6 and LRR7 of LHR-WT and LHR-Del7. LHR-Del7 caused an in-frame deletion of 23 amino acids. The XXLXLXX motif constituted the hydrophobic core for hormone binding. LHR-Del7 destroyed the hydrophobic core in LRR6 and LRR7. (The concave surface of the LRR region was shown in gray).
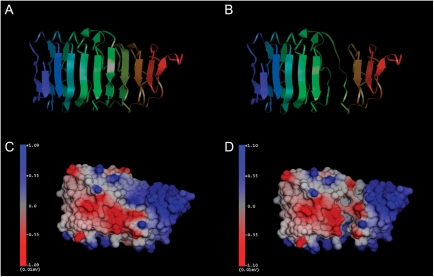
Molecular modeling of LHR-Del7 LRRs
To further understand the structural changes in the LRRs of LHR-Del7, we constructed a model using the recently developed crystal structure of the ECD of human FSH receptor as the major template. In the wild-type LHR, LRRs are arranged regularly (Fig. 2A), while in LHR-Del7, LRRs 6 and 7 were completely destroyed (Fig. 2B). Compared with the wild-type LHR (Fig. 2C), the local solid surface and electrostatic potential of LRR 6 and 7 were greatly changed in LHR-Del7 (Fig. 2D).
Figure 2.
Spatial conformations of the LRR domains in LHR-WT and LHR-Del7. (A) LRRs were arranged regularly in the wild-type LHR protein (view vertical to the β-strands). The model was shown in cartoon ribbon style and colored by index. (B) The sixth and seventh LRRs were completely destroyed in LHR-Del7. (C) A space-filing model of wild-type LHR colored by electrostatic potential. Blue represented positive charge and red represents negative charge. (D) In the LHR-Del7 protein, the local solid surface and electrostatic potential of LRR 6 and 7 were greatly changed.
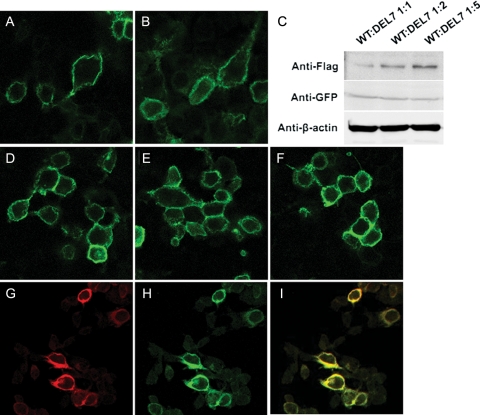
Subcellular location of LHR-Del7 and its influence on wild-type LHR
Cell surface expression of the wild and mutant receptors were analyzed on 293T cells transiently transfected with cDNA of either receptor type. Like wild-type LHR (Fig. 3A and H), the LHR-Del7-EGFP fusion protein was mainly localized to the cell membrane (Fig. 3B). In order to study the interaction between LHR-Del7 and LHR-WT, we co-expressed LHR-WT with LHR-Del7 in different ratios (Fig. 3D 1:1, Fig. 3E 1:2 and Fig. 3F 1:5). By using western blot analysis, we observed that expression of LHR-Del7 was gradually increased when co-transfected with LHR-WT (Fig. 3C). However, there were no detectable changes of LHR-WT-EGFP fusion protein at the cell surface with increasing amounts of LHR-Del7. Cells co-transfected with cDNA of either receptor type were incubated with FLAG monoclonal antibody, then strong cell surface staining was observed (Fig. 3G and H). Confocal microscopy analysis showed that both LHR-WT and LHR-Del7 co-localized in the cell membrane (Fig. 3I) (Fig. 3A, B, D, E, F, G and H, I; ×400). Similar results were obtained using myc-tagged LHR-del7 and immunofluorescent antibodies (Supplementary data, Fig. S1).
Figure 3.
Subcellular location of LHR variant in 293T cells. (A and H) LHR-WT was subcloned into pEGFPN2 and the resulting plasmid was transiently transfected into 293T cells. The expressed LHR-WT protein was localized to the plasma membrane. (B) LHR-Del7 was subcloned and expressed as described for WT; the expressed mutant protein was mainly localized to the plasma membrane. (C) Western blot showed the expression of LHR-Del7 and LHR-WT with anti-FLAG and anti-GFP antibody respectively. (D–F) 293T cells were co-transfected with pGFPN2-LHR-WT and pCDNA3.1-LHR-Del7 at different ratios: 1:1 (D), 1:2 (E) and 1:5. (F) LHR-WT localized to the plasma membrane in all cases. (G) Immunofluorescence analysis detected cell surface expression of LHR-Del7 with anti-FLAG antibody. (I) Co-localization of LHR-Del7 and LHR-WT in the cell membrane (A, B, D, E, F, G, H, I; ×400).
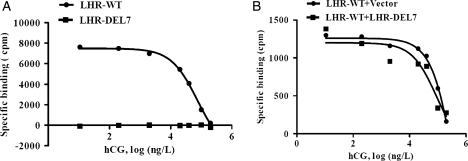
Radioactive ligand-binding assay
We then investigated the hormone-binding ability of LHR-Del7 with [125I]-labeled hCG. The specific binding of [125I]-hCG to detergent-solubilized receptors could be displaced by increasing concentrations of unlabeled hCG in cells transfected with LHR-WT. However, compared with wild-type LHR, LHR-Del7 exhibited almost no hormone-binding activity (Fig. 4A), indicating that LHR-Del7 could not bind to its cognate ligand. Intact cells co-expressing LHR-WT and LHR-Del7 showed similar hCG-binding affinity to cells co-expressing with LHR-WT and empty vector (Fig. 4B).
Figure 4.
Radioactive hCG (ligand) binding of LHR-Del7 and cAMP production of 293T cells stimulated by hCG. (A) 293T cells transiently transfected with LHR-WT and LHR-Del7 were used for this binding assay. [125I]-hCG displacement experiments were performed using detergent-solubilized membrane receptors. Detergent-solubilized receptor was incubated at 4°C with 0.5 ng [125I]-hCG and increasing concentrations of unlabeled hCG. (B) Receptor-binding assays using 293T cells co-transfected with LHR-WT/vector and LHR-WT/ LHR-Del7. Intact cells were incubated at room temperature with constant shaking in the presence of increasing amounts of unlabeled hCG (0.1–l00 ng/tube) and the designated amounts of [125I]-hCG.
hCG-induced cAMP production
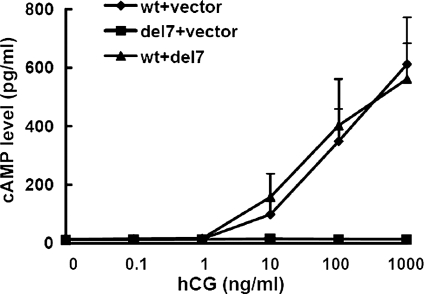
Despite its inability to bind hCG, the LHR-Del7 protein retains its transmembrane domains and C-terminal tail. Thus, we had to investigate whether LHR-Del7 was able to activate downstream signaling, such as cAMP production. 293T cells were stimulated with increasing concentrations of hCG (0–1000 ng/ml) after transient transfection with pCDNA3.1-LHR-WT/vector, pCDNA3.1-LHR-Del7/vector and pCDNA3.1-LHR-WT/pCDNA3.1-LHR-Del7. A robust increase in cAMP concentration was observed in cells transfected with pCDNA3.1-LHR-WT/vector and pCDNA3.1-LHR-WT/pCDNA3.1-LHR-Del7. In contrast, cAMP was not detected in cells transfected with pCDNA3.1-LHR-Del7/vector, suggesting that LHR-Del7 resulted in a dramatic impairment of signal transduction mediated by LHR (Fig. 5).
Figure 5.
cAMP production of 293T cells stimulated by hCG. 293T cells were transiently transfected with LHR-WT/vector, LHR-Del7/vector and LHR-WT/LHR-Del7. Cells were then stimulated with increasing dilutions of hCG and 0.25 mM IBMX for 30 min. The cAMP assay was performed after cell lysis according to the manufacturer's protocol. The values shown are the means ± SD of three independent experiments. There is no significant difference of cAMP production between LHR-WT/vector and LHR-WT/LHR-Del7 (P> 0.05).
Discussion
RNA splicing is carried out by the spliceosome, a complex assembly of small nuclear ribonucleoprotein particles composed of a variety of small nuclear RNAs and associated proteins. Most eukaryotic introns begin with GT and end with AG (GT-AG rule). Splice acceptor sites are pyrimidine-rich, and the C at position –3 is highly conserved in vertebrate genes (Krawczak et al., 1992). The nucleotide sequence of a pre-mRNA and its secondary structure are necessary elements for the spliceosome to recognize the splice donor and splice acceptor sites as well as for cleavage of the intron. Previously, we found that a naturally occurring IVS6-3C>A mutation changes the accessibility of 3′ splicing site as well as its binding with LHR pre-mRNA, leading to the alternative splicing of exon 7 in the LHR mRNA. In this study, we showed that the secondary structure of the RNA is altered by the 3′ splicing site mutation, which might underlie its aberrant splicing. As a result, exon 7 was skipped in the mutant LHR. We then studied the function of this splicing variant protein as well as its interaction with wild-type LHR.
The ECD of LHR, composed of nine LRRs motifs, is involved in the specific recognition and high-affinity ligand binding of the hormone hCG (Vischer et al., 2006). Crystal structure analysis indicated that consecutive LRRs are arranged in a horseshoe-like structure and that each LRR contains a short β-strand composed of a highly conserved X1X2LX3LX4X5 motif. The consecutive β-strands form the concave inner surface of the ECD, to which the cognate ligand can bind with multiple contact points (Vischer et al., 2006). The translated product of the splicing variant has a deletion of 23 amino acids from codon 176 to 199, which comprise part of LRR 6 and LRR 7, disrupting the sixth and seventh β-sheets of the ECD. A three-dimensional structure of LRRs in LHR predicted by molecular modeling showed that LRR 6 and 7 were completely destroyed in LHR-Del7. The local solid surface and electrostatic potential of LRR 6 and 7 were changed significantly in LHR-Del7, which reduced the binding affinity of the receptor to its ligand.
Consistent with the predicted protein structure, LHR-Del7 was localized to the cell membrane, but a radioactive ligand binding test verified that LHR-Del7 was unable to bind hCG. Furthermore, cAMP production was barely detectable in cells transfected with LHR-Del7. At the same time, cells co-expressing LHR-WT and LHR-Del7 generated cAMP in response to hCG. However, there was no significant difference between cells transfected with LHR-WT/vector and LHR-WT/LHR-Del7. This result indicated that LHR-Del7 does not have a dominant negative effect on the cell surface expression and activity of LHR-WT.
Other GPCRs in the LHR family are known to undergo significant alternative splicing, and alternative splicing of LHRs have also been reported in rat, pig, sheep, horse, cow and human corpora lutea (CL). Minegishi et al. (1997) discovered a natural splicing variant of human LHR that lacks exon 9 in a woman with a normal menstrual cycle. Further studies demonstrated that this variant could interact with wild-type LHR and attenuate the expression level of the wild-type LHR receptor (Nakamura et al., 2004). Then, Madhra et al. (2004) found three alternative splice variants of the LHR in human CL and luteinized granulose cells. Among them, a splice variant encoding a protein lacking the transmembrane and carboxyl terminal domains altered the function of wild-type LHR during luteolysis. It was therefore suggested that the expression of different LHR splicing variants may play a role in some physical processes, such as luteolysis (Dickinson et al., 2009). However, a rat LHR variant lacking the transmembrane and C-terminal domains failed to dimerize with full-length LHR in co-immunoprecipitation experiments (Apaja et al., 2006). This may indicate that the truncated variant regulates the function of wild-type LHR through other mechanisms. In this study, we proved that the pathological splicing variant LHR-Del7 has no similar effect on wild-type LHR.
The truncated rat variant was found to localize with the endoplasmic reticulum (ER) chaperones calnexin and calreticulin before being targeted for the ER-associated degradation pathway (Apaja et al., 2006). In vitro expression studies confirmed that the isoform lacking exon 7 mainly localizes to the membrane of 293T cells, unlike the LHR variant lacking exon 9. However, there was some fluorescence detected in the cytoplasm, so we could not exclude the possibility that some of the protein was retained in the ER due to misfolding. We also co-expressed LHR-WT with increasing concentrations of LHR-Del7. Western blot analysis was used to confirm the expression of LHR-WT and LHR-Del7. However, no change in the fluorescence in the plasma could be detected after co-transfection with LHR-WT and LHR-Del7 in different ratios. Our experiment also disclosed the co-localization of LHR-WT and LHR-Del7 in cell membrane, which suggested the splicing variant of LHR-Del7 could be transported to the cell membrane. Furthermore, it was possible that LHR-WT and LHR-Del7 could dimerize together, but since each was normally trafficked to the cell surface, the dimerization would not have an effect on the cell surface expression of LHR-WT.
Until now, two deletion mutants in the ECD of LHR (Del 8, Del 10) were identified in patients with LCH (Laue et al., 1996; Gromoll et al., 2000). A 2- to 3-year-old child with a small penis and severe hypospadias was reported to be a compound heterozygote for LHR gene. He had a deletion of exon 8 on one allele together with a Ser616Tyr substitution in the seventh transmembrane span of the receptor. The deletion of exon 8 abolished hormone binding and cyclase stimulation. Furthermore, a homozygous deletion of ∼5 kb encompassing exon 10 of the LHR gene was revealed in a patient displaying normal male phenotype associated with delayed pubertal development and hypogonadism. However, the properties of LHR lacking exon 10 have not been fully studied. Muller et al. (2003) reported that LHR without exon 10 binds both LH and hCG, and that the second messenger cascade is activated by hCG but not LH. Moreover, LHR without exon 9 is also unable to bind hCG at the cell surface. So, it was assumed that the β-subunit of hCG may be more flexible than that of LH. Based on our experimental result, LHR-Del7 was not able to bind with hCG. Furthermore, from the phenotype of our patient, the testosterone level was low even under the remarkable elevated LH level. Thus, the splicing variant abolished the binding affinity with two hormones. However, further studies will still be needed to fully describe the response of LHR-Del7 to LH. Our patient presented with some degree of masculinization of the external genitalia, which was confirmed to be caused by low levels of endogenous wild-type transcripts whose function was not influenced by aberrant splicing transcripts LHR-Del7.
In conclusion, the splicing variant LHR-Del7 produced in a LCH patient was able to localize to the cell membrane, but failed to bind hCG and had no effect on wild-type LHR. This work also expanded our understanding of the functional properties of LHR. Based on these results and the accumulated knowledge of other forms of LHR splicing variants, we suggest that wild-type LHR might be regulated by physiological splicing variants.
Supplementary data
Supplementary data are available at http://molehr.oxfordjournals.org/.
Authors’ roles
B.H. conducted the cell biology experiments and data analysis. L.-q.X. participated in the western blot. J-h.M., B-l.L. and C.-m.P. supported radioactive ligand-binding experiment. J.-j.W., Z.-q.W. and S.-x.Z. conducted the plasmid construction and partly participated in ELISA. X.C., W.L. and Y.-l.L. conducted data collection and analysis. J.Q. and W.-l.W. partly participated in the experiment design and wrote the paper. H.-d.S. participated in design the experiments and revised the final manuscript.
Funding
This work was supported by the grant from National Natural Science Foundation Program (Grant number: 81070666 and 30900503) and Shanghai Ninth People's Hospital (Grant number: JY2011A13).
Acknowledgements
The authors are very grateful to Prof. Aaron J.W. Hsueh (Division of Reproductive Biology, Department of Obstetrics and Gynecology, Stanford University School of Medicine) for providing the pcDNA3.0 LHR-WT cDNA recombinant plasmid.
References
- Aatsinki JT, Pietila EM, Lakkakorpi JT, Rajaniemi HJ. Expression of the LH/CG receptor gene in rat ovarian tissue is regulated by an extensive alternative splicing of the primary transcript. Mol Cell Endocrinol. 1992;84:127–135. doi: 10.1016/0303-7207(92)90079-l. [DOI] [PubMed] [Google Scholar]
- Apaja PM, Tuusa JT, Pietila EM, Rajaniemi HJ, Petaja-Repo UE. Luteinizing hormone receptor ectodomain splice variant misroutes the full-length receptor into a subcompartment of the endoplasmic reticulum. Mol Biol Cell. 2006;17:2243–2255. doi: 10.1091/mbc.E05-09-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL Workspace: a web-based environment for protein structure homology modeling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Ascoli M, Fanelli F, Segaloff DL. The lutropin/choriogonadotropin receptor, a 2002 perspective. Endocr Rev. 2002;23:141–174. doi: 10.1210/edrv.23.2.0462. [DOI] [PubMed] [Google Scholar]
- Bacich DJ, Rohan RM, Norman RJ, Rodgers RJ. Characterization and relative abundance of alternatively spliced luteinizing hormone receptor messenger ribonucleic acid in the ovine ovary. Endocrinology. 1994;135:735–744. doi: 10.1210/endo.135.2.7518389. [DOI] [PubMed] [Google Scholar]
- Chesler AT, Zou DJ, Le Pichon CE, Peterlin ZA, Matthews GA, Pei X, Miller MC, Firestein S. A G protein/cAMP signal cascade is required for axonal convergence into olfactory glomeruli. Proc Natl Acad Sci USA. 2007;104:1039–1044. doi: 10.1073/pnas.0609215104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson RE, Stewart AJ, Myers M, Millar RP, Duncan WC. Differential expression and functional characterization of luteinizing hormone receptor splice variants in human luteal cells: implications for luteolysis. Endocrinology. 2009;150:2873–2881. doi: 10.1210/en.2008-1382. [DOI] [PubMed] [Google Scholar]
- Elmhurst JL, Xie Z, O'Dowd BF, George SR. The splice variant D3nf reduces ligand binding to the D3 dopamine receptor: evidence for heterooligomerization. Mol Brain Res. 2000;80:63–74. doi: 10.1016/s0169-328x(00)00120-0. [DOI] [PubMed] [Google Scholar]
- Fan QR, Hendrickson WA. Structure of human follicle-stimulating hormone in complex with its receptor. Nature. 2005;433:269–277. doi: 10.1038/nature03206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromoll J, Eiholzer U, Nieschlag E, Simoni M. Male hypogonadism caused by homozygous deletion of exon 10 of the luteinizing hormone receptor: differential action of the luteinizing hormone (LH) and human chorionic gonadotropin (hCG) J Clin Endocrinol Metab. 2000;85:2281–2286. doi: 10.1210/jcem.85.6.6636. [DOI] [PubMed] [Google Scholar]
- Grosse R, Schöneberg T, Schultz G, Gudermann T. Inhibition of gonadotropin-releasing hormone receptor signaling by expression of a splice variant of the human receptor. Mol Endocrinol. 1997;11:1305–1318. doi: 10.1210/mend.11.9.9966. [DOI] [PubMed] [Google Scholar]
- Kawate N, Okuda K. Coordinated expression of splice variants for luteinizing hormone receptor messengerRNAduring the development of bovine corpora lutea. Mol Reprod Dev. 1998;51:66–75. doi: 10.1002/(SICI)1098-2795(199809)51:1<66::AID-MRD8>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- Laue LL, Wu SM, Kudo M, Bourdony CJ, Cutler GB, Hsueh AJ, Chan WY. Compound heterozygous mutations of the luteinizing hormone receptor gene in Leydig cell hypoplasia. Mol Endocrinol. 1996;10:987–997. doi: 10.1210/mend.10.8.8843415. [DOI] [PubMed] [Google Scholar]
- Loosfelt H, Misrahi M, Atger M, Salesse R, Vu Hai-Luu Thi MT, Jolivet A, Guiochon-Mantel A, Sar S, Jallal B, Garnier J, et al. Cloning and sequencing of porcine LH-hCG receptor cDNA: variants lacking transmembrane domain. Science. 1989;245:525–528. doi: 10.1126/science.2502844. [DOI] [PubMed] [Google Scholar]
- Madhra M, Gay E, Fraser HM, Duncan WC. Alternative splicing of the human luteal LH receptor during luteolysis and maternal recognition of pregnancy. Mol Hum Reprod. 2004;10:599–603. doi: 10.1093/molehr/gah076. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minegishi T, Tano M, Abe Y, Nakamura K, Ibuki Y, Miyamoto K. Expression of luteinizing hormone/human chorionic gonadotrophin (LH/HCG) receptor mRNA in the human ovary. Mol Hum Reprod. 1997;3:101–107. doi: 10.1093/molehr/3.2.101. [DOI] [PubMed] [Google Scholar]
- Muller T, Gromoll J, Simoni M. Absence of exon 10 of the human luteinizing hormone (LH) receptor impairs LH, but not human chorionic gonadotropin action. J Clin Endocrinol Metab. 2003;88:2242–2249. doi: 10.1210/jc.2002-021946. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Yamashita S, Omori Y, Minegishi T. A splice variant of the human luteinizing hormone receptor modulates the expression of wildtype human LH receptor. Mol Endocrinol. 2004;18:1461–1470. doi: 10.1210/me.2003-0489. [DOI] [PubMed] [Google Scholar]
- Qiao J, Han B, Liu BL, Chen X, Ru Y, Cheng KX, Chen FG, Zhao SX, Liang J, Lu YL, et al. A splice site mutation combined with a novel missense mutation of LHCGR cause male pseudohermaphroditism. Hum Mutat. 2009;30:E855–E865. doi: 10.1002/humu.21072. [DOI] [PubMed] [Google Scholar]
- Saint-Dizier M, Chopineau M, Dupont J, Daels PF, Combarnous Y. Expression and binding activity of luteinizing hormone/chorionic gonadotropin receptors in the primary corpus luteum during early pregnancy in the mare. Biol Reprod. 2003;69:1743–1749. doi: 10.1095/biolreprod.103.018812. [DOI] [PubMed] [Google Scholar]
- Vischer HF, Granneman JCM, Bogerd J. Identification of follicle-stimulating hormone-selective β-strands in the N-terminal hormone-binding exodomain of human gonadotropin receptors. Mol Endocrinol. 2006;20:1880–1893. doi: 10.1210/me.2005-0202. [DOI] [PubMed] [Google Scholar]