Abstract
The nucleotide excision repair (NER) machinery, the primary defense against cancer-causing bulky DNA lesions, is surprisingly inefficient in recognizing certain mutagenic DNA adducts and other forms of DNA damage. However, the biochemical basis of resistance to repair remains poorly understood. In order to address this problem, we have investigated a series of intercalated DNA-adenine lesions derived from carcinogenic polycyclic aromatic hydrocarbon (PAH) diol epoxide metabolites that differ in their response to the mammalian NER apparatus. These stereoisomeric PAH-derived adenine lesions represent ideal model systems for elucidating the effects of structural, dynamic and thermodynamic properties that determine the recognition of these bulky DNA lesions by NER factors. The objective of this work was to gain a systematic understanding of the relation between aromatic ring topology and adduct stereochemistry with existing experimental NER efficiencies and known thermodynamic stabilities of the damaged DNA duplexes. For this purpose, we performed 100 ns molecular dynamics studies of the lesions embedded in identical double-stranded 11-mer sequences. Our studies show that, depending on topology and stereochemistry, stabilizing PAH-DNA base van der Waals stacking interactions can compensate for destabilizing distortions caused by these lesions that can, in turn, cause resistance to NER. The results suggest that the balance between helix stabilizing and destabilizing interactions between the adduct and nearby DNA residues can account for the variability of NER efficiencies observed in this class of PAH-DNA lesions.
Keywords: polycyclic aromatic hydrocarbon-adenine adducts, nucleotide excision repair, fjord region, bay region, stereochemistry
Nucleotide excision repair (NER) is a key mammalian defense mechanism against promutagenic bulky polycyclic aromatic DNA lesions. In the global genomic repair pathway of NER, the heterodimeric XPC-RAD23B protein factor recognizes the local distortion/destabilization of the DNA caused by the lesion and binds to the damaged site causing local strand separation.1–3 Other factors are subsequently recruited to this site that further enhance the separation of the two strands, and ultimately lead to the excision of oligonucleotides 24 – 32 nucleotides in length that contain the lesion.1–11 A crystal structure of the yeast S. cerevisiae NER recognition factor Rad4/Rad23 (a homologue to the human XPC-RAD23B NER factor) bound to a cyclobutane pyrimidine dimer, revealed novel insights into the NER recognition mechanism.12 While the exact position of the lesion could not be ascertained, a β-hairpin was found to be inserted between the two strands at the lesion site, and the two bases opposite the lesion were flipped out of the duplex and bound to Rad4/Rad23.12 This structure suggests that the local thermodynamic stability at the lesion site play an important role in determining the ease or difficulty of DNA strand separation and concerted base-flipping, which may facilitate or hinder β-hairpin insertion.12–14 However, the relative incision efficiencies for structurally different adducts vary over several orders of magnitude,4, 15 and the molecular origins of these differences are not well understood. It has been shown experimentally that factors which impact the relative excision efficiencies include the chemical structures of the lesions and base sequence contexts in eukaryotic16–20 and prokaryotic21–27 systems. Recently, we reported that the relative excision efficiencies, catalyzed by prokaryotic and eukaryotic NER systems, bear some resemblance to one another for certain polycyclic aromatic and other DNA lesions;28 these results were interpreted in terms of similar β-hairpin insertion mechanisms since a crystal structure of a complex of the prokaryotic UvrB protein with a damaged DNA duplex showed that the β-hairpin plays an analogous role in prokaryotic NER systems.29 Consistent with a thermodynamic probing mechanism of DNA damage, we found that the interactions between polycyclic aromatic carcinogen-DNA lesions can, in addition to inducing destabilizing structural distortions, also generate stabilizing van der Waals stacking interactions.20 We hypothesized that even bulky polycyclic aromatic lesions can be resistant to NER if these stabilizing van der Waals interactions compensate for the destabilizing structural distortions that facilitate β-hairpin insertion. However, incision efficiencies depend also on damage processing events that are downstream of lesion recognition, but are not yet well understood.28
The objectives of the present work were to explore the balance between stabilizing carcinogen–DNA interactions and destabilizing structural distortions associated with structurally related but topologically and stereochemically different sets of polycyclic aromatic hydrocarbon (PAH)-derived N 6-adenine lesions in double-stranded DNA. In order to assess the overall impact of these interactions on NER efficiencies, we elucidated the dynamic, structural and energetic characteristics of six structurally or stereoisomerically different PAH-N 6-dA adducts (Figure 1, A and B) in double-stranded DNA. Our goals were to gain a systematic understanding of the relationships between adduct ring topology and adduct stereochemistry on carcinogen-DNA interactions and their impact on nucleotide excision repair efficiencies.
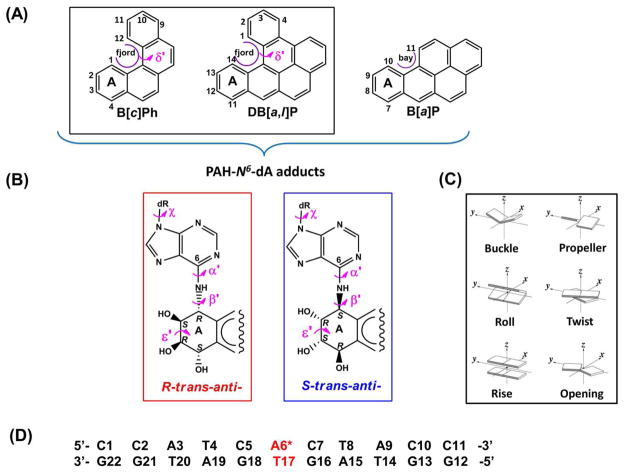
Figure 1.
Chemical structures, torsion angle definitions, and base sequence context. (A) Chemical structures of B[c]Ph, B[a]P, and DB[a,l]P with definitions of the fjord regions of B[c]Ph and DB[a,l]P and the bay region of B[a]P. The fjord region torsion δ′ is shown that defines the out of plane twist of the indicated aromatic ring. (B) Stereochemical structures (absolute configurations) of the 10S (+)- and 10R (−)-trans-anti-B[a]P-N 6 -dA, 1R (+)- and 1S (−)-trans-anti-B[c]Ph-N 6 -dA, and 14R (+)- and 14S (−)-trans-anti-DB[a,l]P-N 6 -dA adducts. The lesion-DNA linkage site torsion angles α′ and β′, glycosidic torsion angle χ, and benzylic ring pucker torsion angle ε′ are also designated. (C) Definitions of helicoidal parameters used to describe the geometry of base pairs and sequential base pair steps. The cartoons are reprinted with permission from 88. Copyright 2003 Oxford University Press. (D) The base sequence context in which the lesions are embedded. A6* designates the modified adenine residue.
Carcinogenicity and Topology of Parent PAHs
The parent PAHs are widespread environmental contaminants that are products of fossil fuel combustion and are therefore present in cigarette smoke, automobile exhaust, urban air and food.30–33 In this study we focused on three PAH compounds, the two fjord benzo[c]phenanthrene (B[c]Ph) and dibenzo[a,l]pyrene (DB[a,l]P), and the bay region benzo[a]pyrene (B[a]P) that have different numbers of aromatic rings (Figure 1A). These compounds belong to the two topologically distinct classes of PAH termed ‘fjord’ and ‘bay’ (Figure 1A). The sterically hindered fjord region compounds are partially nonplanar34 (Figure 1A) due to steric hindrance between the C1 and C12 (B[c]Ph) and C1 and C14 (DB[a,l]P) protons, respectively. By contrast, the bay region is sterically unhindered because the analogous C10 and C11 protons are too far apart and the aromatic ring system is thus fully planar and inflexible. DB[a,l]P is the most potent PAH tumorigen yet identified,33, 35–38 and B[a]P is classified by the IARC (International Agency for Research on Cancer)39 as a human carcinogen. While B[c]Ph is a weak carcinogen in rodents because it is not metabolically activated to the highly tumorigenic fjord B[c]Ph-3,4-dihydrodiol-1,2-epoxide, in humans B[c]Ph is metabolically activated to this diol epoxide and B[c]Ph is therefore suspected to be a potent human carcinogen.40 Overall, the tumorigenicity of the different PAH compounds is a consequence of combinations of factors that include metabolic activation, reactivity with DNA, mutagenicity of the adducts, and their repairability by cellular defense mechanisms such as NER.31, 33, 41, 42
Structures, stabilities, and NER susceptibilities of PAH-DNA adducts
In mammalian cells, PAH compounds are metabolized to highly reactive diol epoxide intermediates that bind predominantly to the exocyclic amino groups of adenine and guanine in DNA in a stereo-selective manner.43–47 The metabolized rings are denoted by “A” in Figure 1, A and B. The stereochemical properties of the DNA lesions formed have a strong impact on their structural features and the associated distortions of the B-DNA structure.48–52 For example, stereochemically different B[a]P diol epoxide -N 6-adenine and -N 2-guanine adducts exhibit variable stereochemistry-dependent conformations in double-stranded DNA.50–55
NMR studies and molecular modeling have provided structural characterization of the 1R (+)- and 1S (−)-trans-anti-B[c]Ph-N6-dA (R and S B[c]Ph-N6-dA), the 14R (+)- and 14S (−)-trans-anti-DB[a,l]P-N 6 -dA (R and S DB[a,l]P-N 6 -dA), and the 10R (−)- and 10S (+)-trans-anti-B[a]P-N 6 -dA (R and S B[a]P-N 6 -dA) adducts.53–62 The structural studies indicate that all adopt classical intercalation conformations in which the bulky aromatic ring systems are inserted between adjacent Watson-Crick base pairs.53, 55–59, 61, 62 The aromatic rings intercalate into the DNA by stretching and unwinding the double helix; the stereoisomeric lesions with S absolute configuration at the PAH-DNA linkage site are intercalated on the 3′-side of the damaged base while the R stereoisomers are intercalated on the 5′-side. For the DB[a,l]P adducts models were created based on the NMR structures of the B[c]Ph adducts57, 58 of identical stereochemistry and fjord region topology, together with spectroscopic data that revealed the intercalative nature of these adducts.63 The current 100 ns simulations for these DB[a,l]P adducts showed that structural, dynamic and energetic features were very similar to our previous 30 ns molecular dynamics (MD) simulations.56
Thermal melting data has shown that the fjord R DB[a,l]P-N 6-dA adduct stabilizes double-stranded DNA relative to the unmodified duplex, while the S stereoisomer is somewhat destabilizing. However, both R and S stereoisomeric adducts derived from B[a]P are destabilizing and the impact of the lesion is much greater for the S than the R adduct64, 65; this greater destabilization has been shown to result from steric hinderance between the benzylic rings of the PAH-DNA adducts and neighboring DNA base or backbone atoms that are more pronounced in the case of the 3′-side S- than the 5′-side R-intercalation.53, 56, 59 We term this phenomenon the “S destabilization effect”. However, in the case of the compact B[c]Ph adducts, the DNA duplex melting profiles show that neither of the two stereoisomeric adducts either stabilizes or destabilizes double-stranded DNA.65, 66 Interestingly, the R and S B[c]Ph-N 6 -dA and DB[a,l]P-N 6-dA adducts are resistant to NER.67 By contrast, the R and S B[a]P-N 6 -dA adducts are substrates of NER with excision efficiencies that depend on base sequence context.59, 62, 67
The overall NER process includes the recognition of the DNA lesions as the key rate-determining step.3, 4, 7 In order to determine how the structural features of the different PAH-N 6-dA lesions might affect this recognition process, we have carried out 100 ns MD studies of the R and S B[c]Ph-N 6-dA, B[a]P-N 6-dA, and DB[a,l]P-N 6-dA adducts embedded in the sequence context shown in Figure 1D, as well as an unmodified control duplex. Our results have elucidated how the different distorting and stabilizing properties together might affect the NER efficiency.
Materials and Methods
Starting structures and force field
The initial models for the R and S B[c]Ph-N 6-dA adducts were the high resolution NMR solution structures (R: 57; S: 58). The initial models for the R and S B[a]P-N 6-dA adducts were the minimized average structures from our prior 2.0 ns unrestrained MD simulations.62 Base sequences were remodeled to match those shown in Figure 1C. Initial models are presented in Figure S1, Supporting Information. The MD simulations of 30 ns for the R and S DB[a,l]PN 6-dA adducts, extended here to 100 ns, were reported previously.56
MD simulations were carried out using SANDER in the AMBER 9 simulation package.68 The Cornell et al. force field,69 parm99.dat parameter set70 modified by parmbsc071 were employed for all simulations. Partial charges and other added parameters for the R and S B[c]Ph-N 6-dA adducts,72 B[a]P-N 6-dA adducts,62 and DB[a,l]P-N 6-dA adducts56 on the nucleoside level were reported previously.
MD computation protocols
Details of the MD protocols are given in Supplementary Materials. The stability of the MD simulation was evaluated for each model. For each sequence context, the root-mean-square deviation (RMSD) of each snapshot in the trajectory relative to its respective starting structure was plotted as a function of time and is shown in Figure S2, Supporting Information. Also given in Figure S2 are the average RMSD values with standard deviations of all atoms, excluding two base pairs at each end, in the current structure against the initial model for the 100 ns MD simulations. For all except the S B[a]P-N 6-dA case, MD achieved good stability, fluctuating around the mean after 10 ns, and we employed the structural ensembles from the 10 ns to 100 ns time frame for further analyses. For the S B[a]P-N 6-dA adduct, MD achieved good stability only after 45 ns; therefore in order to acquire an additional 90 ns of MD for analysis, as for the other cases, we ran the simulation to 135 ns and analyzed the 45 ns to 135 ns time frame.
Structural analyses
The PTRAJ module of the AMBER 9 package68 was employed for structural analyses. The CARNAL module of the AMBER 7 package73 was utilized to compute hydrogen bond occupancies. Frames were selected at 5 ps intervals from the last 90 ns of simulation. DNA duplex helicoidal parameters and groove dimensions were analyzed using MD Toolchest;74, 75 for the groove dimensions, 5.8 Å was subtracted from the pairwise phosphorous-phosphorous distances to account for the van der Waals radius of the P atoms.76 The first and last base pairs were excluded in this analysis due to possible end effects. Visualization and model building were performed with the INSIGHT II 2005 program (Accelrys Software, Inc.). PyMOL (Delano Scientific, LLC.)77 was employed to make molecular images and movies. Computations were carried out on our own cluster of Silicon Graphic Origin and Altix high-performance computers, and at the National Science Foundation Texas Advanced Computing Center.
Stacking interactions
Stacking interactions were estimated by computing the van der Waals interaction energies between the PAH aromatic rings and all the adjacent base pairs, utilizing the ANAL module of the AMBER 9 package.68
Distortion energy
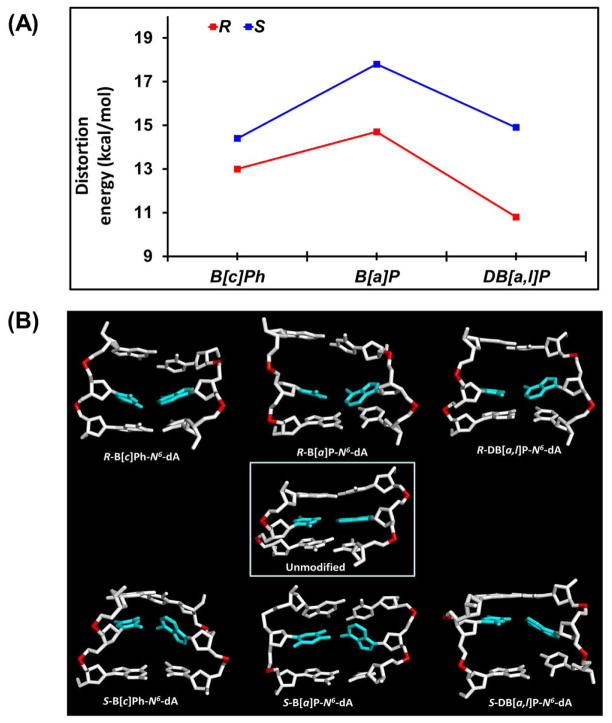
We evaluated a distortion energy due to adduct intercalation for our simulated structures, following the protocol in Wu et al72 from our group. In this approach, we compute the energy required to distort the unmodified duplex to the intercalated conformation adopted by the adduct. First, we truncated the lesion-containing 11-mer to a 3-mer, that is the damaged base pair and the 3′- and 5′-side neighboring base pairs. Then, we substituted the lesion with a hydrogen atom in each modified duplex and obtained a structure which contained the intercalation pocket without the lesion. We then computed the molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) energies78 for each adduct. Next, we subtracted from this energy the MM-PBSA energy of the unmodified duplex, which had also been truncated to the central 3-mer 5′-CAC-3′ level. Therefore, this value determines the energy required to distort the central 3-mer DNA duplex to accommodate the lesion in the intercalation pocket.
Results
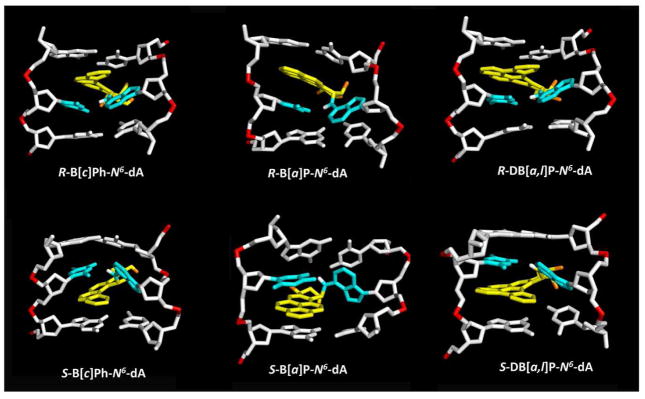
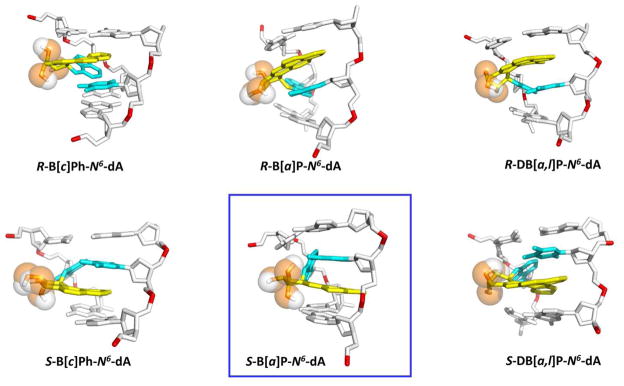
We utilized molecular modeling based on NMR solution structures (See Methods) and 100 ns MD simulations with MM-PBSA energy analyses to investigate classically intercalated conformations of six topologically and stereochemically different PAH-N6-dA adducts in double-stranded DNA, specifically the R and S fjord B[c]Ph-N 6 -dA, R and S bay region B[a]P-N 6 -dA, and R and S fjord DB[a,l]P-N 6 -dA lesions in double-stranded 11-mer oligonucleotides (Figure 1). These adducts are embedded in the identical sequence context 5′-…CA*C…-3′ (Figure 1D). All six PAH-N 6 -dA adducts maintain classical intercalation conformations during the entire MD simulations. The lesion is intercalated from the major groove side in the case of the S stereoisomer on the 3′-side, and in the case of the R stereoisomer on the 5′-side of the damaged base (Figure 2). The stretching and unwinding of the double helix that accompany the insertion of the bulky aromatic ring systems between adjacent base pairs are signature distortions of classical intercalation.79–81 These structural disturbances are accompanied by other correlated and lesion-specific distortions of the DNA structure. As a control, we also investigated the structural features of the corresponding unmodified duplex 5′-…CAC….-3′ (Figure 1D). The opposite orientations of the S and R stereoisomeric adducts are reflected in the opposite values of their structural parameters (Table 1). These include the linkage site torsion angles α′ and β′, as well as the fjord region twist angle δ′ (defined in Figure 1A), benzylic ring pucker ε′ and base pair parameters Buckle and Propeller. Of particular interest is the parameter δ′ that governs the non-planarity of the fjord region: opposite signs of this parameter and the corresponding opposite directionalities reflect directly the fjord region flexibility, which can twist the single aromatic ring protruding into the fjord region in opposite directions to optimize carcinogen-base stacking interactions within the intercalation pocket in each of the stereoisomeric adducts (Figure 2).
Figure 2.
Best representative structures96 from our work. Only the central 3-mers are shown. The view is looking into the minor groove. For the B[c]Ph, B[a]P and DB[a,l]P moieties, the carbon atoms are in yellow and the oxygen atoms in orange. The damaged bases are cyan, and the DNA duplexes are in white, except for the phosphorus atoms, which are in red color. Hydrogen atoms are not displayed for clarity, except for the hydrogen bonding N 6 proton at the lesion modified adenine. Movies S 1–6, showing these structures rotating in both stick and CPK rendering, are provided in the Supporting Information.
Table 1.
Structural parameters of adducts that characterize opposite orientations in S and R stereoisomers.a
| B[c]Ph-N6-dA | B[a]P-N6-dA | DB[a,l]P-N6-dA | Unmodified | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| R | S | R | S | R | S | ||
| Buckle (°) | −30.5 (7.7) | 43.4 (8.2) | −36 (7.5) | 36.4 (8.1) | −26.4 (7.9) | 28.2 (7.5) | 4.4 (11.1) |
| Propeller (°) | 25.1 (8.5) | −36 (10.0) | 36.4 (9.5) | −48.2 (12.5) | 21.4 (8.9) | −24.5 (7.6) | −6.8 (9.1) |
| α′ (°) | −11.6 (8.9) | 11.5 (9.9) | −26.4 (8.8) | 18.4 (10.4) | −13.8 (8.1) | 13.3 (8.1) | N/A |
| β′ (°) | 101.5 (8.1) | −94.9 (10.1) | 101.2 (9.9) | −106.7 (11.5) | 103.0 (7.5) | −100.0 (7.5) | N/A |
| ε′ (°) | −62.0 (5.0) | 60.4 (5.1) | −59.9 (5.2) | −57.8 (5.9) | −62.7(4.9) | 61.5 (5.3) | N/A |
| δ′ (°) | 21.1 (6.6) | −20.1 (8.3) | N/A | N/A | 24.2 (6.2) | −23.3 (6.4) | N/A |
See Figure 1, A and B, for definitions of α ′, β ′, ε′ and δ′. Values given are ensemble averages for the last 90 ns of the MD simulations. Standard deviations are given in parentheses. Helicoidal parameters are defined in Figure 1C. N/A, not applicable.
The “S destabilization effect” is a function of lesion topology
Watson-Crick hydrogen bonding is disrupted in the S B[a]P-N6-dA adduct
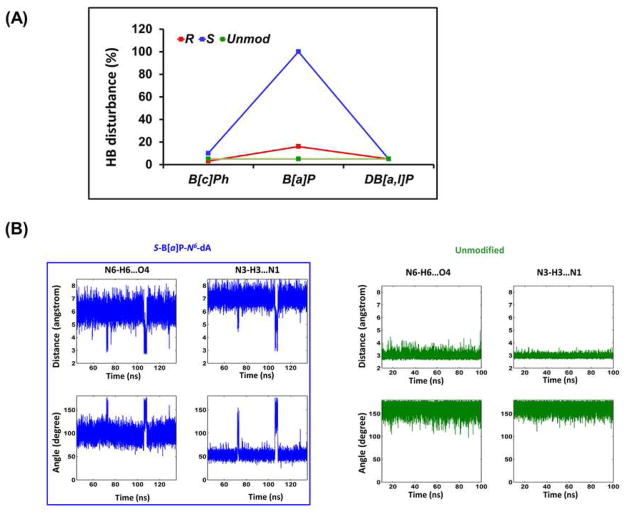
Perturbed Watson-Crick hydrogen bonding has been correlated with enhanced susceptibility to NER in a number of cases.7, 16, 18–20, 82, 83 We evaluated the percent hydrogen bond disruption for all of the six adducts. Only in the case of the S B[a]P-N 6-dA adduct is the Watson-Crick base pair at the lesion step, A6*:T17, severely disturbed with both hydrogen bonds disrupted (Figure 3 and Table S1, Supporting Information). The disruption of the hydrogen bonding at A6*:T17 is due to steric hindrance resulting from the crowding between the benzylic ring (ring A, Figure 1B) and DNA atoms (Figures 1, A and B), combined with the rigidity of the planar B[a]P ring system; together, these factors require rupturing of the A6*:T17 base pair to accommodate the B[a]P moiety in the intercalation pocket. This observation is consistent with the high resolution NMR solution structures that display conformational heterogeneity and strongly disrupted hydrogen bonding.54, 60, 84 However, the hydrogen bonds of the base pair C7:G16 flanking the intercalated aromatic B[a]P ring system on the 3′-side of A6*:T17, as well as all other base pairs are essentially undisturbed (Table S1, Supporting Information). By contrast, the other five lesion-containing duplexes, as well as the unmodified duplex, maintain intact Watson-Crick hydrogen bonding at the lesion site and at the adjacent base pairs, with only 16% (R-B[a]P) or less of the population exhibiting any Watson-Crick hydrogen bond disruption (Table S1A, Supporting Information).
Figure 3.
Watson-Crick hydrogen bonding at the A6*:T17 base pair featuring the complete disruption in the S-B[a]P-N 6-dA adduct case. (A) Percent disruption of hydrogen bonds at A6*:T17. The values for the most disrupted of the two hydrogen bonds in the Watson-Crick base pair are shown. The full data set for the central 3-mer is given in Table S3A, Supporting Information. Our hydrogen bond quality index reveals the same pattern of distortions (Table S3B, Supporting Information). (B) Time dependence of hydrogen bond distances and angles for the A6*:T17 in the S-B[a]P-N 6-dA adduct, and at A6:T17 for the unmodified control, showing disrupted and dynamic distances and angles at the A6*:T17 base pair. The time dependent fluctuations of hydrogen bond distances and angles for all adducts are shown in Figure S3, Supporting Information; these reveal that all are similar to the unmodified control, except for the S-B[a]P-N 6-dA adduct.
Distortions in groove dimensions and helicoidal parameters have also been correlated with susceptibilities of some PAH-DNA lesions to NER;1, 16, 19 however, these properties are generally not independent of one another,85–87 and some parameters, notably the Opening parameter,88 are correlated with Watson-Crick hydrogen bonding. Concomitant with the greatest hydrogen bond disruption, the S B[a]P-N 6-dA adduct has the largest and most dynamic base pair Opening value (Figure 1C) at A6*:T17 (15.4 ± 12.1° vs. 2.2 ± 6.2° in the unmodified case, Table S2, Supporting Information). In addition, the intercalation-induced unwinding is the most dynamic of all cases in the S B[a]P-N 6-dA adduct, as reflected in its largest standard deviation: the Twist angle is 11.2 ± 14.8°, while for the unmodified DNA at the same step it is 32 ± 5.4°; this greater flexibility is attributed to the disrupted hydrogen bonds at the lesion site (Figure 3B).
The S DB[a,l]P-N6-dA adduct is the most severely untwisted
The S DB[a,l]P-N 6-dA adduct has one more aromatic ring than the B[a]P-N 6-dA adduct, which defines the flexible aromatic fjord PAH region. However, in case of the S DB[a,l]P-N 6-dA adduct, the hydrogen bonding does not need to be disrupted to relieve the steric crowding, since this can be achieved by an out-of-plane twisting of the aromatic ring in the fjord region (designated as ‘δ′’ in Figure 1A)34 that helps to relieve the steric hinderance. Consequently, the “S destabilization effect” in the case of the DB[a,l]P-N 6-dA duplex manifests itself mainly in terms of severe unwinding required to accommodate the five aromatic rings in the intercalation pocket (Table S2, Supporting Information). The ensemble average value for the Twist parameter at the lesion site is −13.2° ± 6.0° while this value for the analogous step in the unmodified duplex is 32.0° ± 5.4°, a local unwinding of ~45°. This is the most severe unwinding observed for any adduct (although not the most dynamic, which was observed in the case of the S B[a]P-N 6-dA adduct). Coupled with the greatest unwinding, the S DB[a,l]P-N 6-dA adduct also features the most enhanced Roll value,85–87 with an ensemble average value of 21.8° ± 5.8°; these combined effects generate a bend towards the major groove side, which widens the minor groove (Table S3, Supporting Information). We note that intercalation always causes major and minor groove widening because it forces the phosphate groups further apart, as well as entailing the unwinding of the double helix;81 however, the extent of these effects is modulated by the impact of adduct stereochemistry and topology. Our previous, shorter 30 ns simulation for this adduct conformation uncovered the unwinding effect, as well as changes in other structural parameters and energetic properties; these adduct-induced changes remained nearly identical when the simulation time was extended to 100 ns for both the S and the R DB[a,l]P-N 6-dA adducts.56
The S B[c]Ph-N6-dA adduct manifests only minor structural distortions
The most notable findings are the smaller structural distortions for this adduct, compared to the great unwinding due to the “S destabilization effect” observed for the S DB[a,l]P-N 6-dA adduct. We observe a 10% disruption of the N6-H6…O4 hydrogen bond at the lesion site (Table S3A, Supporting Information) for the S stereoisomer, which allows for a large base pair Buckle (41.8 ± 8.4°) and Opening (19.3 ± 10.1°) (Table S2, Supporting Information). However, this represents a much lesser distortion of the hydrogen bonding than in the case of the bay-region B[a]P-derived adduct, where the hydrogen bonds are entirely ruptured. As in the case of the S DB[a,l]P-N 6-dA adduct, the flexibility and resulting twist of the aromatic ring protruding into the fjord region (designated as ‘δ′’ in Figure 1A) helps to relieve the steric hindrance that causes hydrogen bond rupturing in the bay region S B[a]P-N 6-dA adduct. However, the intercalation of the three aromatic rings in the S B[c]Ph-N 6-dA adduct requires much less unwinding than the five rings of the S DB[a,l]P-N 6-dA adduct. Figure 4B shows that the S stereoisomeric adducts with their various topologies are oriented differently in the intercalation pockets to optimize their stacking interactions; consequently, only the rigid bay region adduct manifests disrupted hydrogen bonding. By contrast, the fjord region adducts do not exhibit this kind of severe structural distortion, because of the flexible fjord region twist.
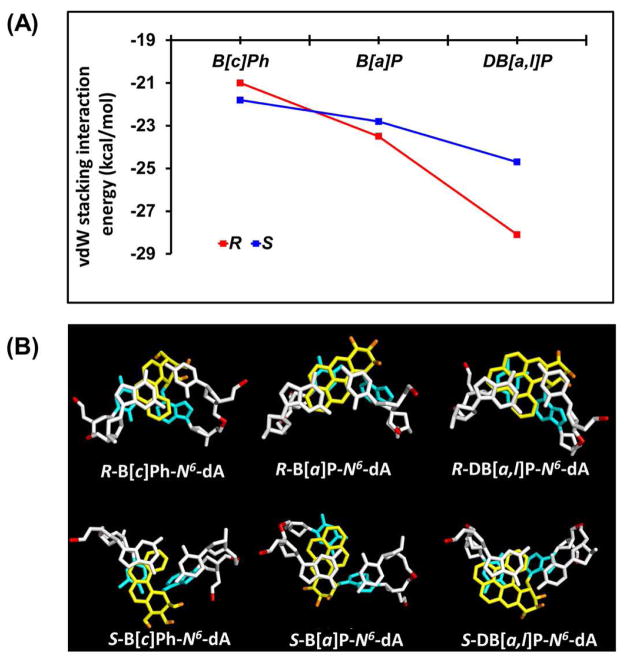
Figure 4.
Stabilizing carcinogen-base stacking interactions. (A) Ensemble average van der Waals interaction energies between aromatic rings and adjacent base pairs. See Table S4, Supporting Information, for values and standard deviations. (B) Views looking down the helix axis of the intercalation pockets showing the stacking interactions. The color code is the same as in Figure 2. Stereoviews are shown in Supporting Information Figure S5.
The rigid bay region topology in the R B[a]P-N6-dA adduct produces distorting enlargement in the Rise parameter
Among the R stereoisomeric adducts, the B[a]P-N 6-dA lesion stands out for manifesting by far the largest Rise of all of the adducts; the ensemble average value for the step at the lesion intercalation pocket (C5:G18-A6*-T17) is 8.8 ± 0.6 Å. Since the base pair at the lesion site is not ruptured as in the S adduct case, the rigid bay region ring system greatly enlarges the Rise to accommodate the intercalated B[a]P residue. Concomitantly, the minor groove is widened more than in any other adduct, a reflection of the enhanced stretching81 (Table S3, Supporting Information). By contrast, in the case of the R fjord region adducts, the flexible aromatic ring twist (Figure 1A) allows for optimal stacking in the intercalation pocket with much less stretching. The major groove is always widened by the intercalation of the N 6-dA adducts from that side; however, the extent of widening is modulated by the number of aromatic rings and their topology: e.g., the rigid, four-ring R B[a]P-N 6-dA adduct with the largest Rise, produces major groove widening similar to that of the five-aromatic ring R DB[a,l]P-N 6-dA adduct (Table S3, Supporting Information). Overall, the R fjord region adducts manifest only the distortions associated with classical intercalation of stretching, unwinding, and groove enlargement,81 while achieving maximal stabilizing stacking interactions; these stacking energies are summarized in Figure 4A.
van der Waals stacking interactions between the lesion aromatic ring systems and neighboring base pairs stabilize the carcinogen-modified duplexes in a topology and stereochemistry-dependent manner
The stabilizing van der Waals stacking interactions have been shown to play an important role in the resistance of polycyclic aromatic lesions to NER.20, 56 In order to evaluate the stabilizing impact of the stacking interactions between the aromatic ring systems and the neighboring base pairs, we computed the ensemble average van der Waals stacking interaction energies (See Methods) (Figure 5A, Table 2 and Table S4, Supporting Information). The stabilizing stacking interactions range from −21 to −28 kcal/mol (the more negative number corresponds to the higher stabilization energy), and are a function of the number of aromatic rings, the topology of the aromatic ring system, and the adduct stereochemistry. There is a general trend showing that stabilization increases with the number of aromatic rings, and that the stacking interactions are stronger in the case of the R than the corresponding S stereoisomeric adducts. However, in the case of the smallest three-aromatic ring B[c]Ph adducts with the least stacking interaction energies, the trend is reversed: in this case, the S stereoisomer’s stacking is slightly better than that of the R adduct; this is most likely due to the existence of some disturbance of the hydrogen bonding interactions, already discussed above, allowing stacking to optimize better. However, for the other S stereoisomeric adducts, the distortions diminish the stacking interactions: in the case of the B[a]P-N 6-dA adduct, the ruptured hydrogen bonds at the lesion site lead to diminished stacking of the B[a]P aromatic ring system with adjacent bases; in the case of the S DB[a,l]P-N 6-dA adduct, the large adduct-induced duplex unwinding, to alleviate the steric hindrance associated with the 3′-side intercalation, causes diminished stacking. Figure 4B and Supporting Information Figure S5 show the stacking views along the axis of the DNA helix for all six adducts. These figures illustrate how optimal stacking is achieved in each case according to the number of aromatic rings, the lesion topology and the adduct stereochemistry.
Figure 5.
Destabilizing lesion-induced duplex distortions. (A) Ensemble average distortion energies. (B) Intercalation pockets with lesions replaced by hydrogen atoms. The counterpart central 3-mer in the unmodified duplex is also shown for comparison. The view is looking into the minor groove. Note the large values of Rise for the R-B[a]P-N 6-dA adduct, the ruptured hydrogen bonding for the S-B[a]P-N 6-dA adduct, and the severe unwinding established in the case of the S-DB[a,l]P-N 6-dA adduct. The color code is the same as in Figure 2.
Table 2.
Summary of topological features, energies, melting temperatures and relative NER susceptibilities for the PAH-N6-dA adducts investigated
| PAH-N6-dA adduct | Topology | Distortion energy (kcal/mol) | VDW stacking interaction energy (kcal/mol) | Distortion + VDW (kcal/mol) | ΔTm (°C)a | NERf |
|---|---|---|---|---|---|---|
| R B[c]Ph | Fjord 3 rings | 13.0 | −21.0 | −8.0 | −0.5 b,c | Resistant |
| S B[c]Ph | Fjord 3 rings | 14.4 | −21.8 | −7.4 | −0.5 b,c | Resistant |
| R B[a]P | Bay 4 rings | 14.7 | −23.5 | −8.8 | −11.8 b,d | Modest |
| S B[a]P | Bay 4 rings | 17.8 | −22.8 | −5.0 | −18.8 b,d | Greatest |
| R DB[a,l]P | Fjord 5 rings | 10.8 | −28.1 | −17.3 | +7.7 b,e | Resistant |
| S DB[a,l]P | Fjord 5 rings | 14.9 | −24.7 | −9.8 | −5.5 b,e | Resistant |
ΔTm = Tm (Lesion-containing duplex) − Tm (Unmodified);
Sequence context: 5′-CTCTCA*CTTCC-3′;
Melting data for the B[c]Ph-modified duplexes are from 66;
melting data for the B[a]P-modified duplexes are from 64;
melting data for the DB[a,l]P-modified duplexes and the unmodified duplex are from 65; the Tm for the unmodified duplex is 43.8 ± 0.5 °C.
Destabilizing distortion energies and stabilizing stacking energies combine to establish local duplex stability
The energy of local distortion of the double-stranded helix caused by the intercalated adducts was computed for each of our simulated structures. This entailed computing the energy required to distort the unmodified duplex central 3-mer to the actual conformation assumed in the intercalated structure, but in the absence of the PAH lesions, as detailed in Material and Methods. The structures of the intercalation pockets of each of the distorted duplexes are shown in Figure 5B, and demonstrate how hydrogen bonding and unwinding depend on adduct topology and stereochemistry. As shown in Figure 5A and Table 2, the distortion energy is 17.8 kcal/mol in the S-B[a]P-N 6 -dA adduct, which is significantly higher than for all the other five fjord PAH-DNA adducts. This high distortion energy is a reflection of the disrupted hydrogen bonding at the lesion site A6*:T17, and the associated diminished stacking energies of the bases with their neighbors (Figures 3 and 5). Furthermore, we observe that S adducts are always more distorting than the R stereoisomers. However, for the R and S stereoisomeric B[c]Ph-N 6 - dA adducts, the distortion energy values are close to one another (13.0 kcal/mol and 14.4 kcal/mol, respectively), in line with the minimal “S destabilization effect” noted above. The least distorting of all the adducts is the R DB[a,l]P-N 6-dA adduct: in this case, the five aromatic rings, the fjord region flexibility, and the absence of the “S destabilization effect”, combine to support the optimally stacked intercalation with minimal distortion (Figure 6). The greater distortion energy of the S DB[a,l]P adduct than the R stereoisomers stems from its larger unwinding effect. However, intercalation is more distorting in the R B[c]Ph- than for the R DB[a,l]P-N 6-dA adduct; the reason is that the better stacking interactions between the five-ringed DB[a,l]P aromatic ring system and the neighboring base pairs in the intercalation pocket lessens the distortion of the A6*:T17 base pair, as manifested by the lower Buckle and Propeller structural parameters (Table 1).
Figure 6.
“S destabilization effect” for the lesion-containing DNA duplexes. The structures are the same as Figure 2 except rotated to highlight the steric hinderance. Steric hindrance between the adduct benzylic rings and the neighboring DNA atoms is associated only with the 3′-side intercalation of the S adducts, while no such crowding is present in the case of the 5′-intercalated R stereoisomeric adducts. The color code is the same as in Figure 2. For the S-B[c]Ph-N6-dA adduct, the steric hinderance is essentially absent.
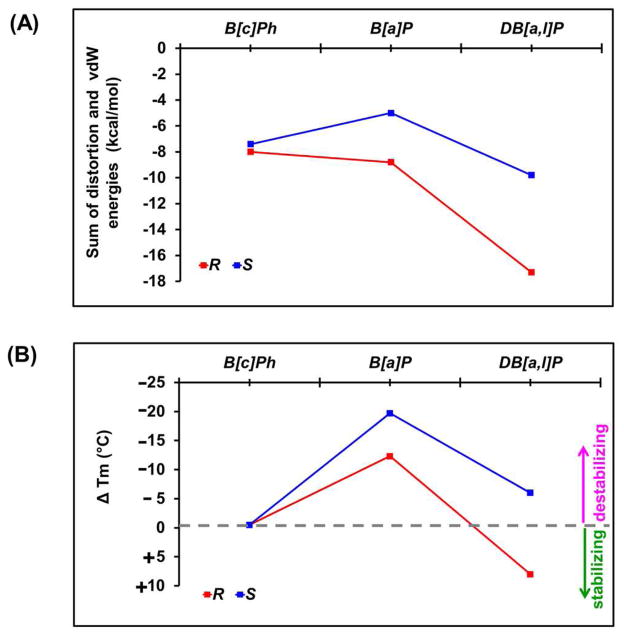
The overall impact of the lesion on the local stabilities of double-stranded DNA molecules may be decomposed into two terms: (1) the “cost” of distorting the DNA duplex to accommodate the intercalated polycyclic aromatic ring systems, and (2) the “gain” associated with the stabilizing van der Waals stacking interaction energies. These two computed terms were summed for each of the six modified duplexes studied. The results are presented in Figure 7A and Table 2, and indicate that, by this measure, the S B[a]P-N 6-dA adduct is the most destabilizing one, while the R DB[a,l]P-N 6-dA adduct is the most stabilizing.
Figure 7.
(A) Sum of ensemble average distortion and van der Waals interaction energies. The more negative energies are more stabilizing. (B) ΔTm = Tm (modified) − Tm (unmodified), the differences between the thermal melting temperatures (Tm) of the modified and unmodified duplexes. Data is given in Table 2. The ΔTm values are derived from experiments with identical concentrations of modified or unmodified DNA duplexes (Table 2).
Discussion
In this work, we explored the structural underpinnings of the resistance to NER of the stereoisomeric R and S fjord PAH-N 6-dA adducts, while the bay region R and S B[a]P-N 6-dA adducts are susceptible to NER (Figure 1, A and B, and Table 2). Our basic hypothesis is that successful NER is initiated by the binding of XPC-RAD23B to the damaged DNA, and depends on the insertion of the XPC β-hairpin between the two DNA strands,12 a mechanism that is facilitated by a sufficient thermodynamic destabilization around the lesion site.
Impact of the DNA lesions on the local stabilities of DNA duplexes
The overall impact of the lesion on the local stability is approximated in terms of the sum of two different effects: (1) the energy required to distort the double-stranded DNA to create the intercalation pocket (Figure 5A), and (2) the energy gained by the dominant van der Waals interactions between the DNA and the polycyclic aromatic ring system inserted into this pocket (Figure 4A). These destabilizing and stabilizing energies, respectively, depend on the number of aromatic rings, their topology, and the stereochemical properties of the adducts. How the sum of these energies (Figure 7A) is correlated with the stabilities of the modified duplexes as ΔTm (Figure 7B), is of particular interest since the global duplex melting point is not the same as the local melting point around the lesion site; these two Tm values may occur at a different temperature, as demonstrated by NMR methods earlier.89 Our hypothesis is that the NER efficiency is related to the local destabilization effect in the vicinity of the lesion which is only crudely and not necessarily quantitatively correlated with the global duplex melting point reflected in the ΔTm values.
The R and S DB[a,l]P-N 6-dA adducts have the greatest number of aromatic rings and also manifest the highest stabilizing stacking interactions, that are favored by the flexible twist of the aromatic ring in the fjord region (Figure 4A and Table 1). The R DB[a,l]P-N 6-dA adduct has a lower distortion energy (+10.8 kcal/mol) than the S DB[a,l]P-N 6-dA (+14.9 kcal/mol distortion energy). On the other hand, the stabilization energy is greater in the case of the R adduct (−28.1 kcal/mol) than the S adduct (−24.7 kcal/mol). The sum of these computed energies is −17.3 (R) and −9.8 (S) kcal/mol, predicting that the R is more stable than the S adduct; this is in agreement with the experimental ΔTm values that show a stabilization of the R duplex by ~ +8° and a relatively small destabilization of the S duplex by −6° (Figure 7B).
The fjord B[c]Ph-N 6-dA adducts possess only three aromatic rings and the van der Waals stacking interactions are smaller. The sum of the distortion and stacking energies is only −0.6 kcal/mol smaller in the case of the S (−7.4 kcal/mol) than the R B[c]Ph-dA adduct (−8.0 kcal/mol), which is consistent with their identical melting points with ΔTm ≈ 0° C in each case (Table 2).
The S B[a]P-N 6-dA adduct is the only one with a significant disruption of the Watson-Crick hydrogen bonding at the lesion site (Figure 3A, and Table S1, Supporting Information). This disruption is also reflected in the largest and most dynamic Opening parameter (Table S2, Supporting Information), the most dynamic untwisting (Table S2, Supporting Information), the largest distortion energy (17.8 kcal/mol), and the smallest total energy (−5.0 kcal/mol). These computational results are consistent with the experimentally observed strong destabilization of the DNA duplex as evidenced by the large negative ΔTm value of −19 °C (Figure 7B and Table 2). By contrast, the R stereoisomeric adduct is considerably less destabilizing (ΔTm = −12 °C) and is repaired only modestly.59 On the other hand, the S DB[a,l]P-N 6-dA adduct, with ΔTm of −5.5 °C, is repair-resistant. Both adducts have very similar distortion energies but the DB[a,l]P-N 6-dA adduct has a modestly greater calculated van der Waals stacking interaction energy by ~1 kcal/mol (Table 2); this calls attention to the importance of the greater number of aromatic rings and flexible fjord region sufficient local stabilization to inhibit NER recognition, which is only approximately manifested in ΔTm values of full duplexes.
The summed energy terms plotted in Figure 7A represent a rough estimate of the overall impact of the different lesions on the stability of the modified DNA duplexes (lesion-induced local distortions plus stabilizing interactions). Figure 7B also shows a plot of the ΔTm values for the different lesions.
The two-state model of association of two complementary DNA strands to form double-stranded DNA predicts that Tm is proportional to the free energy of association of the two strands according to the equation Tm = ΔG 0/(Rln[CT/4]) = ΔH 0 − Tm ΔS 0) (e.g., 90, where CT is the total DNA strand concentration, and ΔG 0, ΔH 0, and ΔS 0 are the equilibrium free energy, enthalpy and entropy terms, respectively, at the temperature Tm. Since our summed energy terms in Figure 7A reflect only contributions to the enthalpy ΔH 0 term, these comparisons are significant only when ΔH 0 > TmΔS 0. Since the computational evaluation of entropy terms is presently the most uncertain,78 we did not here attempt to estimate the ΔS0 terms. We note that the summed energy terms and the ΔTm values follow one another approximately and qualitatively (Figure 7A): the lower Tm values of the S as compared to the R B[a]P- and DB[a,l]P-N 6-dA adducts, as well the similar stabilities of the duplexes with R and S B[c]Ph-N 6-dA adducts are correlated. However, the computed summed energy value for the R B[a]P-N 6-dA adduct is −9 kcal/mol, but should have been about −6 kcal/mol to reflect that this duplex is destabilized as much as it is (ΔTm ~ −12 °C) (Figure 7B). Thus, the computed summed energies reflect trends rather than accurate predictions of modified DNA duplex stabilities, and seem most useful for evaluating the extreme cases with the most stabilizing and the most destabilizing effects of the lesions. As discussed above, the quantity of interest is local destabilization, and it is for this reason we computed our distortion and stabilization energies for the three-nucleotide duplex sequence with the modified adenine base in the center (Figure 7A, Table 2, and Materials and Methods Section). However, distortions beyond the central trimer are certainly worthy of consideration and might partly account for the energetic discrepancy mentioned above.
Impact of stabilizing and destabilizing interactions on susceptibilities of DNA lesions to NER
The relative NER susceptibilities of all six PAH-N 6-dA adducts considered in this work, together with the computed van der Waals stacking and distortion energies, are summarized in Table 2. The computational results provide insights into the structural reasons for the resistance to NER of all fjord PAH-N 6-dA adducts studied up till now,67 as well as into the differences in NER efficiencies of the R and S B[a]P-N 6-dA adducts.
The S B[a]P-N 6-dA adduct stands out as the best NER substrate in this group. It is also the most destabilized one because it is the only one with complete Watson-Crick hydrogen bonding disruption at the lesion site; the rigidity of the polycyclic aromatic B[a]P residue creates distortions in these classically intercalated structures that are not compensated by stacking interactions, thus providing the overall destabilization that would allow XPC β-hairpin insertion and successful NER. In contrast, the R B[a]P-N 6-dA adduct is considerably less destabilizing than the S stereoisomeric adduct, and the NER efficiency of duplexes with the R adduct is significantly smaller59, 67 and dependent on base sequence context.59, 67, 91
In the case of the R and S DB[a,l]P-N 6-dA adducts, the stabilizing carcinogen-base stacking interactions sufficiently compensate for the distortions induced by these bulky intercalated lesions to account for the observed NER resistance (Table 2). Our hypothesis is that carcinogen-base stacking interactions prevent the separation of the two DNA strands that allows for the successful insertion of the XPC β-hairpin into the damaged DNA duplex, with concomitant flipping of the lesion partner bases into the protein.12, 13 There may be a threshold activation energy to this partner base flipping that could be related to the free energy barrier for the flipping process,92 which, however, is not specifically evaluated here. The recently observed repair resistance of a very bulky aristolactam II-dA lesion,93 which intercalates both the damaged adenine and the lesion ring system, while displacing the partner base thymine into the major groove,94 suggests that the very powerful stabilizing stacking interactions largely compensate for the distortions, explaining the observed resistance to NER.93 The observed modest thermal destabilization, which varies from 3 °C to 6.4 °C, depending on the sequence context of the DNA duplex, is similar to that of the repair-resistant S DB[a,l]P-N 6-dA adduct (Table 2). In the case of the smaller, three-aromatic ring system B[c]Ph-N 6-dA adducts, the lesion-induced distortions and van der Waals stacking interactions appear to compensate one another consistent with the ΔTm of ~0 °C stereoisomeric adducts.
Conclusions
Our overall findings show that lesion-induced destabilizing distortions combined with stabilizing van der Waals interactions together determine the overall stabilizing/destabilizing impact of the lesion, and recognition and removal of these same lesions by NER. The overall stabilizing impact of the lesion may stem from various combinations of van der Waals stacking interactions and lesion-induced distortions, that depend on the topological characteristics of specific lesions (number of aromatic rings together with their arrangements), and adduct stereochemistry. Our current results apply to the classically intercalated N 6-dA adducts. Combining the stabilizing stacking interactions with the destabilizing distortions may thus provide a rough estimate of the stabilizing/destabilizing impact of different lesions. This estimate could yield insights into the NER susceptibilities of other intercalating DNA lesions, at least for those that exhibit the most pronounced destabilizing or stabilizing effects, and for suggesting particularly interesting lesions for experimental investigation. More accurate evaluations of the local thermodynamic effects that are relevant to NER but are not quantitatively correlated with global melting stabilities of DNA duplexes derived from ΔTm measurements are amenable to studies by NMR methods.89, 94 We hypothesize that the extensive polycyclic aromatic residue-base stacking interactions provide sufficient local stabilization to prevent the XPC β-hairpin intrusion and base flipping,12 and the subsequent NER processing steps in the repair resistant adducts. For initiation of this process, productive binding of the XPC-RAD23B to the lesion is required.1, 3, 7 In addition, downstream events in the processing of lesions, which are currently of great interest, likely play a role in determining NER efficiencies.28, 95
Supplementary Material
Acknowledgments
We thank Professor Carlos de los Santos, Stony Brook University, for very valuable discussion.
Funding Sources: This research was supported by NIH Grants CA-28038 to S.B. and CA-099194 to N.E.G.. Computational infrastructure and systems management was partially supported by CA-75449 to S.B.. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. We gratefully acknowledge TeraGrid resources provided by the Texas Advanced Computing Center Supported by National Science Foundation.
Abbreviations
- NER
nucleotide excision repair
- PAHs
polynuclear aromatic hydrocarbons
- B[c]Ph
benzo[c]phenanthrene
- B[a]P
benzo[a]pyrene
- DB[a,l]P
dibenzo[a,l]pyrene
- DE
diol epoxide
- MD
molecular dynamics
- RMSD
root mean square deviation
- SASA
solvent accessible surface area
- MM-PBSA
molecular mechanics Poisson-Boltzmann surface area
- ns
nanosecond
Footnotes
Disclosure
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Associated Content
Supporting Information: MD computation protocols, Supporting Data: Supporting Tables S1 to S6, and Figures S1 to S5; Movies S1 to S6 show the best representative structures rotating. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Mocquet V, Kropachev K, Kolbanovskiy M, Kolbanovskiy A, Tapias A, Cai Y, Broyde S, Geacintov NE, Egly JM. The human DNA repair factor XPC-HR23B distinguishes stereoisomeric benzo[a]pyrenyl-DNA lesions. EMBO J. 2007;26:2923–2932. doi: 10.1038/sj.emboj.7601730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riedl T, Hanaoka F, Egly JM. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 2003;22:5293–5303. doi: 10.1093/emboj/cdg489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sugasawa K, Okamoto T, Shimizu Y, Masutani C, Iwai S, Hanaoka F. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 2001;15:507–521. doi: 10.1101/gad.866301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 5.Mocquet V, Laine JP, Riedl T, Yajin Z, Lee MY, Egly JM. Sequential recruitment of the repair factors during NER: the role of XPG in initiating the resynthesis step. EMBO J. 2008;27:155–167. doi: 10.1038/sj.emboj.7601948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scharer OD. In: The Chemical Biology of DNA Damage. Geacintov NE, Broyde S, editors. Wiley-VCH; Weinheim, Germany: 2010. pp. 239–260. [Google Scholar]
- 7.Sugasawa K, Shimizu Y, Iwai S, Hanaoka F. A molecular mechanism for DNA damage recognition by the xeroderma pigmentosum group C protein complex. DNA Repair (Amst) 2002;1:95–107. doi: 10.1016/s1568-7864(01)00008-8. [DOI] [PubMed] [Google Scholar]
- 8.Naegeli H, Sugasawa K. The xeroderma pigmentosum pathway: Decision tree analysis of DNA quality. DNA Repair (Amst) 2011;10:673–683. doi: 10.1016/j.dnarep.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 9.Camenisch U, Nageli H. XPA gene, its product and biological roles. Adv Exp Med Biol. 2008;637:28–38. doi: 10.1007/978-0-387-09599-8_4. [DOI] [PubMed] [Google Scholar]
- 10.Fuss JO, Tainer JA. XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase. DNA Repair (Amst) 2011;10:697–713. doi: 10.1016/j.dnarep.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 12.Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 13.Scharer OD. A molecular basis for damage recognition in eukaryotic nucleotide excision repair. Chembiochem. 2008;9:21–23. doi: 10.1002/cbic.200700619. [DOI] [PubMed] [Google Scholar]
- 14.Cai Y, Patel DJ, Broyde S, Geacintov NE. Base sequence context effects on nucleotide excision repair. J Nucleic Acids. 2010 doi: 10.4061/2010/174252. doi:10.4061/2010/174252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wood RD. DNA damage recognition during nucleotide excision repair in mammalian cells. Biochimie. 1999;81:39–44. doi: 10.1016/s0300-9084(99)80036-4. [DOI] [PubMed] [Google Scholar]
- 16.Cai Y, Patel DJ, Geacintov NE, Broyde S. Differential nucleotide excision repair susceptibility of bulky DNA adducts in different sequence contexts: hierarchies of recognition signals. J Mol Biol. 2009;385:30–44. doi: 10.1016/j.jmb.2008.09.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunz D, Hess MT, Naegeli H. Recognition of DNA adducts by human nucleotide excision repair. Evidence for a thermodynamic probing mechanism. J Biol Chem. 1996;271:25089–25098. doi: 10.1074/jbc.271.41.25089. [DOI] [PubMed] [Google Scholar]
- 18.Hess MT, Schwitter U, Petretta M, Giese B, Naegeli H. Bipartite substrate discrimination by human nucleotide excision repair. Proc Natl Acad Sci U S A. 1997;94:6664–6669. doi: 10.1073/pnas.94.13.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kropachev K, Kolbanovskii M, Cai Y, Rodriguez F, Kolbanovskii A, Liu Y, Zhang L, Amin S, Patel D, Broyde S, Geacintov NE. The sequence dependence of human nucleotide excision repair efficiencies of benzo[a]pyrene-derived DNA lesions: insights into the structural factors that favor dual incisions. J Mol Biol. 2009;386:1193–1203. doi: 10.1016/j.jmb.2008.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reeves DA, Mu H, Kropachev K, Cai Y, Ding S, Kolbanovskiy A, Kolbanovskiy M, Chen Y, Krzeminski J, Amin S, Patel DJ, Broyde S, Geacintov NE. Resistance of bulky DNA lesions to nucleotide excision repair can result from extensive aromatic lesion-base stacking interactions. Nucleic Acids Res. 2011;39:8752–8764. doi: 10.1093/nar/gkr537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoare S, Zou Y, Purohit V, Krishnasamy R, Skorvaga M, Van Houten B, Geacintov NE, Basu AK. Differential incision of bulky carcinogen-DNA adducts by the UvrABC nuclease: comparison of incision rates and the interactions of Uvr subunits with lesions of different structures. Biochemistry. 2000;39:12252–12261. doi: 10.1021/bi0013187. [DOI] [PubMed] [Google Scholar]
- 22.Kow YW, Wallace SS, Van Houten B. UvrABC nuclease complex repairs thymine glycol, an oxidative DNA base damage. Mutat Res. 1990;235:147–156. doi: 10.1016/0921-8777(90)90068-g. [DOI] [PubMed] [Google Scholar]
- 23.Mu D, Bertrand-Burggraf E, Huang JC, Fuchs RP, Sancar A, Fuchs BP. Human and E. coli excinucleases are affected differently by the sequence context of acetylaminofluorene-guanine adduct. Nucleic Acids Res. 1994;22:4869–4871. doi: 10.1093/nar/22.23.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruan Q, Liu T, Kolbanovskiy A, Liu Y, Ren J, Skorvaga M, Zou Y, Lader J, Malkani B, Amin S, Van Houten B, Geacintov NE. Sequence context–and temperature-dependent nucleotide excision repair of a benzo[a]pyrene diol epoxide-guanine DNA adduct catalyzed by thermophilic UvrABC proteins. Biochemistry. 2007;46:7006–7015. doi: 10.1021/bi700294k. [DOI] [PubMed] [Google Scholar]
- 25.Snowden A, Kow YW, Van Houten B. Damage repertoire of the Escherichia coli UvrABC nuclease complex includes abasic sites, base-damage analogues, and lesions containing adjacent 5′ or 3′ nicks. Biochemistry. 1990;29:7251–7259. doi: 10.1021/bi00483a013. [DOI] [PubMed] [Google Scholar]
- 26.Zou Y, Liu TM, Geacintov NE, Van Houten B. Interaction of the UvrABC nuclease system with a DNA duplex containing a single stereoisomer of dG-(+)- or dG-(−)-anti-BPDE. Biochemistry. 1995;34:13582–13593. doi: 10.1021/bi00041a038. [DOI] [PubMed] [Google Scholar]
- 27.Zou Y, Shell SM, Utzat CD, Luo C, Yang Z, Geacintov NE, Basu AK. Effects of DNA adduct structure and sequence context on strand opening of repair intermediates and incision by UvrABC nuclease. Biochemistry. 2003;42:12654–12661. doi: 10.1021/bi034446e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Reeves D, Kropachev K, Cai Y, Ding S, Kolbanovskiy M, Kolbanovskiy A, Bolton JL, Broyde S, Van Houten B, Geacintov NE. Probing for DNA damage with beta-hairpins: Similarities in incision efficiencies of bulky DNA adducts by prokaryotic and human nucleotide excision repair systems in vitro. DNA Repair (Amst) 2011;10:684–696. doi: 10.1016/j.dnarep.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Truglio JJ, Karakas E, Rhau B, Wang H, DellaVecchia MJ, Van Houten B, Kisker C. Structural basis for DNA recognition and processing by UvrB. Nat Struct Mol Biol. 2006;13:360–364. doi: 10.1038/nsmb1072. [DOI] [PubMed] [Google Scholar]
- 30.Baek SO, Field RA, Goldstone PW, Kirk PW, JNL, Perry RA. A review of atmospheric polycyclic aromatic hydrocarbons: Sources, fate and behavior. Water, Air, Soil Pollut. 1991;60:279–300. [Google Scholar]
- 31.Clapp RW, Jacobs MM, Loechler EL. Environmental and occupational causes of cancer: new evidence 2005–2007. Rev Environ Health. 2008;23:1–37. doi: 10.1515/reveh.2008.23.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luch A. Nature and nurture - lessons from chemical carcinogenesis. Nat Rev Cancer. 2005;5:113–125. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- 33.Luch A. On the impact of the molecule structure in chemical carcinogenesis. EXS. 2009;99:151–179. doi: 10.1007/978-3-7643-8336-7_6. [DOI] [PubMed] [Google Scholar]
- 34.Katz AK, Carrell HL, Glusker JP. Dibenzo[a,l]pyrene (dibenzo[def,p]chrysene): fjord-region distortions. Carcinogenesis. 1998;19:1641–1648. doi: 10.1093/carcin/19.9.1641. [DOI] [PubMed] [Google Scholar]
- 35.Cavalieri EL, Higginbotham S, RamaKrishna NV, Devanesan PD, Todorovic R, Rogan EG, Salmasi S. Comparative dose-response tumorigenicity studies of dibenzo[alpha,l]pyrene versus 7,12-dimethylbenz[alpha]anthracene, benzo[alpha]pyrene and two dibenzo[alpha,l]pyrene dihydrodiols in mouse skin and rat mammary gland. Carcinogenesis. 1991;12:1939–1944. doi: 10.1093/carcin/12.10.1939. [DOI] [PubMed] [Google Scholar]
- 36.Cavalieri EL, Higginbotham S, Rogan EG. Dibenzo[a,l]pyrene:The most potent carcinogenic aromatic hydrocarbon. Polycyclic Aromat Compd. 1994;6:177–183. [Google Scholar]
- 37.Hecht SS. Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst. 1999;91:1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 38.Phillips DH. Smoking-related DNA and protein adducts in human tissues. Carcinogenesis. 2002;23:1979–2004. doi: 10.1093/carcin/23.12.1979. [DOI] [PubMed] [Google Scholar]
- 39.IARC. IARC Monogr Eval Carcinog Risks Hum. International Agency for Research on Cancer; Lyon, France: 2010. Some non-heterocyclic polycyclic aromatic hydrocarbons and some related exposures; pp. 1–853. [PMC free article] [PubMed] [Google Scholar]
- 40.Baum M, Amin S, Guengerich FP, Hecht SS, Kohl W, Eisenbrand G. Metabolic activation of benzo[c]phenanthrene by cytochrome P450 enzymes in human liver and lung. Chem Res Toxicol. 2001;14:686–693. doi: 10.1021/tx000240s. [DOI] [PubMed] [Google Scholar]
- 41.Lagerqvist A, Hakansson D, Lundin C, Prochazka G, Dreij K, Segerback D, Jernstrom B, Tornqvist M, Frank H, Seidel A, Erixon K, Jenssen D. DNA repair and replication influence the number of mutations per adduct of polycyclic aromatic hydrocarbons in mammalian cells. DNA Repair (Amst) 2011 doi: 10.1016/j.dnarep.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 42.Luch A. The mode of action of organic carcinogens on cellular structures. EXS. 2006:65–95. doi: 10.1007/3-7643-7378-4_4. [DOI] [PubMed] [Google Scholar]
- 43.Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- 44.Penning TM. In: The Chemical Biology of DNA Damage. Geacintov NE, Broyde S, editors. Wiley-VCH Verlag; Weinheim, Germany: 2010. [Google Scholar]
- 45.Penning TM. In: Chemical Carcinogenesis. Penning TM, editor. Springer; 2011. [Google Scholar]
- 46.Xue W, Warshawsky D. Metabolic activation of polycyclic and heterocyclic aromatic hydrocarbons and DNA damage: a review. Toxicol Appl Pharmacol. 2005;206:73–93. doi: 10.1016/j.taap.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 47.Zhang SM, Chen KM, Aliaga C, Sun YW, Lin JM, Sharma AK, Amin S, El-Bayoumy K. Identification and quantification of DNA adducts in the oral tissues of mice treated with the environmental carcinogen dibenzo[a,l]pyrene by HPLC-MS/MS. Chem Res Toxicol. 2011 doi: 10.1021/tx200188j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cosman M, de los Santos C, Fiala R, Hingerty BE, Singh SB, Ibanez V, Margulis LA, Live D, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the major adduct between the carcinogen (+)-antibenzo[a]pyrene diol epoxide and DNA. Proc Natl Acad Sci U S A. 1992;89:1914–1918. doi: 10.1073/pnas.89.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de los Santos C, Cosman M, Hingerty BE, Ibanez V, Margulis LA, Geacintov NE, Broyde S, Patel DJ. Influence of benzo[a]pyrene diol epoxide chirality on solution conformations of DNA covalent adducts: the (−)-trans-anti-[BP]G.C adduct structure and comparison with the (+)-trans-anti- [BP]G.C enantiomer. Biochemistry. 1992;31:5245–5252. doi: 10.1021/bi00138a002. [DOI] [PubMed] [Google Scholar]
- 50.Geacintov NE, Cosman M, Hingerty BE, Amin S, Broyde S, Patel DJ. NMR solution structures of stereoisometric covalent polycyclic aromatic carcinogen-DNA adduct: principles, patterns, and diversity. Chem Res Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 51.Lukin M, de Los Santos C. NMR structures of damaged DNA. Chem Rev. 2006;106:607–686. doi: 10.1021/cr0404646. [DOI] [PubMed] [Google Scholar]
- 52.Stone MP, Huang H, Brown KL, Shanmugam G. Chemistry and structural biology of DNA damage and biological consequences. Chemistry & biodiversity. 2011;8:1571–1615. doi: 10.1002/cbdv.201100033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schurter EJ, Yeh HJ, Sayer JM, Lakshman MK, Yagi H, Jerina DM, Gorenstein DG. NMR solution structure of a nonanucleotide duplex with a dG mismatch opposite a 10R adduct derived from trans addition of a deoxyadenosine N6-amino group to (−)-(7S,8R,9R,10S)-7,8-dihydroxy-9,10- epoxy-7,8,9,10- tetrahydrobenzo[a]pyrene. Biochemistry. 1995;34:1364–1375. doi: 10.1021/bi00004a031. [DOI] [PubMed] [Google Scholar]
- 54.Yeh HJ, Sayer JM, Liu X, Altieri AS, Byrd RA, Lakshman MK, Yagi H, Schurter EJ, Gorenstein DG, Jerina DM. NMR solution structure of a nonanucleotide duplex with a dG mismatch opposite a 10S adduct derived from trans addition of a deoxyadenosine N6-amino group to (+)- (7R,8S,9S,10R)-7,8-dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene: an unusual syn glycosidic torsion angle at the modified dA. Biochemistry. 1995;34:13570–13581. doi: 10.1021/bi00041a037. [DOI] [PubMed] [Google Scholar]
- 55.Zegar IS, Kim SJ, Johansen TN, Horton PJ, Harris CM, Harris TM, Stone MP. Adduction of the human N-ras codon 61 sequence with (−)-(7S,8R,9R,10S)-7,8-dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a] pyrene: structural refinement of the intercalated SRSR(61,2) (−)-(7S,8R,9S,10R)-N 6-[10-(7,8,9,10-tetrahydrobenzo[a]pyrenyl)]-2′-deoxyadenosyl adduct from 1H NMR. Biochemistry. 1996;35:6212–6224. doi: 10.1021/bi9524732. [DOI] [PubMed] [Google Scholar]
- 56.Cai Y, Ding S, Geacintov NE, Broyde S. Intercalative conformations of the 14R (+)- and 14S (−)-trans-anti-DB[a,l]P-N 6-dA adducts: molecular modeling and MD simulations. Chem Res Toxicol. 2011;24:522–531. doi: 10.1021/tx1004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cosman M, Fiala R, Hingerty BE, Laryea A, Lee H, Harvey RG, Amin S, Geacintov NE, Broyde S, Patel D. Solution conformation of the (+)-trans-anti-[BPh]dA adduct opposite dT in a DNA duplex: intercalation of the covalently attached benzo[c]phenanthrene to the 5′-side of the adduct site without disruption of the modified base pair. Biochemistry. 1993;32:12488–12497. doi: 10.1021/bi00097a029. [DOI] [PubMed] [Google Scholar]
- 58.Cosman M, Laryea A, Fiala R, Hingerty BE, Amin S, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (−)-trans-anti-benzo[c]phenanthrene-dA ([BPh]dA) adduct opposite dT in a DNA duplex: intercalation of the covalently attached benzo[c]phenanthrenyl ring to the 3′-side of the adduct site and comparison with the (+)-trans-anti-[BPh]dA opposite dT stereoisomer. Biochemistry. 1995;34:1295–1307. doi: 10.1021/bi00004a024. [DOI] [PubMed] [Google Scholar]
- 59.Yan S, Wu M, Buterin T, Naegeli H, Geacintov NE, Broyde S. Role of base sequence context in conformational equilibria and nucleotide excision repair of benzo[a]pyrene diol epoxide-adenine adducts. Biochemistry. 2003;42:2339–2354. doi: 10.1021/bi0270081. [DOI] [PubMed] [Google Scholar]
- 60.Zegar IS, Chary P, Jabil RJ, Tamura PJ, Johansen TN, Lloyd RS, Harris CM, Harris TM, Stone MP. Multiple conformations of an intercalated (−)-(7S,8R,9S,10R)-N 6-[10-(7,8,9,10-tetrahydrobenzo[a]pyrenyl)]-2′-deoxyadenosyl adduct in the N-ras codon 61 sequence. Biochemistry. 1998;37:16516– 16528. doi: 10.1021/bi9817616. [DOI] [PubMed] [Google Scholar]
- 61.Schwartz JL, Rice JS, Luxon BA, Sayer JM, Xie G, Yeh HJ, Liu X, Jerina DM, Gorenstein DG. Solution structure of the minor conformer of a DNA duplex containing a dG mismatch opposite a benzo[a]pyrene diol epoxide/dA adduct: glycosidic rotation from syn to anti at the modified deoxyadenosine. Biochemistry. 1997;36:11069–11076. doi: 10.1021/bi971306u. [DOI] [PubMed] [Google Scholar]
- 62.Yan S, Shapiro R, Geacintov NE, Broyde S. Stereochemical, structural, and thermodynamic origins of stability differences between stereoisomeric benzo[a]pyrene diol epoxide deoxyadenosine adducts in a DNA mutational hot spot sequence. J Am Chem Soc. 2001;123:7054–7066. doi: 10.1021/ja0043035. [DOI] [PubMed] [Google Scholar]
- 63.Cai Y, Wang L, Ding S, Schwaid A, Geacintov NE, Broyde S. A bulky DNA lesion derived from a highly potent polycyclic aromatic tumorigen stabilizes nucleosome core particle structure. Biochemistry. 2010;49:9943–9945. doi: 10.1021/bi101560y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krzeminski J, Ni J, Zhuang P, Luneva N, Amin S, Geacintov NE. Total synthesis, mass spectrometric sequencing, and stabilities of oligonucleotide duplexes with single trans-anti-BPDE-N 6-dA lesions in the N-ras codon 61 and other sequence contexts. Polycyclic Aromat Compd. 1999;17:1–10. [Google Scholar]
- 65.Ruan Q, Kolbanovskiy A, Zhuang P, Chen J, Krzeminski J, Amin S, Geacintov NE. Synthesis and characterization of site-specific and stereoisomeric fjord dibenzo[a,l]pyrene diol epoxide-N 6-adenine adducts: unusual thermal stabilization of modified DNA duplexes. Chem Res Toxicol. 2002;15:249–261. doi: 10.1021/tx010157k. [DOI] [PubMed] [Google Scholar]
- 66.Laryea A, Cosman M, Lin JM, Liu T, Agarwal R, Smirnov S, Amin S, Harvey RG, Dipple A, Geacintov NE. Direct synthesis and characterization of site-specific adenosyl adducts derived from the binding of a 3,4-dihydroxy-1,2-epoxybenzo[c]phenanthrene stereoisomer to an 11-mer oligodeoxyribonucleotide. Chem Res Toxicol. 1995;8:444–454. doi: 10.1021/tx00045a017. [DOI] [PubMed] [Google Scholar]
- 67.Buterin T, Hess MT, Luneva N, Geacintov NE, Amin S, Kroth H, Seidel A, Naegeli H. Unrepaired fjord region polycyclic aromatic hydrocarbon-DNA adducts in ras codon 61 mutational hot spots. Cancer Res. 2000;60:1849–1856. [PubMed] [Google Scholar]
- 68.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA. AMBER. Vol. 9. University of California; San Francisco, CA: 2006. [Google Scholar]
- 69.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force-field for the simulation of proteins, nucleic acids, and organic-molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 70.Cheatham TE, Cieplak P, Kollman PA. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J Biomol Struct Dyn. 1999;16:845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- 71.Perez A, Marchan I, Svozil D, Sponer J, Cheatham TE, 3rd, Laughton CA, Orozco M. Refinement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu M, Yan S, Patel DJ, Geacintov NE, Broyde S. Relating repair susceptibility of carcinogen-damaged DNA with structural distortion and thermodynamic stability. Nucleic Acids Res. 2002;30:3422–3432. doi: 10.1093/nar/gkf427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Case DA, Pearlman DA, Caldwell JW, Cheatham TE, III, Wang J, Ross WS, Simmerling CL, Darden TA, Merz KM, Stanton RV, Cheng AL, Vincent JJ, Crowley M, Tsui V, Gohlke H, Radmer RJ, Duan Y, Pitera J, Massova I, Seibel GL, Singh UC, Weiner PK, Kollman PA. AMBER. Vol. 7. University of California; San Francisco, CA: 2002. [Google Scholar]
- 74.Ravishanker G, Beveridge DL. MD Toolchest 2.0. Wesleyan University; Middletown, CT 06459: 1993. [Google Scholar]
- 75.Ravishanker G, Swaminathan S, Beveridge DL, Lavery R, Sklenar H. Conformational and helicoidal analysis of 30 ps of molecular dynamics on the d(CGCGAATTCGCG) double helix: “curves”, dials and windows. J Biomol Struct Dyn. 1989;6:669–699. doi: 10.1080/07391102.1989.10507729. [DOI] [PubMed] [Google Scholar]
- 76.Fratini AV, Kopka ML, Drew HR, Dickerson RE. Reversible bending and helix geometry in a B-DNA dodecamer: CGCGAATTBrCGCG. J Biol Chem. 1982;257:14686–14707. [PubMed] [Google Scholar]
- 77.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]
- 78.Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE., 3rd Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 79.Mukherjee A, Lavery R, Bagchi B, Hynes JT. On the molecular mechanism of drug intercalation into DNA: a simulation study of the intercalation pathway, free energy, and DNA structural changes. J Am Chem Soc. 2008;130:9747–9755. doi: 10.1021/ja8001666. [DOI] [PubMed] [Google Scholar]
- 80.Li S, Cooper VR, Thonhauser T, Lundqvist BI, Langreth DC. Stacking interactions and DNA intercalation. J Phys Chem B. 2009;113:11166–11172. doi: 10.1021/jp905765c. [DOI] [PubMed] [Google Scholar]
- 81.Neidle S, Pearl LH, Herzyk P, Berman HM. A molecular model for proflavine-DNA intercalation. Nucleic Acids Res. 1988;16:8999–9016. doi: 10.1093/nar/16.18.8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cai Y, Kropachev K, Kolbanovskiy M, Kolbanovskiy A, Broyde S, Patel DJ, Geacintov NE. In: Recognition and removal of bulky DNA lesiosn by the nucleotide excision repair system in the The Chemical Biology of DNA Damage. Geacintov N, Broyde S, editors. Chapter 12. Wiley-VCH; Weinheim, Germany: 2010. pp. 261–298. [Google Scholar]
- 83.Cai Y, Patel DJ, Geacintov NE, Broyde S. Dynamics of a benzo[a]pyrene-derived guanine DNA lesion in TGT and CGC sequence contexts: enhanced mobility in TGT explains conformational heterogeneity, flexible bending, and greater susceptibility to nucleotide excision repair. J Mol Biol. 2007;374:292–305. doi: 10.1016/j.jmb.2007.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Volk DE, Rice JS, Luxon BA, Yeh HJ, Liang C, Xie G, Sayer JM, Jerina DM, Gorenstein DG. NMR evidence for syn-anti interconversion of a trans opened (10R)-dA adduct of benzo[a]pyrene (7S,8R)-diol (9R,10S)-epoxide in a DNA duplex. Biochemistry. 2000;39:14040–14053. doi: 10.1021/bi001669l. [DOI] [PubMed] [Google Scholar]
- 85.Dickerson RE. DNA bending: the prevalence of kinkiness and the virtues of normality. Nucleic Acids Res. 1998;26:1906–1926. doi: 10.1093/nar/26.8.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gorin AA, Zhurkin VB, Olson WK. B-DNA twisting correlates with base-pair morphology. J Mol Biol. 1995;247:34–48. doi: 10.1006/jmbi.1994.0120. [DOI] [PubMed] [Google Scholar]
- 87.Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc Natl Acad Sci U S A. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rodriguez FA, Cai Y, Lin C, Tang Y, Kolbanovskiy A, Amin S, Patel DJ, Broyde S, Geacintov NE. Exocyclic amino groups of flanking guanines govern sequence-dependent adduct conformations and local structural distortions for minor groove-aligned benzo[a]pyrenyl-guanine lesions in a GG mutation hotspot context. Nucleic Acids Res. 2007;35:1555–1568. doi: 10.1093/nar/gkm022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marky LA, Breslauer KJ. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 91.Geacintov NE, Broyde S, Buterin T, Naegeli H, Wu M, Yan S, Patel DJ. Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery. Biopolymers. 2002;65:202–210. doi: 10.1002/bip.10239. [DOI] [PubMed] [Google Scholar]
- 92.Zheng H, Cai Y, Ding S, Tang Y, Kropachev K, Zhou Y, Wang L, Wang S, Geacintov NE, Zhang Y, Broyde S. Base flipping free energy profiles for damaged and undamaged DNA. Chem Res Toxicol. 2010;23:1868–1870. doi: 10.1021/tx1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sidorenko VS, Yeo JE, Bonala RR, Johnson F, Scharer OD, Grollman AP. Lack of recognition by global-genome nucleotide excision repair accounts for the high mutagenicity and persistence of aristolactam-DNA adducts. Nucleic Acids Res. 2011 Nov 25; doi: 10.1093/nar/gkr1095. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lukin M, Zaliznyak T, Johnson F, de Los Santos C. Structure and stability of DNA containing an aristolactam II-dA lesion: implications for the NER recognition of bulky adducts. Nucleic Acids Res. 2011 Nov 25; doi: 10.1093/nar/gkr1094. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Scharer OD. Multistep damage recognition, pathway coordination and connections to transcription, damage signaling, chromatin structure, cancer and aging: current perspectives on the nucleotide excision repair pathway. DNA Repair (Amst) 2011;10:667. doi: 10.1016/j.dnarep.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 96.Simmerling C, Elber R, Zhang J. MOIL-View -A program for visualization of structure and dynamics of biomolecules and STO- A program for computing stochastic paths. In: Pullman A, et al., editors. Modeling of Biomolecular Structure and Mechanisms. Netherlands: Kluwer; 1995. pp. 241–265. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.