Abstract
TET2 converts 5-methylcytosine to 5-hydroxymethylcytosine (5-hmC) in DNA and is frequently mutated in myeloid malignancies, including myeloproliferative neoplasms. Here we show that the level of 5-hmC is decreased in granulocyte DNA from myeloproliferative neoplasm patients with TET2 mutations compared with granulocyte DNA from healthy patients. Inhibition of TET2 by RNA interference decreases 5-hmC levels in both human leukemia cell lines and cord blood CD34+ cells. These results confirm the enzymatic function of TET2 in human hematopoietic cells. Knockdown of TET2 in cord blood CD34+ cells skews progenitor differentiation toward the granulomonocytic lineage at the expense of lymphoid and erythroid lineages. In addition, by monitoring in vitro granulomonocytic development we found a decreased granulocytic differentiation and an increase in monocytic cells. Our results indicate that TET2 disruption affects 5-hmC levels in human myeloid cells and participates in the pathogenesis of myeloid malignancies through the disturbance of myeloid differentiation.
Introduction
Tet1, Tet2, and Tet3 convert 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine (5-hmC) in DNA from various mouse tissues.1–3 Acquired mutations in TET2 have been found in a variety of myeloid malignancies, including myeloproliferative neoplasms (MPNs).4–8 These defects are considered to be loss-of-function mutations affecting a HSC, but their actual involvement in malignant hematopoiesis remains unclear. TET2 mutations have been recently associated with impaired hydroxylation of 5-mC in myeloid cancers, and Tet2 has been shown to regulate murine myeloid differentiation.9,10 As in mice, TET2 expression is predominant in hematopoietic cells in humans.5 To test whether TET2 alterations could have consequences in 5-mC hydroxylation and in the biology of human hematopoietic cells, we studied primary cells from MPN patients with or without TET2 mutations, as well as cell lines and normal CD34+ cells where TET2 expression was knocked down by RNA interference.
Methods
Patient samples
The study was approved by the Local Research Ethics Committee of Assistance Publique–Hôpitaux de Paris. Peripheral blood was collected from 58 MPN patients (supplemental Table 2, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) and 31 healthy patients with their informed consent, in accordance with the Declaration of Helsinki. Umbilical cord blood samples were collected from healthy newborns with mothers' consent. CD34+ cells or granulocytes were isolated as described.11
Nucleic acid extraction
DNA and RNA were extracted by the use of QIAGEN kits (QIAGEN).
Quantification of 5-hmC
Cytosine, 5-mC, and 5-hmC were quantified by the use of HPLC coupled to tandem mass spectrometry (HPLC-MS/MS; supplemental Figure 1)12,13 Dot blots were obtained by spotting DNA onto nylon hybond N+ membranes (Amersham). Membranes were washed, air-dried, UV cross-linked, blocked, and incubated with anti–5-hmC antibody (1:10 000; Active Motif) and HRP-conjugated anti–rabbit IgG secondary antibody (Jackson ImmunoResearch Laboratories). To control spotting, blots were stained with 0.02% methylene blue (MB) in 0.3M sodium acetate (pH 5.2). Immunofluorescent staining was performed with the use of anti–5-hmC antibody and Alexa-546–conjugated secondary antibody (Molecular Probes). Nuclei were stained with Hoechst 33342 (Molecular Probes), and cells were examined with a LSM 510 microscope (Zeiss).
Real-time quantitative RT-PCR
PCRs were performed by the use of primers and probes listed in supplemental Table 1 with an ABI Prism GeneAmp 7500 (Applied Biosystems).
TET2 knockdown by lentiviral delivery of shRNA
MO7e, Kasumi-1, TF1, UKE1, HL60, and UT7 cell lines or CD34+ cells were transduced as previously described14 with lentiviruses expressing the green fluorescent protein (GFP) and either shRNA-TET2 (5′-GGGTAAGCCAAGAAAGAAA-3′) or shRNA-scramble (5′-GCCGGCAGCTAGCGACGCCAT-3′) as control. GFP-positive cells were sorted with the use of a MOFLO (Beckman Coulter) cell sorter. TET2 protein knockdown was assessed by Western blot with an anti-TET2 antibody generated in the mouse and an anti-HSC70 antibody (Enzo Life Sciences).
Cell cultures and flow cytometry
CD34+ cells were grown in colony-forming cell (CFC) assays in methylcellulose11 or in liquid cultures stimulating erythroid, granulomonocytic, or monocyte/macrophage differentiation with SCF (50 ng/mL; Immunex), IL-3 (100 IU/mL; Novartis), erythropoietin (EPO; 3 IU/mL; Bellon), Fms-like tyrosine kinase-3–ligand (FLT3-L; 50 ng/mL; Diaclone), G-CSF (10 ng/mL; Peprotech), and M-CSF (100 ng/mL; Miltenyi Biotec; Figure 2 and supplemental Figures 5-7). Cord blood CD34+CD38− cells were seeded at one cell per well in a B-cell/natural killer/granulo-monocytic (B/NK/GM) culture system for 4-6 weeks.15 Morphology was examined after May-Grunwald-Giemsa staining of cytospun cells. For sorting or immunophenotypic analyses, cells were labeled with anti-CD14–PE, anti-CD15–APC, anti-CD11b–PE, anti-CD19–PE, anti-CD34–APC, anti-CD36–APC, anti-CD38–PE (BD Biosciences), anti-CD34–PC7, anti-CD56–PC7 (Beckman Coulter), and anti–glycophorin-A–PE (Caltag Laboratories) antibodies.
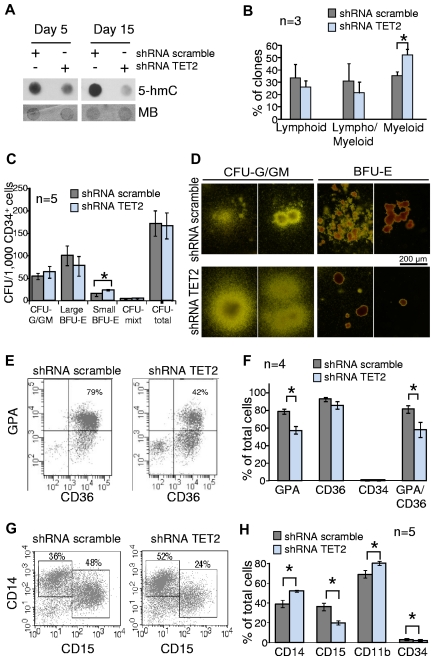
Figure 2.
Knockdown of TET2 disturbs myeloid differentiation of cord blood CD34+ cells in vitro. Cord blood CD34+ cells were transduced by lentiviruses expressing GFP and either shRNA scramble or shRNA TET2. GFP+ cells were sorted 2 days after the end of the transduction procedure. (A) Sorted GFP+ cells were grown in MEM-α medium supplemented with SCF, FLT3-L, IL-3, and G-CSF. After 5-15 days of culture cells were harvested, DNA extracted, and spotted for 5-hmC dot blot assay. Membranes were stained with MB to assess equal spotting. Results are representative of 3 experiments, each performed at 3 time points of culture. (B) Sorted GFP+ CD34+CD38− cells were seeded at one cell per well on a confluent layer of MS-5 cells in a specific medium supporting B/NK/GM differentiation. After 4-6 weeks of culture wells with significant cell growth were harvested and 450 clones from 3 independent experiments were tested for B, natural killer, and myeloid differentiations with the use of anti-CD19, anti-CD56, and anti-CD15 antibodies. Histograms show the percentages of lymphoid (CD15− and CD56+ or CD19+), lympho/myeloid (CD15+CD19+ or CD15+CD56+ or CD15+CD19+CD56+), and myeloid (CD15+CD19−CD56−) clones. *P < .05, unpaired Student t test. Error bars indicate SEM. (C) Transduced CD34+ cells were seeded in methylcellulose in the presence of EPO, IL-3, SCF, and G-CSF. At day 14, colonies derived from shRNA scramble and shRNA TET2 BFU-E and CFU-G/GM were enumerated. Histograms show the number of colonies derived from transduced CD34+ cells (n = 5 independent experiments). *P < .05, unpaired Student t test. Error bars indicate SEM. (D) Photographs showing BFU-E– and CFU-G/GM–derived colonies. Colonies derived from shRNA TET2 BFU-E were smaller than those derived from shRNA scramble BFU-E. In contrast, CFU-G/GM–derived colonies appeared larger. (E) Transduced CD34+ cells were grown in serum-free medium supplemented with SCF, IL-3, and EPO. At day 14, CD34, CD36, and glycophorin-A (GPA) expression were analyzed by flow cytometry. Scattergrams from one representative of 4 experiments are shown. (F) Histograms represent the mean percentages of cells positive for GPA, CD36, CD34, and double-positive for GPA and CD36 antigens in the whole cell suspension after the 14-day culture. *P < .05, unpaired Student t test. Error bars indicate SEM (G) Transduced CD34+ cells were grown in a medium containing SCF, FLT3-L, IL-3, and G-CSF to monitor granulomonocytic differentiation in vitro. CD14 and CD15 immunophenotypic analysis at day 10 in 1 representative of 5 independent experiments is shown. (H) CD14, CD15, CD11b, and CD34 immunophenotypic analysis of cultured cells at day 10. Histograms represent the mean percentages of cells positive for each antigen in the whole cell suspension (5 independent experiments). *P < .05, unpaired Student t test. Error bars indicate SEM.
Results and discussion
We first assessed whether TET2 mutations could influence 5-hmC levels in MPN. CD34+ cells from 2 healthy patients, 2 patients with mutant TET2, and 1 patient with wild-type TET2 were grown in erythroid differentiation culture to obtain sufficient DNA for HPLC-MS/MS experiments. On day 8, 5-mC levels were similar, but the ratio of 5-hmC to 5-mC was lower in DNA from mutant TET2 cells than in wild-type TET2 and control cells (supplemental Figure 1). We next analyzed peripheral blood leukocytes from a series of 19 healthy control patients, 19 patients with wild-type TET2, and 15 patients with mutant TET2 with an HPLC-MS/MS technique that allowed for the use of smaller amounts of DNA (supplemental Figure 1). We observed a trend toward lower 5-hmC values in the group of mutant TET2 patients (median = 0.495) compared with healthy control values (median = 0.838, P = .0686, Mann-Whitney 2-tailed test). We then analyzed the 5-hmC content in purified granulocytes from an additional series of samples from 10 healthy control patients, 14 patients with wild-type TET2, and 9 patients with mutant TET2 by using dot blots with anti–5-hmC antibody. Analysis of 5-hmC spot intensities relative to MB signals revealed that mutant TET2 MPN granulocytes had a reduction in 5-hmC content compared with granulocytes from both control patients and wild-type TET2 MPN patients, with a median normalized value of 0.526 versus 0.986 and 0.819, respectively (P < .05, Mann-Whitney 2-tailed test; Figure 1A-B). These results demonstrate that some patients with MPN have an impaired hydroxylation of 5-mC, especially when they carry a TET2 mutation. However some wild-type TET2 patients also had low 5-hmC levels, as reported in other myeloid malignancies,10 and a few patients had a high 5-hmC content. Known mutations, such as IDH1/2 mutations,9 or unknown alternative genetic lesions affecting TET2 function or DNA hydroxy-methylation may be responsible for these variations in 5-hmC content.
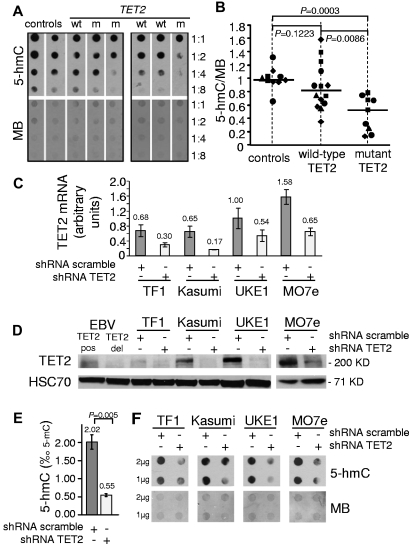
Figure 1.
Mutation or knockdown of TET2 lead to decreased levels of 5-hmC in human hematopoietic cells. (A) 5-hmC content of granulocytes from healthy patients (controls) and wild-type (wt) or mutant (m) TET2 MPN patients was determined by analyzing serial 2-fold dilutions of granulocyte DNA on dot blots with an anti–5-hmC antibody. The membranes were stained by MB to allow quantification of spot intensities by ImageJ software. (B) The ratios of 5-hmC to MB spot intensities from 4 experiments (10 control samples, 14 wild-type TET2 MPN samples, and 9 mutant TET2 MPN samples) were calculated, and individual values were normalized to the average value of control DNA for each experiment. Normalized values are plotted as squares, triangles, diamonds, and circles, each symbol representing one experiment. Horizontal bars indicate median values. P values were obtained with the use of the 2-tailed Mann-Whitney test. (C) TET2 mRNA quantification in TF1, Kasumi-1, UKE1, and MO7e cell lines transduced by lentiviruses expressing GFP and shRNA designed against either TET2 (shRNA TET2) or a control scramble sequence (shRNA scramble). Error bars indicate SEM. (D) Western blot analysis of transduced cell lines with anti-TET2 and anti-HSC70 antibodies. EBV lymphoblastoid cell lines from a patient with no TET2 mutation (EBV TET2 pos) and a patient with a biallelic deletion of TET2 (EBV TET2 del) were used as positive and negative controls. (E) 5-hmC content was determined by HPLC-MSMS in transduced MO7E cells. Histograms show 5-hmC/5-mC ratios (mean of 3 experiments). Error bars indicate SEM. P value was obtained with an unpaired Student t test. (F) A total of 1-2 μg of DNA from transduced TF1, Kasumi-1, UKE1, and MO7e cells was spotted for 5-hmC dot blot assay. Membranes were stained with MB to control spotting.
To confirm that 5-hmC levels were directly dependent on TET2 function in human hematopoietic cells, we used lentiviral delivery of shRNA-TET2 or control shRNA-scramble in leukemic cell lines. Quantitative RT-PCR and Western blot analyses showed efficient TET2 knockdown by shRNA-TET2 (Figure 1C-D, supplemental Figure 2). In MO7e cells, HPLC-MS/MS revealed that TET2 knockdown led to a dramatic reduction of the 5-hmC/5-mC ratio (0.55‰ vs 2.02‰ in control cells, P = .005, unpaired Student t test; Figure 1E, supplemental Figure 3). Decreased 5-hmC levels also were observed in shRNA-TET2–expressing TF1, Kasumi-1, UKE1, MO7e, UT7, and HL60 cells when 5-hmC dot blots were used (Figure 1F, supplemental Figure 3). These data indicate that TET2 knockdown results in decreased 5-hmC levels in human leukemic cell lines, thus arguing for the role of the protein in generating 5-hmC in human hematopoietic cells.
Because TET2 mutations are found in myeloid malignancies, we hypothesized that the disruption of TET2 function may disturb myelopoiesis. Thus, we transduced CD34+ cells from umbilical cord blood with shRNA-TET2 and shRNA-scramble lentiviruses. Transduced cells were analyzed in long-term B/NK/GM culture, CFC assays, and liquid cultures for granulomonocytic or erythroid differentiation. We assessed efficient TET2 knockdown by analyzing both TET2 expression and 5-hmC levels (supplemental Figure 4, Figure 2A). In B/NK/GM conditions, TET2-knockdown CD34+CD38− cells gave rise to 52.4% ± 4.3% of myeloid clones versus 35.5% ± 3.1% for control cells (P = .003, unpaired Student t test; Figure 2B). CFC assays showed that TET2 knockdown had no impact on the total number of colonies. However granulomonocytic colonies were slightly increased in number and size, although erythroid colonies were decreased (Figure 2C-D). These results suggest that the granulomonocytic compartment had an advantage over erythroid differentiation.
Impaired terminal erythroid differentiation was confirmed when we analyzed CD36 and glycophorin-A acquisition of CD34+ cells grown in liquid erythroid differentiation culture for 14 days, with 57% ± 8% of glycophorin-A/CD36–double-positive erythroblasts within shRNA-TET2–expressing cells compared with 81% ± 4% in control cells (P = .024, unpaired Student t test; Figure 2E-F). In a granulomonocytic liquid culture assay, morphologic analysis revealed that shRNA-TET2–expressing cells contained more monocytic and less granulocytic cells than control cells (47% ± 4% vs 37% ± 3%, and 52% ± 4% vs 62% ± 3%, respectively, at day 10, P < .05, unpaired Student t test; supplemental Figure 5). Immunophenotypic analyses confirmed this observation. On day 10 and 15, the percentages of granulocytic CD15+ cells were lower in TET2-knockdown cells (20% ± 2% and 19% ± 2%, respectively) compared with shRNA-scramble–expressing cells (36% ± 4% and 26% ± 2%, respectively), whereas the percentages of monocytic CD14+ cells were greater in TET2-knockdown cell suspension (52% ± 1% and 53% ± 2%, respectively) than in control culture (39% ± 4% and 42% ± 3%, respectively; Figure 2G-H, supplemental Figure 6). In contrast, cultures in the presence of M-CSF revealed no significant difference between shRNA-TET2–expressing cells and control cells (supplemental Figure 7). In these experiments we did not observe any increase in immature myeloid cells as recently reported with a Tet2 shRNA in the mouse.9
Altogether, our results confirm the ability of TET2 to convert 5-mC to 5-hmC in human hematopoietic progenitor cells and demonstrate that TET2 knockdown skews human progenitor differentiation toward the myeloid lineage, with an advantage for monocytic development at the expense of the granulocytic lineage. Moreover, we observed an impairment of erythroid differentiation. These results could explain the high frequency of TET2 mutations in chronic myelomonocytic leukemia, a disease associated with both myelodysplastic features and an expansion of the monocytic compartment.6,16,17 In addition, other lesions such as JAK2V617F could behave as a rescuer of myelopoiesis defects of TET2 mutant cells and lead to MPN7,18 or refractory anemia with ringed sideroblasts associated with marked thrombocytosis.19
The authors of previous studies5,7,9,10,20 suggested that hematopoietic cells with TET2 mutations had defects in the control of both early and late steps of myelopoiesis. How TET2 expression and 5-hmC levels control hematopoietic development is unknown. The mechanism has been proposed to be epigenetic through modifications of DNA methylation.10 Other genes involved in DNA methylation maintenance, such as DNMT1 and DNMT3A, have been implicated in HSC function,21 myeloid differentiation,22 or myeloid malignancies.23 In this regard, preliminary gene expression analyses revealed that the knockdown of TET2 was related to a slightly decreased expression of some genes implicated in granulocyte differentiation such as GFI1, RARA, and RXRA (supplemental Figure 8). Further studies will be required to understand whether they are primary target genes of TET2 function or secondary events amplifying the skewing toward monocytic differentiation.
Supplementary Material
Acknowledgments
The authors thank François Girodon for the recruitment of patients; Annie Robert for statistical analysis; and Corinne Hurtaud, Oliver Bluteau, Yann Lescluse, Thierry Langlois, Dorota Jeziorowska, and Philippe Rameau for helpful discussions and technical support.
This work received funding from the MPN research foundation (F.D.), the Fondation de France (F.D.), the Institut National du Cancer (F.D., N.C., and W.V.), the Cancéropôle Ile de France (Cancer Stem Cell network, W.V. and H.M.), the Agence Nationale pour la Recherche (ANR blanc 2010, I.P. and J.L.R.), the Ligue Nationale Contre le Cancer (équipe labélisée 2009), and National Institute of Health grants CA129831 and CA129831-03S1 (L.A.G). E.P. is a Ministère de la Recherche MNERT recipient.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: E.P. and C.A. performed cellular and molecular experiments, analyzed the data, and wrote the paper; H.M., A.S., Bd.C.R.M.M., and a.m. contributed to cellular and molecular experiments; J.-P.L.C. did molecular analyses; F.P. and O.A.B. contributed to research design and provided cell lines; B.C. and J.L. provided umbilical cord blood samples; A.V., J.-L.R., and L.A.G. performed HPLC-MS/MS and analyzed data; N.C. recruited patients and contributed to research design and writing of the paper; N.D. and E.S. contributed to research design, data analysis, and writing of the paper; W.V. and I.P. designed research and contributed to the writing of the manuscript; and F.D. designed and directed research, performed cellular experiments, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: François Delhommeau, Laboratoire d'Hématologie, Hôpital Saint-Antoine, 184 rue du Faubourg Saint-Antoine, 75012, Paris, France; e-mail: francois.delhommeau@sat.aphp.fr.
References
- 1.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466(7310):1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ficz G, Branco MR, Seisenberger S, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473(7347):398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 4.Tefferi A, Lim KH, Abdel-Wahab O, et al. Detection of mutant TET2 in myeloid malignancies other than myeloproliferative neoplasms: CMML, MDS, MDS/MPN and AML. Leukemia. 2009;23(7):1343–1345. doi: 10.1038/leu.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langemeijer SM, Kuiper RP, Berends M, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41(7):838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- 6.Jankowska AM, Szpurka H, Tiu RV, et al. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood. 2009;113(25):6403–6410. doi: 10.1182/blood-2009-02-205690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delhommeau F, Dupont S, Della Valle V, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360(22):2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 8.Abdel-Wahab O, Mullally A, Hedvat C, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114(1):144–147. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ko M, Huang Y, Jankowska AM, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468(7325):839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupont S, Masse A, James C, et al. The JAK2 617V>F mutation triggers erythropoietin hypersensitivity and terminal erythroid amplification in primary cells from patients with polycythemia vera. Blood. 2007;110(3):1013–1021. doi: 10.1182/blood-2006-10-054940. [DOI] [PubMed] [Google Scholar]
- 12.Marie-Desvergne C, Maitre A, Bouchard M, Ravanat JL, Viau C. Evaluation of DNA adducts, DNA and RNA oxidative lesions, and 3-hydroxybenzo(a)pyrene as biomarkers of DNA damage in lung following intravenous injection of the parent compound in rats. Chem Res Toxicol. 2010;23(7):1207–1214. doi: 10.1021/tx100081p. [DOI] [PubMed] [Google Scholar]
- 13.Song L, James SR, Kazim L, Karpf AR. Specific method for the determination of genomic DNA methylation by liquid chromatography-electrospray ionization tandem mass spectrometry. Anal Chem. 2005;77(2):504–510. doi: 10.1021/ac0489420. [DOI] [PubMed] [Google Scholar]
- 14.Gilles L, Bluteau D, Boukour S, et al. MAL/SRF complex is involved in platelet formation and megakaryocyte migration by regulating MYL9 (MLC2) and MMP9. Blood. 2009;114(19):4221–4232. doi: 10.1182/blood-2009-03-209932. [DOI] [PubMed] [Google Scholar]
- 15.Delhommeau F, Dupont S, Tonetti C, et al. Evidence that the JAK2 G1849T (V617F) mutation occurs in a lymphomyeloid progenitor in polycythemia vera and idiopathic myelofibrosis. Blood. 2007;109(1):71–77. doi: 10.1182/blood-2006-03-007146. [DOI] [PubMed] [Google Scholar]
- 16.Kosmider O, Gelsi-Boyer V, Ciudad M, et al. TET2 gene mutation is a frequent and adverse event in chronic myelomonocytic leukemia. Haematologica. 2009;94(12):1676–1681. doi: 10.3324/haematol.2009.011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohlmann A, Grossmann V, Klein HU, et al. Next-generation sequencing technology reveals a characteristic pattern of molecular mutations in 72.8% of chronic myelomonocytic leukemia by detecting frequent alterations in TET2, CBL, RAS, and RUNX1. J Clin Oncol. 2010;28(24):3858–3865. doi: 10.1200/JCO.2009.27.1361. [DOI] [PubMed] [Google Scholar]
- 18.Schaub FX, Looser R, Li S, et al. Clonal analysis of TET2 and JAK2 mutations suggests that TET2 can be a late event in the progression of myeloproliferative neoplasms. Blood. 2010;115(10):2003–2007. doi: 10.1182/blood-2009-09-245381. [DOI] [PubMed] [Google Scholar]
- 19.Flach J, Dicker F, Schnittger S, Kohlmann A, Haferlach T, Haferlach C. Mutations of JAK2 and TET2, but not CBL are detectable in a high portion of patients with refractory anemia with ring sideroblasts and thrombocytosis. Haematologica. 2010;95(3):518–519. doi: 10.3324/haematol.2009.013631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mullighan CG. TET2 mutations in myelodysplasia and myeloid malignancies. Nat Genet. 2009;41(7):766–767. doi: 10.1038/ng0709-766. [DOI] [PubMed] [Google Scholar]
- 21.Trowbridge JJ, Snow JW, Kim J, Orkin SH. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell. 2009;5(4):442–449. doi: 10.1016/j.stem.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bröske AM, Vockentanz L, Kharazi S, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41(11):1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- 23.Ley TJ, Ding L, Walter MJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.