SUMMARY
Steroid hormones are known systemic regulators of multiple normal and cancerous tissues; however, whether or how they impact the fate and function of adult stem cells is unclear. In the Drosophila ovary, insulin signals modulate the proliferation and self-renewal of germline stem cells (GSCs), yet despite evidence that additional systemic factors control GSC activity, these have remained largely unknown. Here, we report that ecdysone, a steroid hormone structurally related to mammalian sex steroids, directly regulates adult GSC proliferation and self-renewal independently of insulin signaling. Ecdysone controls GSCs through a functional interaction with the chromatin remodeling factors ISWI, an intrinsic epigenetic factor required for GSC fate and activity, and Nurf301, the largest subunit of the ISWI-containing NURF chromatin remodeling complex. Our findings support a link between systemic steroid hormones and the intrinsic chromatin remodeling machinery as a potential mechanism to promote broad transcriptional programs required for adult stem cell self-renewal.
INTRODUCTION
Maintenance and regeneration of many adult tissues are fueled by stem cells that self-renew and create daughters that differentiate. Abnormal behavior of stem cells can lead to their depletion and loss of tissue integrity or, conversely, to overproliferation. Stem cells are therefore tightly regulated by intrinsic factors, local signals from their niche, and systemic hormones, which couple stem cell behavior to physiological status (Drummond-Barbosa, 2008). Many studies have addressed how intrinsic and local regulators maintain stem cell identity and proliferative potential. In contrast, much less is known about the direct actions of systemic hormones on stem cells, despite the therapeutic relevance of exploring this level of regulation.
Steroid hormone levels vary with gender and physiology, and affect proliferation, survival, and cell fate in multiple adult tissues supported by stem cells, such as the brain, mammary epithelium, and hematopoietic system (Asselin-Labat et al., 2010; Pawluski et al., 2009; Ray et al., 2008). In the dentate gyrus, ectopic estrogen administration increases proliferation, while glucocorticoids suppress it (Pawluski et al., 2009); however, it is unclear whether these steroids act directly on neural stem cells. A recent study suggests that estrogen and progesterone stimulate mammary stem cells via an indirect mechanism (Asselin-Labat et al., 2010). Thus, although it has been proposed that steroid hormones exert their effects directly on adult stem cells (Ray et al., 2008), there is a paucity of conclusive experimental evidence to support this model, and potential mechanisms are poorly understood. The link between increased cancer risk and elevated steroid hormone levels (Eliassen and Hankinson, 2008) underscores the importance of understanding the extent to which adult stem cells respond to these hormones.
The Drosophila ovary is a powerful system for dissecting the control of adult stem cells (Kirilly and Xie, 2007). Ovaries are composed of ovarioles that harbor progressively more mature egg chambers, or follicles (Spradling, 1993). Germline stem cells (GSCs) in the anterior of the ovariole, or germarium, reside in a specialized niche that modulates their behavior (Figure 1A). For example, the bone morphogenetic protein (BMP) ligand Decapentaplegic (Dpp) is a key niche signal that stimulates Punt (Put) and Thickveins (Tkv) receptors on GSCs to promote their proliferation and self-renewal (Xie and Spradling, 1998). Asymmetric GSC division yields another GSC and a cystoblast committed to differentiation. The cystoblast divides to ultimately give rise to a germline cyst comprised of one oocyte and 15 nurse cells. GSCs, cystoblasts, and cysts can be easily distinguished by the morphology of the fusome, a membranous, germline-specific organelle (de Cuevas and Spradling, 1998). Somatic follicle cells envelop each cyst to form a follicle that progresses through fourteen developmental stages.
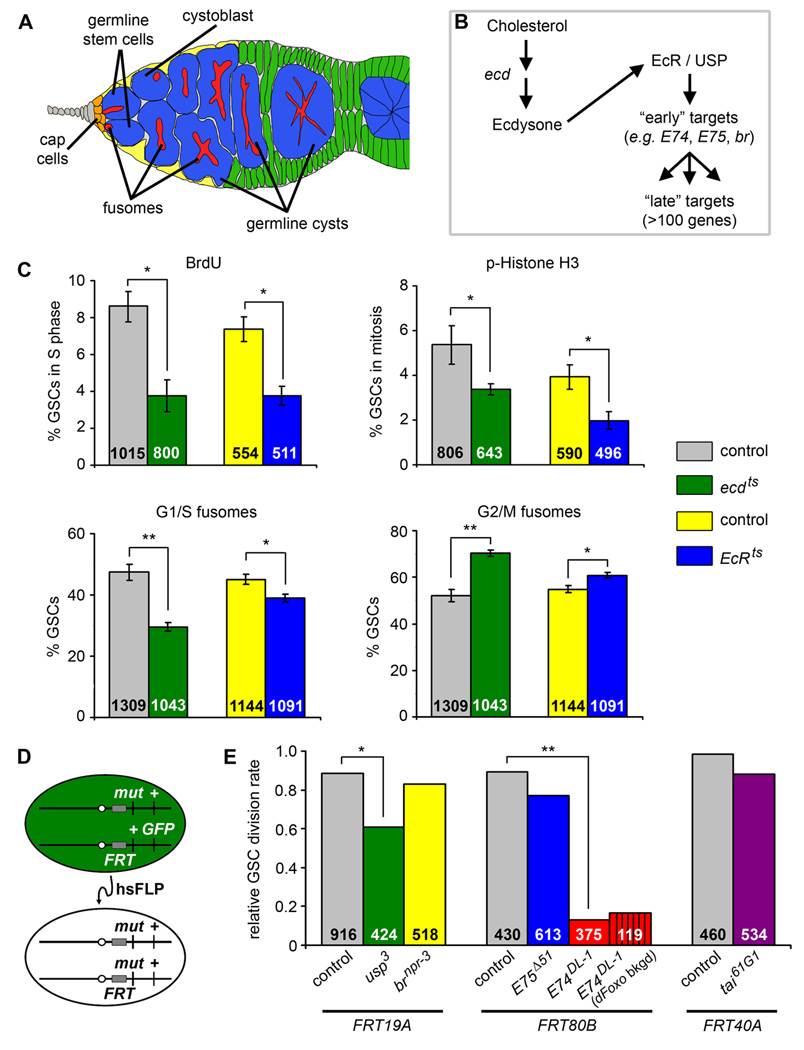
Figure 1. Ecdysone acts directly on GSCs to control GSC proliferation at the level of the G2 cell cycle phase.
(A) Drosophila germarium. GSCs are identifiable by their typical fusome (red) and attachment to the niche (cap cells, orange; terminal filament cells, gray), and associate with a subset of inner germarial sheath cells (yellow). GSC division produces one self-renewing daughter and one cystoblast that divides to form a 16-cell germline cyst encapsulated by somatic follicle cells (green). (B) Diagram of ecdysone pathway showing components relevant to this study. (C) Frequencies of GSCs in S phase (measured by BrdU incorporation), mitosis (measured by phospho-histone H3 labeling, p-Histone H3), or displaying G1/S or G2/M fusome morphologies in temperature-sensitive ecd1/Df(3L)R-G7 (ecdts) and EcRM554fs/EcRA483T (EcRts) mutants versus heterozygous controls at 29°C for five days. The number of GSCs analyzed is shown inside bars. Error bars, mean ± s.e.m. (D) FLP/FRT technique used to generate genetic mosaics. Mitotic recombination is mediated by heat shock-induced expression of flipase (hsFLP). Homozygous mutant (mut) GSCs are identified by the absence of a GFP marker, which is linked to the wild-type allele. (E) Relative GSC division rates (normalized ratio between GFP-negative and -positive progeny) in wild-type control, usp3, brnpr-3, E75Δ51, E74DL-1, and tai61G1 mosaic germaria. E74DL-1 mosaic germaria were also analyzed in the dFOXO21/dFOXO25 mutant background (dFoxo bkgd). The number of germline cystoblasts/cysts analyzed is shown inside bars. *p < 0.005, **p < 0.001. (See also Figure S1 and Table S2.)
Our recent studies uncovered key roles of systemic factors on GSC division and maintenance. Insulin signals directly regulate GSC proliferation (Hsu et al., 2008; LaFever and Drummond-Barbosa, 2005), but have a separate, indirect role in GSC self-renewal via modulation of niche size and adhesion to GSCs (Hsu and Drummond-Barbosa, 2009). Although insulin signals are major GSC regulators, additional unknown factors control the GSC division cycle in response to external cues (Hsu et al., 2008).
The best characterized steroid hormone in Drosophila is ecdysone, which is similar to human sex steroids (Mangelsdorf et al., 1995) and modulates oogenesis. Ecdysone is required for follicle development (Carney and Bender, 2000), and regulates border cell migration (Bai et al., 2000; Jang et al., 2009). Binding of ecdysone to the ecdysone receptor (EcR) initiates a transcriptional cascade that triggers many cellular responses (Figure 1B) (Riddiford et al., 2000). Among EcR downstream targets, the early-response genes E74, E75, and broad (br) are expressed in ovarian germline and somatic cells. As for EcR mutants, E74 or E75 germline loss-of-function results in failure of follicles to develop past early oogenesis (Buszczak et al., 1999; Carney and Bender, 2000). It remains unclear, however, whether GSCs themselves sense and respond to ecdysone.
Here, we demonstrate that ecdysone directly stimulates GSCs to promote their self-renewal and activity independently of insulin signaling. We also show that ecdysone controls GSCs through a functional interaction with the chromatin remodeling factors ISWI, intrinsically required for GSC fate and division, and Nurf301, the largest subunit of the ISWI-containing NURF chromatin remodeling complex. Mutation of components of the ecdysone pathway in GSCs leads to impaired BMP signaling, consistent with known roles of ISWI (Xi and Xie, 2005). We propose that ecdysone produced by more differentiated follicles under favorable conditions acts on GSCs as a positive feedback mechanism. Our findings suggest a potentially widespread mechanism whereby steroid hormones act directly on adult stem cells via a functional interaction with their epigenetic machinery to broadly control the transcription of genes required for stem cell self-renewal and activity. Finally, this study offers new insights into how steroids may promote tumors, as steroid hormone signaling is often altered in cancers thought to arise from cancer stem cells (Britt et al., 2007; Clarke and Fuller, 2006; D'Errico and Moschetta, 2008).
RESULTS
Ecdysone Promotes GSC Proliferation at G2
Ecdysone antagonizes insulin signaling to control larval growth (Colombani et al., 2005), and insulin signals directly promote GSC proliferation (LaFever and Drummond-Barbosa, 2005). We therefore asked if ecdysone antagonizes insulin signaling in GSCs. To test if ecdysone controls GSC division, we measured cell cycle marker frequencies in GSCs of temperature-sensitive mutants of ecdysoneless (ecd; required for normal ecdysone levels) and EcR (Figure 1B) at the restrictive temperature. ecdts and EcRts mutants display reduced frequencies of GSCs positive for bromodeoxyuridine (BrdU) incorporation, an S phase marker, or for the mitosis marker phospho-histone H3 (Figure 1C), indicating that ecdysone normally stimulates GSC proliferation and thus that it does not antagonize insulin signaling.
We also monitored GSC fusome morphology as an indicator of cell cycle phases (Hsu et al., 2008) (see Experimental Procedures), and found increased frequencies of “G2/M” fusomes and decreased frequencies of “G1/S” fusomes under reduced ecdysone signaling (Figure 1C). These results suggest that ecdysone promotes GSC progression through G2, similar to its role in histoblast proliferation during insect metamorphosis (Ninov et al., 2009).
Ecdysone Directly Stimulates GSC Proliferation
The EcR is widely expressed in the ovary (Buszczak et al., 1999); thus, multiple cells can potentially receive the ecdysone signal. We asked whether ecdysone acts directly on GSCs to control their proliferation, or indirectly, via intermediate cell types. Ecdysone signaling requires EcR dimerization with its obligate co-receptor, Ultraspiracle (Usp; Figure 1B), the Drosophila ortholog of the mammalian retinoid X receptor (Riddiford et al., 2000). To render GSCs unable to receive the ecdysone signal, we inactivated usp specifically in GSCs (mutant GSCs and their progeny identified by loss of a GFP marker) and measured their proliferation relative to that of neighboring control GFP-positive GSCs (Fig. 1D). [For technical reasons, we could not perform the equivalent experiment with EcR mutations (see Experimental Procedures)]. Relative division rates were obtained by dividing the number of GFP-negative cystoblasts/cysts by the number of GFP-positive cystoblasts/cysts in mosaic germaria. In control mosaics, GSC relative division rates were close to 1.0, reflecting similar proliferation rates of GFP-positive and -negative GSCs. In contrast, usp3 GSCs had significantly reduced relative division rates (Figure 1E), and there was no increased death of usp3 cystoblasts/cysts (see below and Figure 2). Thus, we conclude that ecdysone is directly received by GSCs to control their proliferation.
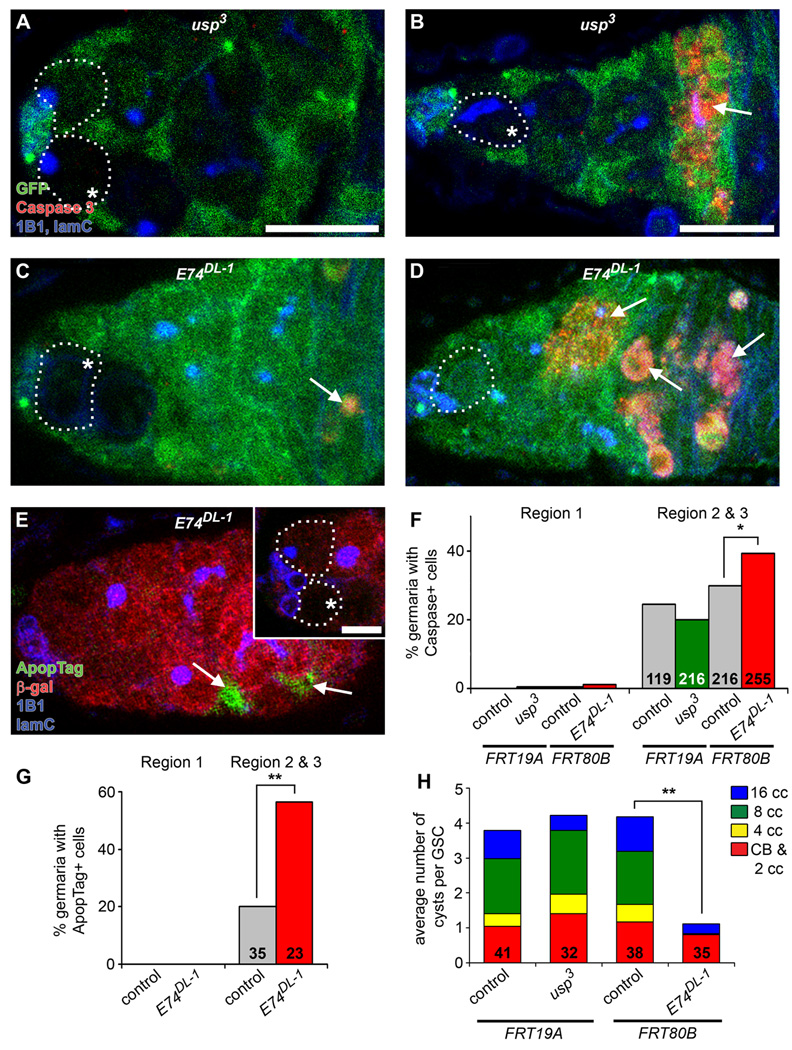
Figure 2. E74DL-1, but not usp3, mosaic germaria display increased cell death of germline cysts in posterior regions of germarium.
(A–D) usp3 (A and B) and E74DL-1 (C and D) mosaic germaria labeled with GFP (green), cleaved Caspase 3 (red, apoptotic cells), 1B1 (blue, fusome) and laminC (blue, nuclear envelope of cap cells) at six days after heat shock (AHS). GSCs (outlined) are identified by their fusome morphology and attachment to cap cells. (E) E74DL-1 mosaic germaria labeled with ApopTag (green, apoptotic cells), β-galactosidase (red), 1B1 (blue) and laminC (blue) at six days AHS. In (A–E), mutant clones are recognized by the absence of GFP (A–D) or β-galactosidase (E) expression in GSCs (asterisks). Arrows indicate apoptotic cells. Scale bar represents 10 µm (A–E) or 5 µm (inset in E, showing GSCs from same germarium in different focal plane). (F and G) Percentages of control, usp3, and E74DL-1 mosaic germaria containing cleaved Caspase 3-positive (F) or ApopTag-positive (G) cells in either region 1 (anterior-most region, containing GSCs, cystoblasts, and dividing cysts) or region 2 and 3 (containing 16-cell cysts and associated follicle cells). Number of germaria (F) or number of germaria with a β-galactosidase-negative clone (G) is shown inside bars. *p < 0.05, **p < 0.005. (H) Distribution of the average number of control (GFP-positive) or mutant (GFP-negative) cystoblasts/2-cell cysts, 4-, 8-, and 16-cell cysts (CB & 2 cc, 4 cc, 8 cc, and 16 cc, respectively) per corresponding GSC in E74DL-1 or usp3 mosaic germaria. The number of germaria analyzed is shown inside bars. **p < 0.001 for 4-cell, 8-cell, and 16-cell cysts.
The EcR Co-Activator tai Is Not Intrinsically Required for GSC Proliferation
Activity of the EcR/Usp receptor can be modified by at least one co-activator, Taiman (Tai), which is required during border cell migration (Bai et al., 2000; Jang et al., 2009). We asked whether tai is required for EcR/Usp activity in GSC proliferation control. The relative division rates of tai61G1 mutant versus control GSCs in mosaic germaria, however, were not statistically different (Figure 1E), suggesting specificity in EcR/Usp signaling in GSCs versus somatic border cells.
The Ecdysone Early-Response Target E74, but not E75 or br, is Required for Proper GSC Proliferation and Survival of Early Cysts
Among the best-characterized ecdysone early response genes are br, E75, and E74 (Figure 1B), all with known roles in later follicles (Buszczak et al., 1999; Deng and Bownes, 1997). To determine whether a specific ecdysone pathway branch controls GSC proliferation, we measured the relative division rates of brnpr-3, E75Δ51 and E74DL-1 mutant GSCs in mosaics. The relative division rates of E75Δ51 and brnpr-3 GSCs were not statistically different from control rates. In contrast, the relative division rate of E74DL-1 GSCs was significantly decreased (Figure 1E), suggesting that E74 specifically mediates the effects of ecdysone on GSC proliferation.
E74DL-1 GSCs displayed a more pronounced decrease in relative division rate than usp3 GSCs (Figure 1E). This difference was likely due to the hypomorphic nature of usp3 because germaria mosaic for the null usp2 allele displayed stronger phenotypes than usp3 mosaics (see Figure S1), consistent with reports that usp3 retains the ability to activate a subset of downstream targets (Ghbeish et al., 2001). We also detected a significant increase in apoptotic cysts and associated somatic cells in more posterior regions of E74DL-1 mosaic germaria, but not in usp3 mosaics (Figure 2A–G). Consistent with increased apoptosis, there was a decrease in E74DL-1 4-, 8-, and 16-cell cysts in mosaic germaria, which was not evident in usp3 mosaics (Figure 2H). This indicates that E74 is required for the viability of cysts as they divide and associate with follicle cells. Thus, the relative division rate of E74DL-1 GSCs is skewed lower due to increased E74DL-1 cyst death.
Ecdysone Controls GSC Proliferation Independently of Insulin Signaling
The analyses of ecdysone pathway mutants suggested that ecdysone signals directly to GSCs via E74 to promote G2 progression. Insulin signals also directly control GSC division at G2 (Hsu et al., 2008); therefore, ecdysone signaling could either converge on the insulin signaling pathway, or control GSC proliferation by a distinct mechanism. The G2 delay caused by mutation of the insulin receptor (InR) gene can be suppressed by mutation of the negative downstream effector dFOXO (Hsu et al., 2008). dFOXO removal, however, had no effect on the relative division rate of E74DL-1 GSCs (Figure 1E). We did not expect a complete suppression because of the contribution of early cyst death to this measurement (see Figure 2). Nevertheless, the absence of a partial suppression indicates that loss of dFOXO does not reverse the E74DL-1 GSC proliferation defect. Although we cannot exclude the possibility that E74 acts downstream of dFOXO, these results suggest that ecdysone and insulin signals control GSC G2 via separate mechanisms.
Ecdysone Signals Directly Promote Maintenance of GSCs
Systemic signals may control not only the proliferation of stem cells, but also their ability to self-renew. Indeed, in addition to directly controlling the proliferation of GSCs, insulin signals also control their self-renewal indirectly, via the niche (Hsu and Drummond-Barbosa, 2009; LaFever and Drummond-Barbosa, 2005). We thus asked whether ecdysone signals are required for GSC self-renewal. Upon switching to the restrictive temperature, ecdts and EcRts females showed a rapid loss of GSCs relative to heterozygous or wild-type controls (Figure 3A and Table S1), indicating that systemic ecdysone signals are required for GSC maintenance.
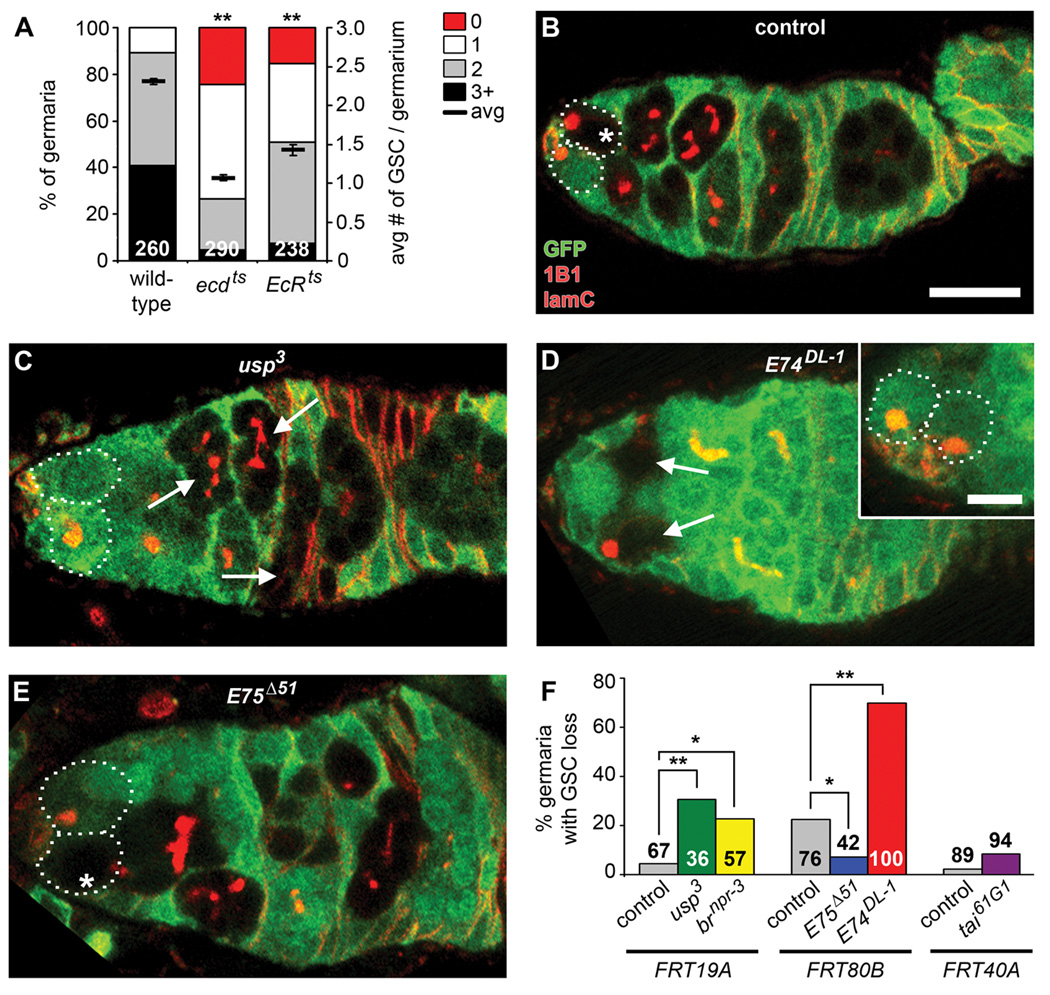
Figure 3. Ecdysone directly controls GSC maintenance predominantly via E74.
(A) Frequencies of germaria containing zero, one, two, or three or more GSCs (left Y-axis), and average number of GSCs per germarium (right Y-axis) in ecdts and EcRts mutant versus wild-type females one week after incubation at the restrictive temperature. See Table S1 for heterozygous controls. The number of germaria analyzed is shown inside bars. Error bars, mean ± s.e.m. (B–E) Control, usp3, E74DL-1, and E75Δ51 mosaic germaria labeled with GFP (green), 1B1 (red, fusome) and laminC (red, nuclear envelope of cap cells) at 10 days AHS (outlines indicate GSCs). Control and mutant clones are recognized by the absence of GFP expression in GSCs (asterisks) and their progeny (B and E) or just in their progeny (arrows), if the GSC has been lost from the niche (C and D). Scale bar represents 10 µm (B–E) or 5 µm (inset in D, showing GSCs from same germarium in different focal plane). (F) Frequencies of germaria with control, usp3, brnpr-3, E75Δ51, E74DL-1, and tai61G1 mosaic germline showing GSC loss at 10 days AHS. Number of germaria with mosaic germline analyzed is shown inside or above bars. *p < 0.05, **p < 0.001. (See also Figure S1 and Tables S1 and S2.)
To test if ecdysone is received directly by GSCs to control their maintenance, we inactivated components of the ecdysone pathway in GSCs (Figure 1D; see Table S2). GSCs homozygous for a given ecdysone pathway mutation and their descendents were recognized by loss of GFP in mosaic germaria. In most control mosaic germaria, GFP-negative GSCs and cystoblasts/cysts were observed (Figure 3B and F). In contrast, a significant percentage of hypomorphic usp3 mosaic germaria contained GFP-negative cystoblasts/cysts in the absence of a GFP-negative GSC, indicating GSC loss (Figure 3C and F). The loss of null usp2 GSCs was considerably more severe than that of usp3 GSCs (Figure S1). These results indicate that GSCs directly receive the ecdysone signal for their maintenance.
The E74 Branch of the Ecdysone Pathway Predominantly Promotes GSC Maintenance
We also found evidence for specificity of EcR signaling during GSC maintenance. While loss of tai61G1 or E75Δ51 GSCs was not significantly increased, E74DL-1 GSCs were lost markedly faster relative to control GSCs in mosaic germaria, suggesting that signaling through E74 is critical for GSC maintenance (Figure 3D–F), as is the case for their proliferation. E74DL-1 GSC loss is likely not due to their death, as increased apoptosis in the GSC region was not evident in E74DL-1 mosaics (see Figure 2C–G). brnpr-3 GSCs also have a less severe, albeit significantly increased rate of loss (Figure 3F), suggesting that an additional ecdysone pathway branch contributes to GSC maintenance. E74 and br have been reported to cooperate to modulate gene expression during metamorphosis (Fletcher and Thummel, 1995); it is thus possible that the contribution of br to the control of GSC maintenance reflects an analogous interaction with E74. A small decrease in E75Δ51 GSC loss was detected (Figure 3F), suggesting that E75 weakly inhibits GSC maintenance. This result may reflect a negative feedback role of E75 in the ecdysone pathway, or an ecdysone-independent role of E75. Together, these results suggest that the E74 branch of the ecdysone pathway has a predominant role in promoting GSC self-renewal.
Ecdysone Acts Via a Functional Interaction with the Intrinsic NURF Chromatin Remodeling Complex to Maintain GSCs
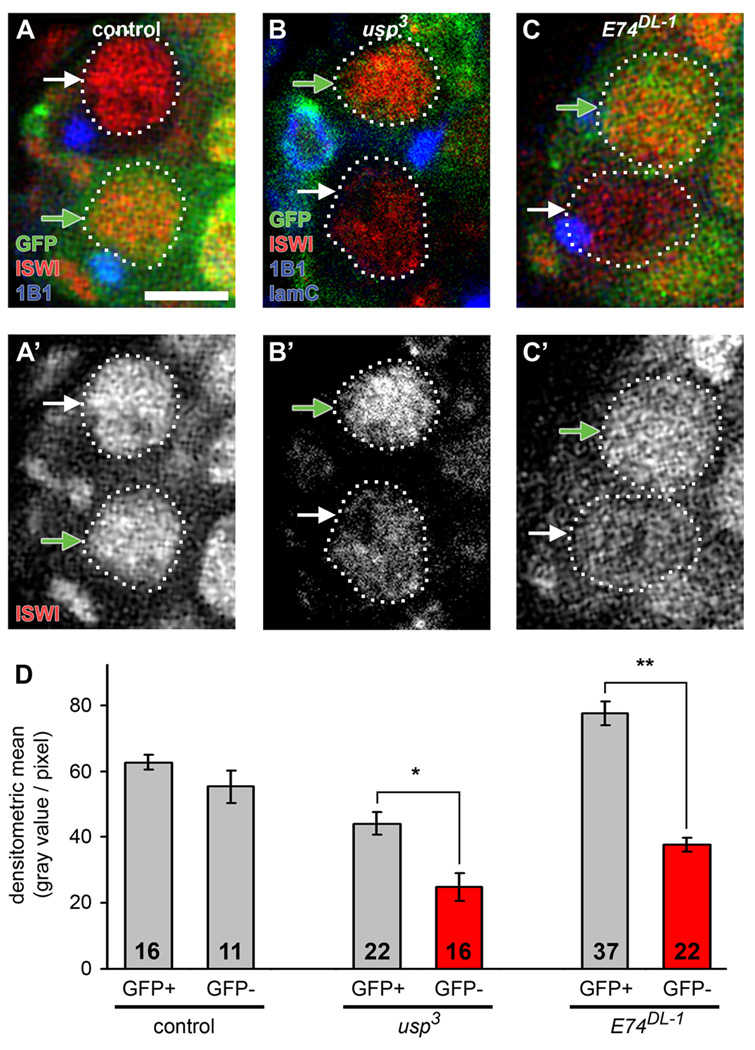
The intrinsic requirement for ecdysone signaling for GSC maintenance implies that the underlying mechanism does not involve insulin signaling because insulin-like peptides control GSC self-renewal indirectly, via the niche (Hsu and Drummond-Barbosa, 2009). Instead, it is conceivable that ecdysone signaling modulates intrinsic factors required for GSC maintenance. ISWI, a member of the SWI/SNF family of chromatin remodeling factors, is intrinsically required both for proliferation and self-renewal of ovarian GSCs (Xi and Xie, 2005). We also found that the Nurf301 subunit of the ISWI-containing NURF complex (Figure S2A) is required for GSC proliferation and maintenance, as nurf3013 GSCs showed impaired proliferation and increased loss in mosaic germaria, comparable to the defects of iswi2 GSCs (Xi and Xie, 2005) (Figure S2B–G). EcR and the NURF complex have been shown to cooperate in the induction of ecdysone-responsive genes during metamorphosis, and also to physically interact in an ecdysone-dependent manner in vitro, leading to the model that NURF is a co-activator of EcR (Badenhorst et al., 2005). Intriguingly, both usp3 and E74DL-1 GSCs, germline cysts, and follicle cells displayed reduced levels of nuclear ISWI protein relative to those of control cells in mosaic germaria (Figure 4 and Figure S3A–B). Reduced ecdysone signaling did not, however, result in alterations in either iswi or nurf301 transcript levels in whole ovaries (Figure S3C–D), suggesting that ecdysone signaling instead might modulate the translation or stability of ISWI. Antibodies against other subunits of NURF were unavailable; therefore, we could not test whether ecdysone affects the stability of the entire NURF complex. Taken together, these results raise the possibility that a functional interaction between ecdysone signaling and the ISWI-containing NURF complex may mediate the effects of ecdysone on GSCs.
Figure 4. Ecdysone signaling affects nuclear ISWI protein levels.
(A–C) Control (A), usp3 (B), and E74DL-1 (C) mosaic germaria analyzed six days AHS and labeled with GFP (green), ISWI (red), 1B1 (blue, fusomes) and laminC (in B, blue, nuclear envelope of cap cells). Control and mutant GSCs (outlined) are recognized by the absence of GFP expression (GFP-negative GSCs, white arrows), while cells that did not undergo mitotic recombination are GFP-positive (GFP-positive GSCs, green arrows). Scale bar represents 5 µm. (D) Quantification of nuclear ISWI protein levels in control and mutant GSCs. The number of GSCs analyzed is shown inside bars. Error bars, mean ± s.e.m. *p < 0.05, **p < 0.001. (See also Figure S3.)
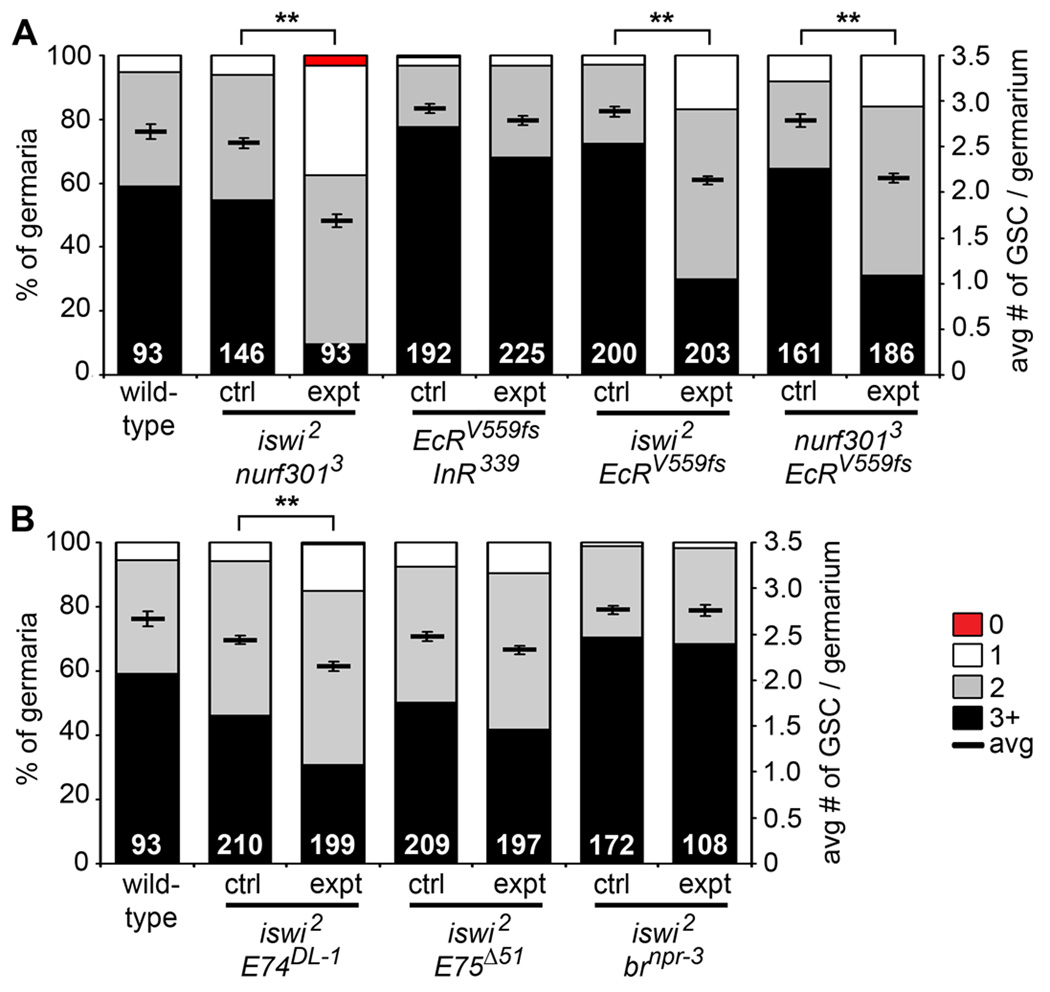
To specifically test this model, we examined genetic interactions between subunits of the NURF complex and ecdysone pathway components (Figure 5 and Table 1; see Experimental Procedures). Single heterozygous females for iswi1, iswi2, nurf3013, E74DL-1, E74neo24, E75Δ51, or brnpr-3 showed very low rates of GSC loss, similar to that of age-matched wild-type females. Double heterozygous iswi2 and nurf3013 females showed a significant increase in GSC loss (Figure 5A and Table 1). This result was not surprising, based on the published evidence that they function in a complex (Badenhorst et al., 2002). As a negative control, we tested double heterozygous females for EcRV559fs and the null InR339 allele, as insulin-like peptides and ecdysone control GSC maintenance via distinct mechanisms (see above). As expected, we found no significant genetic interaction between the insulin and ecdysone pathways in controlling GSC number (Figure 5A and Table 1). Similarly, nurf3013 and InR339 did not show a genetic interaction (Table 1). In contrast, we detected increased GSC loss in double heterozygotes for nurf3013 and EcRV559fs, or iswi2 and EcRV559fs, relative to controls (Figure 5A and Table 1), in support of the model that ecdysone cooperates with the ISWI-containing NURF complex to modulate GSCs. Further, we found strong genetic interactions between iswi2 and E74DL-1, or between iswi2 and E74neo24 (Figure 5B and Table 1). In contrast, we detected no genetic interactions between iswi2 and E75Δ51, or iswi2 and brnpr-3 (Figure 5B and Table 1), consistent with the weaker effects of the E75 and br mutations on GSC maintenance (see Figure 3F). These results are consistent with our findings that E74 predominantly mediates the effects of ecdysone on GSCs, and they strongly suggest that ecdysone functions with the ISWI-containing NURF complex to control GSCs.
Figure 5. Ecdysone signaling functionally interacts with iswi and nurf301 to control the maintenance of GSCs.
(A and B) Frequencies of germaria containing zero, one, two, or three or more GSCs (left Y-axis), and average number of GSCs per germarium (right Y-axis) in seven-day old wild-type females versus single heterozygotes (ctrl) or double heterozygotes (expt) for mutations in iswi, nurf301, EcR, and InR (A), and iswi, E75, E74, and br (B). The same wild-type data are shown in (A) and (B). Additional results are shown in Table 1. The number of germaria analyzed is shown inside bars. Error bars, mean ± s.e.m. **p < 0.001. (See also Figure S2.)
Table 1.
Genetic interactions between ecdysone pathway components, the chromatin remodeling factors iswi and nurf301, and the BMP pathway components dpp and put
| Genotypea | Number of germaria | Average # of GSCsb |
p valuec | |||
|---|---|---|---|---|---|---|
| 3+ GSCs |
2 GSCs |
1 GSC |
0 GSCs |
|||
| y w wild-type controls | 55 | 33 | 5 | 0 | 2.7 ± 0.08 (93)d | -- |
| iswi2/+ OR nurf3013/+ sibling controlse | 80 | 57 | 9 | 0 | 2.5 ± 0.06 (146) | |
| iswi2/+; nurf3013/+ | 9 | 49 | 32 | 3 | 1.7 ± 0.07 (93) | 3.21E-17 |
| EcRV559fs/+ OR InR339/+ sibling controls | 149 | 37 | 5 | 1 | 2.9 ± 0.05 (192) | |
| EcRV559fs/+; InR339/+ | 153 | 65 | 7 | 0 | 2.8 ± 0.05 (225) | 0.069 |
| nurf3013/+ OR EcRV559fs/+ sibling controls | 104 | 44 | 13 | 0 | 2.8 ± 0.07 (161) | |
| EcRV559fs/+; nurf3013/+ | 58 | 98 | 30 | 0 | 2.2 ± 0.05 (186) | 9.79E-12 |
| iswi1/+ OR EcRV559fs/+ sibling controls | 106 | 39 | 4 | 0 | 3.0 ± 0.07 (149) | |
| + iswi1/EcRV559fs + | 61 | 108 | 28 | 0 | 2.2 ± 0.05 (197) | 4.73E-19 |
| iswi2/+ OR EcRV559fs/+ sibling controls | 145 | 49 | 6 | 0 | 2.9 ± 0.05 (200) | |
| + iswi2/EcRV559fs + | 61 | 108 | 34 | 0 | 2.1 ± 0.05 (203) | 3.38E-23 |
| iswi1/+ OR E75Δ51/+ sibling controls | 144 | 70 | 7 | 0 | 2.8 ± 0.05 (221) | |
| iswi1/+; E75Δ51/+ | 151 | 63 | 5 | 0 | 2.8 ± 0.05 (219) | 0.620 |
| iswi2/+ OR E75Δ51/+ sibling controls | 105 | 88 | 16 | 0 | 2.5 ± 0.05 (209) | |
| iswi2/+; E75Δ51/+ | 82 | 96 | 19 | 0 | 2.3 ± 0.05 (197) | 0.035 |
| iswi1/+ OR E74DL-1/+ sibling controls | 128 | 53 | 6 | 0 | 2.7 ± 0.05 (187) | |
| iswi1/+; E74DL-1/+ | 78 | 102 | 24 | 0 | 2.3 ± 0.05 (204) | 4.07E-12 |
| iswi2/+ OR E74DL-1/+ sibling controls | 97 | 101 | 12 | 0 | 2.4 ± 0.05 (210) | |
| iswi2/+; E74DL-1/+ | 61 | 108 | 29 | 1 | 2.2 ± 0.05 (199) | 1.55E-05 |
| iswi1/+ OR E74neo24/+ sibling controls | 122 | 28 | 1 | 0 | 3.0 ± 0.05 (151) | |
| iswi1/+; E74neo24/+ | 75 | 103 | 16 | 5 | 2.3 ± 0.05 (199) | 1.86E-20 |
| iswi2/+ OR E74neo24/+ sibling controls | 132 | 74 | 8 | 0 | 2.7 ± 0.05 (214) | |
| iswi2/+; E74neo24/+ | 39 | 69 | 15 | 1 | 2.2 ± 0.06 (124) | 3.21E-09 |
| iswi1/+ OR brnpr-3/+ sibling controls | 137 | 74 | 5 | 0 | 2.7 ± 0.05 (216) | |
| brnpr-3/+; iswi1/+ | 168 | 57 | 5 | 0 | 2.8 ± 0.04 (230) | 0.140 |
| iswi2/+ OR brnpr-3/+ sibling controls | 121 | 49 | 2 | 0 | 2.8 ± 0.05 (172) | |
| brnpr-3/+; iswi2/+ | 74 | 32 | 2 | 0 | 2.8 ± 0.06 (108) | 0.915 |
| iswi2/+ OR InR339/+ sibling controls | 135 | 61 | 11 | 0 | 2.7 ± 0.05 (207) | |
| iswi2/+; InR339/+ | 102 | 93 | 7 | 0 | 2.5 ± 0.05 (202) | 0.004f |
| nurf3013/+ OR InR339/+ sibling controls | 100 | 85 | 8 | 1 | 2.5 ± 0.05 (194) | |
| nurf3013 +/+ InR339 | 103 | 71 | 15 | 2 | 2.5 ± 0.05 (191) | 0.587 |
| EcRV559fs/+ OR dppe87/+ sibling controls | 122 | 47 | 11 | 0 | 2.8 ± 0.06 (180) | |
| + EcRV559fs/dppe87 + | 99 | 70 | 14 | 2 | 2.5 ± 0.06 (185) | 6.76E-04 |
| EcRV559fs/+ OR dpphr56/+ sibling controls | 78 | 65 | 28 | 2 | 2.3 ± 0.06 (173) | |
| + EcRV559fs/dpphr56 + | 27 | 82 | 49 | 10 | 1.8 ± 0.06 (168) | 6.84E-10 |
| EcRV559fs/+ OR put135/+ sibling controls | 140 | 39 | 4 | 0 | 3.0 ± 0.05 (183) | |
| EcRV559fs/+; put135/+ | 102 | 74 | 9 | 2 | 2.6 ± 0.06 (187) | 8.71E-06 |
Females kept at 22–25°C were analyzed at seven days of age.
Average number of germline stem cells (GSCs) per germarium ± s.e.m.
p values relative to sibling controls. p < 0.001 was considered statistically significant (shown in bold face).
The number of germaria analyzed is shown in parentheses.
All heterozygous sibling controls carry balancer chromosomes containing wild-type alleles (+).
We detect a very weak interaction between InR339 and iswi2, suggesting a possible NURF-independent role of ISWI in the niche, where InR is required for GSC maintenance (Hsu and Drummond-Barbosa, 2009).
Ecdysone signaling positively regulates BMP signaling in GSCs
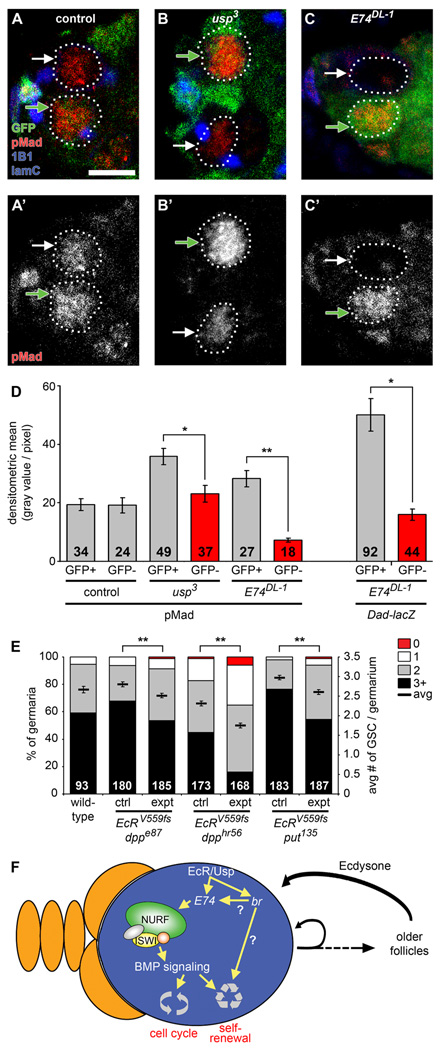
Loss of iswi function in GSCs reportedly leads to aberrant BMP signaling (Xi and Xie, 2005). We thus asked whether BMP signaling was altered in ecdysone pathway mutants by monitoring either phosphorylated Mad (pMad) or Dad-lacZ levels, both of which are well established reporters of BMP signaling in GSCs (Kai and Spradling, 2003). pMad and Dad-lacZ levels are experimentally variable in wildtype GSCs (Figure 6A–D). Nevertheless, we found that BMP signaling is consistently decreased in usp3 and E74DL-1 GSCs relative to neighboring control GFP-positive GSCs within individual mosaic genotypes (Figure 6A–D). These results indicate that ecdysone potentiates BMP signaling in GSCs, and are consistent both with the genetic interactions between the ecdysone pathway and NURF complex components and with the decreased levels of ISWI in E74DL-1 and usp3 GSCs (see Figures 4 and 5).
Figure 6. The ecdysone pathway affects BMP signaling levels in GSCs and genetically interacts with dpp and put to control GSC maintenance.
(A–C) Control (A), usp3 (B), and E74DL-1 (C) mosaic germaria analyzed six days AHS and labeled with GFP (green), pMad (red), 1B1 (blue, fusomes) and laminC (blue, nuclear envelope of cap cells). Control and mutant GSCs (outlined) are recognized by the absence of GFP expression (GFP-negative GSCs indicated by white arrows), while cells that did not undergo mitotic recombination are GFP-positive (GFP-positive GSCs indicated by green arrows). Scale bar represents 5 µm. (D) Quantification of nuclear pMad protein levels in control, usp3 and E74DL-1 GSCs (left) and nuclear Dad-lacZ levels in E74DL-1 GSCs (right). The number of GSCs analyzed is shown inside bars. (E) Frequencies of germaria containing zero, one, two, or three or more GSCs (left Y-axis), and average number of GSCs per germarium (right Y-axis) in seven-day old wild-type females versus single heterozygotes (ctrl) or double heterozygotes (expt) for mutations in EcR, dpp, and put. Wild-type data are the same shown in Figure 5. Additional results are shown in Table 1. The number of germaria analyzed is shown inside bars. Error bars, mean ± s.e.m. *p < 0.005, **p < 0.001. (F) Proposed model for how a GSC (blue) within its niche (orange) is controlled by an interaction between the steroid hormone ecdysone and the intrinsic epigenetic machinery (see text for details).
To ascertain whether decreased levels of BMP signaling observed in E74DL-1 and usp3 GSCs reflect a functional interaction between the ecdysone and BMP pathways, we analyzed double heterozygous females carrying EcRV559fs and either dppe87, dpphr56, or put135. Indeed, we observed remarkably strong genetic interactions between EcRV559fs and each of the BMP signaling components tested (Figure 6E and Table 1). These results indicate that ecdysone positively regulates BMP signaling in GSCs.
DISCUSSION
Although steroid hormones are known to modulate cell proliferation in many adult tissues, it has remained unclear how they affect stem cells. Our studies demonstrate that the steroid hormone ecdysone directly modulates GSC maintenance and proliferation. We also provide evidence suggesting that ecdysone and insulin-like peptides control stem cell activity independently, and thus that stem cells can integrate multiple systemic signals to modulate their behavior. Strikingly, our genetic interaction analyses reveal a specific functional cooperation between ecdysone and the intrinsic chromatin remodeling machinery of GSCs, presumably promoting BMP signaling. These results suggest that steroid hormones may change the epigenetic state of stem cells as a broad mechanism to control their fate and activity, and their ability to respond to signals from the niche.
Ecdyone Regulation of GSCs May Be Part of a Positive Feedback Loop
Several lines of evidence suggest that in the adult female, older follicles are the major site of ecdysteroid production. Ecdysone secretion can be detected in cultured Drosophila ovaries (Schwartz, 1985), and multiple genes encoding steroidogenic enzymes are expressed in more differentiated follicles (Huang et al., 2008). Not surprisingly, levels of circulating ecdysone are reduced in females maintained under nutrient-poor conditions (Schwartz, 1985) and in InR mutants (Tu et al., 2002), both of which have reduced numbers of vitellogenic stages (Drummond-Barbosa and Spradling, 2001; LaFever and Drummond-Barbosa, 2005).
These published data, in combination with our results, suggest that ecdysone may participate in a positive feedback loop. Under favorable physiological conditions, increased production of steroids from older follicles would reinforce self-renewal and proliferation of GSCs (Figure 6F). A similar positive feedback loop may be inferred from the effects of estrogen on folliculogenesis in the mammalian ovary. Estrogen, a potent mitogen for follicular granulosa cells, is produced by antral follicles mid-way through folliculogenesis, but is also necessary for production of proper numbers of both primordial and primary follicles early in follicle development (Britt et al., 2004).
Ecdysone and Insulin Signaling in the Control of GSCs
Taken together, our data describing the effects of both ecdysone (this study) and insulin (Hsu and Drummond-Barbosa, 2009; LaFever and Drummond-Barbosa, 2005) on GSCs demonstrate that these endocrine signals work independently to control GSC proliferation and self-renewal. Parallel regulation by ecdysone and insulin contrasts with studies reporting antagonistic or cooperative relationships between steroid hormones and insulin in both Drosophila and mammalian tissues. In the larval prothoracic gland, insulin signaling stimulates ecdysone production, which signals cells of the larval fat body to retain dFOXO in the nucleus, suppressing insulin-stimulated growth (Colombani et al., 2005). Studies in mammalian neural tissue have demonstrated extensive synergistic cross-talk between estrogen and insulin-like growth factor (Mendez et al., 2006). Nevertheless, although GSCs receive ecdysone and insulin signals independently, production of ecdysone in older follicles appears to be insulin-dependent. Indeed, both InR mutant females and nutrient-deprived females have decreased ecdysone levels (Schwartz, 1985; Tu et al., 2002), and insulin can induce ecdysteroidogenesis in mosquito ovaries (Riehle and Brown, 1999).
Ecdysone Appears to Mediate Specific Cellular Responses During Oogenesis Via a Variety of Targets and Co-Regulators
Intriguingly, our results demonstrate that GSCs have distinct requirements for ecdysone target genes. While E74 is critical for GSC activity, two other early response transcription factors, E75 and br, impact only GSC maintenance, and to a much smaller degree than E74. In contrast, germline inactivation of E74 or E75 reportedly results in similar follicle degeneration during later stages of oogenesis (Buszczak et al., 1999), suggesting that ecdysone regulates different oogenesis processes via distinct mechanisms. Given the divergent properties of these three transcription factors, it is easy to conceive that they might mediate different cell type-dependent outcomes downstream of ecdysone signaling. E75 is itself a nuclear hormone receptor, which is activated not only downstream of ecdysteroids, but also by carbon monoxide and nitric oxide binding to the ligand-binding domain (Marvin et al., 2009). E74 and E75 null mutants die at different developmental stages (Bialecki et al., 2002; Fletcher et al., 1995), suggesting that they may have distinct roles during larval and pupal development. Further, while genetic interactions between E74 and br are well-documented (Fletcher and Thummel, 1995), little evidence suggests that E74 and E75 regulate similar molecular pathways.
Steroid hormone receptor activity is regulated not only by ligand binding, but also by the interaction of the receptor with co-activators and co-repressors that refine the transcriptional response in both the presence and absence of ligand (Privalsky, 2004; Xu, 2005). tai, a co-activator of the EcR/Usp complex required for border cell migration (Bai et al., 2000; Jang et al., 2009), is not required for GSC maintenance or activity, indicating specific requirements for ecdysone signaling in GSCs. It is tempting to speculate that specificity for the ecdysone response in GSCs is at least in part afforded by its functional interaction with NURF, potentially via its proposed role as a co-activator of the EcR/Usp complex (Badenhorst et al., 2005). Several co-activators and co-repressors, however, have been identified in Drosophila (see below), and further studies will be necessary to determine whether or not additional co-regulators play roles in GSC control by ecdysone.
Steroid Hormones and the Chromatin Modifying Machinery
To our knowledge, this study is the first report of a functional interaction between a systemic steroid hormone and intrinsic chromatin remodeling factors in the regulation of adult stem cell biology. Nevertheless, a wide variety of nuclear hormone receptors have potential roles in other stem cell systems (Jeong and Mangelsdorf, 2009), and this may represent a more general mechanism for stem cell regulation. Indeed, many steroid hormone co-activators and co-repressors either possess chromatin-modifying enzymatic functions, or can interact with other proteins that have these functions (Wolf et al., 2008). For example, the EcR/Usp receptor has been reported to interact with several co-regulators, including the co-activator Nurf301 (Badenhorst et al., 2005) and SMRTER, a co-repressor that associates with Sin3A, known to facilitate histone deacetylation (Tsai et al., 1999). Retinoic acid receptors recruit histone acetyltransferases and BAF proteins, subunits of the ATP-dependent SWI/SNF family (Flajollet et al., 2007), which are thought to act in sequence to increase the accessibility of retinoic acid-responsive promoters to general transcription factors (Dilworth et al., 2000). A similar mechanism has been proposed for the estrogen receptor, which is coupled to a co-activator complex with histone acetyltransferase activity and Brahma-related gene 1, another member of the SWI/SNF family (DiRenzo et al., 2000). Therefore, regulatory circuits involving effects of systemic steroids on the intrinsic epigenetic machinery of stem cells may represent a broadly conserved mechanism in the multilayered regulation of stem cells.
Steroids, Insulin, and Cancer
One model of tumor development suggests that tumors are fueled by cancer stem cells derived from adult stem cells that acquired genetic mutations or epigenetic changes conferring altered sensitivity to their microenvironment (Clarke and Fuller, 2006). Similar to normal adult stem cells, cancer stem cells are self-renewing and can give rise to a range of differentiated and proliferative cell types. Many cancer stem cells are likely to be steroid-responsive. For example, the majority of primary breast cancers express both estrogen and progesterone receptors, suggesting that mammary cancer stem cells may arise from a steroid-responsive cell and/or themselves be responsive to steroid signaling (Britt et al., 2007). There is a strong epidemiological correlation between breast cancer risk and prolonged ovarian steroid exposure (Britt et al., 2007; Eliassen and Hankinson, 2008), and obese individuals have increased cancer risk, likely due to alterations in sex hormone levels and to chronic hyperinsulinemia (Fair and Montgomery, 2009). The extensive molecular cross-talk between steroid and insulin/insulin-like growth factors (Lanzino et al., 2008), and perturbations in the epigenetic landscape associated with altered steroid hormone signaling found commonly in cancers (Thorne et al., 2009), suggest striking parallels between how cancer stem cells and Drosophila GSCs respond to their systemic environments. We further speculate that misregulation of steroid signaling may be part of the mechanism by which normal stem cells become cancer stem cells, thereby augmenting the response of these cells to physiological stimuli.
EXPERIMENTAL PROCEDURES
Drosophila Strains and Culture Conditions
Drosophila stocks were maintained at 22–25°C on standard medium containing cornmeal, molasses, yeast, and agar. A nutrient-rich diet, consisting of the standard medium supplemented with wet yeast paste, was used for all experiments described. For GSC analyses in flies defective for ecdysone signaling, we used the following mutant combinations: ecd1 (Garen et al., 1977) in trans to Df(3L)R-G7, which uncovers the ecd locus; and the temperature-sensitive EcRA483T in trans to either null EcRV559fs or null EcRM554fs (Carney and Bender, 2000). These temperature-sensitive genotypes, referred to as ecdts and EcRts, respectively, were analyzed in parallel to y w wild-type or heterozygous controls after incubation at the restrictive temperature of 29°C as described below.
For genetic mosaic analyses, we obtained the following alleles previously recombined onto FRT chromosomes: usp3 FRT19A (Lee et al., 2000); brnpr-3 FRT19A (Schubiger et al., 2005); tai61G1 FRT40A (Bai et al., 2000); and FRT42B iswi2 (Deuring et al., 2000). We also recombined the following alleles individually onto the FRT19A or FRT80B chromosome using standard crosses: usp2 (Ghbeish et al., 2001), E74DL-1 (Fletcher et al., 1995); E75Δ51 (Bialecki et al., 2002); and nurf3013 (Badenhorst et al., 2002). The above FRT alleles were analyzed in trans to the corresponding wild-type FRT chromosomes containing the ubiquitously expressed Ubi-GFP or arm-lacZ markers (www.flybase.org). For generation of E74 genetic mosaics in the dFOXO21/dFOXO25 (Hsu et al., 2008) background, E74DL-1FRT80B recombined with dFOXO21 was analyzed in trans to the Ubi-GFP FRT80B dFOXO25 recombinant chromosome. To monitor BMP signaling in E74 mutant GSCs, E74DL-1 FRT80B was also recombined with Dad-lacZ (Kai and Spradling, 2003) and analyzed in trans to Ubi-GFP FRT80B.
For genetic interaction analyses, the following alleles were used: E74DL-1 (Fletcher et al., 1995), E74neo24 (Fletcher et al., 1995), E75Δ51 (Bialecki et al., 2002); iswi1 (Deuring et al., 2000); iswi2 (Deuring et al., 2000); nurf3013 (Badenhorst et al., 2002); put135 (Xie and Spradling, 1998); dpphr56 (Xie and Spradling, 1998); dppE87 (Irish and Gelbart, 1987); and InR339 (LaFever and Drummond-Barbosa, 2005). Controls (single heterozygotes carrying a balancer) were compared to double heterozygotes. hs-FLP strains, balancer chromosomes and other genetic tools are described in FlyBase (www.flybase.org).
Generation of Genetically Mosaic Germaria and GSC Analyses
To measure GSC loss in ecdts and EcRts mutants, two- or three-day old experimental and control females raised at the permissive temperature (18–22°C) were incubated at the restrictive temperature (29°C) for three, five, or seven days. GSCs were identified based on their juxtaposition to cap cells (recognized by laminC labeling of their nuclear envelopes) and on the morphology and location of their anteriorly anchored fusomes (visualized by 1B1 labeling), as described (Hsu and Drummond-Barbosa, 2009; Hsu et al., 2008). Results were subjected to Student’s t-test. Analysis of fusome morphology as an indicator of GSC cell cycle stage has been previously described (Hsu et al., 2008), and data are displayed as the average of six independent experiments subjected to Student’s t-test.
Genetic mosaics were generated by FLP/FRT-mediated mitotic recombination (Xu and Rubin, 1993) as described (LaFever and Drummond-Barbosa, 2005). Briefly, two- or three-day old females carrying a mutant allele on an FRT chromosome arm in trans to a wild-type allele linked to a Ubi-GFP or arm-lacZ marker on the homologous FRT arm (Figure 1D) were heat-shocked for one hour at 37°C twice daily for three consecutive days. Ovaries were dissected six or 10 days after the last heat shock (AHS) to ensure that non-stem cell clones had exited the germaria (Margolis and Spradling, 1995) and were excluded from our analyses. Unmarked wild-type FRT chromosomes were used for generation of control clones. FLP/FRT-mediated recombination could not be used to create EcR null clones, as the EcR locus lies between the centromere and the FRT42 insertion. GSC relative division rate and maintenance were quantified as previously described (Hsu and Drummond-Barbosa, 2009; LaFever and Drummond-Barbosa, 2005; Xie and Spradling, 1998).
Tissue Preparation, Immunofluorescence, and Microscopy
Ovaries were dissected and ovarioles teased apart in Grace’s medium (BioWhittaker), fixed for 13 minutes at room temperature in 5.3% formaldehyde (Ted Pella) in Grace’s medium (one part 16% formaldehyde to two parts Grace’s), and washed extensively in phosphate buffered saline (PBS, pH 7.0) with 0.1% Triton X-100 (Sigma). Ovaries were blocked in 5% bovine serum albumin (BSA; Sigma), 5% normal goat serum (NGS; Jackson ImmunoResearch), and 0.1% Tween-20 (Sigma) in PBS for three hours at room temperature or overnight at 4°C. The following primary antibodies were incubated in blocking solution overnight at 4°C: mouse anti-Hts (1B1) (DSHB; 1:10), mouse anti-laminC (LC28.26) (DSHB; 1:100 or 1:400), rabbit anti-GFP (Torrey Pines, 1:2500), mouse anti-β-galactosidase (Promega, 1:500), rat anti-BrdU (Accurate Biochemicals, 1:500), rabbit anti-phosphohistone H3 (Upstate Biotechnology/Millipore, 1:250), rabbit anti-pMad (Smad 3, #1880) (Epitomics, 1:100), rabbit anti-cleaved Caspase 3 (#9661S) (Cell Signaling Technology, 1:50), and rabbit anti-ISWI (a gift of John Tamkun, 1:400). AlexaFluor 448-, 568-, or 633-conjugated goat species-specific secondary antibodies (Molecular Probes/Invitrogen, 1:200) were incubated for two hours at room temperature in blocking solution, and counterstained with 0.5 µg/mL DAPI (Sigma, 1:1000 in PBS) to visualize nuclei. Ovaries were mounted in either Vectashield (Vector Labs) or 90% glycerol containing 20.0 µg/mL N-propyl gallate (Sigma). Data were collected using a Zeiss Axioplan-2 or AxioImager-A2 fluorescence microscope, or a Zeiss LSM510, LSM510Meta, or LSM700 confocal microscope. Fluorescence intensity in confocal sections was measured using ImageJ or AxioVision (Zeiss) by manually demarcating individual GSC nuclei and measuring densitometric mean (gray value/pixel) at the largest nuclear diameter. Due to slight variations in pixel intensity among stain sets, an average value was obtained from two to five independent experiments. Statistical analysis was performed using the Student’s t-test.
Apoptosis Assay
To measure the incidence of apoptosis, the ApopTag (Chemicon/Millipore) fluorescein direct in situ detection kit was used as described (Drummond-Barbosa and Spradling, 2001). Briefly, ovaries were dissected, fixed, and washed as described above. Ovaries were then washed twice for five minutes in equilibration buffer, and incubated for one hour at 37°C in terminal deoxynucleotidyl transferase (TdT) solution with occasional mixing. The TdT reaction was quenched by washing for five minutes in stop/wash solution, and ovaries were washed and immunolabeled as described above. Results were subjected to Chi-square analysis.
Supplementary Material
ACKNOWLEDGMENTS
We wish to thank: M. Bender, E. Hafen, T. Lee, E. Matunis and C. Cherry, D. Montell, J. Truman, J. Tamkun, J. Maines, W.-M. Deng, T. Nystul, A. Spradling, the Bloomington Stock Center, and the Developmental Studies Hybridoma Bank for reagents; the Vanderbilt and Johns Hopkins Microscopy Facilities; and V. Culotta, B. Zirkin, H.-J. Hsu, and L. LaFever for critical reading of this manuscript. E.T.A. and D.D.-B. designed experiments, interpreted results, and wrote the manuscript. E.T.A. performed all experiments. This work was supported by National Institutes of Health R01 GM069875 and American Cancer Society RSG-DDC-112316. E.T.A. was supported by training grant T32 DK007563 and National Research Service Award F32 GM086031 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing conflicts of interest.
REFERENCES
- Asselin-Labat ML, Vaillant F, Sheridan JM, Pal B, Wu D, Simpson ER, Yasuda H, Smyth GK, Martin TJ, Lindeman GJ, et al. Control of mammary stem cell function by steroid hormone signalling. Nature. 2010;465:798–802. doi: 10.1038/nature09027. [DOI] [PubMed] [Google Scholar]
- Badenhorst P, Voas M, Rebay I, Wu C. Biological functions of the ISWI chromatin remodeling complex NURF. Genes & Dev. 2002;16:3186–3198. doi: 10.1101/gad.1032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenhorst P, Xiao H, Cherbas L, Kwon SY, Voas M, Rebay I, Cherbas P, Wu C. The Drosophila nucleosome remodeling factor NURF is required for Ecdysteroid signaling and metamorphosis. Genes & Dev. 2005;19:2540–2545. doi: 10.1101/gad.1342605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Uehara Y, Montell DJ. Regulation of invasive cell behavior by taiman, a Drosophila protein related to AIB1, a steroid receptor coactivator amplified in breast cancer. Cell. 2000;103:1047–1058. doi: 10.1016/s0092-8674(00)00208-7. [DOI] [PubMed] [Google Scholar]
- Bialecki M, Shilton A, Fichtenberg C, Segraves WA, Thummel CS. Loss of the ecdysteroid-inducible E75A orphan nuclear receptor uncouples molting from metamorphosis in Drosophila. Dev Cell. 2002;3:209–220. doi: 10.1016/s1534-5807(02)00204-6. [DOI] [PubMed] [Google Scholar]
- Britt K, Ashworth A, Smalley M. Pregnancy and the risk of breast cancer. Endocr-rel Cancer. 2007;14:907–933. doi: 10.1677/ERC-07-0137. [DOI] [PubMed] [Google Scholar]
- Britt KL, Saunders PK, McPherson SJ, Misso ML, Simpson ER, Findlay JK. Estrogen actions on follicle formation and early follicle development. Biol Reprod. 2004;71:1712–1723. doi: 10.1095/biolreprod.104.028175. [DOI] [PubMed] [Google Scholar]
- Buszczak M, Freeman MR, Carlson JR, Bender M, Cooley L, Segraves WA. Ecdysone response genes govern egg chamber development during mid-oogenesis in Drosophila. Dev. 1999;126:4581–4589. doi: 10.1242/dev.126.20.4581. [DOI] [PubMed] [Google Scholar]
- Carney GE, Bender M. The Drosophila ecdysone receptor (EcR) gene is required maternally for normal oogenesis. Genetics. 2000;154:1203–1211. doi: 10.1093/genetics/154.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke MF, Fuller M. Stem cells and cancer: two faces of eve. Cell. 2006;124:1111–1115. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Colombani J, Bianchini L, Layalle S, Pondeville E, Dauphin-Villemant C, Antoniewski C, Carre C, Noselli S, Leopold P. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science. 2005;310:667–670. doi: 10.1126/science.1119432. [DOI] [PubMed] [Google Scholar]
- D'Errico I, Moschetta A. Nuclear receptors, intestinal architecture and colon cancer: an intriguing link. Cell Mol Life Sci. 2008;65:1523–1543. doi: 10.1007/s00018-008-7552-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cuevas M, Spradling AC. Morphogenesis of the Drosophila fusome and its implications for oocyte specification. Dev. 1998;125:2781–2789. doi: 10.1242/dev.125.15.2781. [DOI] [PubMed] [Google Scholar]
- Deng WM, Bownes M. Two signalling pathways specify localised expression of the Broad-Complex in Drosophila eggshell patterning and morphogenesis. Dev. 1997;124:4639–4647. doi: 10.1242/dev.124.22.4639. [DOI] [PubMed] [Google Scholar]
- Deuring R, Fanti L, Armstrong JA, Sarte M, Papoulas O, Prestel M, Daubresse G, Verardo M, Moseley SL, Berloco M, et al. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell. 2000;5:355–365. doi: 10.1016/s1097-2765(00)80430-x. [DOI] [PubMed] [Google Scholar]
- Dilworth FJ, Fromental-Ramain C, Yamamoto K, Chambon P. ATP-driven chromatin remodeling activity and histone acetyltransferases act sequentially during transactivation by RAR/RXR In vitro. Mol Cell. 2000;6:1049–1058. doi: 10.1016/s1097-2765(00)00103-9. [DOI] [PubMed] [Google Scholar]
- DiRenzo J, Shang Y, Phelan M, Sif S, Myers M, Kingston R, Brown M. BRG-1 is recruited to estrogen-responsive promoters and cooperates with factors involved in histone acetylation. Mol Cell Biol. 2000;20:7541–7549. doi: 10.1128/mcb.20.20.7541-7549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond-Barbosa D. Stem cells, their niches and the systemic environment: an aging network. Genetics. 2008;180:1787–1797. doi: 10.1534/genetics.108.098244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond-Barbosa D, Spradling AC. Stem cells and their progeny respond to nutritional changes during Drosophila oogenesis. Dev Biol. 2001;231:265–278. doi: 10.1006/dbio.2000.0135. [DOI] [PubMed] [Google Scholar]
- Eliassen AH, Hankinson SE. Endogenous hormone levels and risk of breast, endometrial and ovarian cancers: prospective studies. Adv Exp Med Biol. 2008;630:148–165. [PubMed] [Google Scholar]
- Fair AM, Montgomery K. Energy balance, physical activity, and cancer risk. Meth Mol Biol. 2009;472:57–88. doi: 10.1007/978-1-60327-492-0_3. [DOI] [PubMed] [Google Scholar]
- Flajollet S, Lefebvre B, Cudejko C, Staels B, Lefebvre P. The core component of the mammalian SWI/SNF complex SMARCD3/BAF60c is a coactivator for the nuclear retinoic acid receptor. Mol Cell Endocr. 2007;270:23–32. doi: 10.1016/j.mce.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Burtis KC, Hogness DS, Thummel CS. The Drosophila E74 gene is required for metamorphosis and plays a role in the polytene chromosome puffing response to ecdysone. Dev. 1995;121:1455–1465. doi: 10.1242/dev.121.5.1455. [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Thummel CS. The ecdysone-inducible Broad-complex and E74 early genes interact to regulate target gene transcription and Drosophila metamorphosis. Genetics. 1995;141:1025–1035. doi: 10.1093/genetics/141.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garen A, Kauvar L, Lepesant JA. Roles of ecdysone in Drosophila development. Proc Nat Acad Sci, U.S.A. 1977;74:5099–5103. doi: 10.1073/pnas.74.11.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghbeish N, Tsai CC, Schubiger M, Zhou JY, Evans RM, McKeown M. The dual role of ultraspiracle, the Drosophila retinoid X receptor, in the ecdysone response. Proc Nat Acad Sci, U.S.A. 2001;98:3867–3872. doi: 10.1073/pnas.061437798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HJ, Drummond-Barbosa D. Insulin levels control female germline stem cell maintenance via the niche in Drosophila. Proc Nat Acad Sci, U.S.A. 2009;106:1117–1121. doi: 10.1073/pnas.0809144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HJ, LaFever L, Drummond-Barbosa D. Diet controls normal and tumorous germline stem cells via insulin-dependent and -independent mechanisms in Drosophila. Dev Biol. 2008;313:700–712. doi: 10.1016/j.ydbio.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Warren JT, Gilbert LI. New players in the regulation of ecdysone biosynthesis. J Genetics Genomics. 2008;35:1–10. doi: 10.1016/S1673-8527(08)60001-6. [DOI] [PubMed] [Google Scholar]
- Irish VF, Gelbart WM. The decapentaplegic gene is required for dorsal-ventral patterning of the Drosophila embryo. Genes & Dev. 1987;1:868–879. doi: 10.1101/gad.1.8.868. [DOI] [PubMed] [Google Scholar]
- Jang AC, Chang YC, Bai J, Montell D. Border-cell migration requires integration of spatial and temporal signals by the BTB protein Abrupt. Nat Cell Biol. 2009;11:569–579. doi: 10.1038/ncb1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y, Mangelsdorf DJ. Nuclear receptor regulation of stemness and stem cell differentiation. Exp Mol Med. 2009;41:525–537. doi: 10.3858/emm.2009.41.8.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai T, Spradling A. An empty Drosophila stem cell niche reactivates the proliferation of ectopic cells. Proc Nat Acad Sci, U.S.A. 2003;100:4633–4638. doi: 10.1073/pnas.0830856100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirilly D, Xie T. The Drosophila ovary: an active stem cell community. Cell Res. 2007;17:15–25. doi: 10.1038/sj.cr.7310123. [DOI] [PubMed] [Google Scholar]
- LaFever L, Drummond-Barbosa D. Direct control of germline stem cell division and cyst growth by neural insulin in Drosophila. Science. 2005;309:1071–1073. doi: 10.1126/science.1111410. [DOI] [PubMed] [Google Scholar]
- Lanzino M, Morelli C, Garofalo C, Panno ML, Mauro L, Ando S, Sisci D. Interaction between estrogen receptor alpha and insulin/IGF signaling in breast cancer. Curr Cancer Drug Targets. 2008;8:597–610. doi: 10.2174/156800908786241104. [DOI] [PubMed] [Google Scholar]
- Lee T, Marticke S, Sung C, Robinow S, Luo L. Cell-autonomous requirement of the USP/EcR-B ecdysone receptor for mushroom body neuronal remodeling in Drosophila. Neuron. 2000;28:807–818. doi: 10.1016/s0896-6273(00)00155-0. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis J, Spradling A. Identification and behavior of epithelial stem cells in the Drosophila ovary. Dev. 1995;121:3797–3807. doi: 10.1242/dev.121.11.3797. [DOI] [PubMed] [Google Scholar]
- Marvin KA, Reinking JL, Lee AJ, Pardee K, Krause HM, Burstyn JN. Nuclear receptors homo sapiens Rev-erbbeta and Drosophila melanogaster E75 are thiolate-ligated heme proteins which undergo redox-mediated ligand switching and bind CO and NO. Biochem. 2009;48:7056–7071. doi: 10.1021/bi900697c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez P, Wandosell F, Garcia-Segura LM. Cross-talk between estrogen receptors and insulin-like growth factor-I receptor in the brain: cellular and molecular mechanisms. Front Neuroendocr. 2006;27:391–403. doi: 10.1016/j.yfrne.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Ninov N, Manjon C, Martin-Blanco E. Dynamic control of cell cycle and growth coupling by ecdysone, EGFR, and PI3K signaling in Drosophila histoblasts. PLoS Biol. 2009;7:e1000079. doi: 10.1371/journal.pbio.1000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawluski JL, Brummelte S, Barha CK, Crozier TM, Galea LA. Effects of steroid hormones on neurogenesis in the hippocampus of the adult female rodent during the estrous cycle, pregnancy, lactation and aging. Front Neuroendocr. 2009;30:343–357. doi: 10.1016/j.yfrne.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Privalsky ML. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Ann Rev Phys. 2004;66:315–360. doi: 10.1146/annurev.physiol.66.032802.155556. [DOI] [PubMed] [Google Scholar]
- Ray R, Novotny NM, Crisostomo PR, Lahm T, Abarbanell A, Meldrum DR. Sex steroids and stem cell function. Mol Med. 2008;14:493–501. doi: 10.2119/2008-00004.Ray. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddiford LM, Cherbas P, Truman JW. Ecdysone receptors and their biological actions. Vita Horm. 2000;60:1–73. doi: 10.1016/s0083-6729(00)60016-x. [DOI] [PubMed] [Google Scholar]
- Riehle MA, Brown MR. Insulin stimulates ecdysteroid production through a conserved signaling cascade in the mosquito Aedes aegypti. Insect Biochem Mol Biol. 1999;29:855–860. doi: 10.1016/s0965-1748(99)00084-3. [DOI] [PubMed] [Google Scholar]
- Schubiger M, Carre C, Antoniewski C, Truman JW. Ligand-dependent de-repression via EcR/USP acts as a gate to coordinate the differentiation of sensory neurons in the Drosophila wing. Dev. 2005;132:5239–5248. doi: 10.1242/dev.02093. [DOI] [PubMed] [Google Scholar]
- Schwartz M, Kelly TJ, Imberski RB, Rubenstein EC. The effects of nutrition and methoprene treatment on ovarian ecdysteroid synthesis in Drosophila Melanogaster. J Insect Phys. 1985;31:947–957. [Google Scholar]
- Spradling A. Developmental Genetics of Oogenesis. In: Bate M, editor. The Development of Drosophila melanogaster. Plainview, N.Y.: Cold Spring Harbor Laboratory Press; 1993. pp. 1–70. [Google Scholar]
- Thorne JL, Campbell MJ, Turner BM. Transcription factors, chromatin and cancer. Int J Biochem Cell Biol. 2009;41:164–175. doi: 10.1016/j.biocel.2008.08.029. [DOI] [PubMed] [Google Scholar]
- Tsai CC, Kao HY, Yao TP, McKeown M, Evans RM. SMRTER, a Drosophila nuclear receptor coregulator, reveals that EcR-mediated repression is critical for development. Mol Cell. 1999;4:175–186. doi: 10.1016/s1097-2765(00)80365-2. [DOI] [PubMed] [Google Scholar]
- Tu MP, Yin CM, Tatar M. Impaired ovarian ecdysone synthesis of Drosophila melanogaster insulin receptor mutants. Aging Cell. 2002;1:158–160. doi: 10.1046/j.1474-9728.2002.00016.x. [DOI] [PubMed] [Google Scholar]
- Wolf IM, Heitzer MD, Grubisha M, DeFranco DB. Coactivators and nuclear receptor transactivation. J Cell Biochem. 2008;104:1580–1586. doi: 10.1002/jcb.21755. [DOI] [PubMed] [Google Scholar]
- Xi R, Xie T. Stem cell self-renewal controlled by chromatin remodeling factors. Science. 2005;310:1487–1489. doi: 10.1126/science.1120140. [DOI] [PubMed] [Google Scholar]
- Xie T, Spradling AC. decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell. 1998;94:251–260. doi: 10.1016/s0092-8674(00)81424-5. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Dev. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Xu W. Nuclear receptor coactivators: the key to unlock chromatin. Biochem Cell Biol. 2005;83:418–428. doi: 10.1139/o05-057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.