Abstract
Background
Non-target-site based resistance to herbicides is a major threat to the chemical control of agronomically noxious weeds. This adaptive trait is endowed by differences in the expression of a number of genes in plants that are resistant or sensitive to herbicides. Quantification of the expression of such genes requires normalising qPCR data using reference genes with stable expression in the system studied as internal standards. The aim of this study was to validate reference genes in Alopecurus myosuroides, a grass (Poaceae) weed of economic and agronomic importance with no genomic resources.
Results
The stability of 11 candidate reference genes was assessed in plants resistant or sensitive to herbicides subjected or not to herbicide stress using the complementary statistical methods implemented by NormFinder, BestKeeper and geNorm. Ubiquitin, beta-tubulin and glyceraldehyde-3-phosphate dehydrogenase were identified as the best reference genes. The reference gene set accuracy was confirmed by analysing the expression of the gene encoding acetyl-coenzyme A carboxylase, a major herbicide target enzyme, and of an herbicide-induced gene encoding a glutathione-S-transferase.
Conclusions
This is the first study describing a set of reference genes (ubiquitin, beta-tubulin and glyceraldehyde-3-phosphate dehydrogenase) with a stable expression under herbicide stress in grasses. These genes are also candidate reference genes of choice for studies seeking to identify stress-responsive genes in grasses.
Background
Differences in gene expression are at the root of plant adaptive response to the environment. Analysing gene expression patterns requires tools enabling sensitive, precise, and reproducible quantification of specific mRNAs. Quantitative real-time polymerase chain reaction (qPCR) is currently the technique of choice for this purpose [1]. However, technical and sample variations usually render absolute quantification of gene expression unreliable. Thus, a normalisation strategy using one, or preferably several, reference gene(s) is generally implemented in qPCR studies [1-4]. A reference gene must be constitutively and constantly expressed in all experimental conditions and samples studied [5,6]. Compared to animals, relatively few studies so far had described validated sets of reference genes in plants [7], and most of them considered species with sequenced genomes (i.e., crop or model species). Exceptions are the grass species Lolium sp. (e.g. [8,9]) and Brachiaria brizantha [10].
Alopecurus myosuroides (black-grass, Poaceae) is a grass weed of major economic importance in winter crops in Europe that is removed from crops by herbicide applications. Resistance to herbicides has evolved in numerous A. myosuroides field populations [11]. Resistance is mostly endowed by a range of mechanisms decreasing the amount of herbicide molecules reaching their target (non target-site-based resistance [11-13]). This type of resistance is a major threat to crop protection, because it can confer unpredictable resistance to herbicides with different modes of action [12]. As it is considered to be endowed by differential regulation of many stress-responsive genes between resistant and sensitive plants [14,15], transcriptomics-based approaches should identify genes involved. Designing a reliable qPCR assay is a prerequisite to adequately validating the gene expression data. Here, we describe the validation of a set of three reference genes in A. myosuroides, a non-model species with no genomic resources. These genes can reliably be used for qPCR normalisation of gene expression data in plants resistant or sensitive to herbicides before and up to at least 6 hours after herbicide application.
Methods
Plant material
All plants used were grown from seeds in a climatic chamber (22°C/18°C day/night; 14-h photoperiod at 450 μmol m-2 s-1). Individual plants with a dozen tillers each were split into ten individual tillers, and every tiller was grown for 3 days in an individual pot before being used for spraying experiments [13]. All tillers from a given plant are clones, i.e., genetically identical plants at the same growth stage (3-4 leaves). To identify genes with stable expression among phenotypes (resistant versus sensitive to herbicides) and among experimental conditions (herbicide-treated versus untreated), a time-course experiment was subsequently conducted. Two clones per plant (= one sample) were collected at each of three different times (before, 2.5 hours and 6 hours after herbicide application) encompassing the most crucial period after herbicide application when expression of non-target-site-based resistance genes enables resistant plants to survive herbicide exposure [14,15]. The basal part of the clones (approximately 10 cm high and 100 mg fresh weight tissue) was cut just above the ground, immediately frozen in liquid nitrogen and stored at -80°C until RNA extraction. The basal section contains the meristematic tissues, where most of the activity of chloroplastic acetyl coenzyme A carboxylase (ACCase; EC 6.1.4.2), the target enzyme of the herbicide used, is located [14]. Two additional clones per plant were used to characterise the phenotype (i.e. resistant or sensitive). The last two clones were sprayed with water ("untreated control"). The herbicide fenoxaprop, a broadly used ACCase inhibitor, was applied using a custom-built, single-nozzle (nozzle 110-04; Albuz, France) sprayer delivering herbicide in 300 L ha-1 water at 400 kPa, at a speed of 6.6 km h-1 [13]. Clones from sensitive plants from a reference population [16] were used as a control for herbicide application efficacy. All clones were grown, sprayed and collected at the same time and in the same conditions. Plant survival was assessed one month after herbicide application. Dead and surviving plants were classified as sensitive and resistant, respectively.
RNA extraction, quantification and quality assessment
Total RNA from every sample was extracted and DNA contamination removed using the RNeasy plant mini kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. Nucleic acid concentration of each sample was measured twice at 260 nm using a NanoDropND-1,000 spectrophotometer (LABTECH, Luton, UK). Measurement was repeated if the two measures differed by more than 3%. Total RNA quality was assessed using the A260/A280 and A260/A230 absorption ratios. Only RNA samples with A260/A280 and A260/A230 ratios between 1.8 and 2.2 and between 2 and 2.2, respectively, were subsequently used. Total RNA integrity was checked by electrophoresis on 0.8% (w/v) agarose gels under denaturing conditions. Finally, RNA quality and genomic DNA (gDNA) contamination were assessed by capillary electrophoresis on an Experion labchip electrophoresis system (Bio-Rad, Hercules, USA). No gDNA contamination was detected. Each step from total RNA extraction to gDNA contamination assessment was performed at the same time for all samples.
Total RNA was extracted from 3 sensitive and 7 resistant plants at each of three time-course points (before, 2.5 and 6 hours after herbicide application), yielding thirty RNA samples. Following quality assessment, 19 of these samples were used for qPCR experiments (Table 1).
Table 1.
RNA samples used for assessment of the stability of the candidate reference genes
| Plant phenotype | Herbicide application | Total | ||
|---|---|---|---|---|
| BEFORE treatment | AFTER treatment | |||
| +2.5H | +6H | |||
| SENSITIVE | 1 | 2 | 2 | 5 |
| RESISTANT | 4 | 5 | 5 | 14 |
| Total | 5 | 7 | 7 | 19 |
Samples were assigned to five subsets according to plant phenotype or to the time after herbicide application. Subsets containing samples collected at the same time (BEFORE, + 2.5H or + 6H) enabled to assess gene stability between phenotypes (i.e. phenotype effect). Subsets containing samples from plants with a same phenotype (SENSITIVE or RESISTANT) enabled to assess the effect of the time after herbicide application on gene stability.
cDNA synthesis
cDNA synthesis was performed in duplicate for every RNA sample that passed the quality controls. Reverse-transcription (RT) reactions were performed simultaneously for all samples. cDNA was synthesized from 5 μg of total RNA using the two-step RT-PCR protocol of the Masterscript RT-PCR System (5 PRIME, Hamburg, Germany). Both the oligo(dT)18 primer (0.5 μg) and the random primers (50 ng) were used with 1 μL reverse-transcriptase in a 20 μL-reaction volume following the manufacturer's instructions. The two-step protocol reduces unwanted primer dimer formation [17], which must be avoided when using SYBR Green as the qPCR quantification dye. Reactions were immediately stored at -20°C until further use. To detect gDNA contamination, all cDNA samples and a sample of A. myosuroides gDNA were simultaneously used in PCRs to amplify a fragment of ACCase [genbank: AM408429] using intron-spanning primers ACVII8 (5'- AGGACACGCAGAGGAACCTCTTTCATTTAC) and ACVII8R (5'- CAACTCTCCAGCTACCACTGGCAGG). Amplicon sizes (340 bp for cDNA and 426 bp for gDNA) were compared on 2.5% (w/v) agarose gels. No gDNA contamination was detected.
Primer design
Eleven genes commonly reported as stable reference genes in grasses and/or in studies investigating plant response to stress were selected. They are involved in biosynthesis pathways [glyceraldehyde-3-phosphate dehydrogenase (GAPDH), sucrose phosphate synthase (SPS) and ribulose biphosphate carboxylase (RUBISCO)], cytoskeletal structure [beta-tubulin (TUB) and actin (ACT)], protein metabolism [elongation factor 1α (EF1), ubiquitin (UBQ) and cyclophilin (CYC)], and ribosomal structure [nucleus-encoded 18S rRNA (18S) and 25S rRNA (25S), and mitochondrial-DNA-encoded 26S rRNA (26S)].
Hardly any genomic data is available for A. myosuroides. Primers for reference genes (Table 2) were therefore designed based on conserved regions in the homologous genes in other grasses (rice, barley, wheat, maize, Lolium sp. and Brachypodium distachyon; Additional file 1: Table S1). Primers were designed using Primer3 [18], using a primer length = 23 ± 3 bp, melting temperature (Tm) = 60°C ± 3°C, a guanine-cytosine content around 50%, and an expected amplicon size of 150 to 250 bp. To detect gDNA contamination, the primers targeting TUB or GAPDH were designed to amplify an intron-containing amplicon. Primers were synthesized by MWG Biotech (Germany, Ebersberg).
Table 2.
Candidate reference genes tested and primer sequences
| Reference gene1 (accession number) | Primer sequences (5'-3')2 | Amplicon lenght (bp) | Tm (°C) | Manual threshold3 | PCR efficiency (%) | Regression coefficient (R2) | Average Cq value |
|---|---|---|---|---|---|---|---|
| CYC | F: AGCTTTGAAGTTGGCAGTAG | ||||||
| R: GATCGCGTATTCATGGACTTTAG | Discarded (aspecific amplification) | ||||||
| SPS | F: CATTGCAAGAACTATTTGTCACG | ||||||
| R: GCAGAGATCAAATGGTTCAAATC | |||||||
| ACT | F: TGTGCTTGACTCTGGTGATG | 220 | 58 | ||||
| R: TTCATAATCAAGGGCAACGTAAGC | Discarded (aspecific amplification) | ||||||
| RUBISCO | F: CATTATCAAGAAGGGCAAGATGTG | 169 | 60 | ||||
| R: TGTTGTACATCCCTGGAAGTTG | |||||||
| GAPDH | F: GTATTGTTGAGGGACTGATGACC | 182 | 57 | 0.047 | 92 | 0.999 | 23.31 |
| (JN599100) | R: AGTAAGCTTGCCATTGAACTCAG | ||||||
| TUB | F: TACTGTGGTTGAGCCATACAATG | 162 | 60 | 0.069 | 98 | 0.993 | 23.87 |
| (JN599101) | R: GCAGAGATCAAATGGTTCAAATC | ||||||
| EF1 | F: CAAGTACTACTGCACCGTCATTG | 199 | 57 | 0.025 | 89 | 0.984 | 25.10 |
| (JN599095) | R: GATCATCTGCTTCACTCCAAGAG | ||||||
| UBQ | F: GCAAGAAGAAGACCTACACCAAG | 225 | 60 | 0.054 | 100 | 0.991 | 19.00 |
| (JN599096) | R: CCTTCTGGTTGTAGACGTAGGTG | ||||||
| 18S | F: GTCCAGACATAGGAAGGATTGAC | 245 | 63 | 0.052 | 106 | 0.995 | 15.08 |
| (JN599097) | R: GAACATCTAAGGGCATCACAGAC | ||||||
| 25S | F: GCATGAATGGATTAACGAGATTC | 165 | 63 | 0.098 | 95 | 0.998 | 17.00 |
| (JN599099) | R: GGCTCCCACTTATCCTACAC | ||||||
| 26S | F: GATAGCGTACAAGTACCGTGAGG | 238 | 63 | 0.102 | 94 | 0.993 | 20.45 |
| (JN599098) | R: GTTTCGGGTCAAATAGGAAGAAC | ||||||
1CYC cyclophilin, SPS sucrose phosphate synthase, ACT actin, RUBISCO ribulose biphosphate carboxylase, GAPDH glyceraldehyde-3-phosphate dehydrogenase, TUB beta-tubulin, EF1 elongation factor 1α, UBQ ubiquitin, 18S 18S ribosomal RNA (nuclear gene), 25S 25S ribosomal RNA (nuclear gene), 26S 26S ribosomal RNA (mitochondrial gene)
2F: forward primer and R: reverse primer
3The threshold for fluorescence detection was set using the logarithmic amplification plot so that it is above the background fluorescence, below the linear region and at the beginning of the region of exponential amplification (i.e. the linear portion of the plot)
PCR
Specific amplification from cDNA and gDNA was checked by PCR followed by electrophoresis on 2% (w/v) agarose gels, and the annealing temperature optimised where necessary. PCR mixes were as described [16]. Amplicons were sequenced to confirm amplification of the targeted gene in A. myosuroides (accession numbers are in Table 2).
QPCR
qPCR was performed in fast optical 0.1 ml, 96-well reaction plates (MicroAmp™, Applied Biosystems, Cheshire, UK) using the ABI PRISM 7,900 HT Sequence Detection System (Applied Biosystems, Foster City, USA). The reaction volume (20 μl) contained 2 μl of a cDNA RT mix diluted 125-fold, 10 μl of ABsolute QPCR SYBR Green ROX mix (ThermoScientific, Epsom, UK) and 0.5 μM of each gene-specific primer. Polymerase activation (95°C for 15 min) was followed by 40 quantification cycles [95°C for 15 s, Tm (Table 2) for 30s and 72°C for 30s]. After 40 cycles, a melting-curve analysis (68°C to 95°C, one fluorescence read every 0.3°C) was performed to check the specificity of the amplifications. Amplicon sizes were checked on 3% (w/v) agarose gels. PCRs with each primer pair were also performed on three samples lacking cDNA template (negative controls).
To assess the amplification efficiency of each candidate gene, identical volumes of all cDNA samples (2 RT reactions per RNA sample) were pooled. The pool was diluted and used to generate five-point standard curves based on a five-fold dilution series (1:5-1:3125). This was performed in duplicate. Each duplicate series was amplified in two independent qPCR runs. Amplification efficiency (E) was computed as: E = 10(-1/a) -1 [19], where a is the slope of the linear regression model (y = a log(x) + b) fitted over log-transformed data of the input cDNA concentration (y) plotted against quantification cycle (Cq) values (x). The four E-values obtained from the dilution series were averaged for each primer pair. E-values for different target genes were considered comparable when included in the range of 100 ± 10% (standard curve slope of -3. 3 ± 0.33) [20].
To assess gene stability, all cDNA samples that passed quality assessment were diluted 125-fold. For each gene, every diluted sample was amplified twice in two independent qPCR runs. As two independent RTs were performed per RNA sample, this yielded four technical replicates per RNA sample.
Data were analysed using SDS 2.3 (Applied Biosystems, Foster City, USA). To generate a baseline-subtracted plot of the logarithmic increase in fluorescence signal (DRn) against cycle number, baseline data were collected between qPCR cycles 3 and 15, so that the amplification curve growth begins at a cycle number greater than the stop baseline cycle. The threshold for fluorescence detection was set using the logarithmic amplification plot so that it is above the background fluorescence, below the linear region and at the beginning of the region of exponential amplification.
Data analysis
To evaluate the stability of candidate reference genes expressed as Cq values, we used BestKeeper [21], geNorm v. 3.5 [4] and NormFinder [22]. NormFinder and geNorm require the transformation of Cq values by the 2-ΔΔCq method [20], using the lowest Cq as a calibrator. BestKeeper first computes the variation in Cq values and its standard deviation (SD) for each gene. Genes with SD > 1 are considered unstable [21]. The remaining genes are ranked based on pairwise correlations between the Cq values of each gene and the geometric mean of the Cq values of all genes (BestKeeper Index). Candidate genes showing the strongest correlation with the BestKeeper Index are considered the most suitable [21]. NormFinder ranks the candidate genes after their Stability Values (SV) based on the variations of their respective transformed-Cq values within and among groups [22]. Reference genes with the lowest SVs are top ranked, and considered the most suitable reference genes. NormFinder subsequently computes the SV of the combination of the two most stable genes. geNorm computes all possible average pairwise variation between the candidate gene transformed-Cq values, and provides a measure of the expression stability (M) of each gene. An M value below 1.5 identifies stable reference genes [4]. geNorm then performs stepwise exclusion of the gene with the highest M-value (least stably expressed gene) and recalculates M values for the remaining genes. This iterative process enables to rank candidate genes based on their stability of expression. As a single reference gene may not allow adequate normalisation, geNorm computes the optimal number of reference genes required for accurate normalisation by calculating pairwise variations Vn/n+1 between consecutively ranked normalisation factors NFn and NFn+1, where n and n+1 are the number of genes considered, and NFi are the geometric means of the i best candidate reference gene transformed Cq values. A pairwise variation of 0.15 is suggested as a cut-off value below which the inclusion of an additional reference gene is not required for reliable normalisation [4].
Application: Expression level of ACCase and GSTL in A Myosuroides
UBQ, GADPH and TUB were used to normalise the expression data of two genes. Expression data were generated using all cDNA samples that passed quality assessment. ACCase that encodes the target of the herbicide used in this study is expected to be stably expressed in herbicide-resistant compared to herbicide-sensitive plants [14,23]. GSTL that encodes a lambda-class glutathione-S-transferase had been shown to be up-regulated in herbicide-resistant plants compared to sensitive plants [24]. Amplification of a 175 bp fragment of ACCase [genbank: AM408429] was performed using primers ACVII 27 (5'-CACAAGATGCAGCTAGATAGTGGCG) and ACVII34R (5'-TTCCAACAGTTCGTCCAGTAACGAATG) with a Tm of 60°C. Amplification of a 156 bp fragment of a GSTL [genbank: FN394979, FN394980, FN394981] was performed using primers VGSTL-3 (5'-GGTTCAGATATACTATTCTCACC) and VGSTL-3R (5'-CTTGAGATGCCTCTTGGCAAC) with a Tm of 60°C. qPCR results were analysed using REST 2009 ver. 2.0.13 [25] that compares the expression level of a target gene in a 'sample' group using a 'control' group as a reference, taking into account the respective amplification efficiencies of each reference gene and target gene computed as described in the 'qPCR' section. Gene expression data analysed consisted into average Cq values computed for two replicates (two independent qPCR runs, each performed from one of two independent RT reactions). REST 2009 implements the 2-ΔΔCq method to the transformation of Cq values [20]. ACCase and GSTL expression was compared in sensitive plants ('control') and in resistant plants ('sample') before, 2.5 hours and 6 hours after herbicide application, using a pairwise fixed reallocation randomisation test (2,000 iterations).
Results
Specificity and efficiency of amplification
PCRs using primers targeting CYC and SPS were aspecific, and melting curve profiles revealed primer dimer formation or aspecific amplification for primers targeting ACT or RUBISCO (Additional file 2: Figure S1A and B). Other sets of primers designed and tested for these four genes did not increase the specificity of amplification (not shown). These four genes were thus not further considered (Table 2).
The single-peak melting curves obtained for the seven remaining candidate genes confirmed the absence of primer dimers or non-specific products (Additional file 2: Figure S1 C-I). The no-template controls (NTCs) yielded Cq values comprised between 31 and 35 cycles. The corresponding melting curves showed a small peak located before the position of the amplicon peak observed in the template samples. As no amplicon peak was detected in the NTCs, the positive Cq values observed were attributed to primer dimer formation, and were ignored. Specific amplification of the expected amplicons was confirmed by agarose gel electrophoresis (Figure 1).
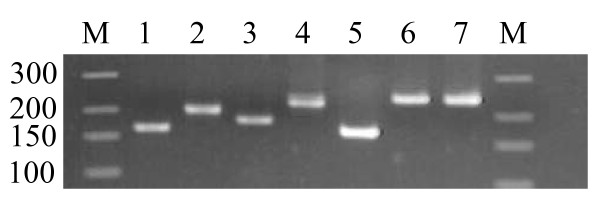
Figure 1.
Agarose gel (3%) electrophoresis showing amplicon size for seven candidate reference genes. These genes all passed the specificity and efficiency assessment steps. 1, TUB; 2, EF1; 3, GAPDH; 4, UBQ; 5, 25S; 6, 26S; 7, 18S. M, DNA ladder (the size of the DNA ladder fragments is given on the left in base pairs).
qPCR efficiency and correlation coefficients computed from the five-fold dilution series are given in Table 2. The amplification efficiency of EF1 (89%) was just below the 90% cut-off value. As amplification was specific for EF1, it was included in the subsequent analyses. All six remaining genes fulfilled our criteria for specific and effective amplification (Table 2).
Stability of candidate reference genes
For every gene, qPCR data consisting into the mean Cq value computed from four technical replicates was generated from each of the 19 RNA samples that passed quality control (Table 1). The technical replicates consisted into two independent qPCR runs performed on each of two independent RT reactions per RNA sample. Expression data was subdivided in five subsets according to the phenotype or to the time after herbicide application (Table 1).
BestKeeper analyses
Gene stability was assessed considering all samples as a single set where all possible combinations of the tested effects (time after herbicide application and phenotype) were present. All genes except EF1 were found suitable for normalisation (SD < 1, Table 3; Additional file 1: Table S2). Among the remaining 6 genes, the highest r values were observed for TUB, 26S, GAPDH and UBQ. TUB was ranked the second most stable gene because of its SD value (1.07). 26S that had the lowest SD value and the second highest r value was ranked the most stable gene (Table 3; Additional file 1: Table S2).
Table 3.
Ranking of the candidate reference genes according to their stability value using BestKeeper and NormFinder
| BestKeeper | NormFinder | |||||||||
| Reference genesa | All samples | All samples | Herbicide effect | Phenotype effect | ||||||
| BEFORE/+2.5H+6H | SENSITIVE/RESISTANT | |||||||||
| SDb | r c | Ranking order | SVd | Ranking order | SV | Ranking order | SV | Ranking order | ||
| 26S | 0.46 | 0.80* | 1 | 0.335 | 1 | 0.148 | 1 | 0.132 | 1 | |
| TUB | 1.07 | 0.88* | 2 | 0.488 | 2 | 0.204 | 4 | 0.156 | 2 | |
| GAPDH | 0.93 | 0.70* | 3 | 0.494 | 3 | 0.188 | 3 | 0.242 | 4 | |
| UBQ | 0.81 | 0.67 | 4 | 0.515 | 4 | 0.175 | 2 | 0.254 | 5 | |
| 25S | 0.50 | 0.53 | 5 | 0.669 | 5 | 0.258 | 5 | 0.279 | 6 | |
| 18S | 0.96 | 0.42 | 6 | 1.039 | 7 | 0.378 | 7 | 0.370 | 7 | |
| EF1 | 1.45 | 0.85* | 7 | 0.813 | 6 | 0.332 | 6 | 0.231 | 3 | |
aThe three reference genes selected are in bold
bStandard deviation of Cq values. SD values higher than the cut-off value (1.00) are underlined
cPearson coefficient of correlation
dStability value
*: associated p value <0.001
Norm Finder analyses
To assess the effect of the plant phenotype and of the time after herbicide application on gene stability, we analysed three different data sets comprising all samples 1- without subset assignment; 2- with every sample assigned to one of the subsets BEFORE, + 2.5H or + 6H and 3- with every sample assigned to one of the two subsets SENSITIVE or RESISTANT. 26S, TUB, GAPDH and UBQ were the most stable genes overall and considering the time after herbicide application (Table 3). 26S, TUB, GAPDH and EF1 were the most stable genes between sensitive and resistant plants (i.e. phenotype effect) (Table 3).
When the samples were not assigned to a subset or assigned to subsets SENSITIVE or RESISTANT, the combination of the two most stable genes was 26S and TUB (Table 3). This combination had a SV value (0.092) lower than that of the most stable gene (26S, SV = 0.132; Table 3). When the samples were assigned to the subsets BEFORE, + 2.5H and + 6 H, the most stable combination of two genes was 26S and UBQ, with a SV value (0.119) lower than that of the most stable gene (26S, SV = 0.148; Table 3).
Overall, 26S, TUB, UBQ and GAPDH were identified as the most stable genes.
geNorm analyses
The subsets BEFORE and SENSITIVE (five replicates per subset, Table 1) did not contain enough biological replicates to allow reliable analysis. Thus, we first used the whole data set to assess gene stability when the two effects (i.e. plant phenotype and time after herbicide application) are combined. We subsequently analysed each of the subsets + 2.5H and + 6H independently to assess phenotype effect, and the subset RESISTANT to assess the effect of time after herbicide application.
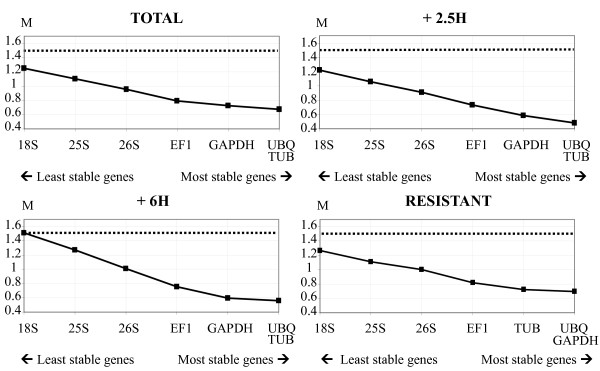
In all analyses, all genes showed M values below the default cutoff value (1.5) (Figure 2). Overall, UBQ, TUB and GAPDH were the three most stable genes, UBQ and TUB being the most stable among all samples and between contrasted phenotypes, and UBQ and GAPDH being the most stable when considering the effect of the time after herbicide application (Figure 2).
Figure 2.
Average expression stability values (M) of seven candidate reference genes computed using geNorm. Ranking was performed for all RNA samples (TOTAL), and for the subsets +2.5H, +6H and RESISTANT. M-values of the remaining genes at each step during stepwise exclusion of the least stable gene are shown. The genes are ranked according to increasing expression stability (decreasing M-values) starting from the least stable gene on the left. The two most stable genes are on the right. The horizontal dotted line indicates the cutoff M-value (1.5).
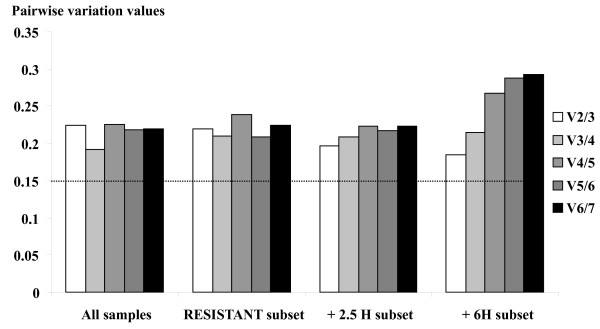
The optimal number of reference genes required for accurate normalisation was computed. All values obtained for the pairwise variation between consecutive normalisation factors (Vi/j values, Figure 3) were higher than the proposed 0.15 cutoff threshold [4]. As recommended in this case [4], we considered the change in the Vi/j values when including additional reference genes, and we used the lowest Vi/j value to determine the number of reference genes adequate for normalisation. Regarding the phenotype effect (assessed from subsets + 2.5 H and + 6H; Figure 3), the inclusion of the third most stably expressed gene yielded the lowest variation of the normalisation factor (V2/3), indicating that using three genes (UBQ, TUB and GAPDH) was adequate for normalisation. Overall (TOTAL subset) and when considering the effect of the time after herbicide application (assessed from subset RESISTANT; Figure 3), the inclusion of the fourth most stably expressed gene yielded the lowest variation of the normalisation factor (V3/4), indicating that using four genes (UBQ, TUB, GAPDH and EF1) was adequate for normalisation.
Figure 3.
Determination of the optimal number of reference genes for accurate normalisation. geNorm calculates pairwise variations (Vn/n+1) between consecutively ranked normalisation factors NFn and NFn+1 (NFi is the geometric mean of the expression values for the i first-ranked candidate reference genes). Each pairwise variation value is compared to the cutoff value (0.15, horizontal dotted line). For example, V2/3 corresponds to the pairwise variation between the normalisation factors of the two first-ranked (2) and the three first-ranked (3) reference genes.
Expression level of ACCase and GSTL in A. Myosuroides
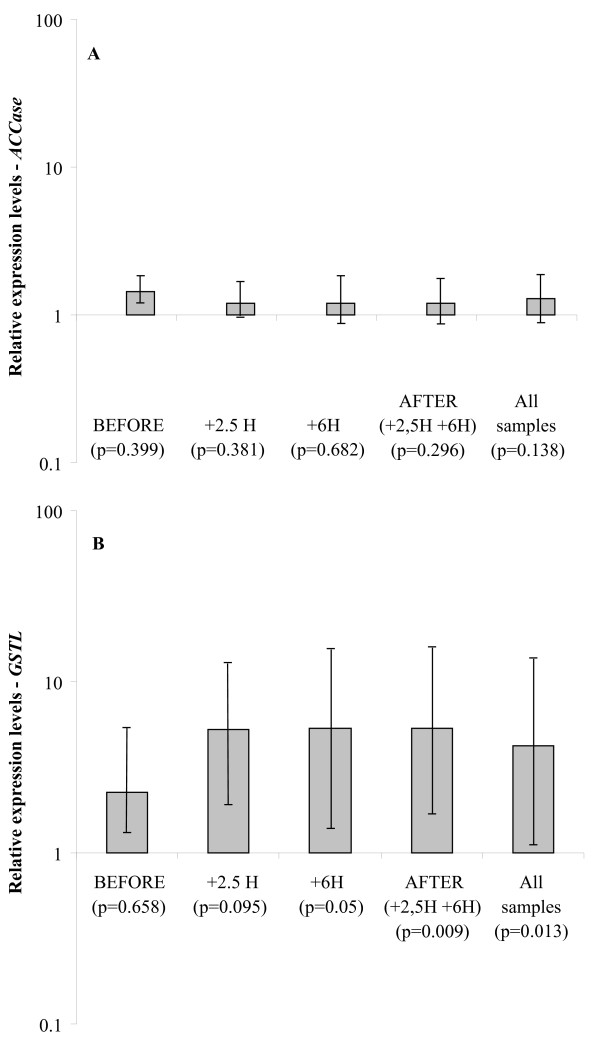
The expression of ACCase and GSTL was normalised using UBQ, TUB and GAPDH, and compared in herbicide-resistant or sensitive plants overall and in each of the subsets BEFORE, +2.5H, +6H and AFTER (Table 1). ACCase showed an average Cq value of 25.88 (PCR efficiency = 105%; R2 = 0.997; manual threshold = 0.235). No significant differences in the expression of ACCase were observed between resistant and sensitive plants for all conditions tested (Figure 4a). GSTL showed an average Cq value of 30.11 (PCR efficiency = 109%; R2 = 0.980; manual threshold = 0.249). GSTL was found to be 4.3-fold up-regulated in resistant plants compared to sensitive ones when considering all samples (Figure 4b). GSTL was 5.4-fold and 5.3-fold up-regulated when considering the two subsets + 6H and AFTER (+2.5H and +6H), respectively (Figure 4b). No significant differences in the expression of GSTL were observed when comparing resistant and sensitive plants before and 2.5 hours after treatment (Figure 4b).
Figure 4.
Relative expression levels of ACCase (a) and GSTL (b) in resistant versus sensitive plants. UBQ, TUB and GADPH were used for normalisation. Each measurement consists of two repetitions. BEFORE, before herbicide application; +2.5H and +6H, 2.5 and 6 h after herbicide application, respectively; AFTER, after herbicide application (2.5H and +6H); TOTAL, all samples. p values for differences in gene expression are given in brackets.
Discussion
The aim of this study was to identify suitable reference genes for the normalisation of gene expression data in A. myosuroides plants with contrasted phenotypes (sensitive or resistant) that had been submitted or not to herbicide stress. As no single method is generally accepted to test for the stability of candidate reference genes, we used the algorithms implemented by three different programs.
BestKeeper is a useful first approach generating descriptive statistics and coefficients of correlation. geNorm is considered one of the best methods to determine the most stable genes in plant studies (e.g. [7,26,27]). geNorm also indicates the optimal number of genes required for normalisation in a given experimental dataset [4]. However, it has a tendency to assign close ranks to co-regulated genes, which expression ratios show less pairwise variation than those of independently regulated genes [4]. Therefore, we also used NormFinder [22] that ranks candidate reference genes according to the variation of their expression within and among experimental modalities, and is less sensitive to possible bias arising from co-regulation [2].
The results yielded by the three programs were very similar (Table 3, Figure 2), and indicated that the most stable genes in our system were UBQ, TUB and GAPDH, which were always ranked among the four most stable genes. geNorm indicated that four genes would be most adequate for normalisation, also including EF1. However, because EF1 was ranked among the two least stable genes by the two other programs, we did not include it in the reference gene set. Furthermore, in most cases, using the three 'best' reference genes is a valid normalisation strategy [4].
26S was top-ranked by NormFinder and BestKeeper, but ranked fifth by geNorm (Figure 2). This could be explained by the sensitivity of geNorm to the co-regulation of the genes tested. It is indeed reasonable to consider that 26S, a ribosomal RNA gene, may be co-regulated with 18S and 25S, ribosomal RNA genes that were consistently ranked among the least stable genes (Table 3). The high abundance of ribosomal RNAs compared with mRNAs makes it difficult to accurately subtract the baseline value in qPCR data analysis [4]. Furthermore, ribosomal RNA seems less affected than mRNA by partial degradation, which may introduce bias in normalisation [28]. Considering all these points, and because UBQ, TUB and GAPDH used together enabled reliable normalisation (Figure 4a-b), we did not include 26S in the reference gene set. The combination of UBQ, TUB and GAPDH was thus selected to normalise gene expression data in herbicide resistance studies in A. myosuroides.
To check the accuracy of this set of reference genes, we investigated the expression profiles of two genes of interest in resistant and sensitive plants. Differential regulation of ACCase had never been shown to confer resistance to ACCase inhibitors [14,23]. Thus, ACCase is expected to have a similar expression level in plants sensitive or resistant to the ACCase inhibitor fenoxaprop, which was observed (Figure 4a). In contrast, GSTL was expected to be up-regulated after herbicide application in resistant plants compared to sensitive ones [24], which was observed considering all samples. The expression level of GSTL was not significantly different between resistant and sensitive plants before and 2.5 hours after treatment, but was up-regulated in resistant plants six hours after treatment (Figure 4b). This suggests an herbicide-induced up-regulation of GSTL in resistant plants compared to sensitive plants, which is consistent with a previous study [24]. These results thus confirm that UBQ, TUB and GAPDH can reliably be used as a set of reference genes to normalise qPCR data in studies on herbicide action in A. myosuroides.
Herbicides are an abiotic stress. Previous works identified reference genes with stable expression under different abiotic or biotic stresses in a range of plant species. UBQ had been shown to be stable under abiotic stresses in grasses [8,26]. UBQ was also stable in the broadleaf Arabidopsis thaliana under herbicide stress [29]. TUB was among the most stable genes reported in several cereal (grasses) species under different stresses [30]. GAPDH showed a stable expression during stress in grasses [30-32], although some stresses induced a high variability in its expression in wheat [33].
EF1 and 18S were among the least stably expressed genes in our study. They are among the most commonly used reference genes in plant studies, with a stable expression reported in grasses under different stresses in several studies (e.g. [8,26,31,32]). However, other studies reported an unstable expression of these genes in grasses under biotic stress (e.g., [30,32]). This was also observed for herbicide stress in our work (Table 3). Our results and the literature thus clearly confirm the need for a thorough validation of the stability of expression of candidate reference genes in the system considered prior to any gene expression study.
Conclusions
This is the first study describing a set of reference genes in a non-model grass plant species that is also a weed of economic and agronomic importance. To the best of our knowledge, this is also the first study conducted on a non-model plant species demonstrating the stability of expression of a set of reference genes under herbicide stress and in contrasted herbicide-related phenotypes (sensitive and resistant).
In grass weeds, herbicide resistance is mostly endowed by non-target-site-based mechanisms [11,12,23], an adaptive trait under polygenic control endowed by constitutive and/or induced differential expression of numerous genes in resistant versus sensitive plants [15]. Hardly any such genes have been identified so far in a weed species [14,23]. The reference gene set validated for A. myosuroides will be of immense help to identify genes involved in non-target-site-based resistance. Given that the expression of these reference genes seems stable in grasses, they are clearly candidate reference genes of choice for studies seeking to identify herbicide-responsive genes in other grasses with no associated genomic resources (i.e., most grass weeds), and also very likely to identify stress-responsive genes in this taxon.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
All authors participated in the design of the study. CP and FP carried out the experiments and performed the analyses. CP drafted the manuscript. JMH participated to the analyses of the study and helped to draft the manuscript. CD participated in the coordination of the study and critically revised the manuscript. All authors contributed to, read and approved the final manuscript.
Supplementary Material
Table S1. Accession numbers of the sequences used for primer design. Table S2. Descriptive statistics of reference gene expression in black-grass based on the BestKeeper approach.
Figure S1. Primer specificity test. - melting curve generated for RUBISCO. Melting curves generated for RUBISCO (A), ACT (B), UBQ (C), EF1 (D), GAPDH (E), TUB (F), 25S (G), 26S (H) and 18S (I).
Contributor Information
Cécile Petit, Email: cecilepetit1982@yahoo.fr.
Fanny Pernin, Email: fpernin@dijon.inra.fr.
Jean-Marie Heydel, Email: jean-marie.heydel@u-bourgogne.fr.
Christophe Délye, Email: delye@dijon.inra.fr.
Acknowledgements
The authors thank David Bru (INRA Dijon) for his helpful advice on quantitative PCR experiments and the MSE and FLAVIC laboratories (INRA Dijon) for allowing us to use their facilities.
References
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol. 2002;29:23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005;6:279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034.1–0s034.11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E. Housekeeping genes as internal standards: use and limits. J Biotechnol. 1999;75:291–295. doi: 10.1016/S0168-1656(99)00163-7. [DOI] [PubMed] [Google Scholar]
- Stürzenbaum SR, Kille P. Control genes in quantitative molecular biological techniques: the variability of invariance. Comp Biochem Physiol B Biochem Mol Biol. 2001;130:281–289. doi: 10.1016/S1096-4959(01)00440-7. [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, Van Wuytswinkel O. The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. 2008;6:609–618. doi: 10.1111/j.1467-7652.2008.00346.x. [DOI] [PubMed] [Google Scholar]
- Dombrowski JE, Martin RC. Evaluation of reference genes for quantitative RT-PCR in Lolium temulentum under abiotic stress. Plant Sci. 2009;176:390–396. doi: 10.1016/j.plantsci.2008.12.005. [DOI] [Google Scholar]
- Lee JM, Roche JR, Donaghy DJ, Thrush A, Sathish P. Validation of reference genes for quantitative RT-PCR studies of gene expression in perennial ryegrass (Lolium perenne L.) BMC Mol Biol. 2010;11:8. doi: 10.1186/1471-2199-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira ED, Alves-Ferreira M, Guimarães LA, Rodrigues da Silva F, Tavares de Campos Carneiro V. Selection of reference genes for quantitative real-time PCR expression studies in the apomictic and sexual grass Brachiaria brizantha. BMC Plant Biol. 2009;9:84. doi: 10.1186/1471-2229-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Délye C, Michel S, Berard A, Chauvel B, Brunel D, Guillemin JP, Dessaint F, Le Corre V. Geographical variation in resistance to acetyl-coenzyme a carboxylase-inhibiting herbicides across the range of the arable weed Alopecurus myosuroides (black-grass) New Phytol. 2010;186:1005–1017. doi: 10.1111/j.1469-8137.2010.03233.x. [DOI] [PubMed] [Google Scholar]
- Petit C, Bay G, Pernin F, Délye C. Prevalence of cross- or multiple resistance to the acetyl-coenzyme A carboxylase inhibitors fenoxaprop, clodinafop and pinoxaden in black-grass (Alopecurus myosuroides Huds.) in France. Pest Manag Sci. 2010;66:168–177. doi: 10.1002/ps.1851. [DOI] [PubMed] [Google Scholar]
- Petit C, Duhieu B, Boucansaud K, Délye C. Complex genetic control of non-target-site-based resistance to herbicides inhibiting acetyl-coenzyme a carboxylase and acetolactate-synthase in Alopecurus myosuroides Huds. Plant Sci. 2010;178:501–509. doi: 10.1016/j.plantsci.2010.03.007. [DOI] [Google Scholar]
- Délye C. Weed resistance to acetyl coenzyme A carboxylase inhibitors: an update. Weed Sci. 2005;53:728–746. doi: 10.1614/WS-04-203R.1. [DOI] [Google Scholar]
- Yuan JS, Tranel PJ, Stewart JCN. Non-target-site herbicide resistance: a family business. Trends Plant Sci. 2007;12:6–13. doi: 10.1016/j.tplants.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Délye C, Matéjicek A, Michel S. Cross-resistance patterns to ACCase-inhibiting herbicides conferred by mutant ACCase isoforms in Alopecurus myosuroides Huds. (black-grass), re-examined at the recommended herbicide field rate. Pest Manag Sci. 2008;64:1179–1186. doi: 10.1002/ps.1614. [DOI] [PubMed] [Google Scholar]
- Caldana C, Scheible WR, Mueller-Roeber B, Ruzicic S. A quantitative RT-PCR platform for high-throughput expression profiling of 2500 rice transcription factors. Plant Methods. 2007;3:7. doi: 10.1186/1746-4811-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. In: Bioinformatics Methods and Protocols: Methods in Molecular Biology. Krawetz SA, Misener S, editor. Totowa, Humana Press; 2000. Primer3 on the www for general users and for biologist programmers; pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Radstrom P, Lofstrom C, Lovenklev M. In: PCR Primer: A Laboratory Manual. Dieffenbach CW, Dveksler GS, editor. Cold Spring Harbor, Cold Spring Harbor Laboratory Press; 2003. Strategies for overcoming PCR inhibition; pp. 149–161. [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- Andersen L, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Powles SB, Yu Q. Evolution in action: plants resistance to herbicides. Annu Rev Plant Biol. 2010;178:317–347. doi: 10.1146/annurev-arplant-042809-112119. [DOI] [PubMed] [Google Scholar]
- Cummins I, Bryant DN, Edwards R. Safener responsiveness and multiple herbicide resistance in the weed black-grass (Alopecurus myosuroides) Plant Biotech J. 2009;7:807–820. doi: 10.1111/j.1467-7652.2009.00445.x. [DOI] [PubMed] [Google Scholar]
- REST 2009 V2.0.13. http://www.REST.de.com
- Jain MNA, Tyagi AK, Khurana JP. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun. 2006;345:646–651. doi: 10.1016/j.bbrc.2006.04.140. [DOI] [PubMed] [Google Scholar]
- Schmidt GW, Delaney SK. Stable internal reference genes for normalization of realtime RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol Genet Genomics. 2010;283:233–241. doi: 10.1007/s00438-010-0511-1. [DOI] [PubMed] [Google Scholar]
- Brunner AM, Yakovlev IA, Strauss SH. Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol. 2004;4:14. doi: 10.1186/1471-2229-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M, Reichman JR, Haberer G, Welzl G, Aceituno FF, Mader MT, Watrud LS, Pfleeger TG, Gutiérrez RA, Schäffner AR, Olszyk DM. A composite transcriptional signature differentiates responses towards closely related herbicides in Arabidopsis thaliana and Brassica napus. Plant Mol Biol. 2010;72:545–556. doi: 10.1007/s11103-009-9590-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarošová J, Kundu JK. Validation of reference genes as internal control for studying viral infections in cereals by quantitative real-time RT-PCR. BMC Plant Biol. 2010;10:146. doi: 10.1186/1471-2229-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Seo PJ, Yang MSs, Xiang F, Park CM. Exploring valid reference genes for gene expression studies in Brachypodium distachyon by realtime PCR. BMC Plant Biol. 2008;8:112. doi: 10.1186/1471-2229-8-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faccioli P, Ciceri GP, Provero P, Stanca AM, Morcia C, Terzi V. A combined strategy of "in silico" transcriptome analysis and web search engine optimization allows an agile identification of reference genes suitable for normalization in gene expression studies. Plant Mol Biol. 2007;63:679–688. doi: 10.1007/s11103-006-9116-9. [DOI] [PubMed] [Google Scholar]
- Long XY, Wang JR, Ouellet T, Rocheleau H, Wei YM, Pu ZE, Jiang QT, Lan XJ, Zheng YL. Genome-wide identification and evaluation of novel internal control genes for q-PCR based transcript normalization in wheat. Plant Mol Biol. 2010;74:307–311. doi: 10.1007/s11103-010-9666-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Accession numbers of the sequences used for primer design. Table S2. Descriptive statistics of reference gene expression in black-grass based on the BestKeeper approach.
Figure S1. Primer specificity test. - melting curve generated for RUBISCO. Melting curves generated for RUBISCO (A), ACT (B), UBQ (C), EF1 (D), GAPDH (E), TUB (F), 25S (G), 26S (H) and 18S (I).